Abstract
Individuated finger and wrist movements can be decoded using random subpopulations of neurons that are widely distributed in the primary motor (M1) hand area. This work investigates (i) whether it is possible to decode dexterous finger movements using spatially-constrained volumes of neurons as typically recorded from a microelectrode array; and (ii) whether decoding accuracy differs due to the configuration or location of the array within the M1 hand area. Single-unit activities were sequentially recorded from task-related neurons in two rhesus monkeys as they performed individuated movements of the fingers and the wrist. Simultaneous neuronal ensembles were re-created by constraining these activities to the recording field dimensions of conventional microelectrode array architectures. Artificial Neural Network (ANN) based filters were able to decode individuated finger movements with greater than 90% accuracy for the majority of movement types, using as few as 20 neurons from these ensemble activities. Furthermore, for the large majority of cases there were no significant differences (p < 0.01) in decoding accuracy as a function of the location of the recording volume. The results suggest that a Brain-Machine Interface (BMI) for dexterous control of individuated fingers and the wrist can be implemented using microelectrode arrays placed broadly in the M1 hand area.
I. INTRODUCTION
Recent advances in Brain-Machine Interfaces (BMIs) have enabled direct neural control of computer cursors and robotic arms in monkeys [1]–[3] and humans [4]. In these systems, simultaneous activity from multiple neurons is obtained using implanted microelectrode arrays, and decoded in real-time to actuate external devices. One of the emerging applications of BMIs is for the control of prosthetic limbs. Trained monkeys have been able to control basic limb functions such as reach, grasp and 3-D trajectories of robotic and prosthetic devices using their neural signals [2], [3]. However, to mimic the full range of natural limb motions, including movements of individual fingers, BMIs for multi-channel dexterous control need to be developed.
In order to develop a BMI for control of individual finger and wrist movements, one needs to determine the nature of the control signal; more specifically, the exact site for deriving such signals and also the appropriate schemes for real-time decoding. The primary motor (M1) hand area seems to be an ideal candidate for such an implantation, since previous work using sequentially recorded single-unit activities from this region has demonstrated the existence of task-related neurons [5], [6]. It has also been shown that individuated finger and wrist movements can be decoded using random subpopulations of neurons widely distributed within this region [7]–[9]. However, it is unclear if it would be possible to decode these movements using ensembles of neurons that are spatially-constrained to volumes typically recorded using implanted microelectrode arrays.
The M1 hand area has been long shown to have functional subdivisions in non-human primates [10] and humans [11], with the individual fingers and wrist representations arranged in a lateral to medial fashion over an approximate area of 36 mm2 [12]. This raises practical difficulties when attempting to record from a single microelectrode array in this region to decode movements related to all the fingers and the wrist. For example, the commonly used Utah Electrode Array [13], which has a footprint of 4 × 4 mm2, would only cover a fraction of the M1 hand area.
Recent empirical evidence, however, has shown that the demarcation of individual sub-regions within the M1 hand area is not as rigid as was previously believed [14]–[17]. This suggests that decoding of all movement types may in fact be possible using a subpopulation of neurons in this region. However, there remains the possibility that, depending on the specific site of implantation, a subset of finger movements may have a negative bias in decoding accuracy or, worse still, not be decodable at all. Therefore, decoding all finger movements with high accuracy may necessitate implanting multiple microelectrode arrays, or a single wide-area electrode array, to adequately cover the M1 hand area.
The other important issue that needs to be investigated is the number of neurons required to decode dexterous movements with high accuracy, and whether this number fluctuates based on the placement of the microelectrode array within the M1 hand area. For example, an implant in the ‘thumb area’ might require fewer neurons to decode thumb movement, and might not have sufficient information to decode wrist movements (the corresponding area for which lies at the opposite end of the M1 hand area). These issues have practical implications on the feasibility of a BMI for dexterous neuroprosthetic control.
In this study, we address two questions: 1) whether confined volumes of neurons from the M1 hand area are sufficient to decode individuated movements of all the fingers and the wrist, and 2) whether any decoding biases are introduced due to the site of implantation of a microelectrode array within this region. Single-unit activities from task-related neurons in the M1 hand area of two trained rhesus monkeys were constrained to contiguous volumes of interest, dubbed voxels, which corresponded to dimensions of the recording fields of commonly used multi-channel microelectrode arrays. The neuronal ensemble activities from these voxels were then used to asynchronously decode individuated finger and wrist movements, which mimics the operation of a self-cued BMI. Statistical inferences were made about the nature of differences between voxel locations and configurations.
II. METHODS
A. Experimental Protocol
This work uses data collected from two male rhesus (Macaca mulatta) monkeys - C and K - that have been the subject of previous studies [8], [17]. The methods used to train the monkeys and the actual experimental protocols have been described in detail in these studies, and are summarized here briefly. The experiments complied with USPHS policy on Humane Care and Use of laboratory animals, and were approved by the institutional Animal Care and Use Committee.
