Abstract
Limb-girdle muscular dystrophy type 2D (LGMD2D) is caused by autosomal recessive mutations in the α-sarcoglycan gene. An R77C substitution is the most prevalent cause of the disease, leading to disruption of the sarcoglycan-sarcospan complex. To model this common mutation, we generated knock-in mice with an H77C substitution in α-sarcoglycan. The floxed neomycin (Neo)-cassette retained at the targeted H77C α-sarcoglycan locus caused a loss of α-sarcoglycan expression, resulting in muscular dystrophy in homozygotes, whereas Cre-mediated deletion of the floxed Neo-cassette led to recovered H77C α-sarcoglycan expression. Contrary to expectations, mice homozygous for the H77C-encoding allele expressed both this mutant α-sarcoglycan and the other components of the sarcoglycan-sarcospan complex in striated muscle, and did not develop muscular dystrophy. Accordingly, conditional rescued expression of the H77C protein in striated muscle of the α-sarcoglycan-deficient mice prevented the disease. Adding to the case that the behavior of mutant α-sarcoglycan is different between humans and mice, mutant human R77C α-sarcoglycan restored the expression of the sarcoglycan-sarcospan complex when introduced by adenoviral vector into the skeletal muscle of previously created α-sarcoglycan null mice. These findings indicate that the α-sarcoglycan with the most frequent missense mutation in LGMD2D is correctly processed, is transported to the sarcolemma, and is fully functional in mouse muscle. Our study presents an unexpected difference in the behavior of a missense-mutated protein in mice versus human patients, and emphasizes the need to understand species-specific protein quality control systems.
INTRODUCTION
Muscular dystrophies are hereditary diseases characterized by the progressive degeneration of skeletal muscle (1). Limb-girdle muscular dystrophy (LGMD) comprises a heterogeneous subset of muscular dystrophies that present with predominantly proximal muscular weakness of the pelvic or shoulder girdles (2, 3). Sarcoglycanopathies are a subgroup of autosomal recessive type 2 LGMD with causative mutations in genes encoding components of the sarcoglycan complex of striated muscle (4). These genes include the α-, β-, γ- and δ-sarcoglycans, defects in which lead to LGMD2D, LGMD2E, LGMD2C, and LGMD2F, respectively (4). Among the sarcoglycanopathies, LGMD2D has the highest frequency, ranging from 35% to 60% (5–8). Clinically, LGMD2D is highly variable in severity (6, 9, 10). Genetically, LGMD2D is heterogeneous with various null and missense mutations reported (11). Among these, a missense mutation resulting in an arginine-to-cysteine substitution at the 77th amino acid (R77C) is the most frequent mutation found worldwide, accounting for 32% – 64% of LGMD2D (6, 11).
The sarcoglycan complex is part of the dystrophin-glycoprotein complex (DGC) that links extracellular ligands such as laminins and perlecan to the subcellular cytoskeleton through the α-dystroglycan - β-dystroglycan - dystrophin axis (4, 12). The DGC is thought to protect muscle cells from contraction-induced damage (13, 14), and its disruption—even if only at the level of the sarcoglycan complex—leads to muscular dystrophy. How a loss of the sarcoglycan component leads to muscular dystrophy is not clear, but disturbance of the DGC axis is a possible explanation that has gained support from the discovery that several lines of sarcoglycan null mice are characterized by a destabilization of binding between α- and β-dystroglycan, and between β-dystroglycan and dystrophin (15–19). Another possibility is a disturbance in intracellular signaling pathways implicated from the association of signaling molecules, such as calmodulin, Grb2 and nNOS, with the DGC (20).
One molecular peculiarity of the sarcoglycan complex is that a disruption of any single component can affect the membrane localization of the other components (21). Sarcospan is an integral membrane protein that associates tightly with sarcoglycans to form a sarcoglycan-sarcospan subcomplex within the DGC (22), and its expression appears to be especially sensitive to the disruption of any sarcoglycan component (22). However, the converse is not true; a lack of sarcospan does not lead to loss of the sarcoglycan complex (23). This could explain the fact that none of the unclassified muscular dystrophies have been linked to this gene (22), and could also account for the finding that sarcospan null mice do not develop muscular dystrophy (23).
Mouse models with targeted gene disruption of α-sarcoglycan (15), β-sarcoglycan (16, 17), γ-sarcoglycan (24, 25), or δ-sarcoglycan (26, 27) have been generated to help us understand the pathophysiology of LGMD2C-2F. Loss of any one of these sarcoglycans was sufficient to obliterate the expression of all four sarcoglycans and sarcospan, and to cause muscular dystrophy. These mouse models provide relevant insights for patients with the respective LGMD, but not for all patients. For example, in human patients the mutated sarcoglycan is often expressed at some reduced level when disease-causing mutations are of the missense type (6, 11). Accordingly, interference with the expression of the other components of the complex is often incomplete (6, 22).
In light of the discrepancies between the sarcoglycanopathies that have been generated in mice and their human counterparts, we reasoned that it may be possible to learn more about the mechanism underlying the human disease by reproducing a missense sarcoglycan protein product in a mouse model. Such an approach could enable us to search for pharmaceutical agents that are capable of counteracting a structural defect in the mutated sarcoglycan, which might lead to recovery of the complex as a whole.
With the above factors in mind, we created a missense knock-in mutation that leads to a histidine-to-cysteine substitution at the codon for the 77th amino acid (H77C). A mouse H77C mutation was expected to mimic the R77C form of human LGMD2D due to the conserved nature of mouse histidine and human arginine (both basic amino acids) in the wild-type proteins. Gene targeting by introduction of the H77C coding region and a floxed Neo cassette in the α-sarcoglycan locus generated an insertional disruption, leading to a complete inactivation of H77C α-sarcoglycan gene expression. Deletion of the floxed Neo cassette led to recovery of α-sarcoglycan mRNA that carries the missense mutation. To our surprise, the H77C mutant protein was expressed at normal levels at the sarcolemma, and no muscle pathology developed. In addition, adenovirus-mediated introduction of the human R77C α-sarcoglycan into previously generated α-sarcoglycan null (SgcaNull/Null) mice (15) led to the expression of human mutant α-sarcoglycan protein as well as recovery of the other components of sarcoglycan-sarcospan complex at the sarcolemma. Overall, our findings demonstrate that the missense mutations leading to human R77C and mouse H77C substitutions are not themselves sufficient to cause a loss of α-sarcoglycan protein expression in these mice. Our data suggest that mice can process these missense-mutated α-sarcoglycans to be folded correctly and transported to the sarcolemma to serve their required function.
RESULTS
Generation of Neo/Neo and H77C/H77C mice
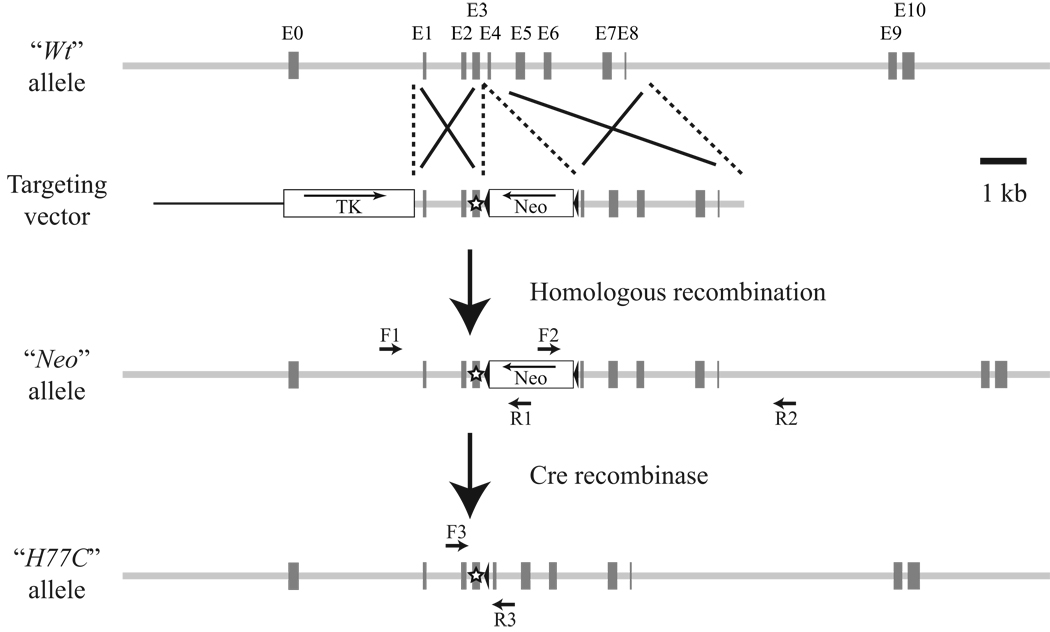
We designed a targeting vector encoding the H77C substitution in exon 3, with a Neo resistance cassette flanked by two loxP sequences (floxed) for positive selection of ES cells (Fig. 1). Homologous recombination was confirmed by PCR analysis over the 5’-and 3’-homologous arms, with one primer in each reaction matching sequence in the Neo cassette and the other matching sequence outside the targeting vector. The presence of coexisting random integrations of the targeting vector in recombinants was ruled out by southern blot analysis, with a Neo-encoding sequence probe detecting only a single band on two independent restriction enzyme digests (NsiI and SpeI; data not shown). ES cell clones were then injected into blastocysts to obtain germline-competent chimeric mice. The chimeric mice were backcrossed to C57BL/6J or 129S6/SvEvTac mice to obtain Neo/Wt heterozygous mice, and these were subsequently used to generate Neo/Neo, Neo/Wt and Wt/Wt mice. (We are going to use these terms, instead of SgcaNeo/Neo etc., for describing genotypes of the mice that were newly developed in this paper.) The Wt/Wt littermates of the Neo/Neo mice will be described as Wt/Wt (Neo) to distinguish them from the Wt/Wt littermates of H77C/H77C mice, which will be referred to as Wt/Wt (H77C) (see below).
