SUMMARY
Ubiquitin-mediated inactivation of caspases has long been postulated to contribute to the regulation of apoptosis. However, detailed mechanisms and functional consequences of caspase ubiquitylation have not been demonstrated. Here we show that the Drosophila Inhibitor of Apoptosis 1, DIAP1, blocks effector caspases by targeting them for polyubiquitylation and nonproteasomal inactivation. We demonstrate that the conjugation of ubiquitin to drICE suppresses its catalytic potential in cleaving caspase substrates. Our data suggest that ubiquitin conjugation sterically interferes with substrate entry and reduces the caspase’s proteolytic velocity. Disruption of drICE ubiquitylation, either by mutation of DIAP1’s E3 activity or drICE’s ubiquitin-acceptor lysines, abrogates DIAP1’s ability to neutralize drICE and suppress apoptosis in vivo. We also show that DIAP1 rests in an “inactive” conformation that requires caspase-mediated cleavage to subsequently ubiquitylate caspases. Taken together, our findings demonstrate that effector caspases regulate their own inhibition through a negative feedback mechanism involving DIAP1 “activation” and nondegradative polyubiquitylation.
INTRODUCTION
The covalent attachment of ubiquitin (Ub) to target proteins critically influences cellular processes such as NF-κB signaling, DNA repair, and cell survival (Haglund and Dikic, 2005). At the molecular level, Ub conjugation can alter a protein’s conformation and/or binding properties, changing its activity, localization, or half-life.
Ub chains are assembled in a stepwise process that involves Ub-activating enzymes (E1), Ub-conjugating enzymes (E2), and Ub-protein ligases (E3) (Hershko et al., 2000). E3s confer substrate specificity and enable the transfer of Ub from an E2 to lysine (K) residues on target substrates (Hershko et al., 2000). Ub is conjugated either as a single moiety (mono-Ub) or as poly-Ub chains (Haglund and Dikic, 2005) that are generally linked through K48 or K63 of Ub. The different types of poly-Ub chains have diverse effects, K48-linked modifications promoting degradation through recognition by the 26S proteasome and mono-Ub, as well as K63-linkages, contributing to a variety of nondegradative signaling processes (Hoeller et al., 2006).
A large body of evidence demonstrates the involvement of Ub conjugation in apoptosis regulation (Hoeller et al., 2006; Vaux and Silke, 2005). Protein levels of many key regulatory molecules, including p53 (Brooks and Gu, 2006), IAPs (Vaux and Silke, 2005), Bim (Ley et al., 2003), Mcl1 (Zhong et al., 2005), and c-FLIP (Chang et al., 2006), are controlled by Ub-dependent degradation. With regards to caspase regulation, the picture is less clear. Caspases are regulated by members of the IAP protein family, and IAP-mediated mono- and polyubiquitylation of caspases has been reported for a number of IAPs and caspases (Vaux and Silke, 2005). However, biochemical insights into the actual molecular and functional consequences of caspase ubiquitylation are missing.
Caspases are expressed as zymogens consisting of a prodomain and a large (p20) and small subunit (p10) (Riedl and Shi, 2004). Proteolytic activation is initiated by the activation of initiator caspases that cleave and activate downstream effector caspases. In a positive feedback loop, effector caspases activate other caspases and amplify the proteolytic activity (Berger et al., 2006; Denault et al., 2006). During activation, their prodomains are removed by cleavage. The position of the initial cleavage differs among caspases; while caspase-3 is first cleaved at the interdomain-linker position, caspase-7 and the Drosophila effector caspases drICE and DCP-1 are first processed at the prodomain site generating ΔN-caspases (Denault and Salvesen, 2003; Riedl and Shi, 2004; Tenev et al., 2005).
Certain IAPs inhibit caspases and can function as a last line of defense against caspase-mediated damage. IAPs are classified by the presence of the Baculovirus IAP repeat (BIR) domain (Vaux and Silke, 2005). Drosophila DIAP1 and mammalian XIAP bind to both initiator and effector caspases via their BIR domains and adjacent flanking regions. In most cases, binding to BIR domains is mediated by an IAP-binding motif (IBM) present in IAP antagonists (Shi, 2002), such as Reaper (Rpr), or caspases such as drICE, DCP-1, and caspase-7 and -9 (Denault and Salvesen, 2003; Srinivasula et al., 2001; Tenev et al., 2005). Generally, IBMs carry an NH2-terminal Alanine (A) residue at position 1 that anchors this motif into the BIR domain (Wu et al., 2001). Upon processing, the IBMs of drICE and DCP-1 become exposed, allowing their detection by IAPs (Tenev et al., 2005). Although required for DIAP1 binding, fully active effector caspases associate with DIAP1 through a bimodular interaction involving both the IBM and the catalytic pocket.
DIAP1 reportedly inhibits caspases in vitro (Kaiser et al., 1998; Yan et al., 2004). However, physical interaction alone is insufficient to maintain cell viability in vivo. Mutations of DIAP1’s RING-finger domain, which abrogate its E3 activity but not caspase binding, cause a loss-of-function phenotype (Lisi et al., 2000; Wilson et al., 2002). Following binding, the RING is required to ubiquitylate and inactivate Drosophila initiator caspase DRONC (Chai et al., 2003; Wilson et al., 2002). Moreover, effector caspases are also neutralized in a RING-dependent manner (this study and Herman-Bachinsky et al. [2007] and Lisi et al. [2000]). However, while necessary, the RING is not sufficient to inhibit caspases as recruitment of the NH2-end-rule Ub-associated machinery is also required (Ditzel et al., 2003). Effector caspases cleave DIAP1 at its NH2 terminus, which exposes a binding motif for UBR-domain-bearing class of NH2-end-rule E3s (UBR-E3s) (Ditzel et al., 2003; Herman-Bachinsky et al., 2007; Yan et al., 2004; Yokokura et al., 2004). Although UBR recruitment and DIAP1’s RING are required for DIAP1’s antiapoptotic activity (Ditzel et al., 2003; Muro et al., 2005), the mechanism and consequence of caspase ubiquitylation remain unclear. IAP-mediated ubiquitylation of caspases is not restricted to Drosophila, as mammalian XIAP reportedly polyubiquitylates caspase-3, targeting it for degradation (Suzuki et al., 2001). Moreover, cIAP2 monoubiquitylates caspase-3 and -7 in vitro (Huang et al., 2000). However, the relevance of these observations remains unresolved, since XIAP knockout animals display limited cell death phenotypes (Harlin et al., 2001), and the functional outcome of caspase monoubiquitylation has not been determined. All in all, there are numerous reports that link Ub conjugation to caspase regulation; however, no clear picture has emerged that might help to explain how Ub conjugation inactivates these proteases.
Here we have addressed the molecular mechanism and functional consequence of DIAP1-mediated ubiquitylation of effector caspases. Surprisingly, we found that full-length DIAP1 rests in an “inactive” configuration that precludes caspase binding. Following caspase-mediated cleavage, DIAP1 becomes fully competent to bind caspases and function as an E3. DIAP1-mediated drICE ubiquitylation suppresses its catalytic ability to cleave the substrates PARP and DEVD-AMC. Disruption of drICE ubiquitylation, either by loss of DIAP1’s E3 activity or generation of a nonubiquitylatable form of drICE, renders it resistant to DIAP1-mediated inactivation. Our data provide a likely explanation of how cells maintain buffered levels of active caspases without losing cell viability. It appears that caspases, through cleaving DIAP1, initiate their own inhibition. Such a scenario would result in a buffered “oscillating cycle” of caspase activity that would establish a continuous presence of sublethal levels of active caspases.
