Abstract
The region of human chromosome 11p15.5 is linked with Beckwith–Wiedemann syndrome that is associated with susceptibility to Wilms’ tumor, rhabdomyosarcoma and hepatoblastoma. TSSC5 (tumor-suppressing subchromosomal transferable fragment cDNA; also known as ORCTL2/IMPT1/BWR1A/SLC22A1L) is located in the region. The expression of TSSC5 and other genes in the region is regulated through paternal imprinting. Mutations and/or reduced expression of TSSC5 have been found in certain tumors. TSSC5 encodes an efflux transporter-like protein with 10 transmembrane domains, whose regulation may affect drug sensitivity, cellular metabolism and growth. Here, we present evidences indicating that RING105, a novel conserved RING-finger protein with a PA (protease-associated) domain and a PEST sequence, is a ubiquitin ligase for TSSC5 that can function in concert with the ubiquitin-conjugating enzyme UbcH6. The polyubiquitin target site on TSSC5 was mapped to a region in the 6th hydrophilic loop. Ectopic expression of RING105 in HeLa cells caused an accumulation of cells during G1 that was not observed with the expression of a form of RING105 in which a residue within the RING finger was mutated to inactivate its ligase activity. UbcH6-RING105 may define a novel ubiquitin–proteasome pathway that targets TSSC5 in mammalian cells.
Keywords: TSSC5, protease associated, RING finger, ubiquitin ligase, UbcH6, 11p15.5
Introduction
Deletion of human chromosome 11p15.5 is associated with Beckwith–Wiedemann syndrome that confers susceptibility to Wilms’ tumor, rhabdomyosarcoma and hepatoblastoma (Feinberg, 1994). This region is thus designated as WT2 (Wilms’ tumor 2) region and is suspected of harboring at least one tumor suppressor that remains unidentified. Overlapping P1 artificial chromosomes constructed from the region were used in an attempt to isolate the genes responsible for the tumors. Introduction of the artificial chromosome (STF: subchromosomal transferable fragment) into rhabdomyosarcoma cells suppressed tumor growth, indicating that the region contains one or more tumor suppressor or suppressors (Koi et al., 1993). The TSSC5 (tumor-suppressing subchromosomal transferable fragment cDNA 5;also known as ORCTL2/IMPT1/BWR1A/SLC22A1L) gene is found in the ~1 Mb region (Cooper et al., 1998;Lee et al., 1998;Scwienbacher et al., 1998). The region is heavily imprinted, leading to suggestions that epigenetic imprinting may be involved in the regulation of cellular growth (Feinberg, 1999). The gene cluster is conserved between human and mouse (Paulsen et al., 1998). Among the genes located in the region, mutation of the IGF2 (insulin-like growth factor 2) gene was shown to have a high correlation with Wilms’ tumor (Ravenel et al., 2001), and loss of imprinting in IGF2 is associated with personal/family history of colorectal neoplasia (Cui et al., 2003) and with hepatocarcinoma (Poirier et al., 2003). However, these effects may be due to global effect on epigenetic imprinting, since IGF2 locus serves as an imprinting center (Lewis and Murrell, 2004). It is not yet established which gene in the 11p15.5 region is directly responsible for tumor suppression in human.
There is evidence suggesting that TSSC5 may play a role as a tumor suppressor, or the loss may serve as an additional factor for tumor formation. Normally TSSC5 is imprinted in embryo. Imprinting is lost in adults allowing TSSC5 protein expression in a variety of tissues (Lee et al., 1998;Feinberg, 1999). Imprinting in TSSC5 and loss of the expression was observed in hepatocarcinoma cells (Schwienbacher et al., 2000). Although mutations within the TSSC5 gene were rarely found, reduction of TSSC5 mRNA expression level was observed in 11 out of 14 hepatoblastoma-derived cells (Albrecht et al., 2004). A TSSC5 germline mutation and frequent aberrant splicing were found in Wilms’ tumor, and somatic mutations were found in lung cancer and breast cancer cells (Lee et al., 1998, Schwienbacher et al., 1998).
The TSSC5 gene encodes a putative integrated membrane protein with 10 membrane-spanning domains and shows homology to efflux transporters (e.g. tetracycline efflux transporter) and to organic cation transporters. TSSC5 mRNA expression is observed in the fetal kidney and liver, and in multiple adult tissues such as the heart, liver, kidney, spleen, thymus, prostate, testis, small intestine, colon and peripheral blood leukocytes (Dao et al., 1998;Lee et al., 1998). The protein is found on the apical membrane surfaces of the proximal tubules in human kidney (Reece et al., 1998). However, how TSSC5 protein functions and how it might be involved in growth regulation are poorly understood. Expression of TSSC5 in bacteria confers drug resistance, and it has been proposed to be similarly involved in drug metabolism in mammalian cells (Reece et al., 1998). Since TSSC5 belongs to the multimembrane-spanning transporter family protein that include MDR1 (multidrug resistance 1;capab le of transporting organic anions as well as small drug molecules), it has been speculated that it may be involved in resistance to chemotherapy drugs (Dao et al., 1998) and/or in export of genotoxic substances whose retention may increase the risk of tumor formation (Reece et al., 1998).
How TSSC5 protein levels are regulated is uncertain. One major pathway is the regulation of protein amount through ubiquitin-dependent proteolysis (Pagano, 1997; Hershko, 1997;Hershko and Chiechanover, 1998; Almond and Cohen, 2002). The ubiquitin-dependent proteolysis system requires a set of enzymes as follows: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3). These enzymes in concert covalently attach multiple ubiquitins to the target. The resulting polyubiquitin chain on the target is recognized by a large protease complex, the proteasome, which degrades the target. Only one E1 for ubiquitin is found in the human genome. In contrast at least 17 E2’s and over 100 putative E3’s have been found. Specific combinations of E2 and E3 enzyme are thought to determine target specificity. Identification of a set of enzymes involved in TSSC5 regulation is of great importance in understanding the regulation of this protein and may provide novel mechanisms for understanding malfunctions of growth control in certain human tumors.