Figure 1. Generation of α-sarcoglycan-targeted Neo/Neo and knock-in H77C/H77C mice.
Schematic representation of the wild-type α-sarcoglycan allele (Wt), the targeting vector, the homologously recombined Neo allele, and the Neo-deleted H77C allele. Exons 0–10 (E0–E10) are depicted by filled boxes, and the two loxP sites are shown as filled arrowheads. The star in exon 3 (E3) designates the H77C mutation. The pgk-Neo and pgk-thymidine kinase cassettes are illustrated as boxed Neo and TK, respectively. Arrows designated F1–F3 and R1–R3 represent the forward and reverse PCR primers, respectively, that were used for genotyping (see Materials and Methods).
To remove the floxed Neo cassette from the targeted allele, Neo/Wt heterozygous mice were crossed with EIIa-Cre mice (28) on the C57BL/6J background or protamine 1-Cre mice (29) on the 129 background; H77C/Wt heterozygotes were thus generated. These mice were further backcrossed to C57BL/6J or 129S6/SvEvTac mice to eliminate the Cre transgene. Heterozygous H77C/Wt mice were subsequently used to produce the H77C/H77C, H77C/Wt and Wt/Wt (H77C) mice.
Neo/Neo mice do not express α-sarcoglycan and develop muscular dystrophy
The presence of a Neo cassette often interferes with the expression of a gene at the targeted locus (30, 31), and may result in total inactivation of a gene (32). This effect occurred in our mice; we were unable to amplify a transcript for the full α-sarcoglycan coding sequence by RT-PCR of skeletal muscle from the Neo/Neo mouse (Fig. 2A). A sample from the previously described SgcaNull/Null mouse (15) was included in this analysis, producing a larger transcript that had previously been demonstrated not to produce functional α-sarcoglycan protein (15). The absence of α-sarcoglycan protein from the skeletal muscle of Neo/Neo mice was confirmed by western blot analysis (Fig. 2B) as well as by immunofluorescence staining (Fig. 2C). Other sarcoglycans and sarcospan were analyzed by immunofluorescence staining in skeletal muscle and heart, and they were not detected in the Neo/Neo mice either (Fig. 2C and data not shown).
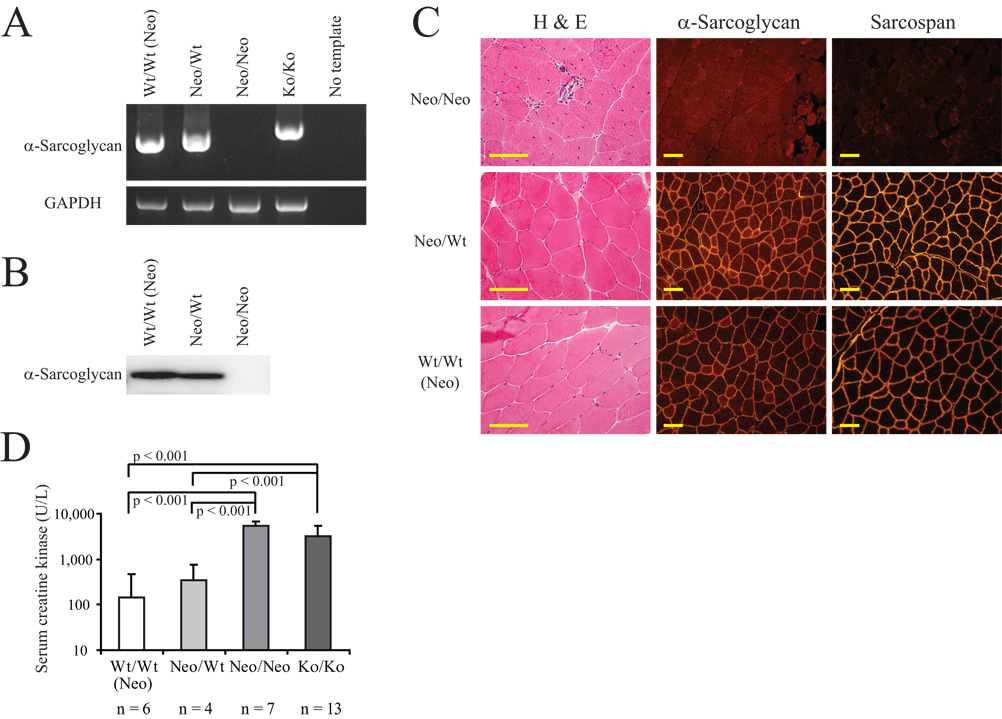
Figure 2. Neo/Neo mice develop a muscular dystrophy phenotype.
(A) RT-PCR analysis of skeletal muscle from a Neo-targeted homozygous mouse (Neo/Neo), heterozygous (Neo/Wt) and wild-type (Wt/Wt (Neo)) littermate controls at 5 weeks of age. cDNA template from a previously generated SgcaNull/Null mouse was also included (“Ko/Ko” lane). RT-PCR products covering the full-coding sequence of α-sarcoglycan (top panel) and a GAPDH internal control (bottom panel) are shown. The Neo/Neo mouse lacked expression of the α-sarcoglycan mRNA. A slightly larger product in the Ko/Ko mouse is described in the text.
(B) Western blot analysis of α-sarcoglycan protein in skeletal muscle from a Neo/Neo mouse and littermate controls at 37 weeks of age. Note a lack of α-sarcoglycan expression in the Neo/Neo mouse.
(C) Quadriceps from a Neo/Neo mouse and its littermate Neo/Wt and Wt/Wt (Neo) control counterparts at 37 weeks old analyzed by H&E staining, and by immunofluorescence staining for α-sarcoglycan and sarcospan. H&E panels demonstrate that the Neo/Neo mouse exhibits a large number of centrally nucleated cells and great variation in fiber size, as well as having necrotic fibers. α-Sarcoglycan and sarcospan panels show significantly reduced expression of these proteins in the Neo/Neo mouse. Scale bars: 100um.
(D) Serum CK levels in Neo/Neo, Neo/Wt, and Wt/Wt (Neo) littermate sets compared to those in SgcaNull/Null (Ko/Ko) mice at 10–12 weeks of age. Values observed in Neo/Neo and Ko/Ko mice were significantly higher than those in Wt/Wt (Neo) and Neo/Wt controls. Error bars indicate the SEM of log-CK values, and n equals the number of mice of each genotype. Statistically significant differences are depicted, and p values are indicated.
Hematoxylin and eosin (H&E) analysis revealed that Neo/Neo mouse muscle features several pathologic characteristics commonly found in muscular dystrophy: fiber size variation, fiber degeneration and regeneration, and central location of nuclei within muscle fibers (Fig. 2C). These features were not observed in control littermates. Muscle pathology can also be assessed based on leakage of muscle-specific intracellular proteins, such as creatine kinase (CK), into the blood indicating a disruption of membrane integrity in dystrophic muscle. Whereas wild type and heterozygous animals exhibited normal low serum CK levels, significantly higher levels were present in Neo/Neo mice (Fig. 2D). SgcaNull/Null mice were also analyzed as a positive control, and their serum CK values were not significantly different from the Neo/Neo mice.
Overall, our Neo/Neo mice recapitulated the phenotype of the SgcaNull/Null mice reported previously (15).
H77C/H77C knock-in mice express α-sarcoglycan at the sarcolemma and do not develop muscular dystrophy
The complete inactivation of H77C gene expression at the “Neo” allele was reversed by Cre-mediated removal of the Neo cassette, as revealed by restoration of the H77C α-sarcoglycan transcript in H77C/H77C mice (Fig. 3A). The presence of mutant mRNA in these animals was confirmed using restriction enzyme Fnu4HI digestion of the RT-PCR product, which cuts the wild-type locus but not the mutated site. The efficiency of transcription from the H77C-encoding allele was comparable to that from the wild-type allele, as demonstrated by similar amounts of RT-PCR product in wild-type and homozygote mutant mice. This was further supported by Fnu4HI digests from the two loci present in heterozygotes (350 bp vs. 218bp+132 bp) (Fig. 3A). In contrast to Neo/Neo mice, H77C/H77C mice expressed equivalent to wild-type levels of α-sarcoglycan protein, as revealed by Western blot analysis (Fig. 3B). Immunofluorescence staining of α-sarcoglycan and sarcospan in these mice also failed to detect differences relative to those in littermate controls, in either skeletal muscle (Fig. 3C) or heart (Supplementary Material, Fig. S1A–D). Additionally, expression of β-, γ-, and δ-sarcoglycans in skeletal muscle and heart was analyzed by immunofluorescence staining, all of which showed equal expression among H77C/H77C mice and their littermate controls (data not shown).