RESULTS
Alteration of DIAP1’s Physical Properties through NH2-Terminal Cleavage
To investigate the biochemical basis for caspase inhibition by cleaved DIAP1 (DIAP121–438), we performed binding assays with DRONC, drICE, DCP-1, and Rpr (Figure 1). Nontagged full-length or truncation mutants of DIAP1 were expressed along with V5-tagged caspases or IAP antagonists. α-V5 antibodies were used to purify caspases and IAP antagonist from cellular extracts, and the presence of copurified DIAP1 was detected using an α-DIAP1 antibody. Mammalian cells were chosen to avoid potential interference by endogenous DIAP1 and because wild-type DIAP1 remains largely unprocessed in 293 cells (Figure 1B). Interestingly, DIAP121–438 interacted with DRONC, drICE, and DCP-1 far better than full-length DIAP1, which bound caspases only weakly (Figures 1B–1D). Notably, drICE enriched the small amount of cleaved DIAP1 over its unprocessed form (Figure 1C, lane 1). Under these conditions, drICE barely bound to noncleaved DIAP1 or DIAP1D20A, which remains full-length due to a mutation in the caspase cleavage site. Removal of DIAP1’s N peptide, but not generation of a particular neo-NH2 terminus, was required for caspase binding. Both, N- and M-DIAP121–438, in which N21 was replaced by M21, and therefore does not bind NH2-end-rule E3s (see below), associated with caspases equally well (Figures 1B–1D). As expected, Rpr equally bound to DIAP1wt, DIAP1D20A, and DIAP121–438 (Figure 1E), a finding that is consistent with its epistatic position upstream of DIAP1 (Wang et al., 1999).
Figure 1. DIAP1 Cleavage Enhances Caspase Binding.

(A) Model depicting DIAP1’s hypothetical “closed” and “open” configuration. A vertical arrow marks the site of caspase cleavage.
(B–E) Coimmunoprecipitation (IP) assays with the indicated constructs. Lysates and V5 immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. The two arrows on the DIAP1 panels indicate the positions of full-length and cleaved DIAP1.
Since DIAP1 bears Alanine at the second position (A2), it was previously suggested that A2 mediates intramolecular binding to the BIR domains (Yan et al., 2004), thereby locking DIAP1 into a “closed” conformation that prevents caspase binding. However, DIAP1A2G, in which A2 was replaced by glycine (G) that does not allow BIR binding (Wu et al., 2001), also failed to interact with DCP-1 (see Figure S1 available online). This indicates that A2 does not contribute to the “closed” configuration of DIAP1.
The Neo-NH2 Terminus of Cleaved DIAP1 Binds to the UBR Box Motif of E3 Ligases
DIAP1 cleavage exposes a previously hidden “docking” site for E3 ligases with a UBR domain. UBR domains generally recognize target proteins via NH2-terminal residues (Tasaki et al., 2005). For N21 of DIAP121–438 to bind to UBR domains, it must be converted to Aspartate (D) by NH2-terminal amidohydrolases and conjugated with Arginine (R) to create the mature NH2-terminal RD21 motif (Figure 2A; Varshavsky et al., 2000). Because of their size (200 kDa for UBR1), we used isolated UBR domains to determine which of the six Drosophila UBR-bearing E3s bind DIAP1. The UBR domains of UBR1, -3, -5, -6, and -7 bound to DIAP121–438, whereas UBR4 showed no detectable interaction (Figure 2B). The binding between DIAP121–438 and UBR1 was strictly dependent on a compatible NH2-terminal. M-DIAP121–438 in which N21 was replaced by methionine (M), an Aa that does not allow UBR binding (Tasaki et al., 2005), failed to copurify with UBR1 (Figure 2C). Under the same conditions, UBR1 readily bound to both N-DIAP121–438 and RD-DIAP121–438, which mimics a fully matured NH2-terminal UBR-binding motif. The observation that several UBRs bind to DIAP121–438 suggests significant redundancy among UBRs, a view that is supported by knockout studies in mice (Kwon et al., 2003).
Figure 2. Cleaved DIAP1 Binds to UBR Domains and Promotes Ubiquitylation of drICE and DCP-1.
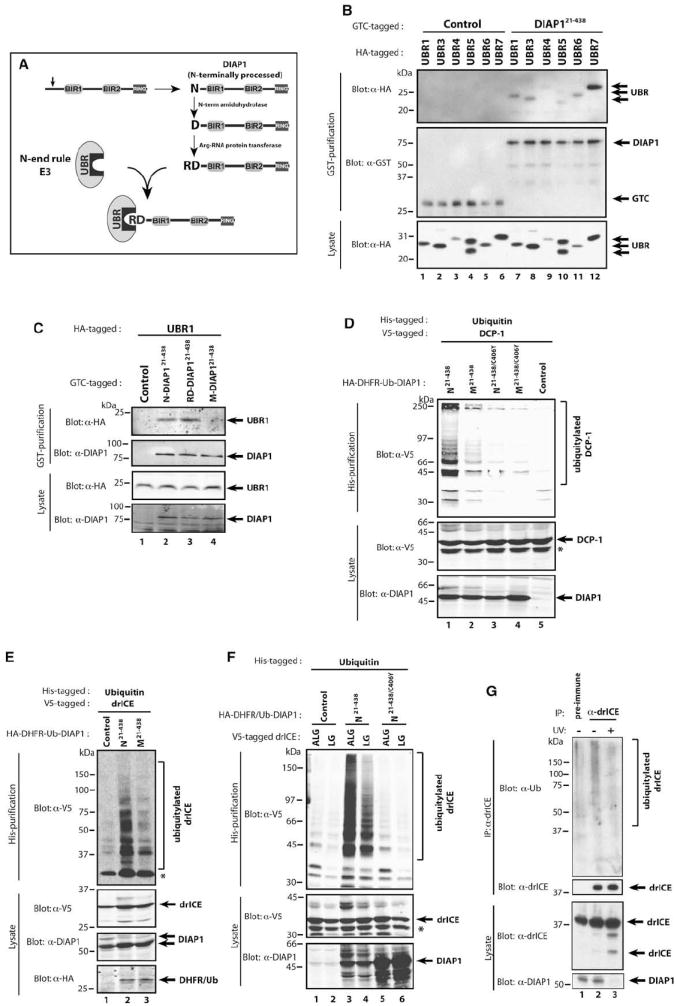
(A) Schematic representation of caspase-mediated DIAP1 cleavage, maturation, and UBR binding. See text for details.
(B) Co-IP assays with DIAP121–438 and various UBR domains. S2 cells were cotransfected with GTC-tagged DIAP121–438 and HA-tagged UBR domains of the indicated UBR proteins. Copurified proteins were analyzed by immunoblotting with the indicated antibodies.
(C) Co-IPs were performed as in (B) with N-, RD-, and M-DIAP121–438 and the UBR domain of UBR1.
(D and E) Requirement of DIAP1’s RING domain and N-exposure in mediating effector caspase ubiquitylation. 293T (D) or S2 (E) cells were cotransfected with the indicated DIAP1 constructs together with Ub-AKG-DCP-1C>A (D) or Ub-ALG-drICEC>A (E) and His-Ub. Cells were lysed under denaturing conditions and ubiquitylated proteins isolated with Ni2+ columns. The presence of ubiquitylated caspases was identified by immunoblotting with the indicated antibodies.
(F) DIAP1 and drICE bind through a bimodular interaction. Mutation of the IBM weakens but does not abrogate DIAP1 binding. Ubiquitylation assays with the indicated constructs were performed as in (D).
Asterisks mark a nonspecific band ([D] and [F]) and unmodified drICE (E) that is due to a nonspecific drICE:matrix association.
(G) Endogenous drICE is ubiquitylated in a DIAP1-dependent manner. S2 cells were either left untreated or exposed to UV, and α-drICE or preimmune serum was used to immunoprecipitate drICE from cellular extracts. The presence of ubiquitylated endogenous drICE was determined by immunoblotting with an α-Ub antibody (top panel). Note, treatment with UV causes depletion of DIAP1 (bottom panel, lane 3).