Results
Isolation of RING105
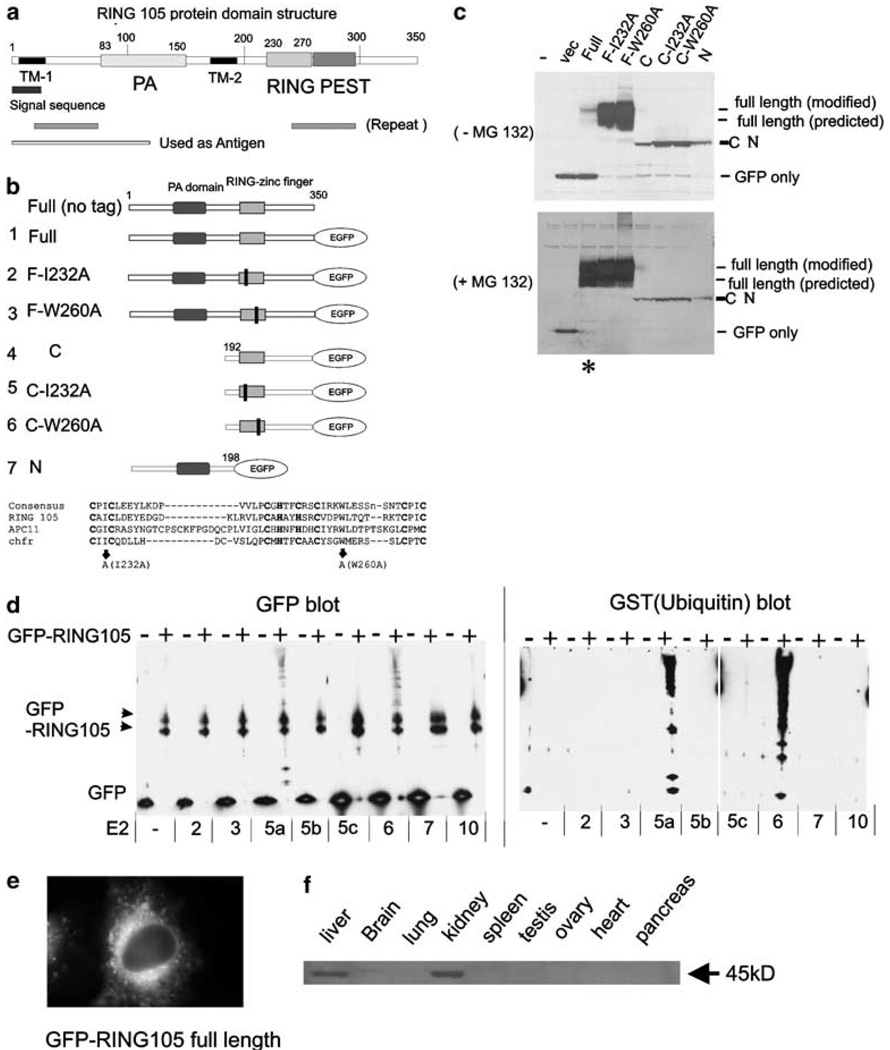
The ubiquitin-conjugating enzyme hCdc34/UbcH3 normally functions with the ubiquitin ligase SCF to facilitate the G1-to-S-phase transition. We found that overexpression of hCdc34/UbcH3 in mitotic cells led to defects in chromosome alignment and progression in mitosis (Topper et al., 2001). Using bacterial two-hybrid screening with hCdc34/UbcH3 as bait, we found a positive clone that is identical in BLAST searches to a human open-reading frame in the database from sequencing projects, DKFZp566H073 (Accession Number NM_015528). Hereafter, we call the protein RING105, as it contains a RING-finger domain and was isolated as 105th clone. RING105 is a 350-aminoacid protein, which contains a PA (protease-associated) domain in the N-terminal region and a RING zinc-finger domain in its C-terminal region that is followed by a putative PEST sequence (Figure 1a). RING105 also contains two transmembrane domains (TM1 and TM2). The PA domain was originally found as an insertion domain in various proteases and in proteins involved in vacuolar sorting pathways, and is proposed to function as a protein–protein interaction domain (Mahon and Bateman, 2000). The RING-finger domain is a putative ubiquitin ligase motif (Lovering et al., 1993;Pagano, 1997). PEST sequences are frequently observed in proteins that are rapidly degraded, such as the targets of SCF ubiquitin ligase complex, cyclins D and E (Rechsteiner and Rogers, 1996). Putative homologs of RING105 are found in mouse, chicken, Xenopus, Caenorhabditis elegans, Drosophila and Arabidopsis. Thus, the protein is widely conserved in metazoans. However, the function has not been identified for any of the putative homologs. Consistent with the presence of the PEST sequence, ectopically expressed GFP-RING105 was degraded in HeLa and in 293 cells, and the degradation was inhibited by proteasome inhibitor MG132 or by point mutations in the RING-finger domain (constructs, F-I232A and F-W260A) (Figure 1b and c). RING105 showed autoubiquitylation activity in vitro with UbcH5a and UbcH6 (Figure 1d). Thus, RING105 is a ubiquitin ligase that is degraded in a proteasome- and its own RING-finger-dependent manner. Ectopically expressed GFP-tagged RING105 targeted to cytoplasmic membranes (Figure 1e). To test RING105 expression in tissues, we raised an antibody to the RING105 N-terminus region and probed extracts from a variety of human tissues. We observed the strongest protein expression in the kidney and liver (Figure 1f, Supplementary Information 1).
Figure 1. RING105 is a RING-finger-containing ubiquitin ligase.
(a) RING105 protein domain structure. RING105 protein is 350 amino acids and contains a PA (protease-associated) domain, a RING-finger domain and a PEST domain. Two TM (transmembrane domain) are also highlighted. (b) Plasmid constructs. RING105 was truncated, or point mutations (represented as black bars) were introduced in conserved positions within the RING-finger domain. Mutations at corresponding sites were previously shown to inactivate ubiquitylation activity in the chfr protein (Kang et al., 2002). The protein sequence alignment comparing RING-finger domains for RING105, Apc11 and chfr is also shown. The residues that were mutated to inactivate the RING finger are indicated by arrows. The cysteine and histidine residues that form zinc-chelating ‘fingers’ are shown in bold. (c) RING105 is degraded by the proteasome in a manner that is dependent on its own RING-finger domain. 293T cells were transfected with plasmids encoding the indicated GFP-tagged RING105 constructs and cell extracts were prepared for immunoblotting with anti-GFP. In the lower blot, the cells were treated with proteasome inhibitor MG132 (10 µm) 2 h before preparation of cell extracts. Note that mutants containing point mutations within the RING finger are stable (compare lane 3 with lanes 4 and 5 in the upper blot). Note also that MG132 treatment results in stabilization of the full-length RING105 protein (compare lane 3 (asterisk) in the upper and lower blots). (d) RING105 has autoubiquitylation activity in vitro when tested with UbcH5a and UbcH6. Immunoprecipitated GFP- or GFP-tagged RING105 were incubated with indicated E2 (ubiquitin-conjugating enzyme), GST-ubiquitin, E1 and ATP for 60 min at 30°C. After the reaction, the beads were rinsed extensively, then treated with SDS sample buffer, separated by PAGE and transferred to membranes. They were then probed with anti-GFP (left panel) and anti-GST (ubiquitin) (right panel). The high molecular weight bands appearing above the GFP-RING105 doublet (black arrowheads) when RING105 is incubated in conjunction with UbcH5a and UbcH6 reflect polyubiquitylated proteins. (e) GFP-tagged RING105 associated with cytoplasmic membranes. HeLa cells were transfected with GFP-RING105 (full) (construct 1). After 18 h, cells were treated with 10 µm MG132 for 2 h and observed. (f) RING105 is enriched in the liver and kidney. Extracts from human tissues were probed with anti-RING105 antibody. Further description of the antibody is shown in Supplementary Information 1.