Each monkey was trained to perform visually-cued individuated finger and wrist movements using a pistol-grip manipulandum that separated each finger into a different slot. The monkey was instructed to flex or extend a single digit until a microswitch was closed. A total of 12 distinct movements were performed - flexion and extension of the right five fingers and wrist.
Single-unit activities were recorded from 312 task-related neurons in M1 for monkey C and 115 task-related neurons for monkey K. The depth of the recording sites from the cortical surface ranged between 250 µm and 7 mm for both monkeys.
B. Virtual Microelectrode Arrays
Virtual neuronal ensembles were created from the single-unit recordings by spatially constraining the neurons to contiguous volumes of interest, or voxels, in the M1 hand area. Temporally, simultaneous multi-unit activity was re-created by event-locking each trial to the time of switch closure. This is summarized in Fig. 1, which shows the approximate location of the M1 hand area in a rhesus monkey (Fig. 1a) and the recording space for monkeys C (Fig. 1b) and monkey K (Fig. 1c), respectively. Four different voxel configurations were investigated, which correspond to the typical recording fields of commonly used microelectrode arrays (Fig. 1d–g).
Fig. 1.
Neural recording from the M1 hand area. (a) Approximate location of the M1 hand area, with respect to the central sulcus (dashed line). Actual locations of neurons recorded from (b) monkey C and (c) monkey K, where the depth is with respect to the cortical surface. Recording volumes for the different electrode arrays are shown in the bottom from left to right: (d) Caltech probe, (applied to monkey C), footprint size: 0.5×3×0.5 mm3; (e) Michigan array (monkey C), footprint size: 1×3×1 mm3; (f) modified Utah array, (monkey C), footprint size: 4×4×0.25 mm3; (g) Caltech array (monkey K), footprint size: 3×3×2 mm3.
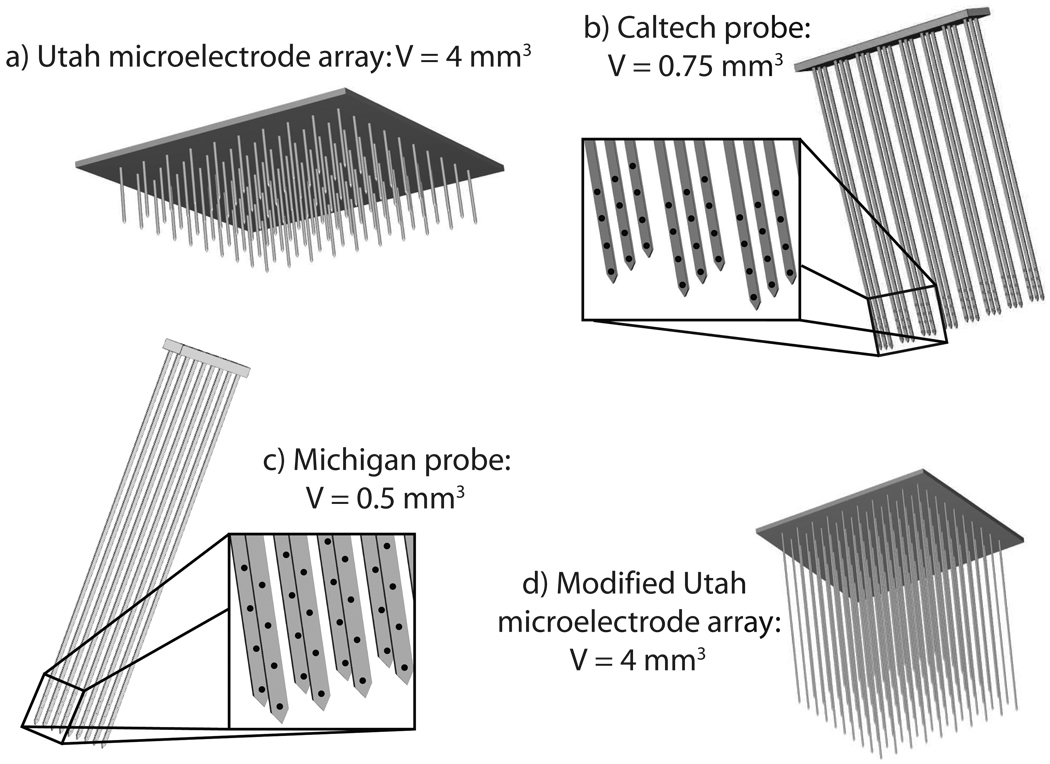
Schematics of the different microelectrode array architectures used in this study are illustrated in Fig. 2. One of the earliest microelectrode arrays developed is the Utah Electrode Array (Fig. 2a) [13]. The electrodes are configured in a 10 × 10 array, with an inter-shank distance of 400 µm and surface footprint of 16 mm2. Each electrode has one recording site per shank and a 125 µm recording radius, which leads to an approximate recording volume of 4 mm3. With shank lengths of 1.5 mm, the Utah array allows recordings exclusively from neurons that are relatively close to the cortical surface. However, since the recording depths for the two monkeys range between 250 µm and 7 mm, it was necessary to modify the recording field of the Utah array by increasing the shank lengths (Fig. 2d). This is in accordance with the Utah group’s intentions to increase the shank length to at least 3 mm [18].