DIAP121–438 Targets Effector Caspases for Polyubiquitylation in a RING and UBR Binding-Dependent Manner
Next, we examined the ability of DIAP1 to promote caspase ubiquitylation (Figures 2D–2F). Nontagged DIAP121–438 was coexpressed with V5-tagged DCP-1 or drICE and His-tagged Ub. Ubiquitylated proteins were affinity purified under denaturing conditions using nickel columns, and the presence of ubiquitylated caspases was assessed by immunoblotting the eluate with α-V5 antibodies. Under these conditions, DIAP121–438 readily ubiquitylated DCP-1 and drICE (Figures 2D–2F). In contrast, M-DIAP121–438 and the DIAP1 RING mutant DIAP121–438/C406Y were impaired in their ability to ubiquitylate DCP-1 and drICE (Figures 2D–2F). Mutation of drICE’s IBM (ALG- to LG-drICE), which weakens DIAP1 binding (Tenev et al., 2005), resulted in less efficient ubiquitylation (Figure 2F). Consistent with the notion that DIAP1 ubiquitylates drICE, we found that, in healthy S2 cells, endogenous drICE was ubiquitylated in a DIAP1-dependent manner (Figure 2G). This is evident because exposure to UV or treatment with a small pharmacological inhibitor of IAPs that mimics SMAC caused DIAP1 depletion and resulted in reduced levels of ubiquitylated drICE (Figure 2G; M.B. and P.M., unpublished data). These results indicate that DIAP1 ubiquitylates effector caspases and that both exposure of a UBR-binding motif and the presence of a functional RING finger are required for maximal ubiquitylation of effector caspases.
Exposure of an UBR-Binding Motif and DIAP1’s Own RING Finger Are Both Required for DIAP1’s Antiapoptotic Activity In Vivo
To address the contribution of DIAP1’s NH2-terminal UBR-binding domain for its antiapoptotic activity in vivo, we generated a number of transgenic fly lines expressing various DIAP1 mutants under the control of the tubulin promoter. We used the Ub-protein reference technique (Varshavsky, 2000) (Figure 3A) and quantitative, infrared fluorescence-based measurement (Odyssey, LI-COR) of V5-DHFR/Ub to select lines with similar transgene expression levels (Figure 3B).
Figure 3. DIAP1 Requires Cleavage and a Functional RING to Suppress Apoptosis In Vivo.
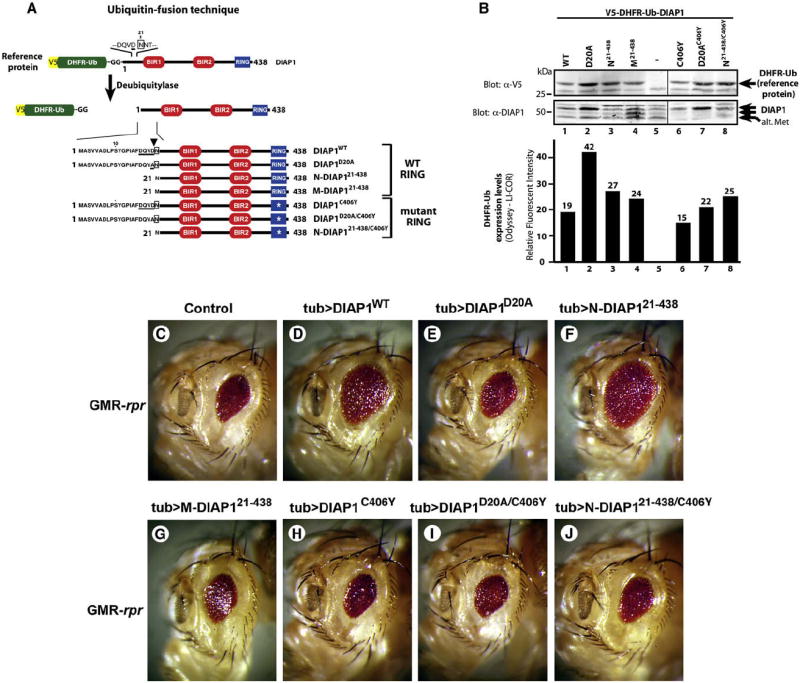
(A) Overview of the Ub fusion technique. Tubulin-driven DIAP1 variants were expressed as V5-DHFR/Ub-DIAP1 fusion in which the reference protein DHFR/Ub is cotranslationally cleaved off by DUBs. Expression of the reference protein indirectly indicates the expression level of the protein of interest. DIAP1’s NH2-terminal sequence and the caspase cleavage site (DQVD, arrowhead) are indicated.
(B) Selection of transgenic lines with “near” equivalent expression levels of the reference protein (upper panel). Quantitative, infrared fluorescence-based measurement of V5-DHFR-Ub expression levels (lower panel).
(C–J) The ability of DIAP1 to suppress the eye phenotypes of Rpr expression. Rpr expression results in a small-eye phenotype (C) that is rescued by tubulin-driven coexpression of DIAP1wt (D) and N-DIAP121–438 (F), but not DIAP1D20A (E), M-DIAP121–438 (G), or RING mutants (H–J). Representative phenotypes are shown.
Eye-specific (GMR) expression of Rpr causes apoptosis in the developing retina that results in a small-eye phenotype (White et al., 1996) (Figure 3C). Tubulin-driven expression of DIAP1wt and N-DIAP121–438 suppressed this eye phenotype (Figures 3D and 3F). In contrast, DIAP1D20A and M-DIAP121–438 failed to provide any significant protection (Figures 3E and 3G), despite DIAP1D20A being expressed 1.5-fold above the levels of N-DIAP121–438 and 2.2-fold above that of DIAP1wt (Figure 3B). Notably, all DIAP1 variants showed comparable binding to Rpr (Figure 1E; Zachariou et al., 2003). Interestingly, N-DIAP121–438/C406Y, which lacks a functional RING, failed to inhibit Rpr-mediated cell death (Figure 3J). Thus, both exposure of an UBR-binding motif and the presence of a functional RING are essential for DIAP1’s antiapoptotic activity. Either mechanism alone fails to protect from Rpr killing.
DIAP1-Mediated Inhibition of drICE Depends on Binding- and Ub-Dependent Activities
To quantitate DIAP1’s ability to suppress caspases, we used the fluorometric caspase substrate Ac-DEVD-AMC in a cell-based assay. In Drosophila tissue culture cells (S2), like in the developing eye, induced expression of Rpr triggers apoptosis that is predominantly mediated by drICE (Fraser et al., 1997; Leulier et al., 2006; Xu et al., 2006) and is efficiently blocked by coexpression of DIAP1wt. Notably, in this system, DIAP1 acts through inhibiting effector caspases (Figure S2; Tenev et al., 2007; and data not shown). DIAP1 efficiently suppressed DEVDase activity induced by Rpr expression (Figure 4A). Cleavable and E3-proficient DIAP1wt and N-DIAP121–438 were most effective at suppressing DEVDase activity. In contrast, DIAP1D20A, M-DIAP121–438, or the E3 mutants DIAP121–438/C406Y, DIAP121–438/CΔ6, and DIAP121–438/F437A were strongly impaired in suppressing DEVDase activity. DIAP121–438/CΔ6 and DIAP121–438/F437A are defective in their E3 activity because they either lack the last 6 Aa (CΔ6) or carry a mutation in F437 essential for E3 activity (Silke et al., 2005). Consistently, DIAP121–438/CΔ6, DIAP121–438/F437A, and DIAP121–438/C406Y failed to polyubiquitylate drICE (Figures 2D, 2F, and 4B). Importantly, these E3 mutants bound to drICE as efficiently as DIAP121–438 (data not shown), indicating that DIAP1’s E3 activity is required to suppress caspase activity. The small, but measurable, residual ability of these mutants to suppress DEVDase activity (Figures 4A and 4C) is due to mere physical interaction (Yan et al., 2004). Therefore, DIAP1-mediated regulation of effector caspases results from a binding-and Ub-dependent activity.
Figure 4. DIAP1-Mediated Ubiquitylation of drICE Correlates with Its Inactivation.