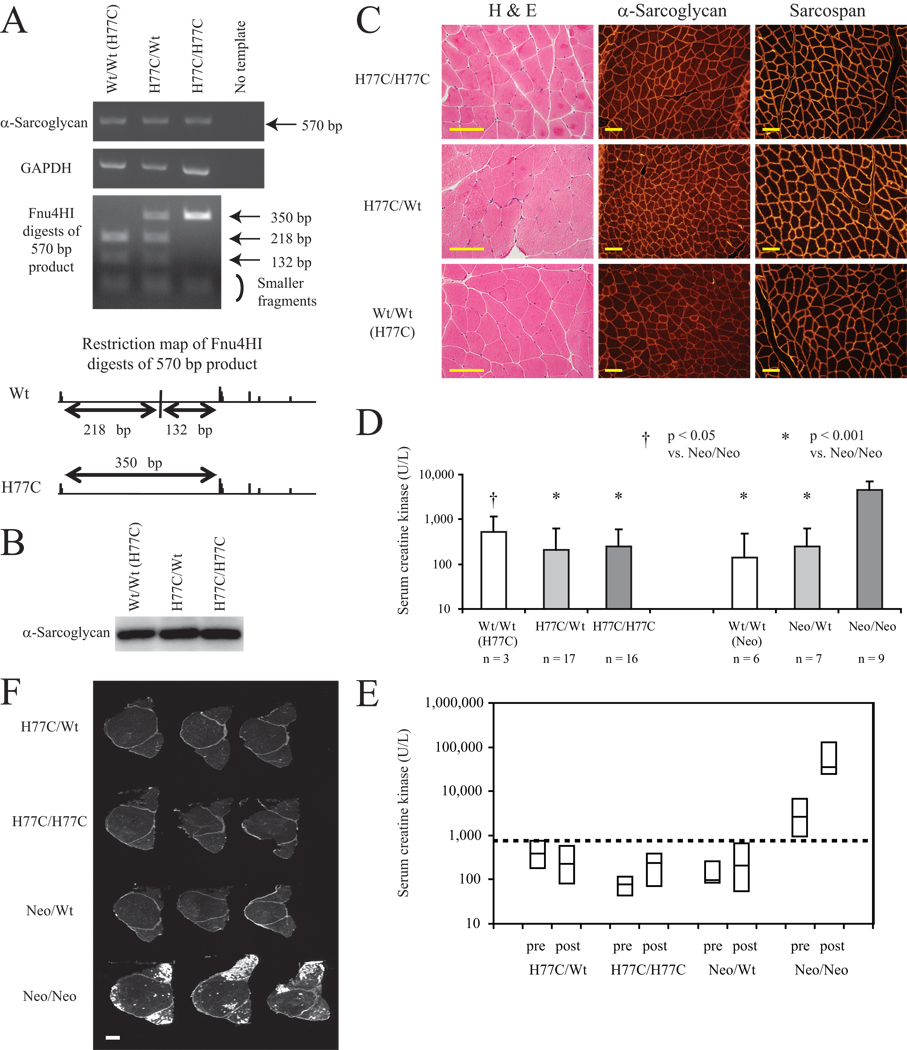
Figure 3. H77C/H77C mice do not develop a muscular dystrophy phenotype.
(A) RT-PCR analysis of skeletal muscle from an H77C/H77C mouse and littermate controls at 15 weeks of age. 570 bp RT-PCR products representing the α-sarcoglycan exon 1 – exon 5 region are shown (top panel), along with a GAPDH internal control (middle panel). Fnu4HI digests of the 570 bp products (bottom panel) indicate that Wt and H77C alleles were equally transcribed. The restriction map of the Fnu4HI sites present in the Wt and H77C 570 bp products is also shown.
(B) Western blot analysis of α-sarcoglycan expression in skeletal muscle from an H77C/H77C mouse and its littermate controls at 55 weeks of age. Note α-sarcoglycan expression in the H77C/H77C mouse.
(C) Quadriceps from an H77C/H77C mouse and its littermate controls H77C/Wt and Wt/Wt (H77C) at 55 weeks of age were analyzed by H&E staining, and by immunofluorescence staining for α-sarcoglycan and sarcospan. H&E panels demonstrate that the H77C/H77C mouse does not exhibit features of muscular dystrophy pathology. α-Sarcoglycan and sarcospan panels show that these proteins are expressed at normal levels and are also localized to the sarcolemma in H77C/H77C mouse as in its littermate controls. Scale bars: 100 µm.
(D) Serum CK levels of H77C littermate sets and Neo littermate sets of mice at 10–21 weeks old. Although the CK level was abnormally high in Neo/Neo mice, this was not the case for H77C/H77C mice. Error bars indicate the SEM of log-CK values, and n equals the number of different mice of each genotype. Statistically significant differences in comparison with Neo/Neo are indicated (†: p<0.05, *: p<0.001).
(E) Box plot of serum CK levels before (‘pre’) and after (‘post’) treadmill exercise. Dotted line: CK value of 750 U/L. Neo/Neo mice showed high basal serum CK levels that increased further after exercise, whereas mice of all other genotypes had serum CK values less than 750 U/L both before and after exercise. n equals 3 for each group.
(F) EBD uptake images of quadriceps samples after treadmill exercise. Each slice was obtained from a different individual mouse. EBD uptake, which is shown as white signal, is clearly visible in Neo/Neo mice but is hardly detected in H77C/H77C mice. n equals 3 for each group. Scale bar: 1 mm.
H&E staining of H77C/H77C mice did not reveal any morphological differences from their wild-type or heterozygous littermate controls (Fig. 3C). In case the onset of muscular dystrophy is delayed more in a missense mutation than in a null mutation, serum CK was measured at a wide range of ages up to 12 months, but H77C/H77C mice were indistinguishable from littermate controls at all time points (Fig. 3D and data not shown). In order to test for more subtle fragility of sarcolemmal integrity, we also measured serum CK following exercise-induced mechanical stress. However, we found serum CK values in H77C/H77C mice before and after exercise settled within a normal range (< 750 U/L) (Fig. 3E), as was the case for H77C/Wt and Neo/Wt mice. This is in stark contrast with the results for Neo/Neo mice, in which the high basal levels before exercise increased further, by more than 3-fold, after exercise (Fig. 3E). Evans blue dye (EBD) is a membrane-impermeant molecule that binds to serum albumin, and is physically restricted from fibers unless the sarcolemmal membrane is damaged (33, 34). EBD uptake in quadriceps after exercise was apparent in Neo/Neo mice, but was observed in no more than a few fibers per cryosection in the other 3 genotypes (H77C/H77C, H77C/Wt, Neo/Wt) (Fig. 3F). These data indicate that H77C/H77C mice do not share the muscular dystrophy phenotype seen in SgcaNull/Null mice (15).
Conditional expression of H77C protein in striated muscle prevents muscular dystrophy in Neo/Neo mice
To validate the functionality of the mutant H77C protein, its ability to rescue α-sarcoglycan-deficient muscle was analyzed. As described above, Neo/Neo mice lack α-sarcoglycan protein expression resulting in the development of muscular dystrophy. Cre-mediated deletion of the floxed Neo cassette, however, converts the “Neo” alleles to the “H77C” alleles, thus restoring expression of both the H77C mRNA and protein. MCK-Cre(+) mice express Cre recombinase in striated muscle (35) to drive high-efficiency deletion of a floxed region by 2–4 weeks of age (36). Breeding of MCK-Cre(+) mice and Neo/Neo mice resulted in doubly heterozygous animals, which were further bred with Neo/Neo mice to obtain 4 genotypes: MCK-Cre(−);Neo/Neo, MCK-Cre(+);Neo/Neo, MCK-Cre(−);Neo/Wt, and MCK-Cre(+);Neo/Wt. The first genotype should lack α-sarcoglycan expression, whereas the second should have restored expression of H77C protein in striated muscle. The remaining genotypes serve as α-sarcoglycan heterozygote littermate controls.
As predicted, MCK-Cre(−);Neo/Neo mice lacked both α-sarcoglycan and sarcospan expression (as shown by immunofluorescence staining), developed a muscular dystrophy pathology (as shown by H&E staining) (Fig. 4A, top panels, and Fig. S1G–H in the Supplementary Material), and had increased serum CK levels (Fig. 4B); in contrast, Neo/Wt heterozygotes expressed both proteins, failed to develop pathology (Fig. 4A, bottom panels) and had normal serum CK values (Fig. 4B). MCK-Cre(+);Neo/Neo mice, in whose striated muscle we expected to observe restored H77C mutant α-sarcoglycan expression, indeed expressed α-sarcoglycan and sarcospan at normal levels, in both skeletal muscle (Fig. 4A, middle panels) and heart (Supplementary Material, Fig. S1E–F). In addition, the serum CK values in these animals were normalized (Fig. 4B). We also assessed the percentage of muscle fibers with centrally located nuclei, and found it to be normalized in MCK(+);Neo/Neo mice to the levels of Neo/Wt littermate controls, leaving only MCK(−);Neo/Neo mice showing increased central nuclei (Fig. 4C).