Fig. 2.
Schematic representations of different microelectrode array architectures used for intra-cortical recording that have been developed at (a) University of Utah (Utah Array), (b) California Institute of Technology, and (c) University of Michigan. A modified version of the Utah Electrode Array with longer shanks is also shown (d). The insets show locations of the recording sites for each of the array architectures that have more than one recording site per shank.
Two additional probes are under development at the University of Michigan [19], [20] and the California Institute of Technology [21]. Each probe design has shank lengths up to 8 mm with multiple (4–8) recording sites per shank, and are thus capable of recording from a deeper 3-D volume. A typical Caltech probe consists of three rows of electrodes, with eight shanks per row and four recording sites per shank separated by 0.125–0.5 mm each (Fig. 2b). The probe’s recording volume of 0.5×3×0.5 mm3 is actually smaller than the Utah array. However, since the shanks can be as long as 8 mm, this probe design is suitable for recording deep within the sulcus where the M1 hand area is located [22]. Similarly, probes from the University of Michigan typically consist of two rows of electrodes with eight shanks per row and four to eight recording sites per shank separated by 0.125 mm (Fig. 2c). This leads to a recording volume of 1 × 0.25 × 0.5 mm3.
Monkey C had over 300 recording sites, which could have been encompassed by either the Caltech probe (Fig. 1d), an array of six Michigan probes (‘Michigan array’, Fig. 1e), or a modified version of the Utah array with longer shank lengths (Fig. 1f). Monkey K had a sparser and more discrete recording space (neural recording sites were typically separated by 1 mm in each direction), which could have been encompassed by an array of six Caltech probes (‘Caltech array’, Fig. 1g). Each voxel configuration, corresponding to different arrays, were placed at five distinct locations within the recording space. Different subpopulations of neurons from within these voxels were randomly selected to mimic the reality of recording from actual electrode arrays placed in the target area.
C. Asynchronous Decoding of Movements
Non-linear decoding filters were designed using multilayer, feedforward Artificial Neural Networks (ANNs). Unlike previous studies [9], [23] that have focused on decoding finger movements in a synchronous fashion using a priori knowledge of the movement events, a novel hierarchical classification scheme was used for asynchronous decoding where cues indicating the onset of movement are not known. This algorithm, which decodes both the onset of movement and movement type, was described in detail in [7], [8] and is outlined here briefly.
First, a Gating Classifier was designed as a committee of N neural networks, and trained to distinguish baseline activity from the onset of movement. Each ANN in the committee was trained using the neuronal firing rate over a 100 ms temporal window shifting every 20 ms, and assigned a fuzzy output label, P{I(tk)}, at discrete times tk, that described the temporal evolution of spiking activity and onset of movement. The output of each individual gating network, g(tk), was thresholded at T1 to produce a discrete binary variable, where an output of 0 corresponded to rest and 1 corresponded to movement. A majority voting rule was used to determine the committee output, and additional threshold parameters T2 and tj were optimized to eliminate spurious gating classifications and maximize classifier accuracy. The final classifier output, G(tk), was,
| (1) |
where
| (2) |
Next, a Movement Classifier was designed as a committee of N neural networks, and trained to distinguish amongst each movement type. Each ANN was trained using the neuronal firing rate during the 100 ms window preceding switch closure, and assigned a binary output label for each movement type, P{Mi(tk)} at discrete times tk. For the ith movement type, the ith output neuron was assigned an output of 1 during this window and all other neurons were assigned an output of 0. The output of each individual movement network, s(tk), was selected based on the output neuron that had the greatest output activity, and a majority voting rule was then used to determine the final committee output of the movement classifier, S(tk),
| (3) |
Where
The final decoded output, F(tk), was obtained from the product of the output of the two committee networks:
| (4) |
D. Statistics
Non-parametric tests were performed to test for differences in the median decoding accuracies of neuronal populations from different voxels. Voxel placement in the M1 hand area was treated as the primary random effect, and the following null hypotheses were tested:
the median overall decoding accuracy was independent of voxel placement;
-
the median decoding accuracies for movements of each of the individual fingers (1–5) and the wrist (W) were not biased by voxel placement;
The following additional hypothesis was tested for monkey C:
the median overall decoding accuracy was independent of the dimensions of the recording volume.
When these null hypotheses were rejected, post hoc tests were performed to identify the pair-wise differences.
III. RESULTS
For each voxel configuration, asynchronous decoding accuracy was evaluated for neuronal ensembles from five voxels placed in varying locations, parallel to the central sulcus, in the M1 hand area. In order to determine the effects of voxel location on decoding accuracy, and minimize the overlap of neuronal subpopulations, voxels were chosen to be maximally separated (in Euclidean distance). Results were calculated for 100 trials per movement type, and the decoding accuracy was calculated for six random repetitions of any given number of neurons.