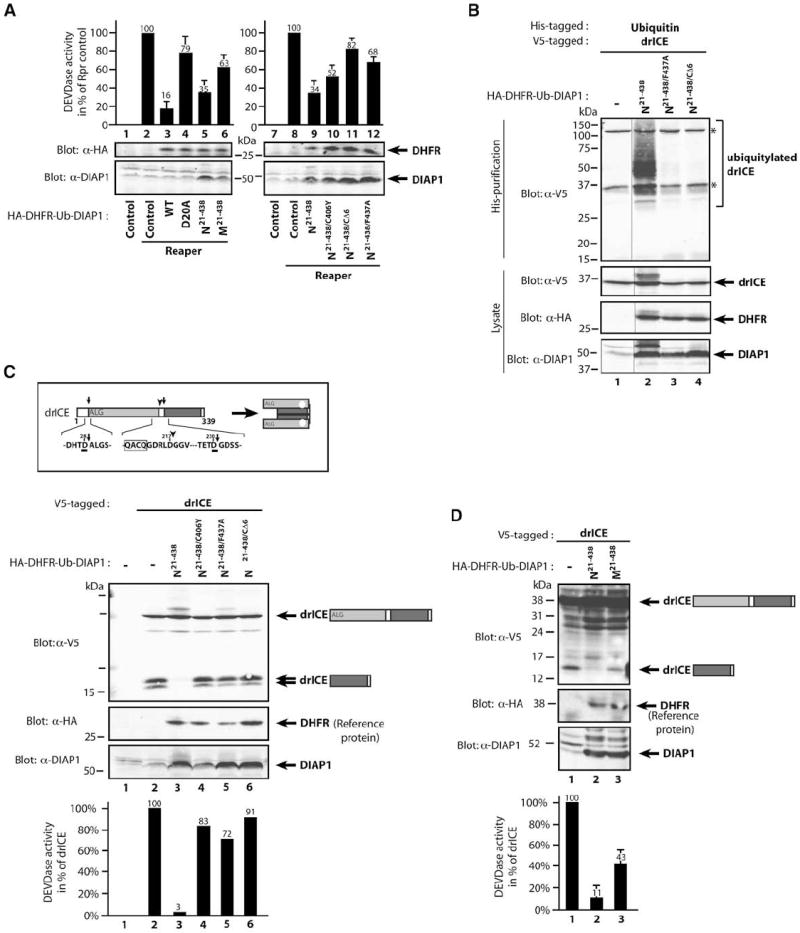
(A) The E3 ligase activity of DIAP1 is required to suppress effector caspase activity induced by Rpr. S2 cells were cotransfected with the indicated constructs, and cell lysates were assayed for DEVDase activity. Values of Rpr-expressing controls were set to 100% (lanes 2 and 8). Immunoblot analysis of the lysates indicates near-equivalent expression levels of the DHFR reference protein (upper panels).
(B) RING-dependent ubiquitylation of drICE. Ubiquitylation assays were performed as in Figure 2E. Asterisks represent nonspecific bands.
(C) DIAP1’s E3 activity is required to suppress appearance of processed drICE (p10) and DEVDase activity. Schematic representation of cleavage-mediated activation of drICE (upper panel). Arrows indicate cleavage sites. DIAP1 coexpression, but not the indicated range of ubiquitylation-deficient DIAP1 mutants, efficiently prevents detection of drICE’s small subunit. S2 cells were cotransfected with the indicated constructs. Following a 6 hr induction of drICE, cell lysates were simultaneously analyzed for DEVDase activity and by immunoblotting with the indicated antibodies. Values were normalized to drICE in vector controls (lane 2). DEVDase values and immunoblot analysis of a representative experiment are shown.
(D) M-DIAP121–438 is less efficient than N-DIAP121–438 in suppressing drICE activation and activity, as indicated by the presence of the p10 subunit of drICE and significant amounts of DEVDase activity. Experiments were performed as in (C). DEVDase values and immunoblot analysis of a representative experiment are shown. Error bars denote SD from three independent experiments.
drICE is activated through cleavage at D230 (Figure 4C), which occurs upon apoptotic insult or induced expression of drICE. A short 6 hr pulse of drICE induction resulted in its proteolytic activation, as measured by the appearance of the p10 in immunoblot analysis and DEVDase activity (Figure 4C). Of note, under these conditions, no apoptosis or morphological changes are detectable, and the majority of drICE is activated by autocleavage. Expression of catalytically inactive drICEC>A did not result in its cleavage (data not shown). The observation that effector caspases can feed back on themselves is consistent with activation of caspase-7, whose processing is by and large mediated by mature caspase-7 (Berger et al., 2006; Denault et al., 2006). When drICEwt was coexpressed with N-DIAP121–438, processed drICE was undetectable, and very little DEVDase activity was measured (Figure 4C). In contrast, processed drICE and DEVDase activity were detectable in the presence of N-DIAP121–438/C406Y, N-DIAP121–438/CΔ6, N-DIAP121–438/F437A, or M-DIAP121–438 (Figure 4C and 4D). Thus, DIAP1 that fails to ubiquitylate drICE efficiently is impaired in suppressing it.
DIAP1 Targets drICE for Polyubiquitylation and Inactivation through a Nondegradative Mechanism
drICE may be inactivated through Ub-mediated proteasomal degradation. According to such a scenario, proteasome inhibition should result in accumulation of ubiquitylated drICE, and appearance of its small subunit despite the presence of DIAP1. However, Lactacystin treatment did not cause accumulation of ubiquitylated drICE when compared to untreated controls (Figure 5A). Moreover, exposure to cycloheximide and/or MG132, another proteasome inhibitor, did not change the ubiquitylation pattern of drICE (Figure S3) or restore the appearance of the p10 (Figure 5B). Further, clonal overexpression of DIAP1 in the developing eye did not result in drICE depletion (Figure 5C), indicating that DIAP1 does not target drICE for proteasomal destruction. Moreover, in vitro ubiquitylation assays suggest that DIAP1 is not only capable, but better at promoting nondegradative K63-linked polyUB chains than degradative K48-linked chains (Herman-Bachinsky et al., 2007). Although DIAP1 can promote K48- and K63-linked polyUB chains on drICE, DIAP1 makes longer K63-Ub chains (Figure S4, compare lane 2 with 4 and 5).
Figure 5. Ubiquitylation of drICE Suppresses Its Catalytic Potential.
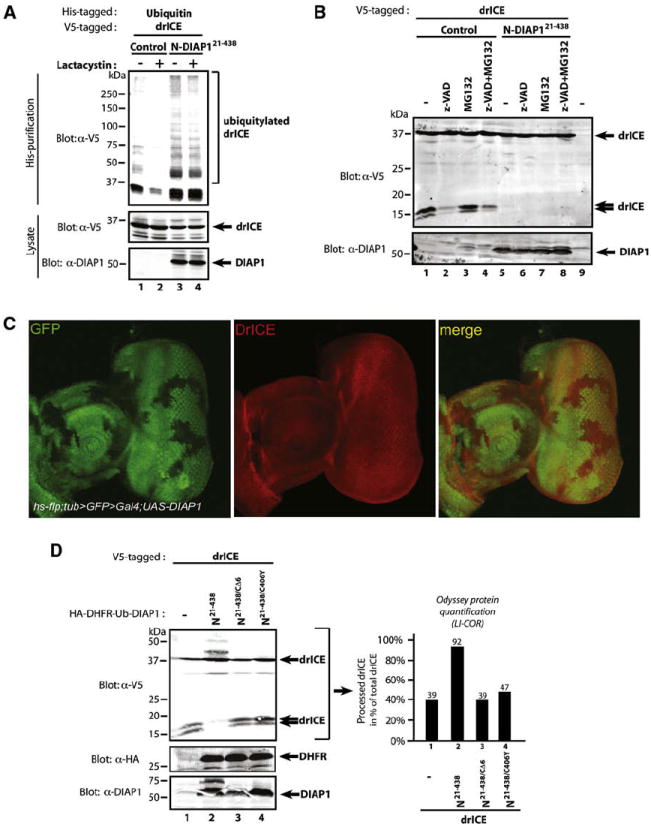
(A) Inhibition of the proteasome does not alter the ubiquitylation pattern of drICE. 293T cells were cotransfected with the indicated constructs and treated with Lactacystin or DMSO (control). The presence of ubiquitylated drICE was assayed as in Figure 2E.