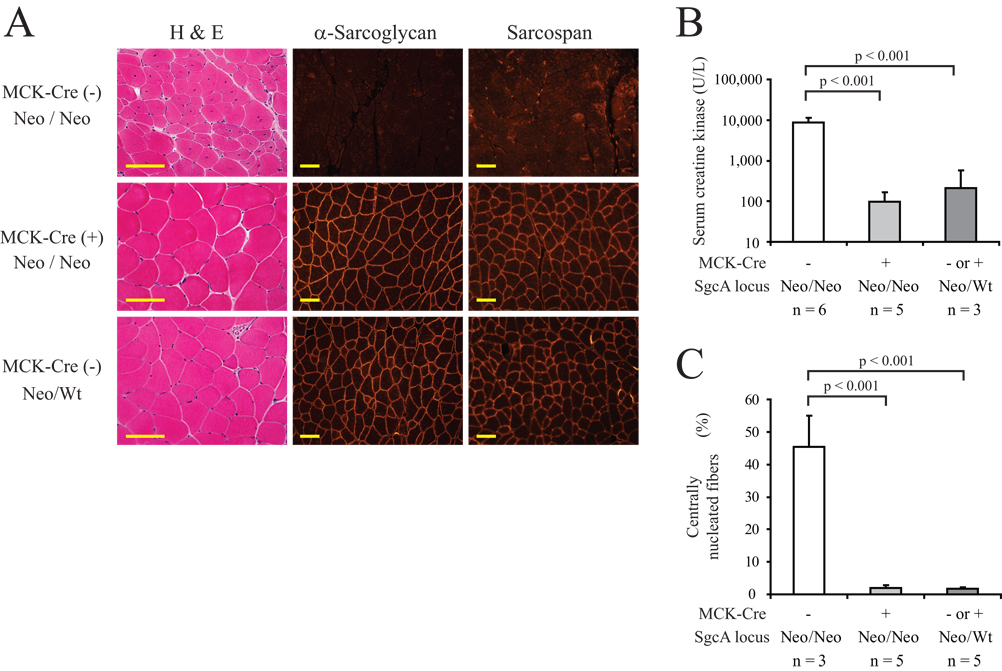
Figure 4. Expression of H77C protein by conditional rescue prevents development of a muscular dystrophy phenotype in Neo/Neo mice.
(A) Quadriceps from an MCK-Cre(+);Neo/Neo mouse and its littermate controls at 55 weeks of age analyzed by H&E staining, as well as by immunofluorescence staining for α-sarcoglycan and sarcospan. The MCK-Cre(+);Neo/Neo mouse (middle panels) lack a muscular dystrophy-like pathology and express both α-sarcoglycan and sarcospan at normal levels at the sarcolemma. Note the contrast to the MCK-Cre(−);Neo/Neo mouse (upper panels). Scale bars: 100 µm.
(B) Serum CK levels in littermate sets of mice at 10–13 weeks of age. Elevated levels were observed in MCK-Cre (−);Neo/Neo but not in MCK-Cre (+);Neo/Neo mice. Error bars indicate the SEM of log-CK values.
(C) The ratio of fibers with centrally located nucleus was examined in quadriceps of littermate sets of mice at 46–56 weeks old. MCK-Cre (+);Neo/Neo mice did not have elevated levels of the centrally nucleated fibers. Error bars indicate the SEM of the ratios.
In (B) and (C), SgcA stands for α-sarcoglycan, and n equals the number of mice in each genotype. Statistically significant differences are depicted, and p values are indicated.
Overall, our results indicate that the presence of H77C protein in α-sarcoglycan-deficient mouse muscle is sufficient to prevent the development of a muscular dystrophy phenotype.
Adenovirally introduced human R77C mutant α-sarcoglycan in SgcaNull/Null mouse muscle is expressed at the sarcolemma and restores the expression of the sarcoglycan-sarcospan complex
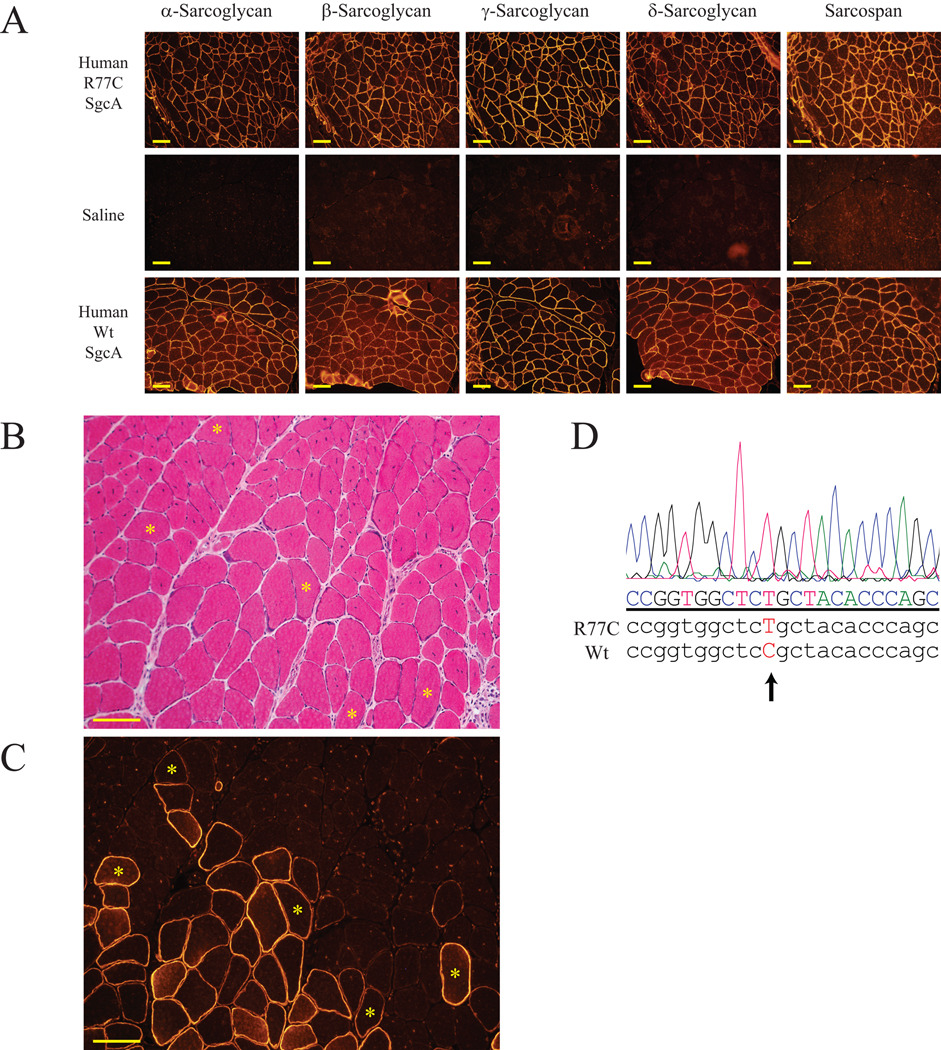
Although human and mouse α-sarcoglycans are highly conserved (88.9% amino acid identity), it is possible that the mouse H77C substitution is not sufficient to reproduce the pathologic effect mediated by the human R77C substitution. To assess the behavior of human R77C mutant α-sarcoglycan in a mouse in vivo system, we injected Ad5CMV-R77C-SgcA-myc adenovirus expressing the human α-sarcoglycan with the R77C-encoding missense mutation into hindlimb muscles of previously reported SgcaNull/Null mice (15). To generate positive controls, we also injected Ad5RSV-SgcA adenovirus carrying the wild-type human α-sarcoglycan into littermate SgcaNull/Null mice. Negative controls in each case were provided by injecting the contralateral leg of each mouse with saline.
As expected based on previous reports (15, 37), wild-type human α-sarcoglycan was expressed at the sarcolemma of the SgcaNull/Null skeletal muscle, and expression of other components of the sarcoglycan-sarcospan complex was restored (Fig. 5A, bottom panels). Saline-injected muscle expressed none of the components (Fig. 5A, middle panels). To our surprise, human R77C α-sarcoglycan was also expressed at the sarcolemma, together with other sarcoglycans and sarcospan (Fig. 5A, top panels). This restored expression of the sarcoglycan-sarcospan complex also provided a protective role against the development of muscular dystrophy pathology (Fig. 5B and C). In order to rule out the possibility of any contamination, RT-PCR of α-sarcoglycan was performed on total RNA purified from muscle sections located next to the region positive for R77C α-sarcoglycan staining. Although SgcaNull/Null mice express a defective α-sarcoglycan transcript as described above, these mice lack exons 2 and 3 (15), and a primer pair that amplifies human α-sarcoglycan from exon 2 to exon 5 can detect only the exogenous human α- sarcoglycan transcript (both wild-type and R77C mutant). DNA sequencing of the amplified products confirmed that the human R77C α-sarcoglycan-transduced muscle samples expressed only human mutant mRNA (Fig. 5D).
Figure 5. Adenovirus-mediated transduction of human wild-type and R77C-mutant α-sarcoglycan protein into skeletal muscle of SgcaNull/Null mice.