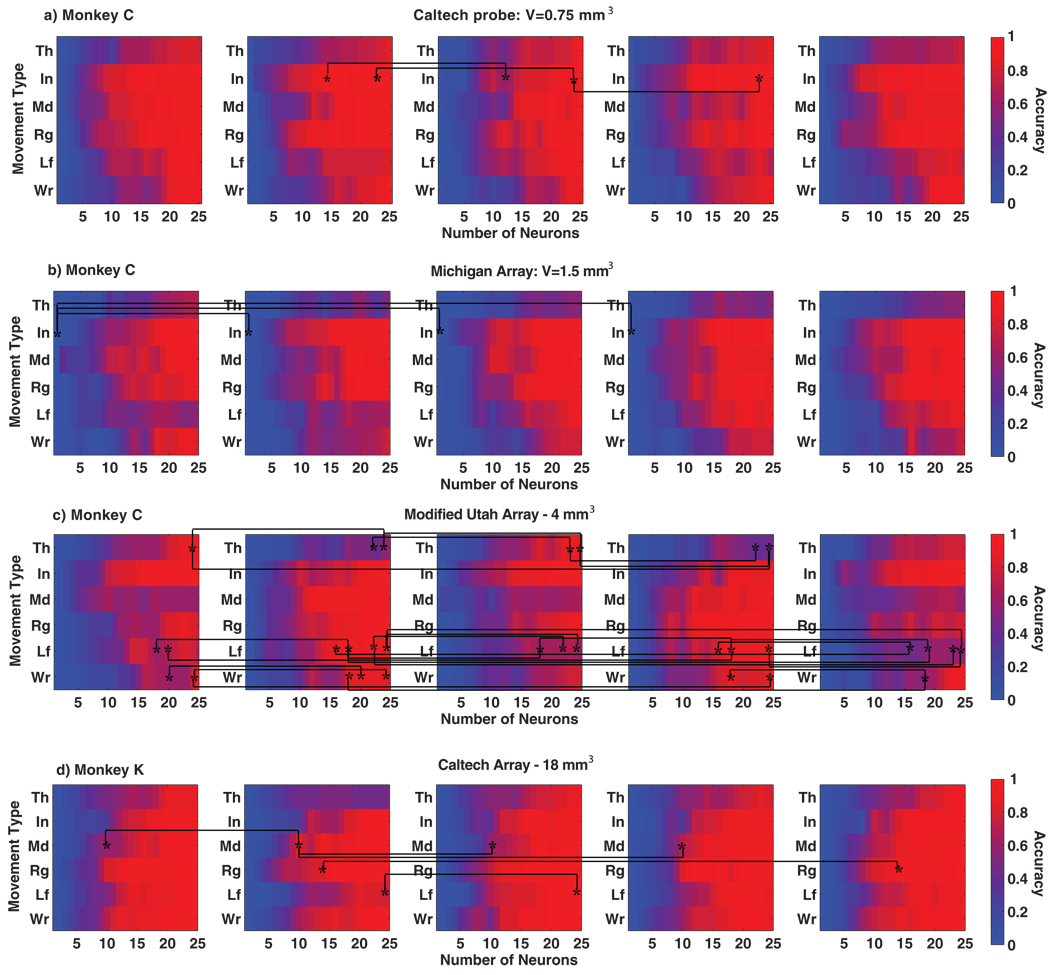
The overall asynchronous decoding accuracies for both monkeys are summarized in Fig. 3. Table I shows the average accuracies and standard errors for 20 and 25 neurons. As can be seen, the average decoding accuracy was approximately greater than 80% with as few as 25 neurons in monkey C and greater than 85% with as few as 20 neurons in monkey K, irrespective of the voxel configuration and placement. The asynchronous decoding accuracies for both monkeys across each movement type are depicted in Fig. 4. For the majority of movement types, decoding accuracy was greater than 90% using as few as 20 neurons. The following sections describe the statistical differences in decoding accuracies between the different voxel configurations and their location across the M1 hand area.
Fig. 3.
Overall asynchronous decoding accuracy for both monkeys and different voxel configurations. Each subplot depicts accuracies from five distinct voxel placements within the M1 hand area. (a) monkey C: Caltech probe; (b) monkey C: Michigan array (c) monkey C: Modified Utah array probe (d) monkey K: Caltech array.
Fig. 4.
Overall decoding accuracy for the different voxel configurations, as a function of the movement type and the number of neurons. For each voxel configuration, results are shown for five distinct voxel placements within the M1 hand area. Significant pair-wise differences in decoding accuracy due to voxel placement are depicted with an asterisk (a) monkey C: Caltech probe (b) monkey C: Michigan array (c) monkey C: Modified Utah array (d) monkey K: Caltech array.