(B) Proteasome inhibition failed to restore the appearance of the small subunit of drICE. S2 cells were cotransfected with the indicated constructs. Following a 2 hr drICE induction, cells were treated for a further 4 hr with MG132 or DMSO.
(C) Clonal expression of DIAP1 in the developing eye does not reduce the levels of drICE. Eye discs overexpressing DIAP1 in clones (marked by the absence of GFP, first and last panel) were stained using an α-drICE antibody (red, middle panel).
(D) S2 cells were cotransfected with the indicated constructs. Immunoblot analysis (left panel) revealed that zymogenic ALG-drICE accumulated at the expense of processed drICE (p10) in the presence of DIAP1. LI-COR Odyssey quantification (right panel) of the drICE signal.
Consistent with the notion that DIAP1 blocks drICE processing, we found that drICE remained predominantly unprocessed (92%) when DIAP1 was coexpressed (Figure 5D). However, in the absence of DIAP1 most drICE was cleaved, with only 35% of total drICE remaining full-length. drICE was also activated (processed) in the presence of DIAP121–438/CΔ6 and DIAP121–438/C406Y. Since cleavage of drICE requires an input from active drICE, it appears that DIAP1-mediated ubiquitylation of drICE results in its inactivation. Similarly, z-VAD-fmk also suppressed the appearance of the small subunit of drICE (Figure 5B).
Ub Conjugation Inhibits Active drICE
To test whether ubiquitylation directly affects the proteolytic activity of drICE, we devised an in vitro ubiquitylation assay followed by a cleavage reaction using PARP as the caspase substrate (Figures 6A and 6B). Several slow migrating drICE species were detected when recombinant, active drICE was incubated with E1, E2/UbcD1, Ub, and DIAP1, which were not detected in the presence of the E3 mutant DIAP1F437A—indicating that they are ubiquitylated products (Figure 6A). The mixture containing either ubiquitylated or nonubiquitylated drICE was subsequently incubated with PARP (Figures 6A and 6B) or DEVD-AMC (Figure 6C). While recombinant drICE cleaved 89% of PARP, the mixture containing ubiquitylated drICE was significantly less active in processing PARP, with only 42% of PARP being cleaved (Figure 6A). In contrast, nonubiquitylated drICE incubated with E3-defective DIAP1F437A cleaved PARP as efficiently as controls (80% cleaved) (Figure 6A). Moreover, time course analysis and concentration-dependent cleavage assays with PARP and DEVD-AMC further corroborate the notion that nonubiquitylated and ubiquitylated drICE significantly differ in their KM and Vmax (Figures 6B–6D). Since DIAP1 and DIAP1F437A bind to drICE equally well, but DIAP1F437A fails to ubiquitylate drICE, this result indicates that ubiquitylation significantly reduces the catalytic potential of drICE. This suggests that DIAP1-mediated drICE ubiquitylation leads to its nondegradative suppression, possibly through steric interference with substrate entry (see below and Figure S5). Since ubiquitylation affects KM and Vmax, it seems possible that it functions as a mixed inhibitor, whereby Ub sterically occludes substrate entry and causes a conformational change of the caspase reducing its catalytic processivity.
Figure 6. Conjugation of Ub to drICE Acts as a “Mixed” Inhibitor.
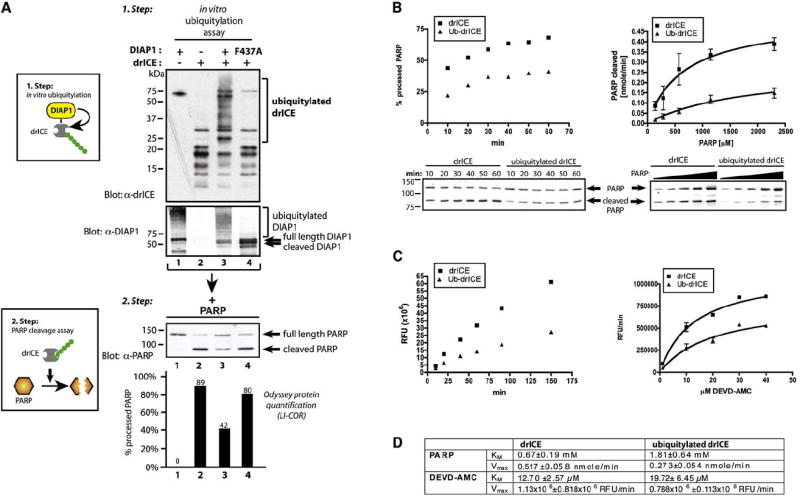
(A) In vitro ubiquitylation of recombinant drICE suppresses its catalytic ability to cleave PARP. Schematic representation of the assay procedure (left panels). In vitro ubiquitylation assay of active drICE with DIAP1 or DIAP1F437A. Arrows on the DIAP1 panel indicate DIAP1 cleavage. (Bottom panel) Step 2 depicting in vitro PARP cleavage assay: the ubiquitylation reactions from step 1 were incubated with recombinant PARP. Shown is immunoblot analysis with α-PARP antibodies. Graphs indicate LI-COR Odyssey quantification of the signal expressed as % of processed PARP in relation to total PARP (lower panel). Values and immunoblot analysis of a representative experiment are shown.
(B and C) Ubiquitylation changes kinetic parameters of active drICE. drICE was incubated in an in vitro ubiquitylation reaction with DIAP1 or DIAPF437A as in Figure 5D. (B) 10.3 nmole of PARP (left panel, a representative experiment is shown) or increasing concentrations of PARP (2.1–34.5 nmole, 15 min, right panel) was incubated with drICE or ubiquitylated drICE. PARP cleavage was monitored by immunoblot analysis. Western blot data were analyzed by LI-COR Odyssey quantification as in Figure 5D. Nonlinear regression using the Michaelis-Menten equation was applied to obtain a curve fit (right panel). (C) drICE was incubated in an in vitro ubiquitylation reaction with DIAP1 or DIAPF437A as in Figure 5D. drICE and ubiquitylated drICE were incubated with 20 μM DEVD-AMC, and DEVDase activity was analyzed at the indicated time points (left panel, shown is a representative experiment). (Right panel) Increasing substrate concentrations were used, and DEVDase activity was measured at 20 min. Curve fit was performed as in (B). RFU, relative fluorescence units.
(D) KM and Vmax (±SE) were determined from the curves shown in (B) and (C) using Prism software.
Nonubiquitylatable drICE Is Refractory to DIAP1-Mediated Inhibition
If ubiquitylation of drICE results in inactivation, then drICE mutants that cannot be ubiquitylated should be resistant to DIAP1-mediated inhibition. To test this, we generated a nonubiquitylatable mutant of drICE. Structural prediction of drICE revealed nine surface exposed K residues (Figures 7A–7E). In vitro ubiquitylation assays with a Ub mutant that lacks all seven lysine residues (K0) and therefore can only form mono-Ub conjugates indicated that DIAP1 can ubiquitylate drICE at five K residues simultaneously (Figure S6A). Sequential, additive rounds of mutagenesis generated a series of K>R mutants (Figure 7F) and revealed that mutation of all nine K residues in the p20 (drICE9K>R) was required to abrogate drICE ubiquitylation (Figure 7G). Under the same conditions, drICEwt, drICE8K>R, and drICEK127R were readily ubiquitylated. Mass spectrometric analysis of ubiquitylated drICE identified at least three ubiquitylated lysines, namely K72, K76, and K142 (Figure S6B). Due to limited coverage, no data were obtained on the ubiquitylation status of other surface-exposed lysine residues.
Figure 7. Nonubiquitylatable drICE Is Refractory to DIAP1-Mediated Inhibition.