Isolation of TSSC5 as an interactor with RING105
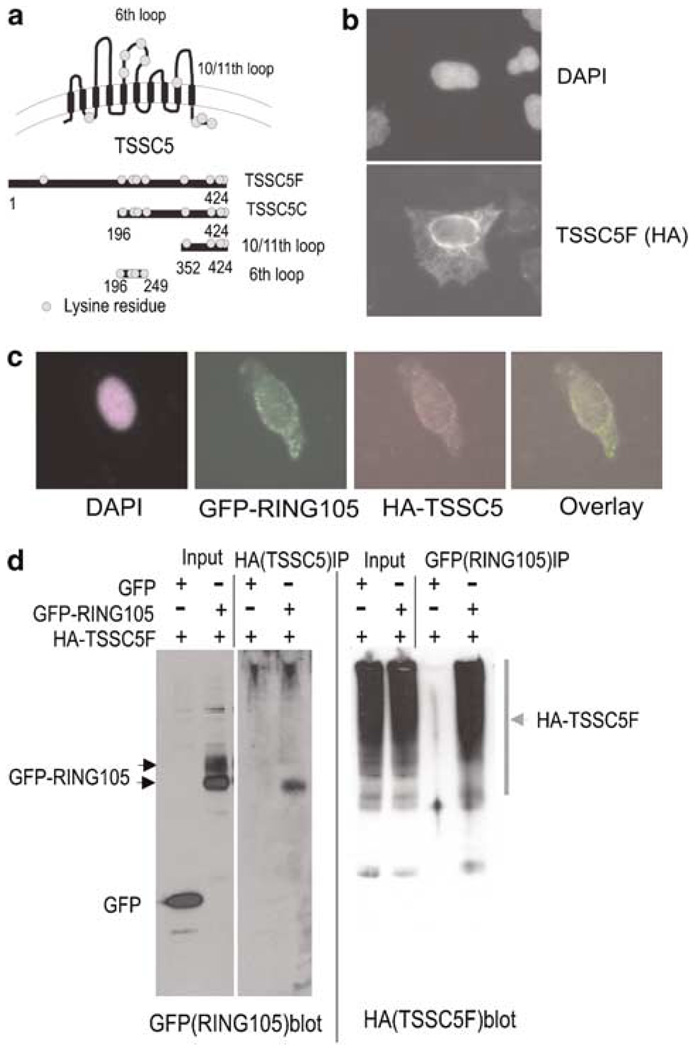
Using bacterial two-hybrid screening with RING105 as bait, we isolated the full-length TSSC5 as positive clones (TSSC5: Accession Number: NP_899056). TSSC5 protein is a 424-amino-acid protein, and protein structure analysis (Dao et al., 1998;or available from www.expasy.com) predicted that it is an integral membrane with 10 membrane-spanning domains (Figure 2a). We generated a series of truncated TSSC5 constructs to use for further analysis. Possible ubiquitin acceptor site lysine (K) residues are marked as circles in Figure 2a.
Figure 2. RING105 binds TSSC5 by co-immunoprecipitation.
(a) Schematic representation of TSSC5 protein and TSSC5 plasmid constructs used for testing ubiquitin targeting. TSSC5 is predicted to be an integral membrane protein with 10 transmembrane domains. (The orientation of TSSC5 with respect to its insertion into cytoplasmic membranes such as ER has not been determined.) Potential ubiquitin acceptor lysine residues are presented as circles. (b) Ectopically expressed TSSC5 was associated with cytoplasmic membranes. HeLa cells were transfected with HA-tagged TSSC5F (full length) and prepared for immunofluorescence with anti-HA antibody. DNA was stained with DAPI (upper panel). (c) Limited colocalization of GFP-RING105 and HA-TSSC5F. GFP-RING105 and HA-TSSC5F (full length) were cotransfected to HeLa cells. At 14 h after transfection, cells were treated with MG132 for 2 h to prevent RING105 degradation, and fixed for localization analysis. Panels represent DAPI (purple), GFP-RING105 (green) and HA-TSSC5F (red) and GFP-HA overlay. (d) HA-tagged TSSC5 co-immunoprecipitated with GFP-tagged RING105. HeLa cells were transfected with HA-TSSC5F (full length) and GFP or GFP-RING105. After 18 h, cells were treated with MG132 for 2 h and then extracted. The extracts were immunoprecipitated with anti-HA and anti-GFP. Samples from the whole extract (input) and the immunoprecipitate (IP) were probed with anti-HA and anti-GFP.
TSSC5 localizes to cytoplasmic membranes
We performed immunofluorescence with anti-HA antibody to observe the localization of HA-tagged TSSC5F (full length). The immunofluorescence signal appeared at the nuclear periphery and to an anastamosing network in the cytoplasm consistent with a localization of TSSC5F-HA to the nuclear envelope and other cytoplasmic membranes (Figure 2b). TSSC5F tagged with GFP instead of HA showed the same cytoplasmic membrane localization (not shown). Both HA-TSSC5 and GFP-RING105 appeared to localize on or to be associated with cytoplasmic membranes (Figure 1e and Figure 2b). However, when TSSC5-HA and GFP-RING105 were coexpressed, we found that colocalization was limited (Figure 2c).
Physical association between TSSC5 and RING105
To test the association between RING105 and TSSC5 in vivo, we cotransfected HeLa cells with HA-TSSC5F and GFP-RING105, treated the cells with MG132 to prevent their degradation and carried out immunoprecipitation (Figure 2d). HA-TSSC5F ran as a smear in the high molecular weight region of the SDS–PAGE gels. Such smeared SDS–PAGE patterns have been preciously noted for some membrane proteins and may be caused by the membrane-spanning domains forming SDS-resistant aggregates during SDS–PAGE sample preparation (Sagne et al., 1996). GFP-RING105 and HA-TSSC5F co-immunoprecipitated reciprocally, which indicates they form a complex in vivo. Ectopically expressed RING105 is modified in vivo, and runs as doublet in SDS–PAGE. Phosphatase treatment did not affect the SDS–PAGE pattern (data not shown). Interestingly, only the faster-running band of RING105 co-immunoprecipitated with HA-TSSC5 (Figure 2d). The existence of a modified form of RING105 that does not associate with TSSC5 may also account for the limited colocalization seen in Figure 2c. These results suggest a subpopulation of RING105 and TSSC5 associate in vivo.