A. Differences in Overall Decoding Accuracy Due to Voxel Location
Kruskal-Wallis tests were conducted (p <0.01) to test the hypothesis that the median overall decoding accuracies were not significantly different due to differences in voxel location within the M1 hand area. This was repeated for the four voxel configurations (as described previously) and for different sizes of neuronal subpopulations sampled from them. Each voxel configuration was placed in five different locations, with six random samples for each neuronal subpopulation. The p-values are reported in Table II. Note that for the large majority of cases, no significant differences were detected in the overall decoding accuracies due to voxel placements.
TABLE II.
P-VALUES FOR STATISTICAL TESTS OF DIFFERENCE IN OVERALL DECODING ACCURACY DUE TO VOXEL LOCATION
| Neurons | Monkey C Caltech probe | Monkey C Mich array | Monkey C Mod Utah array | Monkey K Caltech array |
|---|---|---|---|---|
| 2 | 0.38 | 0.12 | 0.61 | 0.30 |
| 5 | 0.03 | 0.28 | 0.24 | 0.72 |
| 8 | 0.32 | 0.28 | 0.04 | 0.65 |
| 10 | 0.05 | 0.94 | 0.18 | 0.11 |
| 12 | 0.25 | 0.63 | 0.01 | 0.76 |
| 14 | 0.63 | 0.93 | 0.25 | 0.33 |
| 16 | 0.22 | 0.03 | 0.14 | 0.31 |
| 18 | 0.85 | 0.91 | 0.00* | 0.17 |
| 20 | 0.32 | 0.42 | 0.02 | 0.47 |
| 23 | 0.17 | 0.40 | 0.03 | 0.16 |
| 25 | 0.04 | 0.88 | 0.03 | 0.21 |
denotes p< 0.01
B. Biases in Decoding Accuracies of Movements of the Fingers and Wrist due to Voxel Location
Kruskal-Wallis tests were conducted (p <0.01) to test the hypothesis that the median decoding accuracies for movements of each of the individual fingers and the wrist were not significantly biased due to differences in voxel location. Flexions and extensions were combined to form a single group for each finger (1–5) and the wrist (W). This was repeated for the different sizes of neuronal subpopulations sampled from each voxel. The tests were performed for each voxel configuration for both monkeys. Fig. 4 highlights the significant pair-wise differences (Tukey-Kramer method) between different voxel locations with an asterisk. The p-values for a sample number of neurons (n=25) are reported in Table III (six samples for each of the five voxel placements). Note that for a large majority of cases, no significant differences were found in the decoding accuracies of any given finger or the wrist due to voxel location.
TABLE III.
P-VALUES FOR STATISTICAL TESTS OF DIFFERENCE IN DECODING ACCURACY OF INDIVIDUAL DIGITS AND WRIST MOVEMENTS DUE TO VOXEL LOCATION (USING 25 NEURONS)
| Digit | Monkey C Caltech probe | Monkey C Mich array | Monkey C Mod Utah array | Monkey K Caltech array |
|---|---|---|---|---|
| Thumb | 0.49 | 0.70 | 0.00* | 0.69 |
| Index | 0.12 | 0.54 | 0.08 | 0.06 |
| Middle | 0.18 | 0.58 | 0.35 | 0.05 |
| Ring | 0.49 | 0.81 | 0.07 | 0.09 |
| Pinky | 0.37 | 0.39 | 0.00* | 0.00* |
| Wrist | 0.63 | 0.73 | 0.48 | 0.84 |
denotes p< 0.01
C. Differences in Decoding Accuracy Due to the Dimensions of the Recording Volume
This analysis could only be performed for monkey C, since the distribution of the neurons recorded from monkey K did not allow for the simulation of voxel configurations enclosing smaller volumes. Friedman’s 2-way ANOVA was performed to test the hypothesis that the median overall decoding accuracies were not different due to the dimensions of the recording volume. The three different voxel configurations were taken as the primary treatment effect, and the number of sampled neurons was the second treatment effect. Due to the low decoding accuracies and high variability associated with lesser number of neurons, the second treatment effect (number of neurons) was restricted to neuron populations that had a decoding accuracy of at least 60% (i.e. ≥ 14). Significant differences were present between the median ranks of the groups. Post hoc tests revealed that the modified Utah probe configuration has had a lower decoding accuracy than the Caltech probe and Michigan array configurations, but significantly so only with the Caltech probe.
IV. DISCUSSION
A. Differences with Simultaneous Recordings
It should be noted that the data used for this study was generated using sequentially recorded single-unit activities from task-related neurons. These neurons were typically selected based on high firing rates, and possibly contain more information than those that would have been recorded using implanted microelectrode arrays. Additionally, factors such as correlations between neurons could affect the decoding performance favorably or unfavorably depending upon the patterns recorded [24], [25]. Recent comparative studies have shown that the effect of these correlations is negligible when recording from a few neurons, but can be significant when recording from a population of 1000 neurons or more [26].