(A) SgcaNull/Null pups were injected with human wild-type α-sarcoglycan (Wt SgcA) or human R77C α-sarcoglycan-myc (R77C SgcA) adenovirus in one leg, and with saline in the contralateral leg. Calf muscle was harvested 25 weeks after injection, and analyzed by immunofluorescence staining for each component of the sarcoglycan-sarcospan complex. Note that the expression of every component of the sarcoglycan-sarcospan complex at the sarcolemma is efficiently rescued by the injection of R77C α-sarcoglycan-myc adenovirus (top panels) but not by saline injection (middle panels) into the same animal. The results were reproduced in 2 independent experiments.
H&E (B) and α-sarcoglycan immunofluorescence staining (C) at the boundary between R77C α-sarcoglycan-myc adenovirus transduced and non-transduced fibers show that transduced region is associated with a lack of centrally nucleated fibers. Asterisks indicate the same muscle fibers in serial sections. Scale bars: 100 µm.
(D) DNA chromatograms of an α-sarcoglycan transcript expressed in skeletal muscle of an SgcaNull/Null mouse injected with the human R77C-encoding α-sarcoglycan-myc adenovirus. The DNA sequences surrounding the 77th codon of human R77C- and Wt-encoding α-sarcoglycan are also presented for comparison. The introduced missense mutation of C229T is clearly present.
DISCUSSION
Since the discovery of dystrophin, great progress has been made in the field of muscular dystrophy, mainly due to the analysis of the molecules associated with dystrophin (e.g. dystroglycan and sarcoglycans) and the positional cloning of other genes (e.g. calpain-3 and dysferlin) (38). Fourteen genes have so far been shown to cause LGMD. These discoveries have led to the generation of animal models for various forms of LGMD by gene targeting strategies. Although such models serve well as mimics of null mutations (for defects due to nonsense, frameshift or splicing mutations that lead to a failure to produce a full-length protein product), they do not necessarily represent human diseases that result from missense mutations. Therefore, we generated a missense mouse model of sarcoglycanopathy based on the premise that such a model might reveal the effects of a partial reduction in protein expression, or possibly of a misfolding problem that affects the sarcoglycan complex more generally. We reasoned that if either were the case, such a model would also be useful for the screening of drugs capable of correcting the relevant defect. We chose to model the form of LGMD2D caused by the R77C-encoding allele because of its high frequency among sarcoglycanopathies. The arginine at the 77th amino acid position of the protein is identical among humans, rabbits and hamsters (39) and, although the mouse sequence instead bears a histidine at this position, the function of this amino acid is likely the same in that they are both basic amino acids. We thus expected that this approach would generate an accurate mouse model of R77C-associated LGMD2D.
The presence of the Neo cassette in a targeted gene often interferes with the expression of that gene (30, 31). In our case, the effect was a complete inactivation of gene expression, as demonstrated by the fact that neither transcript nor protein could be detected in the Neo/Neo mice. This effect was negated after deletion of the floxed Neo cassette, with easily detectable expression of the missense mRNA. Contrary to our expectation, however, the H77C/H77C mice did not develop muscular dystrophy. α-Sarcoglycan and other components of the sarcoglycan-sarcospan complex were both expressed at normal levels and were localized at the sarcolemma (Fig. 3C and data not shown). Although we examined α-sarcoglycan expression at various ages between 4 and 88 weeks after birth, there was no reduction in α-sarcoglycan expression at any time point (Fig. 3C and data not shown). One potential explanation for this would be that mouse H77C is not, after all, an exact counterpart of the human R77C mutation. This possibility, however, is contradicted by a second unexpected finding—that introduction of the human R77C-encoding mutant α-sarcoglycan into the skeletal muscle of the SgcaNull/Null mice also restored the expression of α-sarcoglycan as well as other components of the sarcoglycan-sarcospan complex in this tissue. Thus, it appears that a single missense mutation that causes an H77C or R77C substitution in the α-sarcoglycan protein is not sufficient to cause a loss of α-sarcoglycan protein expression in mouse skeletal muscle.
Our results might cast doubt on the pathogenic role of human R77C in LGMD2D. However, it is unlikely because i) the R77C mutation cosegregated with the disease trait in each family of LGMD2D with that mutation (11, 40); ii) it has not been found in 200 (11) or 348 (40) normal chromosomes, ruling it out as a common polymorphism; iii) it is found worldwide and is associated with at least 3 distinct haplotypes (40), disfavoring the presence of another causative locus that is linked to the R77C mutation. The frequent occurrence of R77C itself is well explained by the fact that it is located at a CpG site which is a mutational hot spot (11). Instead, our finding suggests that other factors, that are most likely not present in mice, are involved in the pathogenesis of this particular form of LGMD2D.
This hypothesis is consistent with the large variation in clinical severity among LGMD2D cases (6, 9, 10). Even family members with the same mutation can show a significant variability in clinical progression of the disease; this is the case not only for individuals homozygous for the R77C substitution (40, 41), but also for patients with other mutations affecting the α-sarcoglycan protein (42). In addition to our examination of H77C/H77C mice on a mixed background of C57BL/6J and 129 with predominance of the former (75% – 96.9%), we have also analyzed H77C/H77C mice on a 129-pure background. These animals likewise expressed α-sarcoglycan normally, without any sign of the development of muscular dystrophy (data not shown), indicating that the lack of a muscular dystrophy phenotype in our study appears not to be peculiar to a single mouse genetic background.
Because there is no reported loss of human R77C α-sarcoglycan transcript in LGMD2D patients (11, 40, 43, 44), the cause of the loss of R77C α-sarcoglycan expression is more likely ascribed to a failure of the R77C protein to reach the sarcolemma. Missense mutations often impair the propensity of the affected protein to fold to the correct conformation. A typical measure of cellular system against misfolded proteins is to promote its refolding by molecular chaperones, and proteins that failed to be refolded in a timely manner will be degraded by proteases (45). Although we do not have experimental evidence for the role of the protein quality control system on mutated α-sarcoglycan, failure of efficient refolding is an attractive explanation for the disrupted expression of R77C α-sarcoglycan in LGMD2D patients. Aging-associated decline of endoplasmic reticulum chaperones has been reported in rat liver (46) or human cultured fibroblasts (47). If such protein quality control system also exists in skeletal muscle, species difference in its activity, or much shorter life-span of mice than the preclinical period in LGMD2D patients (typically ranging 3–15 years with a mean of 8.5 years (10), but not rarely as long as decades (48)) can explain the different behavior of R77C/H77C α-sarcoglycan between in human patients and in our mice. In the latter case of aging effect, one possibility is that the sarcoglycan-sarcospan complex might be normally expressed long before the onset of clinical manifestation in some patients with missense-caused LGMD2D. Existence of sibling patient cases with different expression levels of α-sarcoglycan with the same mutation (42) favors such possibility. However, there have been no reports of change in sarcoglycan expression at different time points in the same patient. More availability of genetic testing in these days that enables preclinical diagnosis might lead to identification of such cases in future.
While our paper was under review, another manuscript was submitted by Bartoli et al. in which H77C α-sarcoglycan knock-in mice were independently generated and analyzed (49). Those mice also did not develop muscular dystrophy, confirming our result. They report rescue studies in the same SgcaNull/Null mice that we generated (15) and used here, but treated with human R77C α-sarcoglycan using a different promoter and a different virus vector that show aggregation of the transgene products in the endoplasmic reticulum. This same group previously published intracellular accumulation of human Wt α-sarcoglycan transgene product in their gene therapy system (50). In contrast, neither the mutant R77C α-sarcoglycan nor the Wt human α-sarcoglycan accumulated in muscle cells in our rescue experiments. The artificial muscle-specific C5–12 promoter used by Bartoli et al. (49) may be stronger than our CMV and RSV promoters (51–53). A possible explanation for the discrepancies between our two studies is that a greater amount of α-sarcoglycan transgene is produced with the C5–12 promoter, thus promoting intracellular accumulation. Interestingly, Bartoli et al. (49) also show that inhibition of mannosidase I directs more of the R77C α-sarcoglycan to the sarcolemma suggesting that blocking a single component of the protein quality control system can divert mutant polypeptides away from a degradation pathway. This is an important finding that furthers our understanding of the role that trafficking of mutant proteins may play in the pathogenesis of LGMD2D.
The absence of a muscle pathology phenotype in the H77C knock-in mice is not readily explained. However, an in vitro system used by Bartoli et al. (49) to test sarcoglycan expression at the cell surface membrane may provide some insight. This heterologous cellular system was used to determine that a basic amino acid at the 77th residue appears to be necessary for successful trafficking of human α-sarcoglycan. Analysis of the mouse H77C protein in a similar system using both human and murine cells might help determine whether the species difference is intrinsic to the α-sarcoglycan protein (R77C in humans vs. H77C in mice), or extrinsic to α-sarcoglycan and perhaps a function of differences in protein quality control.
Another interesting question is if successful expression of mutant R77C α-sarcoglycan in SgcaNull/Null mice could extend to other mutations of α-sarcoglycan as well as of β-, γ-, and δ-sarcoglycans in respective null mice. If the rescuing function is observed in a number of disease-causing missense mutations of other sarcoglycans, it suggests that the supposed disease-modifying factors would have broader interaction with the sarcoglycan-sarcospan complex instead of one particular mutation of α-sarcoglycan, which could include components of the above mentioned protein quality control system.