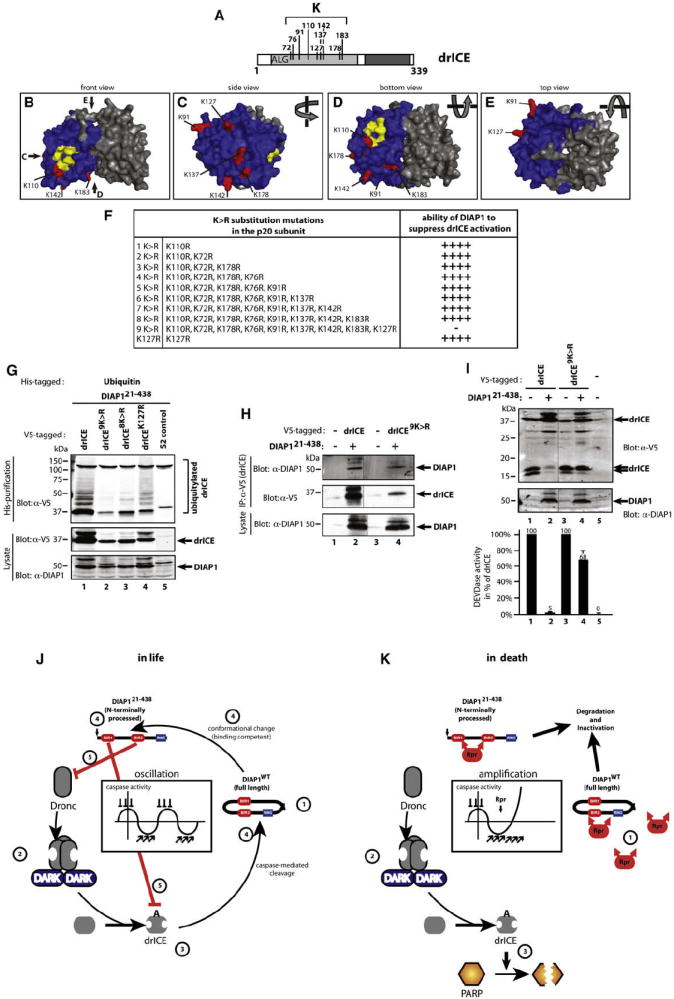
(A) Schematic representation showing the position of surface-exposed K residues in the large subunit.
(B–E) 3D model of drICE dimers. drICE monomers are colored in blue and gray, and surface-exposed K residues are highlighted in red. Residues involved in catalysis are highlighted in yellow. Four different viewpoints of drICE are shown.
(F) K>R mutation schedule. Additive rounds of K>R mutations were carried out, and their effects on DIAP1-mediated regulation of drICE were assessed in cell-based DEVDase assays (as in Figure 4C). “++++” reflects no change and “−” reflects loss of DIAP1-mediated inhibition of drICE.
(G) drICE9K>R is refractory to DIAP1-mediated ubiquitylation. S2 cells were cotransfected with DIAP121–438, His-Ub, and the indicated drICE-V5/FLAG constructs, and cell lysates were analyzed for the presence of ubiquitylated caspases.
(H) drICE9K>R readily binds to DIAP1. Experiment performed as in Figure 1D.
(I) drICE9K>R is refractory to DIAP1-mediated inhibition. drICE9K>R was examined as in Figure S6C. The mean values of triplicate experiments and their SD are shown. Immunoblot analysis of a representative experiment is shown.
(J and K) Model of caspase regulation in living (J) and dying cells (K). See Discussion for details.
Importantly, drICE9K>R interacted with DIAP1 as efficiently as drICEwt (Figure 7H), indicating that the lack of ubiquitylation was not due to impaired DIAP1-binding. Next, we tested the ability of DIAP1 to regulate drICE9K>R. In the absence of DIAP1, expression of all drI-CEK>R mutants resulted in their proteolytic activation, as measured by the appearance of the p10 and DEVDase assays (Figures 7I and S6C). Importantly, proteolytic activation and activity of all these mutants was comparable to the ones of drICEwt (Figures 7I and S6C; and data not shown) indicating that the K>R substitutions preserved the overall caspase structure required for catalytic activity. Coexpression of DIAP121–438 readily suppressed drICEK>R mutants that lacked up to eight K residues (Figure S6C). In contrast, drICE9K>R, which lacks all nine surface-exposed K residues, was significantly refractory to DIAP1-mediated inhibition and efficiently processed its subunits despite the presence of DIAP121–438 (Figure 7I, compare lanes 2 and 4). Under these conditions, drICE9K>R retained nearly 70% of its catalytic activity (Figure 7I), while drICEwt was strongly inhibited, exhibiting merely 5% of its potential activity.
DISCUSSION
Ub-mediated inactivation of caspases has long been postulated to contribute to the regulation of apoptosis. However, detailed biochemical insights into the actual molecular and functional consequences of caspase ubiquitylation have not been demonstrated. Here we provide mechanistic insight into DIAP1’s Ub-dependent ability to inactivate drICE. Unexpectedly, we find that the covalent attachment of Ub to drICE suppresses its catalytic potential through a nondegradative mechanism. Although DIAP1 readily promotes polyubiquitylation of drICE, this does not result in its proteasomal destruction. Several lines of evidence support the notion that drICE is regulated through polyubiquitylation. First, Ub conjugation of drICE in vitro significantly reduces its proteolytic activity toward PARP. Second, drICE9K>R that cannot be ubiquitylated by DIAP1 escapes DIAP1-mediated inhibition, even though DIAP1 binds to drICE9K>R as efficiently as wild-type drICE. Finally, DIAP1 point mutations that abrogate its E3 activity toward drICE, or impair its ability to bind drICE, also fail to suppress drICE in vivo.
The importance of DIAP1’s E3 activity in regulating caspases and cell death is highlighted by diap1 alleles bearing RING mutations. For DIAP1 to maintain cell viability, it needs to synchronously neutralize initiator and effector caspases (Kornbluth and White, 2005). DIAP1 mutations that selectively abrogate binding or ubiquitylation of either initiator or effector caspases cause loss of DIAP1 function. While DIAP1 targets both DRONC and drICE for polyubiquitylation and inactivation, the direct consequences of DRONC ubiquitylation remain to be elucidated (Chai et al., 2003; Wilson et al., 2002). Nevertheless, it is clear that DIAP1 has to interact with DRONC to regulate it. Surprisingly, we find that DIAP1’s ability to bind to DRONC depends on DIAP1 cleavage. Removal of DIAP1’s first 20 Aa residues radically changes DIAP1’s properties, as it interacts with DRONC far better than full-length, noncleaved DIAP1. Thus, full-length DIAP1 seems to reside in an inactive, “closed” configuration that precludes caspase binding (Yan et al., 2004). Only when it is cleaved can it bind tightly to DRONC, or effector caspases. In this respect, DIAP1 may sense caspase activity through caspase-mediated cleavage. Intriguingly, XIAP and cIAP1 reportedly are also cleaved by caspases (Clem, 2001; Deveraux et al., 1999), and it will be interesting to examine whether such cleavages also alter the functional properties of mammalian IAPs.
NH2-terminal cleavage of DIAP1 also substantially changes DIAP1’s E3 activity, as it exposes a new docking site for UBR-containing E3s. Heteromeric E3 protein complexes are frequent, as seen in BRCA1/BARD (Brzovic et al., 2001), cIAP1/TRAF2, cIAP2/TRAF2 (Rothe et al., 1995), and MDM2/MDMX (Tanimura et al., 1999). Our observations indicate that DIAP1 can heterodimerize with several UBR-E3s (Figure 2A) and that its disruption severely impairs DIAP1’s ability to ubiquitylate drICE (Figure 2E) and regulate apoptosis in vivo (Figure 3; Ditzel et al., 2003; Tenev et al., 2007). Hence, DIAP1’s two ubiquitylation-associated activities—UBR-E3 recruitment and DIAP1’s own RING finger—contribute to its antiapoptotic activity.