When using implanted microelectrode arrays, one also does not have the luxury of recording exclusively from task-related neurons. This results in only a fraction of the recorded neurons being related to the movements that are being decoded. Leading groups developing microelectrode arrays have conducted studies to estimate the number of units recorded by such arrays, and the number of task-related neurons as a fraction of that population. In a long term study, the Utah array has been reported to record activity from an average of 1.3 units per recording site [27]. With 100 recording sites, this results in activity from an average of 130 neurons being recorded simultaneously. Furthermore, for neuromotor prosthetic applications using the Utah array, it has been shown that approximately 66% of this recorded population was task-related. Similar results have been reported for the Michigan arrays where activity was recorded from an average of 0.7 units per recording site [19], [28], and where more than 50% of the neurons were reported as being task-related for a specific motor task [29].
These reports suggest that a Michigan array or Utah array (of appropriate depth) with around 100 recording sites would pick up activity from at least 30–40 task-related neurons (by conservative estimates). Since we are able to decode the majority of movement types with greater than 90% accuracy using as few as 20 neurons, this implies that the analysis presented in this work should carry over directly to future closed-loop BMI studies using microelectrode arrays.
B. Decoding Variability within a Voxel
In situations where long-term recordings are made by an implanted microelectrode array, it is a common observation that the neuron population being recorded from is dynamic and could change between recording sessions or even within a given recording session [28]. Both scenarios would require a recalibration of the decoding algorithm. As shown in Table I, the fact that randomly-selected subpopulations of any given number of neurons within a voxel did not have marked variability in decoding accuracy, suggests that such a recalibration for this particular BMI application would indeed be possible.
TABLE I.
SUMMARY OF ASYNCHRONOUS DECODING ACCURACIES FOR ALL VOXEL CONFIGURATIONS AND LOCATIONS (%)
| Neurons | Array Type | Voxel 1 | Voxel 2 | Voxel 3 | Voxel 4 | Voxel 5 |
|---|---|---|---|---|---|---|
| 20 | Monkey C - Caltech probe | 89.8±1.4 | 86.9±3.2 | 83.4±2.4 | 76.1±5.4 | 81.1±2.2 |
| 25 | 94.5±2.1 | 94.2±0.8 | 96.3±0.6 | 90.2±2.4 | 86.7±3.0 | |
| 20 | Monkey C - Mich array | 76.7±5.3 | 77.6±1.7 | 77.2±3.1 | 83.2±2.2 | 76.5±2.8 |
| 25 | 81.3±2.4 | 81.8±2.8 | 84.0±2.9 | 81.5±3.1 | 84.9±1.2 | |
| 20 | Monkey C - Mod Utah array | 70.8±2.6 | 85.1±3.0 | 72.4±2.2 | 80.6±2.9 | 71.5±3.3 |
| 25 | 78.7±3.3 | 89.2±2.8 | 81.4±1.2 | 87.1±2.2 | 76.9±2.5 | |
| 20 | Monkey K - Caltech array | 94.0±0.9 | 86.0±1.9 | 92.2±1.3 | 94.2±1.1 | 91.1±1.7 |
| 25 | 96.9±0.5 | 86.5±2.8 | 97.8±0.5 | 92.9±2.2 | 95.9±1.6 | |
C. Decoding Biases due to Voxel Location
Typically, arrays are implanted in an approximate region using anatomical landmarks and decoding algorithms are trained with whatever population of neurons that the array ends up recording from. In the presence of strong somatotopic organization, placement of an array could result in a decoding bias, against movements that are not represented at the site of implantation. Our results clearly show that for a large majority of cases, decoding accuracies for a given neuronal subpopulation are not significantly different as a function of voxel placement. This implies that it is possible to operate a BMI, for dexterous control of individual fingers, using a single microelectrode array placed anywhere in the broadly-defined M1 hand area, without encountering any movement-specific decoding bias.
D. Choosing an Appropriate Electrode Array Design
The M1 hand area lies buried, in large part, in the anterior bank of the central sulcus [12]. Due to the depth at which task-related neurons for finger movements are found, specialized array types with recording lengths of 4 mm or greater would be needed for reliable decoding [22]. Our results suggest that most of the currently available electrode arrays that can reach to such depths would suffice for the purpose of operating a neuroprosthetic for control of individual fingers. However, the overall decoding accuracy obtained using the modified Utah array seems to be lower than those arrays with a deeper 3-D span. We do not have an explanation for this observation, and it should be noted that this is only the case with a single monkey. Further investigation would be needed before making general inferences about the suitability of one type of array over another. With this particular dataset, however, we observe that microelectrode arrays that are able to record from deeper 3-D volumes result in a better decoding accuracy and may be better suited for this application.