Various knock-out mouse strains have already been generated in efforts to model human disease, and they have contributed immensely to our understanding of the pathogenic processes of many disorders. However, it is important to keep in mind that they typically model null mutations, and do not necessarily accurately represent diseases caused by missense mutations. As shown here, LGMD2D serves as one example of muscular dystrophy for which a knock-in approach was able to provide new insights that might have been missed if knock-out models had been the only available research tool.
MATERIALS AND METHODS
Animals
C57BL/6J and 129S6/SvEvTac mice were purchased from The Jackson Laboratory and Taconic Farms, respectively. EIIa-Cre mice (The Jackson Laboratory) were backcrossed to C57BL/6J at least 10 times before experimental crosses. Protamine 1-Cre mice on the 129 background were purchased from The Jackson Laboratory. SgcaNull/Null mice were generated previously (15) and are available from Mutant Mouse Regional Resource Center (MMRRC) under the stock number of 000425-MU/H. MCK-Cre mice were described previously (35). All animals were housed in the animal care unit of the University of Iowa College of Medicine according to the University of Iowa animal care guidelines. All animal studies were authorized by the Institutional Animal Care and Use Committee of the University of Iowa.
Antibodies
Antibodies for western blots and immunofluorescence were: Mouse monoclonal antibody 20A6 against α-sarcoglycan (15); goat polyclonal antibody Goat26 against β-sarcoglycan (15); rabbit polyclonal antibodies Rbt98 against α-sarcoglycan (54), Rbt245 against γ-sarcoglycan (55), Rbt215 against δ-sarcoglycan (15), Rbt256 against sarcospan (23), and anti-laminin (#L9393, Sigma). Cy3- and HRP-conjugated secondary antibodies were purchased from Jackson Immunoresearch Laboratories and Chemicon, respectively.
Targeting constructs
Genomic fragments of the mouse α-sarcoglycan gene were isolated by PCR from genomic DNA prepared from mice of the 129/Sv genetic background. TGC-to-CAC transition in exon 3 (which generates the H77C missense substitution) was introduced by sequential PCR steps (56). This culminated in the assembly of a 1.5-kb fragment for a 5’-homologous arm, a 3.5-kb fragment for a 3’-homologous arm, a pgk-Neo cassette flanked by two loxP sequences, and a pgk-thymidine kinase cassette within a pBlueScript KS plasmid (Stratagene). The whole sequence of the assembled genomic α-sarcoglycan fragments within the targeting vector, including intronic regions, was checked for a lack of mutations outside that generating the H77C substitution.
Generation of Neo/Neo and H77C/H77C gene-targeted mice
The SalI-linealized construct was electroporated into W4/129S6 ES cells (Taconic Farms). Successful homologous recombination was confirmed by PCR over the 5’- and 3’-sides of the insertion, using the primers described below. Two ES cell clones were microinjected into C57BL/6J blastocysts, and chimeric mice were obtained for each line. The Neo insertional allele was confirmed in the germline-transmitted F1 and 129-pure pups. The floxed Neo cassette was removed by breeding with EIIa-Cre mice on the C57BL/6J background and protamine 1-Cre mice on the 129 background, generating the H77C allele on the C57BL/6J-dominant and 129-pure backgrounds, respectively. Mice of the C57BL/6J-dominant background were used after backcrossing to C57BL/6J 2–5 times, and it is data from these animals that are presented in the figures in this paper. All data from homozygous mice were collected in parallel with data from their littermate controls. Because no differences were detected for the 2 lines of mice generated from 2 independent ES cell clones, the data were combined in this paper.
PCR genotyping
Genotyping of each locus was performed by PCR under standard conditions, with the following primers: 5’-homologous recombination of the Neo allele: F1, GCCACTCAACCGCGCCTGTCTGTAAC and R1, CGCATCGCCTTCTATCGCCTTCTTG; the 3’-homologous recombination of the Neo allele: F2, GCGGCCGGAGAACCTGCGTGCAATCCATCT and R2, AGCTGAGGGGCACAGGACTGGGGGCATTGG; the H77C-encoding allele: F3, CTGCGCCTTCCAGAACACGTTGGTAAGA and R3, TCTAGGGGGAAGCTGACAAGGCACACTT. In successfully recombined clones, the F1 & R1 and F2 & R2 primer pairs yielded bands of 2304 bp and 4347 bp, respectively. The F3 & R3 primer pair produced bands of 490 bp and 442 bp from the H77C-encoding and wild-type alleles, respectively. For genotyping of mice for EIIa-Cre, protamine 1-Cre and MCK-Cre, a Cre-genotyping protocol from The Jackson Laboratory was followed.
RT-PCR
Total RNA from skeletal muscle was extracted using RNAzol B (Tel-Test Inc.). Equal amounts of total RNA from littermates were reverse-transcribed with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) using oligo(dT) primers. The following primers were used for RT-PCR: mouse α-sarcoglycan from 5’- to 3’-untranslated regions, GGAGCCCCTGTCCCTGTCACTCACTGG and AATGCCCTCAGCCCTCCCTACACCATCTGT; mouse α-sarcoglycan from exon 1 to exon 5, ATGGCAGCAGCAGTAACTTGGATA and CTCAATAGGAAGAGGGACTCGG; mouse GAPDH from exon 4 to exon 7, CCATGTTTGTGATGGGTGTGAACC and TGTGAGGGAGATGCTCAGTGTTGG; human α-sarcoglycan from exon 2 to exon 5, CCGTGTCTTTGTGCACACC and AAGCGGCTGGCAGGTGTTGAGG. For determination of DNA sequence, amplified products were purified using a PCR purification kit (Qiagen) and analyzed at the University of Iowa DNA Core Facility.
Western blotting
KCl-washed membranes from skeletal muscle were prepared as previously described (15). 50 µg of protein samples were resolved by SDS-PAGE on 3–15% linear gradient or 10% single-concentration gels and transferred to Immobilon-P membranes (Millipore). Equal protein loading and blotting efficiency among lanes were checked by Ponceau-S staining (#P7170, Sigma) of the blots. Immunoblot staining with the 20A6 antibody was performed as previously described (57), with the following modifications: use of Prestained SDS-PAGE Standards (#161-0318, Bio-Rad); washing solution of 50mM sodium phosphate, pH 7.4, 150 mM sodium chloride, 0.1% Tween-20; chemiluminescence development using SuperSignal West Pico or Dura (Pierce). Images were digitally detected using an Alpha Innotech imaging system.
Immunofluorescence and histological analyses
7 µm (skeletal muscle) or 5 µm (heart) axial cryosections were prepared and analyzed by immunofluorescence or H&E staining. For immunofluorescence staining with the Rbt98, Goat26, Rbt245, Rbt215, Rbt256, or anti-laminin antibody, sections were: washed with phosphate-buffered saline (PBS), blocked with 3% bovine serum albumin (BSA) in PBS for 30 minutes at room temperature, incubated with a primary antibody in PBS+1% BSA overnight at 4°C, washed with PBS, incubated with a Cy3-conjugated secondary antibody in PBS+1% BSA for 1 hour at room temperature, washed with PBS, and mounted with PermaFluor (#IM0752, Beckman Coulter). Microscope images were photographed using a Leica DM RXA microscope equipped with an Olympus DP70 digital camera.
Determining the percentage of muscle fibers with centrally located nuclei
Cryosections of quadriceps were stained with anti-laminin antibody to delineate each muscle fiber, and 4',6-diamidino-2-phenylindole dihydrochloride (#D9542, Sigma) was included in the secondary antibody solution, at a final concentration of 200 ng/ml, for the detection of nuclei. All fibers, except those in direct contact with fascia, were analyzed for the location of their nuclei. For each sample group, the number of fibers with centrally localized nuclei relative to the total number of fibers was recorded. For each individual mouse, about 2500–3500 fibers were counted.
Measurement of serum CK levels
Blood collected from the tail of individual mice was separated by centrifugation (at 10,000 g for 2 min), following which CK levels in the serum were measured using a Creatine Kinase (NAC) Assay kit (#310–56, Diagnostic Chemicals Limited) according to the manufacturer’s instruction. Samples from each mouse were measured in triplicate using Microtest 96-well plates (#353261, BD Falcon) and a SpectraMax 190 microplate reader (Molecular Devices Corp.).
Injection of recombinant adenovirus
An adenovirus construct that expresses wild-type human α-sarcoglycan (Ad5RSV-SgcA) was described previously (15). The C229T nucleotide transition (corresponding to R77C amino acid replacement) in exon 3 of the human a α-sarcoglycan gene was introduced by sequential PCR steps (56) into pcDNA3-human SgcA-myc (21). The R77C-SgcA-myc sequence was then subcloned into a pAd5CMVpA shuttle vector. The Ad5CMV-R77C-SgcA-myc adenovirus was produced at the University of Iowa Gene Transfer Vector Core, by standard methods as previously described (15). Adenovirus injections into 3- to 4-day-old SgcaNull/Null pups were performed as previously described (37), with some modification: the hamstring, quadriceps, calf, and tibialis anterior muscles of one leg were each injected with 2.5 × 109 particles in a 10 µl volume; the same muscles of the contralateral leg were each injected with an equal volume of saline.