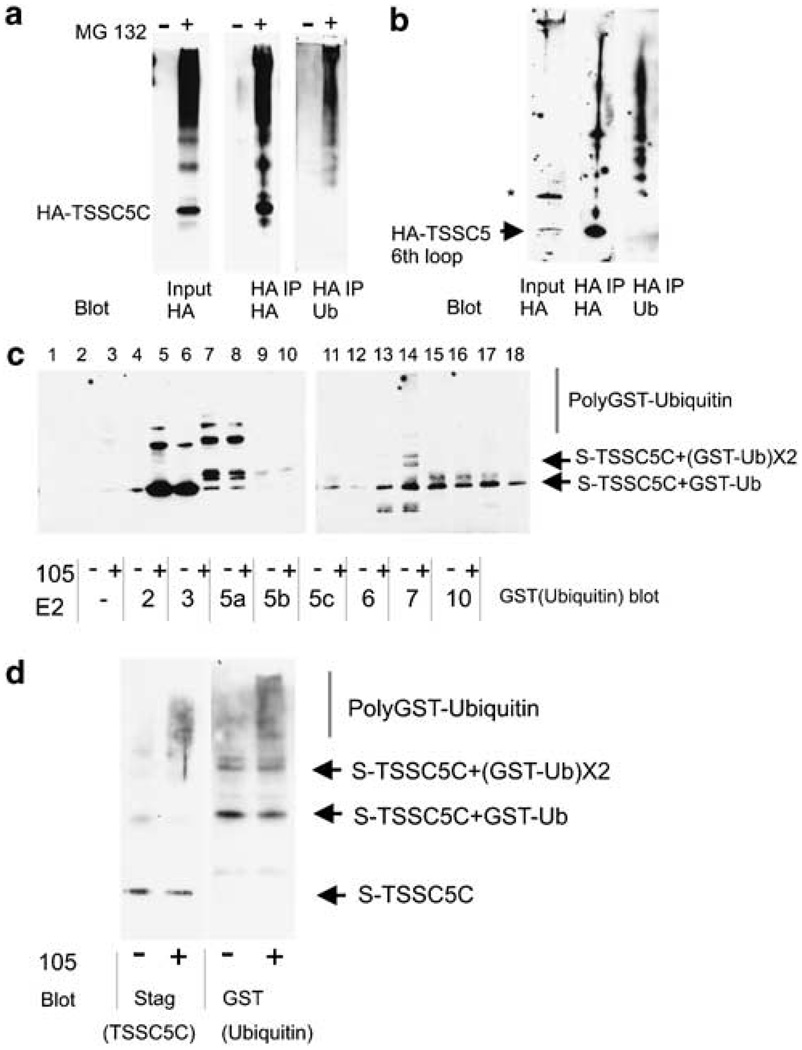
TSSC5 is polyubiquitylated on its 6th loop
Association with an ubiquitin ligase raised a possibility that the levels of TSSC5 protein are regulated through ubiquitin-mediated proteolysis. To test the possibility, firstly we transfected HeLa cells with a plasmid encoding HA-tagged TSSC5 C-terminus region (amino acid 194–424) (hereafter referred to as HA-TSSC5C), treated the cells with a proteasome inhibitor MG132 and tracked HA-TSSC5C ubiquitylation by immunoblot (Figure 3a, input). We used the C-terminus construct because TSSC5 full-length constructs TSSC5F showed smeared SDS–PAGE patterns, making it very difficult to visualize ubiquitylation ladders. In the transfectants, HA-TSSC5C protein accumulated only when treated with MG132, suggesting TSSC5C is subjected to proteasome-dependent degradation. Immunoblotting for TSSC5C showed a ladder-like pattern. To determine whether the ladder-like modification was due to ubiquitylation, we immunoprecipitated with HA antibody after denaturing the sample and blotted the immunoprecipitates with anti-HA and anti-ubiquitin antibodies (Figure 3a, HA IP). The immunoprecipitates were recognized by anti-ubiquitin antibody in a ladder-like pattern, indicating that HA-TSSC5C is polyubiquitylated (Figure 3a, HA IP/Ub blot). The upper bands also labeled with an antibody specific for polyubiquitin chains, confirming that HA-TSSC5C is polyubiquitylated (data not shown). This result is consistent with the notion that TSSC5 is polyubiquitylated at its C-terminus (amino acids 196–424) and degraded through the proteasome pathway. To map the ubiquitylation site(s) more precisely, we expressed shorter fragments of TSSC5. Immunoprecipitation and immunoblot experiments indicated that the 6th loop of TSSC5 (amino acids 196–249) was polyubiquitylated when expressed in HeLa cells that were treated with MG132 (Figure 3b). In contrast, the 10th–11th loop (amino acids 352–424) did not show such polyubiquitylated bands (negative data not shown). Thus, at least one or more of the four lysines in the 6th loop serve as ubiquitin acceptor sites in TSSC5.
Figure 3. TSSC5 ubiquitylation.
(a) TSSC5 C-terminus is polyubiquitylated in vivo. 293T cells were transfected with plasmid encoding HA-tagged TSSC5 C-terminus (TSSC5C). After 18 h, cells were incubated with or without 10 µm MG132 for 2 h. Cell extracts were denatured to break up protein complexes and then HA-tagged proteins were immunoprecipitated with anti-HA antibody. The immunoprecipitates were blotted with anti-HA or antiubiquitin. (b) TSSC5 6th loop is the smallest fragment that is polyubiquitylated efficiently in vivo. Plasmid encoding HA-tagged TSSC5 6th loop was transfected into 293T cells. At 18 h after transfection, cells were treated with MG132 for 2 h and then extracts were prepared for immunoprecipitation with anti-HA antibody in denaturing conditions. Immunoprecipitates were blotted with anti-HA and anti-ubiquitin. Only fragments of TSSC5 containing the 6th loop exhibited polyubiquitylation, while other regions containing lysines (such as the TSSC5 10th loop) did not exhibit polyubiquitylaion (negative data, not shown). (c) Only the combination of UbcH6 and RING105 facilitated polyubiquitylation of TSSC5C in vitro. Immunoprecipitated GFP- or GFP-RING105 was incubated with E1, the indicated E2, GST-ubiquitin, ATP and S-tagged TSSC5C for 2 h at 30°C. Reactions were terminated with 1% SDS and S-tagged protein was collected with S-protein beads. Ubiquitylation was probed with anti-GST. Although several of the E2’s result in the accumulation of high molecular weight bands on the S-tag-precipitated protein, only UbcH6 results in bands that are generated only in the presence of RING105. (d) More detailed examination of the UbcH6/RING105-mediated ubiquitylation reaction shows that high molecular weight bands contain both TSSC5 (labeled with anti-S tag) and GST-ubiquitin (labeled with anti-GST), demonstrating that the high molecular weight bands are poly(GST)ubiquitylated forms of TSSC5C. Note that in the presence of RING105, majority of TSSC5 was polyubiquitylated. In contrast, without RING105 polyubiquitin chain was rarely observed.