Proteins that are ubiquitylated are frequently targeted for proteasomal degradation (Pines and Lindon, 2005). Thus, one possible scenario is that ubiquitylation inactivates drICE through proteasomal degradation. While our findings provide a strong link between drICE ubiquitylation and its activity, we could not detect any Ub-mediated degradation of ubiquitylated drICE. Several lines of evidence indicate that ubiquitylated drICE is not degraded. First, inhibition of the proteasome, through treatment with Lactacystin, cycloheximide, and/or MG132, did not result in any detectable accumulation of ubiquitylated forms of drICE when compared to untreated controls. Second, the nonubiquitylatable drICE mutant drICE9K>R displays a comparable protein half-life as its wild-type counterpart (Figure S7). Third, although coexpression of DIAP1 prevented the detection of the cleaved, activated form of drICE in immunoblot analysis, this was not due to its degradation. Inhibition of the proteasome by MG132 did not rescue the appearance of the p10. Fourth, ectopic expression of DIAP1 in the developing fly eye does not result in the depletion of drICE. Together, our observations are in agreement with the view that DIAP1-mediated ubiquitylation of drICE inactivates it through a nondegradative mechanism. Most likely, the conjugation of Ub to drICE results in steric interference with substrate entry and, in addition, allosteric conformational impairment of the caspase’s catalytic pocket. In this respect, ubiquitylation acts as a “mixed” inhibitor displaying both competitive (increased KM) and noncompetitive (decreased Vmax) inhibitory properties. This notion is consistent with 3D modeling suggesting that ubiquitylation of several surface-exposed K residues can occlude substrate entry (Figure S5). Ultimately, crystal structure analysis of ubiquitylated drICE will be required to validate this model. Allosteric regulation is also seen in caspase-1 (Scheer et al., 2006), indicating that caspases can be regulated through modification of regulatory residues outside the catalytic pocket.
Although ubiquitylation can affect protein function directly, the most common mode of regulation by Ub conjugation involves specific “Ub receptors.” This class of protein recognizes and translates the Ub message into cellular phenotypes (Haglund and Dikic, 2005). Ub receptors carry specialized Ub-binding domains (UBD) through which they interact with Ub-modified proteins (Haglund and Dikic, 2005). Although Ub conjugation of active drICE directly suppresses its proteolytic potential, it is likely that, in vivo, Ub-binding proteins contribute to regulating ubiquitylated caspases, perhaps by spatial sequestration. Moreover, Ub-binding proteins may also prevent further processing of ubiquitylated and partially cleaved caspase intermediates. We would like to speculate that Ub conjugation of caspases might provide cells with a mechanism of restraining the caspase’s proapoptotic potency, allowing the redeployment of the caspase’s residual catalytic activity toward nonapoptotic signaling events (Lamkanfi et al., 2007).
Recent studies indicate that healthy cells harbor significant amounts of active caspases (Ribeiro et al., 2007; Rodriguez et al., 2002). However, little is known about how such cells establish a buffered threshold of active caspases. Our observations are consistent with a model (Figures 7J and 7K) in which DIAP1 acts as a “sensor” for the presence of active effector caspases. While in its full-length form (1), DIAP1 resides in a “closed” inactive configuration that precludes caspase binding. DRONC activation via the scaffold protein DARK (2) (Kornbluth and White, 2005) leads to drICE activation (3) and subsequent cleavage and activation of DIAP1 (4), which converts it into the caspase-binding competent E3 that targets caspases for ubiquitylation and inactivation (5). In this respect, caspases appear to regulate their own inhibition through a negative feedback mechanism that involves DIAP1. Increasing levels of active caspases would hence lead to DIAP1 “activation” that is followed by caspase inhibition. DIAP1’s short half-life of 30 min (Wilson et al., 2002; Yoo et al., 2002) may ensure that once caspase activity levels subside the system returns to the resting state in which full-length DIAP1 resides in a “closed” inactive configuration. In effect, such a scenario would result in a buffered “oscillating cycle” of caspase activity that ensures the continuous presence of sublethal levels of caspase activity (Figure 7J, central panel).
While the above-mentioned oscillating cycle may operate in living cells, under apoptotic conditions (Figure 7K) this system is derailed by IAP antagonists (1), or IKKε, that target DIAP1 for inactivation and/or degradation (Kuranaga et al., 2006; Martin, 2002). Under such conditions, DIAP1 is eliminated, leading to nonubiquitylated and therefore fully active caspases (2), cleavage of cellular proteins (3), and apoptosis.
EXPERIMENTAL PROCEDURES
Constructs, Antibodies, and Recombinant Proteins
Constructs were generated by PCR (Easy-A Polymerase, Stratagene); cloned into pAc, pMT, pMT-GTC (Zachariou et al., 2003), pN-21 (Tenev et al., 2000), or pcDNA3 (Invitrogen); and verified by sequencing. Site-directed mutagenesis was performed using Pfu Turbo (Stratagene). DHFR-based DIAP1 constructs, full-length DRONC-V5/FLAG, drICE-V5/FLAG, Ub-ALG-drICEwt or C>A-V5/FLAG, Ub-AKG-DCP-1C>A-V5/FLAG, and pMT-rpr-FLAG constructs were described previously (Ditzel et al., 2003; Tenev et al., 2007). DIAP1 was expressed untagged throughout, except for in Figures 2B and 2C, where DIAP1-GST was used. HA-UBR domains were as follows: UBR159–223 (CG9086), UBR3174–339 (CG1530), UBR41746–1908 (CG14472), UBR51169–1330 (CG9484), UBR61040–1182 (CG9461), and UBR71–169 (CG15141). Dm Ub-(CG32744) was cloned into His/HA-pAC. Constructs for transgenesis: the dhfr-diap1 coding region was cloned into pKB (Basler and Struhl, 1994) containing the tubulin promoter. The tubulin-promoter-dhfr-diap1 was subcloned into pop-118 (Basler and Struhl, 1994). For the generation of recombinant proteins, the coding regions of drICE, DIAP1, DIAP1F437A, and Effete (Dm UbcD1) were cloned into pGEX6p1 (GE-Healthcare) and purified from bacteria as previously described (Zachariou et al., 2003). GST tags were removed by PreScission cleavage (GE-Healthcare). Recombinant Ub and E1 were a gift from D. Komander (ICR, London). The following antibodies were used: α-V5 (AbD Serotec), α-GST (GE Healthcare), a-HA (Roche), a-PARP1 (Santa Cruz), α-Ub (FK2, Biomol), mouse- and rat-derived secondary HRP-conjugated antibodies (Santa Cruz), and guinea pig-derived secondary HRP-conjugated antibody (Rockland). α-DIAP1 and α-drICE antibodies were described previously (Zachariou et al., 2003).
LI-COR Odyssey Analysis
To quantitate expression levels, immunoblots were incubated with the indicated Odyssey-compatible secondary antibodies (mouse IR680 (Invitrogen), rat IR680 (Invitrogen), and guinea pig IR800 (Rockland) and analyzed at 700 nm excitation. Odyssey software was used to determine the integrated intensities.
Fly Stocks
GMR-rpr was described previously (Wilson et al., 2002). pop118-based transgenic lines expressing tubulin-driven DIAP1-wt, -D20A, -C406Y, and -D20A/C406Y; N-DIAP121–438, -C406Y; and M-DIAP121–438 were generated by Bestgene, USA. Crosses were performed at 25°C throughout. Images of adult heads were acquired using a stereo microscope (Leica) equipped with a Plan Apo 1.0× lens, a digital camera (Nikon), and image capture software (Nikon). Selection of transgenic lines expressing equivalent reference proteins: ten heterozygous female flies were lysed in boiling 1 × SDS sample buffer and analyzed by western blot to detect V5-DHFR expression. To induce DIAP1-overexpressing clones, males of hs-flp;tub>GFP>Gal4 genotype (provided by H.D. Ryoo; > denotes FRT) were crossed to UAS-diap1 females (provided by B. Hay). Clones were induced by 1 hr heat shock at 37°C during first-instar larval stage. third-instar larval eye discs were dissected and labeled with anti-drICE antibody.