Treadmill exercise and analysis of EBD uptake
Animals at 32–37 weeks of age were exercised on an Omnipacer treadmill (Model LC4/M-MGA/AT, Accuscan Instruments) set at a 15° downward angle, with the pace starting at 5 m/min between 0–3 minutes, moving to 10 m/min between 3–6 minutes, and increasing in increments of 2.5 m/min every 3 minutes thereafter (e.g. 12.5 m/min between 6–9 minutes, and so on) until a maximal speed of 30 m/min was reached (no further speed increment after 27 minutes); animals were exercised until exhausted. All mice were intraperitoneally injected with 250 µl of EBD (#E2129, purchased from Sigma, was dissolved in PBS at 10 mg/ml and passed through 0.22 µm filter) 16 hours before exercise. Tail blood was collected both before EBD injection and 2 hours after the exercise for serum CK measurement, and the mice were sacrificed 9 hours after exercise for harvesting of the skeletal muscles. EBD uptake images of 7 µm sections of fresh-frozen quadriceps on glass slides were taken with an Odyssey imaging system (Li-Cor Biosciences) using a 680 nm laser line.
Digital Images
RT-PCR, western blot, immunofluorescence and H&E images were digitally acquired and processed for size, contrast and brightness in Photoshop CS2 (Adobe). To ensure accurate visual comparison among littermates, the same image acquisition conditions and the same processing parameters were applied to each image.
Statistical analysis
Because the distribution of serum CK levels is skewed toward higher values and better matches a log-normal distribution than a normal distribution (58), log conversion was performed prior to analysis. Log-converted CK values, as well as the ratio of centrally nucleated to total muscle fibers, were assessed by one-way ANOVA and multiple comparisons were performed using Bonferroni adjustment. P values less than 0.05 were considered statistically significant, and all significant differences were indicated on the corresponding graph.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to past and present members of the Campbell laboratory for insightful comments and scientific contributions, and thank Drs. Michael G. Anderson and Roger Williamson for critical evaluations of the manuscript. We would especially like to thank Dr. Valérie Allamand for her help on the adenoviral experiments, Dr. Franck Duclos for his advice on the construction of the gene targeting vector, and Dr. C. Ronald Kahn who generously provided us with MCK-Cre mice. We also wish to thank the associates at the Gene Transfer Vector Core Facility of the University of Iowa Center for Gene Therapy of Cystic Fibrosis and Other Genetic Diseases (supported by NIH/NIDDK P30 DK 54759) for their assistance with viral vector production. This work was supported in part by a Paul D. Wellstone Muscular Dystrophy Cooperative Research Center Grant, the Muscular Dystrophy Association, and an NIH Research Grant (1R01AR051199). K.P.C. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
The following abbreviations were used in this paper:
- BSA
bovine serum albumin
- CK
creatine kinase
- DGC
dystrophin-glycoprotein complex
- EBD
Evans blue dye
- ES cell
embryonic stem cell
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- H&E
hematoxylin and eosin
- LGMD
limb-girdle muscular dystrophy
- Neo
neomycin
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RT-PCR
reverse transcriptase-polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Wt
wild-type
Footnotes
Conflict of Interest statement
The authors have no conflict of interest.
REFERENCES
- 1.Gardner-Medwin D, Walton JN. The muscular dystrophies. In: Walton JN, Karpati G, Hilton-Jones D, editors. Disorders of Voluntary Muscle. 6th ed. New York, NY: Churchill Livingstone; 1994. pp. 543–594. [Google Scholar]
- 2.Walton JN, Nattrass FJ. On the classification, natural history and treatment of the myopathies. Brain. 1954;77:169–231. doi: 10.1093/brain/77.2.169. [DOI] [PubMed] [Google Scholar]
- 3.Bushby KM. Diagnostic criteria for the limb-girdle muscular dystrophies: report of the ENMC Consortium on Limb-Girdle Dystrophies. Neuromuscul. Disord. 1995;5:71–74. doi: 10.1016/0960-8966(93)e0006-g. [DOI] [PubMed] [Google Scholar]
- 4.Durbeej M, Campbell KP. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 2002;12:349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 5.Moore SA, Shilling CJ, Westra S, Wall C, Wicklund MP, Stolle C, Brown CA, Michele DE, Piccolo F, Winder TL, et al. Limb-girdle muscular dystrophy in the United States. J. Neuropathol. Exp. Neurol. 2006;65:995–1003. doi: 10.1097/01.jnen.0000235854.77716.6c. [DOI] [PubMed] [Google Scholar]
- 6.Moreira ES, Vainzof M, Suzuki OT, Pavanello RC, Zatz M, Passos-Bueno MR. Genotype-phenotype correlations in 35 Brazilian families with sarcoglycanopathies including the description of three novel mutations. J. Med. Genet. 2003;40:E12. doi: 10.1136/jmg.40.2.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanin M, Duggan DJ, Mostacciuolo ML, Martinello F, Freda MP, Soraru G, Trevisan CP, Hoffman EP, Angelini C. Genetic epidemiology of muscular dystrophies resulting from sarcoglycan gene mutations. J. Med. Genet. 1997;34:973–977. doi: 10.1136/jmg.34.12.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duggan DJ, Manchester D, Stears KP, Mathews DJ, Hart C, Hoffman EP. Mutations in the delta-sarcoglycan gene are a rare cause of autosomal recessive limb-girdle muscular dystrophy (LGMD2) Neurogenetics. 1997;1:49–58. doi: 10.1007/s100480050008. [DOI] [PubMed] [Google Scholar]
- 9.Piccolo F, Roberds SL, Jeanpierre M, Leturcq F, Azibi K, Beldjord C, Carrie A, Recan D, Chaouch M, Reghis A, et al. Primary adhalinopathy: a common cause of autosomal recessive muscular dystrophy of variable severity. Nat. Genet. 1995;10:243–245. doi: 10.1038/ng0695-243. [DOI] [PubMed] [Google Scholar]
- 10.Eymard B, Romero NB, Leturcq F, Piccolo F, Carrie A, Jeanpierre M, Collin H, Deburgrave N, Azibi K, Chaouch M, et al. Primary adhalinopathy (alpha-sarcoglycanopathy): clinical, pathologic, and genetic correlation in 20 patients with autosomal recessive muscular dystrophy. Neurology. 1997;48:1227–1234. doi: 10.1212/wnl.48.5.1227. [DOI] [PubMed] [Google Scholar]
- 11.Carrie A, Piccolo F, Leturcq F, de Toma C, Azibi K, Beldjord C, Vallat JM, Merlini L, Voit T, Sewry C, et al. Mutational diversity and hot spots in the alpha-sarcoglycan gene in autosomal recessive muscular dystrophy (LGMD2D) J. Med. Genet. 1997;34:470–475. doi: 10.1136/jmg.34.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weller B, Karpati G, Carpenter S. Dystrophin-deficient mdx muscle fibers are preferentially vulnerable to necrosis induced by experimental lengthening contractions. J. Neurol. Sci. 1990;100:9–13. doi: 10.1016/0022-510x(90)90005-8. [DOI] [PubMed] [Google Scholar]
- 15.Duclos F, Straub V, Moore SA, Venzke DP, Hrstka RF, Crosbie RH, Durbeej M, Lebakken CS, Ettinger AJ, van der Meulen J, et al. Progressive muscular dystrophy in alpha-sarcoglycan-deficient mice. J. Cell Biol. 1998;142:1461–1471. doi: 10.1083/jcb.142.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbeej M, Cohn RD, Hrstka RF, Moore SA, Allamand V, Davidson BL, Williamson RA, Campbell KP. Disruption of the beta-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol. Cell. 2000;5:141–151. doi: 10.1016/s1097-2765(00)80410-4. [DOI] [PubMed] [Google Scholar]
- 17.Araishi K, Sasaoka T, Imamura M, Noguchi S, Hama H, Wakabayashi E, Yoshida M, Hori T, Ozawa E. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum. Mol. Genet. 1999;8:1589–1598. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- 18.Iwata Y, Nakamura H, Mizuno Y, Yoshida M, Ozawa E, Shigekawa M. Defective association of dystrophin with sarcolemmal glycoproteins in the cardiomyopathic hamster heart. FEBS Lett. 1993;329:227–231. doi: 10.1016/0014-5793(93)80227-l. [DOI] [PubMed] [Google Scholar]
- 19.Roberds SL, Ervasti JM, Anderson RD, Ohlendieck K, Kahl SD, Zoloto D, Campbell KP. Disruption of the dystrophin-glycoprotein complex in the cardiomyopathic hamster. J. Biol. Chem. 1993;268:11496–11499. [PubMed] [Google Scholar]
- 20.Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24:1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 21.Holt KH, Campbell KP. Assembly of the sarcoglycan complex. Insights for muscular dystrophy. J. Biol. Chem. 1998;273:34667–34670. doi: 10.1074/jbc.273.52.34667. [DOI] [PubMed] [Google Scholar]