RING105 is a ubiquitin ligase for TSSC5 in vitro
The results reported above were consistent with the possibility that TSSC5 is a substrate of RING105 ubiquitin ligase. We tested the potential of RING105 for catalysing polyubiquitylation of TSSC5 in vitro. Bacterially produced, S-tagged TSSC5C protein was mixed and incubated with E1, one of a set of E2 proteins, RING105 immunopurified from HeLa cell extracts, ATP and GST-ubiquitin (Figure 3c). After incubation, S-tagged TSSC5C protein was collected with S-protein beads in denaturing conditions, and probed with anti-GST (ubiquitin). In the in vitro system, some of E2’s (UbcH3, UbcH5a, UbcH6, UbcH7, UbcH10) alone were capable of transferring ubiquitin to TSSC5C at a low rate. UbcH2, UbcH5b and UbcH5c showed little activity toward TSSC5. Among the E2’s that showed ubiquitin transfer activity, RING105 increased polyubiquitylation on TSSC5C only in combination with UbcH6 (Figure 3c, lane 14 and Figure 3d). Thus, RING105 accelerates UbcH6-mediated ubiquitylation of TSSC5 in vitro and meets the criteria of an E3 ubiquitin ligase.
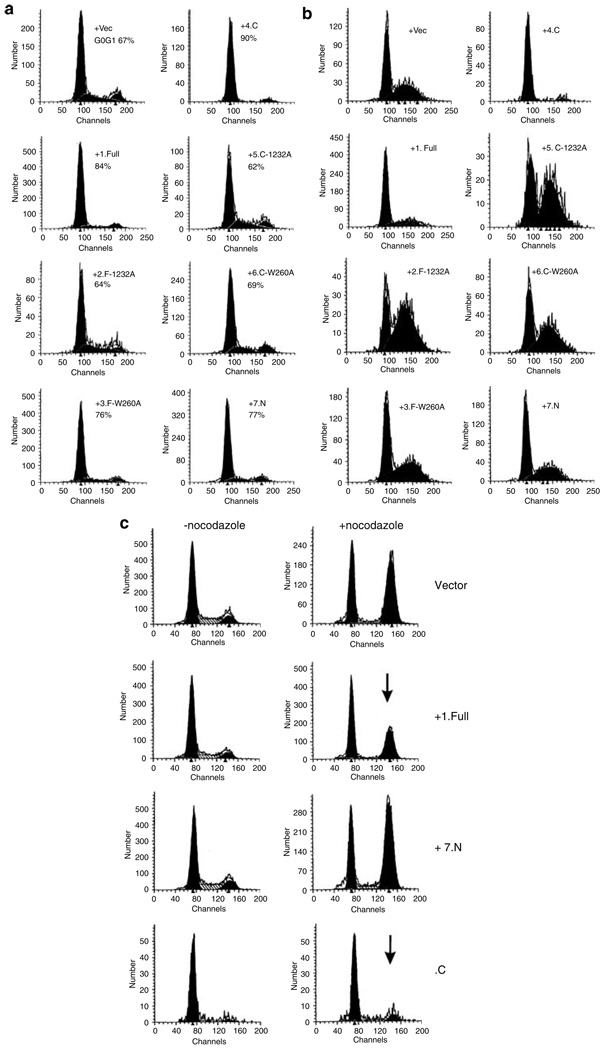
RING105 expression delays the G1-to-S transition in HeLa cells
TSSC5 is suspected to regulate growth. To test whether RING105 expression affects the cell cycle and/or growth, we transfected HeLa cells with plasmids encoding GFP-RING105 and performed cell cycle analysis using FACS. Only GFP-positive cells were analysed. To test the significance of the RING-finger domain, point mutations were introduced into the domain (I232A and W260A, Figure 1b). Homologous changes in other RING-finger ubiquitin ligases lead to loss of ligase activity (Kang et al., 2002), and we confirmed reduction in autoubiquitylation activity of RING105 by I232A mutation (Supplementary Figure 2). Figure 4a shows the effects of expression of wild-type and various mutant forms of RING105 in cycling HeLa cells. Expression of the full-length (construct 1, full) caused a significant increase in the G1 population. Expression of full-length RING105 with point mutations in the RING-finger domain resulted in no change in the G1 population (construct 2, F-I232A) or to a lesser G1 increase (construct 3, F-W260A). Expression of the RING-finger domain region without the rest of the protein (construct 4, C) caused a very large increase in the G1 population, and introduction of the RING-finger point mutations (constructs 5 and 6, C-I232A, C-W260A) abolished the effect. Expression of the N-terminal region of RING105 (construct 7, N) resulted in a slight increase in the G1 population. The results suggested that expression of RING105 delays the G1-to-S transition and that this cell cycle delay is largely dependent on the active RING-finger domain. We also performed the FACS experiments in combination with drugs affecting cell cycle progression. VM26 (teniposide) is a topoisomerase-II inhibitor and causes DNA double-strand breaks. VM26 treatment leads to an accumulation of cells in late S and/or G2 due to the DNA damage checkpoint (Li and Liu, 2001). We transfected HeLa cells with constructs expressing various forms of RING105, and then treated them with VM26 (Figure 4b). When full-length (+1.Full) or RING-finger domain (+4.C) was expressed, fewer cells were accumulated in S/G2. In contrast, point mutant (+2.F-I232A, +3.F-W260A, +5.C-I232A, +6C-W260A)-expressing cells showed the normal, high S/G2 population, supporting the notion that full-length RING105 protein delays/arrests cells in G1. Nocodazole is a spindle poison, activates the mitotic checkpoint and arrests cells in mitosis (Jordan and Wilson, 2004). As before, in the absence of drugs (Figure 4c, left panel), expression of RING105 full-length and of RING-finger domain in HeLa cells showed an increase in G1 population. In the presence of 100 ng/ml nocodazole to induce M-phase arrest, cells transfected by +1.full length or +4.C (RING-finger domain) showed lesser accumulation at G2/M. In contrast, expression of the N-terminus of RING105 did not affect accumulation at G2/M. These results are also consistent with the notion that full-length and RING-finger domain expression delays/arrests cells in G1 and blocks or delays further cell cycle progression. All these results suggest that RING105 may play a role in growth regulation involved in the G1/S transition.