DEVDase Assays and drICE Western Analysis
S2 and HEK293T cells were cultured and transfected as described previously (Tenev et al., 2005). For the cell-based DEVDase assays in S2 cells, expression of Rpr (pMT-rpr) or drICE (pMT-drice) was induced with 350 μM CuSO4 for 4 or 6 hr. Cells were lysed as described previously (Tenev et al., 2005), and 20 μl of cleared supernatant was added to 350 μl of DEVDase assay mix (20 μM DEVD-AMC (Calbiochem), 2 mM DTT, 10 mM Tris (pH 7.5), 150 mM NaCl, 0.1% TX-100, and 5% glycerol). The reaction was incubated at RT for 10 min and analyzed at 380 nM excitation/460 nM emission. Extract from the same lysates were also analyzed for drICE by immunoblot analysis. For Figure 5B, cells were treated as above with DMSO, 40 μM z-VAD (BioMol), 50 μM MG132 (Sigma), or a combination of z-VAD and MG132. For Figure S7, 6 hr after drICE induction, S2 cells were treated with 40 μM z-VAD and lysed at the indicated time points post-z-VAD-addition.
Immunoprecipitation and Immunoblot Analysis
Coimmunoprecipitation and immunoblot assays were performed as described previously (Tenev et al., 2005). For Figure 1E, GTC-tagged Reaper was expressed in S2 cells, bound to GSH-Sepharose, and incubated with DIAP1-containing lysates from transfected 293T cells. For Figure 2G, S2 cells were left untreated or irradiated with 150K μJ UV (UV Stratalinker, Stratagene) and lysed after 4 hr in lysis buffer containing 50 mM Chloroacetamide. Endogenous drICE was immunoprecipitated using guinea-pig-derived α-drICE antibody and the Seize-X Protein A Immunoprecipitation Kit (Pierce). To detect ubiquitylated drICE, the membrane was incubated for 1 hr in 6 M GuHCl, 20 mM Tris (pH 7.5), 1 mM PMSF, and 5 mM β-ME at 4°C.
Ubiquitylation Assays
Cells were cotransfected with the indicated constructs at equal ratios. For Figures 5A and S3, cells were incubated for 6 hr with 10 μM Lactacystin (Calbiochem), 50 μg/ml cycloheximide (Sigma), and/or 50 μM MG132 (Sigma) prior to lysis. After 48 hr, cells were harvested and lysed under denaturing conditions, and ubiquitylated proteins were purified using Ni2+-NTA agarose beads (QIAGEN) as described previously (Rodriguez et al., 1999).
In Vitro Ubiquitylation and PARP Cleavage Assay
Active drICE (4 μg) was incubated in the presence or absence of 1 μg DIAP1, 1 μg DmUbcD1, 80 ng E1, and 2 μg Ub in a final volume of 30 μl reaction buffer (2 mM ATP, 40 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.6 mM DTT). Reactions were carried out at 37°C for 90 min. For immunoblot analysis, 15 μl of the total reaction was used, while the remaining 15 μl were tested for drICE activity on PARP. For PARP cleavage assays, 1.2 μg of recombinant PARP (R&D Systems) was added to the drICE ubiquitylation reactions and incubated for a further 25 min at 37°C. Samples were analyzed by immunoblot analysis and quantified using the LI-COR Odyssey technology.
Enzyme Kinetics
Experiments for determining KM and Vmax were performed in triplicates and analyzed with Prism software. Results of the independent DEVDase assay were standardized to the values obtained for 40 μM substrate of ubiquitylated drICE.
Modeling of drICE
The 3D structure of drICE was modeled using the X-ray structure of human caspase-7 (PDB code 1I4O) as a template. Note, drICE is 40% identical to caspase-7. An initial model of drICE was built using MolIDE (Canutescu and Dunbrack, 2005). The model was refined using QuantaTM and subjected to dynamics and minimization using YASARA (Krieger et al., 2002).
Supplementary Material
Acknowledgments
We would like to thank F. Leulier, D. Xirodimas, D. Komander, A. Varshavsky, F. Levy, Hyung Don Ryoo, Bruce Hay, and B. Seraphin for reagents and advice. M.B. is supported by a fellowship of the Deutsche Forschungsgemein-schaft, and A.B. acknowledges support by the NIH (R01 GM068016) and The Welch Foundation (G-1496).
Footnotes
SUPPLEMENTAL DATA The Supplemental Data include seven figures and can be found with this article online at http://www.molecule.org/supplemental/S1097-2765(08)00761-2.
References
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- Canutescu AA, Dunbrack RL., Jr MollDE: a homology modeling framework you can click with. Bioinformatics. 2005;21:2914–2916. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Clem RJ. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 2001;8:137–143. doi: 10.1038/sj.cdd.4400821. [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS. Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem. 2003;278:34042–34050. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- Denault JB, Bekes M, Scott FL, Sexton KM, Bogyo M, Salvesen GS. Engineered hybrid dimers: tracking the activation pathway of caspase-7. Mol Cell. 2006;23:523–533. doi: 10.1016/j.molcel.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Fraser AG, McCarthy NJ, Evan GI. drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J. 1997;16:6192–6199. doi: 10.1093/emboj/16.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB. Characterization of XIAP-deficient mice. Mol Cell Biol. 2001;21:3604–3608. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitinprotein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 1998;440:243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126:583–596. doi: 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, Sheng J, Xie Y, Varshavsky A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, Zachariou A, Pichaud F, Ueda R, Meier P. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 2006;13:1663–1674. doi: 10.1038/sj.cdd.4401868. [DOI] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ. Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell. 2002;109:793–796. doi: 10.1016/s0092-8674(02)00802-4. [DOI] [PubMed] [Google Scholar]
- Muro I, Means JC, Clem RJ. Cleavage of the apoptosis inhibitor DIAP1 by the apical caspase DRONC in both normal and apoptotic Drosophila cells. J Biol Chem. 2005;280:18683–18688. doi: 10.1074/jbc.M501206200. [DOI] [PubMed] [Google Scholar]
- Pines J, Lindon C. Proteolysis: anytime, any place, anywhere? Nat Cell Biol. 2005;7:731–735. doi: 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–1480. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JMP, Lain S, Midgey CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Chen P, Oliver H, Abrams JM. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 2002;21:2189–2197. doi: 10.1093/emboj/21.9.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. A conserved tetrapeptide motif: potentiating apoptosis through IAP-binding. Cell Death Differ. 2002;9:93–95. doi: 10.1038/sj.cdd.4400957. [DOI] [PubMed] [Google Scholar]
- Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, Huang DC, Vaux DL. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci USA. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bohmer SA, Kaufmann R, Frese S, Bittorf T, Beckers T, Bohmer FD. Perinuclear localization of the protein-tyrosine phosphatase SHP-1 and inhibition of epidermal growth factor-stimulated STAT1/3 activation in A431 cells. Eur J Cell Biol. 2000;79:261–271. doi: 10.1078/S0171-9335(04)70029-1. [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- Tenev T, Ditzel M, Zachariou A, Meier P. The antiapoptotic activity of insect IAPs requires activation by an evolutionarily conserved mechanism. Cell Death Differ. 2007;14:1191–1201. doi: 10.1038/sj.cdd.4402118. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Ubiquitin fusion technique and its descendants. Methods Enzymol. 2000;327:578–593. doi: 10.1016/s0076-6879(00)27303-5. [DOI] [PubMed] [Google Scholar]
- Varshavsky A, Turner G, Du F, Xie Y. Felix Hoppe-Seyler Lecture 2000. The ubiquitin system and the N-end rule pathway. Biol Chem. 2000;381:779–789. doi: 10.1515/BC.2000.101. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A. The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 2006;13:1697–1706. doi: 10.1038/sj.cdd.4401920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, White K. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