- 22.Crosbie RH, Lim LE, Moore SA, Hirano M, Hays AP, Maybaum SW, Collin H, Dovico SA, Stolle CA, Fardeau M, et al. Molecular and genetic characterization of sarcospan: insights into sarcoglycan-sarcospan interactions. Hum. Mol. Genet. 2000;9:2019–2027. doi: 10.1093/hmg/9.13.2019. [DOI] [PubMed] [Google Scholar]
- 23.Lebakken CS, Venzke DP, Hrstka RF, Consolino CM, Faulkner JA, Williamson RA, Campbell KP. Sarcospan-deficient mice maintain normal muscle function. Mol. Cell. Biol. 2000;20:1669–1677. doi: 10.1128/mcb.20.5.1669-1677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, McNally EM. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J. Cell Biol. 1998;142:1279–1287. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaoka T, Imamura M, Araishi K, Noguchi S, Mizuno Y, Takagoshi N, Hama H, Wakabayashi-Takai E, Yoshimoto-Matsuda Y, Nonaka I, et al. Pathological analysis of muscle hypertrophy and degeneration in muscular dystrophy in gamma-sarcoglycan-deficient mice. Neuromuscul. Disord. 2003;13:193–206. doi: 10.1016/s0960-8966(02)00220-1. [DOI] [PubMed] [Google Scholar]
- 26.Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, et al. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 27.Hack AA, Lam MY, Cordier L, Shoturma DI, Ly CT, Hadhazy MA, Hadhazy MR, Sweeney HL, McNally EM. Differential requirement for individual sarcoglycans and dystrophin in the assembly and function of the dystrophin-glycoprotein complex. J. Cell Sci. 2000;113(Pt 14):2535–2544. doi: 10.1242/jcs.113.14.2535. [DOI] [PubMed] [Google Scholar]
- 28.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwan KM. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32:49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- 31.Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 32.Shehee WR, Oliver P, Smithies O. Lethal thalassemia after insertional disruption of the mouse major adult beta-globin gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3177–3181. doi: 10.1073/pnas.90.8.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda R, Nishikawa A, Tanaka H. Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J. Biochem. (Tokyo) 1995;118:959–964. doi: 10.1093/jb/118.5.959. [DOI] [PubMed] [Google Scholar]
- 34.Straub V, Rafael JA, Chamberlain JS, Campbell KP. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J. Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruning JC, Michael MD, Winnay JN, Hayashi T, Horsch D, Accili D, Goodyear LJ, Kahn CR. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 37.Allamand V, Donahue KM, Straub V, Davisson RL, Davidson BL, Campbell KP. Early adenovirus-mediated gene transfer effectively prevents muscular dystrophy in alpha-sarcoglycan-deficient mice. Gene Ther. 2000;7:1385–1391. doi: 10.1038/sj.gt.3301247. [DOI] [PubMed] [Google Scholar]
- 38.Bushby KM. Making sense of the limb-girdle muscular dystrophies. Brain. 1999;122(Pt 8):1403–1420. doi: 10.1093/brain/122.8.1403. [DOI] [PubMed] [Google Scholar]
- 39.Roberds SL, Campbell KP. Adhalin mRNA and cDNA sequence are normal in the cardiomyopathic hamster. FEBS Lett. 1995;364:245–249. doi: 10.1016/0014-5793(95)00395-p. [DOI] [PubMed] [Google Scholar]
- 40.Bueno MR, Moreira ES, Vainzof M, Chamberlain J, Marie SK, Pereira L, Akiyama J, Roberds SL, Campbell KP, Zatz M. A common missense mutation in the adhalin gene in three unrelated Brazilian families with a relatively mild form of autosomal recessive limb-girdle muscular dystrophy. Hum. Mol. Genet. 1995;4:1163–1167. doi: 10.1093/hmg/4.7.1163. [DOI] [PubMed] [Google Scholar]
- 41.Hackman P, Juvonen V, Sarparanta J, Penttinen M, Aarimaa T, Uusitalo M, Auranen M, Pihko H, Alen R, Junes M, et al. Enrichment of the R77C alpha-sarcoglycan gene mutation in Finnish LGMD2D patients. Muscle Nerve. 2005;31:199–204. doi: 10.1002/mus.20267. [DOI] [PubMed] [Google Scholar]
- 42.Angelini C, Fanin M, Menegazzo E, Freda MP, Duggan DJ, Hoffman EP. Homozygous alpha-sarcoglycan mutation in two siblings: one asymptomatic and one steroid-responsive mild limb-girdle muscular dystrophy patient. Muscle Nerve. 1998;21:769–775. doi: 10.1002/(sici)1097-4598(199806)21:6<769::aid-mus9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Duggan DJ, Fanin M, Pegoraro E, Angelini C, Hoffman EP. alpha-Sarcoglycan (adhalin) deficiency: complete deficiency patients are 5% of childhood-onset dystrophin-normal muscular dystrophy and most partial deficiency patients do not have gene mutations. J. Neurol. Sci. 1996;140:30–39. doi: 10.1016/0022-510x(96)00028-7. [DOI] [PubMed] [Google Scholar]
- 44.Duggan DJ, Gorospe JR, Fanin M, Hoffman EP, Angelini C. Mutations in the sarcoglycan genes in patients with myopathy. N. Engl. J. Med. 1997;336:618–624. doi: 10.1056/NEJM199702273360904. [DOI] [PubMed] [Google Scholar]
- 45.Bross P, Corydon TJ, Andresen BS, Jorgensen MM, Bolund L, Gregersen N. Protein misfolding and degradation in genetic diseases. Hum. Mutat. 1999;14:186–198. doi: 10.1002/(SICI)1098-1004(1999)14:3<186::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 46.Erickson RR, Dunning LM, Holtzman JL. The effect of aging on the chaperone concentrations in the hepatic, endoplasmic reticulum of male rats: the possible role of protein misfolding due to the loss of chaperones in the decline in physiological function seen with age. J. Gerontol. A. Biol. Sci. Med. Sci. 2006;61:435–443. doi: 10.1093/gerona/61.5.435. [DOI] [PubMed] [Google Scholar]
- 47.Choi BH, Kim JS. Age-related decline in expression of calnexin. Exp. Mol. Med. 2004;36:499–503. doi: 10.1038/emm.2004.63. [DOI] [PubMed] [Google Scholar]
- 48.Beckmann JS, Brown RH, Muntoni F, Urtizberea A, Bonnemann C, Bushby KM. 66th/67th ENMC sponsored international workshop: The limb-girdle muscular dystrophies, 26–28 March 1999, Naarden, The Netherlands. Neuromuscul. Disord. 1999;9:436–445. doi: 10.1016/s0960-8966(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 49.Bartoli M, Gicquel E, Barrault L, Soheili T, Malissen M, Malissen B, Vincent-Lacaze N, Perez N, Udd B, Danos O, et al. Mannosidase I inhibition rescues the human alpha-sarcoglycan R77C recurrent mutation. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn029. in press. [DOI] [PubMed] [Google Scholar]
- 50.Fougerousse F, Bartoli M, Poupiot J, Arandel L, Durand M, Guerchet N, Gicquel E, Danos O, Richard I. Phenotypic Correction of alpha-Sarcoglycan Deficiency by Intra-arterial Injection of a Muscle-specific Serotype 1 rAAV Vector. Mol. Ther. 2007;15:53–61. doi: 10.1038/sj.mt.6300022. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Eastman EM, Schwartz RJ, Draghia-Akli R. Synthetic muscle promoters: activities exceeding naturally occurring regulatory sequences. Nat. Biotechnol. 1999;17:241–245. doi: 10.1038/6981. [DOI] [PubMed] [Google Scholar]
- 52.Lee AH, Suh YS, Sung JH, Yang SH, Sung YC. Comparison of various expression plasmids for the induction of immune response by DNA immunization. Mol. Cells. 1997;7:495–501. [PubMed] [Google Scholar]
- 53.Chen P, Tian J, Kovesdi I, Bruder JT. Promoters influence the kinetics of transgene expression following adenovector gene delivery. J. Gene Med. 2007 doi: 10.1002/jgm.1127. 10.1002/jgm.1127. [DOI] [PubMed] [Google Scholar]
- 54.Roberds SL, Anderson RD, Ibraghimov-Beskrovnaya O, Campbell KP. Primary structure and muscle-specific expression of the 50-kDa dystrophin-associated glycoprotein (adhalin) J. Biol. Chem. 1993;268:23739–23742. [PubMed] [Google Scholar]
- 55.Durbeej M, Campbell KP. Biochemical characterization of the epithelial dystroglycan complex. J. Biol. Chem. 1999;274:26609–26616. doi: 10.1074/jbc.274.37.26609. [DOI] [PubMed] [Google Scholar]
- 56.Cormack B. Mutagenesis of cloned DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York, NY: John Wiley & Sons, Inc.; 1997. pp. 8.5.7–8.5.9. [Google Scholar]
- 57.Ohlendieck K, Ervasti JM, Snook JB, Campbell KP. Dystrophin-glycoprotein complex is highly enriched in isolated skeletal muscle sarcolemma. J. Cell Biol. 1991;112:135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Percy ME, Chang LS, Murphy EG, Oss I, Verellen-Dumoulin C, Thompson MW. Serum creatine kinase and pyruvate kinase in Duchenne muscular dystrophy carrier detection. Muscle Nerve. 1979;2:329–339. doi: 10.1002/mus.880020503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.