Figure 4. RING105 expression delays the G1-to-S transition in HeLa cells.
(a) RING105 transfection to cycling HeLa cells. HeLa cells were transfected with plasmids expressing RING105-GFP constructs (see Figure 1b) and the effect on the cell cycle was analysed by flow cytometry 24–40 h after transfection. Experiments were repeated at least twice and typical results from GFP-positive cells are shown. Numbers indicate estimated G0/G1 percentage. Cells transfected with plasmids expressing full-length (+1) or the C-terminal region containing the RING-finger domain (+4) showed a substantial accumulation of G1 cells. Point mutations of the RING-finger domain in full-length TSSC5 (+2, +3) or in the C-terminal construct (+5, +6) significantly reduced accumulation of cells in G1. (b) RING105 transfection and DNA-damaging agent VM26 (teniposide) treatment. Transfected HeLa cells were treated with the topoisomerase II inhibitor VM26 (0.2 µg/ml, 12 h) leading to DNA damage. This treatment led to an accumulation of cells in S phase/G2 phase. In cells transfected with plasmids expressing full-length (+1.Full) and C-terminus RING-finger domain (+4.C) of RING105, the S/G2 peaks were reduced, suggesting that the transfection causes delay in the cell cycle progression before S phase. (c) RING105 transfection and spindle inhibitor nocodazole treatment. HeLa cells were transfected with empty plasmid (vector) or with plasmids encoding full-length RING105 (+1.Full), N-terminus (+7.N) and C-terminus (+4.C) of RING105 in the absence (left column) or presence (right column) of nocodazole (100 ng/ml, 16 h). Nocodazole induces mitotic-checkpoint-dependent G2/M arrest. Populations transfected with empty vector or with plasmids expressing the N-terminus showed the normal increase in the G2/M peak. Populations expressing the full-length RING105-GFP (+1) showed a diminished degree of accumulation, while cells expressing the C-terminus RING-finger domain (+4.C) showed almost no accumulation in G2/M (arrows).
Discussion
Here, we identify a conserved E3 ubiquitin ligase RING105 and show that (a) RING105 has autoubiquitylation activity with UbcH5a or UbcH6, (b) TSSC5 and RING105 physically interact in vivo, (c) TSSC5 is polyubiquitylated in vivo at the 6th hydrophilic loop (amino acid 196–249) that contains four lysine residues, and (d) TSSC5 polyubiquitylation is facilitated by RING105 and UbcH6 in vitro. This evidence suggests that RING105 is an E3 ubiquitin ligase for TSSC5 that acts in concert with ubiquitin-conjugating enzyme UbcH6. UbcH6 is unique among E2’s, since it has a C-terminal hydrophobic tail that allows for post-translational anchoring to the endoplasmic reticulum (ER) membrane. In vivo, UbcH6 interacts with a soluble E2 UbcH7, which is recruited to ER membrane by interaction with membrane-anchored proteins, such as Cue1p (Biederer et al., 1997). UbcH6 is reported to be involved in the ERAD (ER-associated degradation) pathway, in which integrated membrane proteins and misfolded proteins are transported from ER to the cytoplasm via the membrane transport complex, the Sec61 translocon, and degraded by the ubiquitin–proteasome pathway. The ERAD pathway serves in stress response and protein quality control, and the pathway is involved in regulating some disease-related proteins including cystic fibrosis transmembrane conductance regulator (Bonifacino and Weissman, 1998; Hampton, 2002;McCracken and Brodsky, 2003; Hirsch et al., 2004, Romisch, 2005). Only limited numbers of E3’s have been identified to work in concert with UbcH6. TSSC5 and RING105 may define a new substrate–E3 combination working with UbcH6. Whether the site for UbcH6- and RING105-mediated ubiquitylation and degradation of TSSC5 is cytoplasmic side of the ER, and thus whether this pathway is part of novel ERAD E3/substrate combination is not yet certain. Some ERAD target proteins have signal sequences for UbcH6/UbcH7-mediated degradation, such as the ACKNWFSSLSHFVIHL sequence in Ura3 protein (Gilon et al., 2000). TSSC5 does not appear to have a matching sequence. Identification of the sequence in TSSC5 required for UbcH6/RING105-mediated degradation will be of interest to answer questions such as whether there is a conserved structure for substrate recognition by UbcH6, and how different E3’s regulate substrate recognition for UbcH6.
We isolated RING105 as a two-hybrid positive clone with hCdc34/UbcH3. However, RING105 did not show strong ubiquitin ligase activity on itself or on TSSC5 with hCdc34/UbcH3 in vitro. This apparent discrepancy may be caused by our two-hybrid bait construction. For technical reasons, we removed the C-terminal region from the bait (see Materials and methods). This alteration may have rendered broader specificity to the bait construct. E2 specificity can be altered by mutation. For example, mutation of the Rad6/Ubc2 catalytic core creates a pluripotent ubiquitin-conjugating enzyme that can complement functions of UBC4 and CDC34 proteins (Ptak et al., 2001). Alternatively, hCdc34/UbcH3 may interact with RING105 but may require additional cofactors for ubiquitylation activity. Finally, hCdc34/UbcH3 and RING105 may be active on yet unidentified substrates in vivo. We reported that expression of RING105 in HeLa cells causes a delay in G1/S transition. The delay might be caused by the RING105 activity on a yet undiscovered substrate, perhaps one whose ubiquitylation involves hCdc34/UbcH3. Multiplicity of substrates may explain why overexpression of a ubiquitin ligase that targets a potential tumor suppressor can also result in the inhibition cell cycle progression.
In tissues such as adult kidney and liver, both TSSC5 and RING105 are abundant. Our coexpression experiment indicated only a fraction of these proteins interact. Only the faster migrating form of RING105 co-immunoprecipitated with TSSC5 (Figure 2d). The nature of the modification on RING105 that results in the two forms remains to be determined, but it may participate in regulating the interaction with TSSC5. Downregulation of TSSC5 is observed in certain tumors. Our results suggest that RING105 is involved in TSSC5 regulation, and manipulation of RING105 may assist restoring TSSC5 function and normal growth control in cancer.
Materials and methods
Bacterial two-hybrid assay with hCdc34/UbcH3 as bait
We followed the manufacturer’s instructions (Stratagene, La Jolla, CA, USA). The N-terminal two-third region of Cdc34, corresponding to amino acid 1–143 and containing catalytic core, was amplified by PCR using primers with engineered restriction enzyme sites and a spacer amino-acid sequence. The primer sequences were as follows: HYYOL-58, 5′-cggcgcggccgcaggaggaggaagcggaatggctcggccgctagtg-3′; and HYYOL-10, 5′-gcggaattctcagctcacgtttgcgggcga-3′.