E. Implications for Closed-Loop Control
Most current BMIs rely on operant conditioning, for successful operation, whereby, with training, users learn to continuously adapt their neural signals to achieve closed-loop control of the end effectors [30]. Therefore, it could be argued that decoding the native state of the neurons with very high accuracy is not as relevant. While this may be true for the first generation of BMIs, the focus on neurally-controlled prosthetic devices with multiple degrees of freedom has brought forth the need for ‘intuitive’ neural control. During actual closed-loop experiments, some degree of operant conditioning will certainly occur; however, starting from a baseline that is close to the natural state of the neurons will make the operation of neuroprosthetic devices seem natural, without necessitating counter-intuitive mental strategies to actuate the desired end-effectors. The results of this study indicate that by using neuronal spike activities as typically recorded by microelectrode arrays, it is possible to decode the native intent of individual finger movements and wrist. This suggests that ‘natural’ dexterous closed-loop control of multi-fingered hand prostheses is indeed feasible.
V. CONCLUSIONS
We have demonstrated that it is possible to decode individual finger and wrist movements from a confined volume of neurons in the M1 hand area typically with decoding accuracy of greater than 90% for the majority of movement types using as few as 20 neurons. Furthermore, the location of the sampled neurons within the M1 hand area did not significantly affect the decoding accuracy, which indicates that a BMI for control of individual fingers would be possible using microelectrode arrays placed anywhere within this cortical region. Ongoing work is directed towards recording simultaneous activity using microelectrode arrays, and subsequently training monkeys to operate a closed-loop BMI for dexterous control of individual fingers and the wrist.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Florian Solzbacher for our discussions about the next generation of Utah microelectrode arrays, Drs. Dan Rizzuto and Jeremy Emken for the information provided on the Caltech arrays, and Dr. Rio Vetter for his updates on the new Michigan arrays.
This work was funded and supported by the Defense Advanced Research Project Agency (DARPA) Revolutionizing Prosthetics 2009 program, and by R01/R37 NS27686 from the National Institute of Neurological Disease and Stroke (NINDS).
REFERENCES
- 1.Serruya MD, Hatsopoulos N, Paninski L, Fellows MR, Donoghue JP. Instant neural control of a movement signal. Nature. 2002;vol. 416(no 6877):141–142. doi: 10.1038/416141a. [DOI] [PubMed] [Google Scholar]
- 2.Carmena JM, Lebedev MA, Crist RE, O’Doherty JE, Santucci DM, Dimitrov DF, Patil PG, Henriquez CS, Nicolelis MA. Learning to control a brain-machine interface for reaching and grasping by primates. Public Library of Science Biology. 2003;vol. 1(no 2):E42. doi: 10.1371/journal.pbio.0000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapin JK, Moxon KA, Markowitz RS, Nicolelis MA. Real-time control of a robot arm using simultaneously recorded neurons in the motor cortex. Nature Neuroscience. 1999;vol. 2:664–670. doi: 10.1038/10223. [DOI] [PubMed] [Google Scholar]
- 4.Hochberg LR, Serruya MD, Friehs GM, Mukand JA, Saleh M, Caplan AH, Branner A, Chen D, Penn RD, Donoghue JP. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature. 2006;vol. 442:164–171. doi: 10.1038/nature04970. [DOI] [PubMed] [Google Scholar]
- 5.Schieber MH. Individuated finger movements of rhesus monkeys. Journal of Neurophysiology. 1991;vol. 65(no 6):1381–1391. doi: 10.1152/jn.1991.65.6.1381. [DOI] [PubMed] [Google Scholar]
- 6.Poliakov AV, Schieber MH. Limited functional grouping of neurons in the motor cortex hand area during individuated finger movements: A cluster analysis. Journal of Neurophysiology. 1999;vol. 82(no 6):3488–3505. doi: 10.1152/jn.1999.82.6.3488. [DOI] [PubMed] [Google Scholar]
- 7.Acharya S, Aggarwal V, Tenore F, Shin HC, Etienne-Cummings R, Schieber MH, Thakor NV. Towards a brain-computer interface for dexterous control of a multi-fingered prosthetic hand. IEEE EMBS Conference on Neural Engineering. 2007:200–203. [Google Scholar]
- 8.Aggarwal V, Acharya S, Tenore F, Shin HC, Etienne-Cummings R, Schieber MH, Thakor NV. Asynchronous decoding of dexterous finger movements using M1 neurons. submitted to IEEE Transactions on Neural Systems and Rehabilitation Engineering. doi: 10.1109/TNSRE.2007.916289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamed SB, Schieber MH, Pouget A. Decoding M1 neurons during multiple finger movements. Journal of Neurophysiology. vol. 13:1027–1034. doi: 10.1152/jn.00760.2006. [DOI] [PubMed] [Google Scholar]