The PCR product was digested by NotI–EcoRI and ligated into the pBT vector. The resulting construct was used as bait (pBT-34delC). (Constructs containing full-length Cdc34 weakly activated reporter cassettes on their own and thus were not used for screening.) For prey constructs, we used a commercial expression library prepared from a human HeLa S3 cells (Stratagene). Among a few hundred carbenicillin-resistant colonies that emerged on sixty-seven 15 cm selection plates, we picked 192 colonies. Of these, 97 colonies were beta-Gal positive. We recovered 87 plasmids from the blue colonies and sequenced them. A clone containing the 24–350 amino-acid region of RING105 was identified. A cDNA containing full-length RING105 was obtained from American Type Culture Collection (ATCC) for further manipulation (ATCC number 6232371).
RING105 plasmids
We prepared constructs encoding C-terminally GFP-tagged RING105 by ligating XhoI–SacII-treated PCR fragments into pEGFP-N (Clontech, Palo Alto, CA, USA). Point mutations were introduced by a site-directed mutagenesis kit (Strata-gene). We made a mammalian expression construct for RING105 without GFP by deleting the GFP coding region with SacII–NotI and religating the DNA after T4 DNA polymerase treatment.
Cell Culture and transfection
Cell culture was performed as described previously (Kallio et al., 1998). We used FuGene 6 reagent (Roche Biochemicals, Indianapolis, IN, USA) for DNA transfection following the manufacturer’s instructions. Occasionally, we used 293T cells because of their high transfection efficiency.
RING105 antibody production and purification
The N-terminal region of RING105 cDNA, corresponding to amino acids 1–134, was amplified by PCR using the ATCC cDNA as template and ligated into XhoI site in pET28C vector (Novagen, Madison, WI, USA). The primer sequences were as follows: HYYOL-82, 5′-cggctcgagttagaccaggataacagggga-3′; and HYYOL-74: 5′-ccggctcgagcgccaccatgcaccctgcagccttcccg-3′
Bacterially expressed protein was purified by Talon Ni affinity beads (Clontech) and sent to Covance Inc. to immunize two rabbits (numbers 98 and 100). For affinity purification, each rabbit anti-RING105 sera was preincubated with CNBr beads (Pharmacia, Rockville, MD, USA) coupled with bacterial lysate containing His-tagged Uba1 to remove background. Sera were then incubated with amino-link beads bound to RING105 (1–134) and eluted following the manufacturer’s protocol (Pierce, Rockford, IL, USA). For further characterization of the antibody, see Supplementary Information 1.
Cell extraction, immunoprecipitation and immunoblotting
For immunoprecipitation samples, cultured cells at 50–70% confluency cells on 10 cm plates were trypsinized, harvested and frozen at −80°C. The cells were thawed, extracted in 600 µl RIPA buffer, supplemented with 10mm PMSF, 10 µm MG132, 5 µg/ml protease inhibitor cocktail (Sigma, St Louis, MO, USA) and 400 nm microcystin LR to preserve phosphorylations. The extract was cleared by centrifugation at 15 800 g for 15 min at 4°C. The supernatant was incubated with antibody-bound protein A beads for 1.5 to 3 h at 4°C. The beads were washed with the same buffer five times. The samples were boiled for 6 min in LDS sample buffer and loaded onto SDS (4–20%)–polyacrylamide gels (Invitrogen, Carlsbad, CA, USA). Proteins were transferred to BioTrace PVDF (Pall Corporation, East Hills, NY, USA) or Immunobilon P membrane (Millipore, Billerica, MA, USA) using an electrophoretic blotting device. For TSSC5 blotting, cell extracts were mixed with SDS–Hepes sample buffer (final concentration: 50mm Hepes, pH 8.0, 1% (w/v) SDS, 5% glycerol, 0.025% bromophenolblue, 1mm DTT) and loaded onto SDS–PAGE gels without heating to reduce protein aggregation occasionally observed in membrane proteins extracted in conventional SDS sample buffer (Sagne et al., 1996).
We used rabbit anti-GFP antibody (BabCo, Richmond, CA, USA), mouse anti-GFP antibody (Clontech), rabbit anti-HA Y-11 (Santa Cruz, Santa Cruz, CA, USA), rabbit anti-S tag (Gorbsky lab) and mouse anti-HA F7 (Santa Cruz) for immunoprecipitation (0.3 µg/sample) and/or immunoblot (2–5 µg/ml in 5% skim milk in PBS). For human tissue blotting, we used a commercial human multiple tissue blot (Oncogene Research, San Diego, CA, USA).
In vitro ubiquitylation assay
Bacterially expressed and affinity-purified E1 enzyme (human Uba1) and GST-tagged ubiquitin were gifts from Dr T Kamura (Kyushu University, Japan). We used a set of commercially available E2 enzymes: UbcH2, UbcH3, UbcH5a, UbcH5b, UbcH5c, UbcH6, Ubc7 and UbcH10 (Calbiochem, San Diego, CA, USA). We immunoprecipitated GFP-RING105 for 2–3 h using rabbit anti-GFP (AbCam, Cambridge, MA, USA) or mouse anti-GFP (BabCo) with extracts prepared from transfected 293T or HeLa cells that were treated with 10 µm MG132 (AbCam).
For the in vitro RING105 autoubiquitylation assay in Figure 2d, E1 (150 ng), E2 (400 ng) and GST-tagged ubiquitin (40 ng) were mixed in a total 10 µl of reaction buffer (50mm Tris-Cl (pH 8.0), 10mM MgCl2, 50mm NaCl, ATP 2mm, DTT 1mm). Anti-rabbit GFP immunoprecipitates (GFP-RING105) were added and incubated in 30°C for 60 min with occasional agitation. After the reaction, the anti-GFP beads were rinsed extensively with PBS to collect only GFP- or GFP-tagged RING105. Then, we added 4 × LDS buffer to samples and heated them to 95°C for 6 min. The samples were separated by SDS–PAGE and transferred to membranes. GFP-RING105 was detected with mouse anti-GFP (Clontech). GST-ubiquitin conjugates were detected with mouse anti-GST (B-14) (Santa Cruz).