- 10.Woolsey C, Settlage P, Meyer D, Sencer W, Hamuy T, Travis A. Patterns of localization in precentral and ”supplementary” motor areas and their relation to the concept of a premotor area. Res. Publ. Assoc. Res. Nerv. Ment. Dis. 1952;vol. 30:238–264. [PubMed] [Google Scholar]
- 11.Penfield W, Rasmussen T. The cerebral cortex of man. New York, NY: Macmillan; 1950. [Google Scholar]
- 12.Park M, Belhaj-Saïf A, Gordon M, Cheney P. Consistent features in the forelimb representation of primary motor cortex in rhesus macaques. Journal of Neuroscience. 2001;vol. 21(no 8):2784–2792. doi: 10.1523/JNEUROSCI.21-08-02784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard EM, Nordhausen CT, Normann RA. The Utah intracortical Electrode Array: A recording structure for potential brain-computer interfaces. Electroencephalogr. Clin. Neurophysiol. 1997;vol. 102:228–239. doi: 10.1016/s0013-4694(96)95176-0. [DOI] [PubMed] [Google Scholar]
- 14.Dechent P, Frahm J. Functional somatotopy of finger representations in human primary motor cortex. Human Brain Mapping. vol. 18:272–283. doi: 10.1002/hbm.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Indovina I, Sanes JN. On somatotopic representation centers for finger movements in human primary motor cortex and supplementary motor area. Neuroimage. vol. 13:1027–1034. doi: 10.1006/nimg.2001.0776. [DOI] [PubMed] [Google Scholar]
- 16.Schieber MH. Constraints on somatotopic organization in the primary motor cortex. Journal of Neurophysiology. 2001;vol. 86(no 5):2125–2143. doi: 10.1152/jn.2001.86.5.2125. [DOI] [PubMed] [Google Scholar]
- 17.Schieber MH, Hibbard LS. How somatotopic is the motor cortex hand area? Science. 1993;vol. 261:489–492. doi: 10.1126/science.8332915. [DOI] [PubMed] [Google Scholar]
- 18.Discussions with Prof. Florian Solzbacher, University of Utah.
- 19.Vetter RJ, Williams JC, Hetke JF, Nunamaker EA, Kipke DR. Chronic neural recording using silicon-substrate microelectrode arrays implanted in cerebral cortex. IEEE Trans. Biomed. Eng. 2004;vol. 51:896–904. doi: 10.1109/TBME.2004.826680. [DOI] [PubMed] [Google Scholar]
- 20.Written communication with Dr. Rio Vetter, University of Michigan.
- 21.Written communication with Drs. Dan Rizzuto and Jeremy Emken, California Institute of Technology.
- 22.Schwartz AB, Cui XT, Weber DJ, Moran D. Brain-controlled interfaces: Review movement restoration with neural prosthetics. Neuron. 2006;vol. 52:205–220. doi: 10.1016/j.neuron.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Georgopoulos AP, Pellizzer G, Poliakov A, Schieber M. Neural coding of finger and wrist movements. Journal of Computational Neuroscience. 1999;vol. 6(no 3):279–288. doi: 10.1023/a:1008810007672. [DOI] [PubMed] [Google Scholar]
- 24.Abbott L, Dayan P. The effect of correlated variability on the accuracy of a population code. Neural Computation. 1999;vol. 11:91–101. doi: 10.1162/089976699300016827. [DOI] [PubMed] [Google Scholar]
- 25.Sompolinsky H, Yoon H, Kang K, Shamir M. Population coding in neuronal systems with correlated noise. Phys Rev E. vol. 64:051904. doi: 10.1103/PhysRevE.64.051904. [DOI] [PubMed] [Google Scholar]
- 26.Zohary E, Shadlen MN, Newsome WT. Correlated neuronal discharge rate and its implications for psychophysical performance. Nature. 1994;vol. 370:140–143. doi: 10.1038/370140a0. [DOI] [PubMed] [Google Scholar]
- 27.Suner S, Fellows MR, Vargas-Irwin C, Nakata GK, Donoghue JP. Reliability of signals from a chronically implanted, silicon-based electrode array in non-human primate primary motor cortex. IEEE Trans. Neural Syst. Rehabil. Eng. 2005;vol. 13:524–541. doi: 10.1109/TNSRE.2005.857687. [DOI] [PubMed] [Google Scholar]
- 28.Kipke D, Vetter R, Williams J, Hetke J. Silicon-substrate intracortical microelectrode arrays for long-term recording of neuronal spike activity in cerebral cortex. Transactions on Neural Systems and Rehabilitation Engineering. 2003;vol. 11(no 2):151–154. doi: 10.1109/TNSRE.2003.814443. [DOI] [PubMed] [Google Scholar]
- 29.Jog M, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Building neural representations of habits. Science. 1999;vol. 286:1745–1749. doi: 10.1126/science.286.5445.1745. [DOI] [PubMed] [Google Scholar]
- 30.Lebedev MA, Carmena JM, O’Doherty JE, Zacksenhouse M, Henriquez CS, Principe JC, Nicolelis MAL. Cortical ensemble adaptation to represent velocity of an artificial actuator controlled by a brain-machine interface. Journal of Neuroscience. vol. 25(no 19):4681–4693. doi: 10.1523/JNEUROSCI.4088-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]