For the in vitro TSSC5 ubiquitylation assay in Figure 3c and d, we mixed E1-expressing bacterial lysate (10 µl), a Ubc (850 ng), GFP-RING105 (rabbit anti-GFP immunoprecipitates), S-tagged TSSC5-C-expressing bacterial extract (25 µl), 1mm DTT, GST-tagged ubiquitin (100 ng), 5mm ATP, 20mm ZnCl2, 50 µm MG132, 50mm Tris-Cl (pH 8.0), 10mm MgCl2 and 50mm NaCl in a total of 50 µl.We incubated the samples for 2 h at 30°C, then stopped the reaction and denatured the proteins by adding 1% SDS. We then diluted the mixture 10-fold to reduce the SDS concentration to 0.1% and removed his-tagged proteins with Talon beads and GFP-tagged proteins with protein A-beads-bound anti-GFP. We took the remaining supernatant and pulled down S-tagged protein with S-protein beads (Novagen/EMD Biosciences Inc., Madison, WI, USA). Samples were separated by SDS–PAGE and transferred to membranes. We visualized ubiquitylated protein with anti-GST antibody.
FACS analysis for GFP-expressing cells
We transfected HeLa cells with plasmids encoding a variety of GFP-tagged RING105 constructs. After 24–40 h, we suspended the cells by typsinization and fixed them with ice-cold 0.5% paraformaldehyde in PBS for 20 min. Fixed cells were rinsed once with PBS and suspended in 80% ethanol (−20°C) for at least 30 min. The permeabilized cells were rehydrated for 5 min and resuspended in PBS, then treated with RNase (0.1 mg/ml) and propidium iodide (20 µg/ml) in room temperature for at least 1 h. The samples were analysed with a FACScalibur and the cell cycle profile was estimated by ModFit software with the aid of The Flow and Image Cytometry Laboratory (University of Oklahoma Health Sciences Center).
Microscopic analysis
GFP-RING105-expressing cells were observed either as living cells or after fixation. For fixation, we used 1% paraformaldehyde (Sigma) in PHEM buffer (details described in Kallio et al., 2002). The samples were analysed with a ZEISS Axioplan IIi microscope equipped with a Hamamatsu Orca II camera and Metamorph Imaging system (Universal Imaging Corp., Downingtown, PA, USA). For live cell observation, a planapochromat × 60 (NA 1.4) objective (Nikon USA) was used with a SenSys CCD camera (Photometrics Ltd, Tucson, AZ, USA) connected to a Nikon Diaphot microscope and imaged with Metamorph software.
Supplementary Material
Acknowledgements
This research is supported by a fellowship from the Breast Cancer Research Program of the US Department of Defense to HY Yamada (DAMD17-02-1-0532), and by a grant from the National Institute of General Medical Science (RO1-GM50412) to GJ Gorbsky. We thank our laboratory members for support, Dr Takumi Kamura (Kyushu University, Japan) for reagents for ubiquitylation assays and Dr Scott Plafker (University of Oklahoma Health Sciences Center) for suggestions.
Footnotes
Supplementary Information accompanies the paper on Oncogene website (http://www.nature.com/onc).
References
- Albrecht S, Hartmann W, Houshdaran F, Koch A, Gartner B, Prawitt D, et al. Int J Cancer. 2004;111:627–632. doi: 10.1002/ijc.20280. [DOI] [PubMed] [Google Scholar]
- Almond JB, Cohen GM. Leukemia. 2002;16:433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper PR, Smilinich NJ, Day CD, Nowak NJ, Reid LH, Pearsall RS, et al. Genomics. 1998;49:38–51. doi: 10.1006/geno.1998.5221. [DOI] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Dao D, Frank D, Qian N, O’Keefe D, Vosatka RJ, Walsh CP, et al. Hum Mol Genet. 1998;7:597–608. doi: 10.1093/hmg/7.4.597. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. J Cell Sci Suppl. 1994;18:7–12. doi: 10.1242/jcs.1994.supplement_18.2. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Cancer Res. 1999;59(Suppl):1743s–1746s. [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Mol Cell Biol. 2000;20:7215–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY. Curr Opin Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- Hershko A, Chiechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Jarosch E, Sommer T, Wolf DH. Biochim Biophys Acta. 2004;1695:215–223. doi: 10.1016/j.bbamcr.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Wilson L. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Chen J, Wong J, Fang G. J Cell Biol. 2002;156:249–259. doi: 10.1083/jcb.200108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koi M, Johnson LA, Kalikin LM, Little PF, Nakamura Y, Feinberg AP. Science. 1993;260:361–364. doi: 10.1126/science.8469989. [DOI] [PubMed] [Google Scholar]
- Lee MP, Reeves C, Schmitt A, Su K, Connors TD, Hu RJ, et al. Cancer Res. 1998;58:4155–4159. [PubMed] [Google Scholar]
- Lewis A, Murrell A. Curr Biol. 2004;14:R284–R286. doi: 10.1016/j.cub.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Li TK, Liu LF. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- Lovering R, Hanson IM, Borden KL, Martin S, O’Reilly NJ, Evan GI, et al. Proc Natl Acad Sci USA. 1993;90:2112–2116. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon P, Bateman A. Protein Sci. 2000;9:1930–1934. doi: 10.1110/ps.9.10.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- Pagano M. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Davies KR, Bowden LM, Villar AJ, Franck O, Fuermann M, et al. Hum Mol Genet. 1998;7:1149–1159. doi: 10.1093/hmg/7.7.1149. [DOI] [PubMed] [Google Scholar]
- Poirier K, Chalas C, Tissier F, Couvert P, Mallet V, Carrie A, et al. J Pathol. 2003;201:473–479. doi: 10.1002/path.1477. [DOI] [PubMed] [Google Scholar]
- Ptak C, Gwozd C, Huzil JT, Gwozd TJ, Garen G, Ellison MJ. Mol Cell Biol. 2001;21:6537–6548. doi: 10.1128/MCB.21.19.6537-6548.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenel JD, Broman KW, Perlman EJ, Niemitz EL, Jayawardena TM, Bell DW, et al. J Natl Cancer Inst. 2001;93:1698–1703. doi: 10.1093/jnci/93.22.1698. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Reece M, Prawitt D, Landers J, Kast C, Gros P, Housman D, et al. FEBS Lett. 1998;433:245–250. doi: 10.1016/s0014-5793(98)00907-7. [DOI] [PubMed] [Google Scholar]
- Romisch K. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Sagne C, Isambert MF, Henry JP, Gasnier B. Biochem J. 1996;316(Part 3):825–831. doi: 10.1042/bj3160825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienbacher C, Gramantieri L, Scelfo R, Veronese A, Calin GA, Bolondi L, et al. Proc Natl Acad Sci USA. 2000;97:5445–5449. doi: 10.1073/pnas.090087497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienbacher C, Sabbioni S, Campi M, Veronese A, Bernardi G, Menegatti A, et al. Proc Natl Acad Sci USA. 1998;95:3873–3878. doi: 10.1073/pnas.95.7.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper LM, Bastians H, Ruderman JV, Gorbsky GJ. J Cell Biol. 2001;154:707–717. doi: 10.1083/jcb.200104130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.