Abstract
To identify risk variants for lung cancer, we conducted a multistage genome-wide association study. In the discovery phase, we analyzed 315,450 tagging SNPs in 1,154 current and former (ever) smoking cases of European ancestry and 1,137 frequency-matched, ever-smoking controls from Houston, Texas. For replication, we evaluated the ten SNPs most significantly associated with lung cancer in an additional 711 cases and 632 controls from Texas and 2,013 cases and 3,062 controls from the UK. Two SNPs, rs1051730 and rs8034191, mapping to a region of strong linkage disequilibrium within 15q25.1 containing PSMA4 and the nicotinic acetylcholine receptor subunit genes CHRNA3 and CHRNA5, were significantly associated with risk in both replication sets. Combined analysis yielded odds ratios of 1.32 (P < 1 × 10−17) for both SNPs. Haplotype analysis was consistent with there being a single risk variant in this region. We conclude that variation in a region of 15q25.1 containing nicotinic acetylcholine receptors genes contributes to lung cancer risk.
Lung cancer is frequently cited as a malignancy attributable solely to environmental exposures—primarily cigarette smoke. However, evidence that genetic factors influence lung cancer susceptibility has been provided by numerous studies, beginning with the landmark study of Tokuhata and Lilienfeld1, which demonstrated a 2.5-fold higher risk in smoking first-degree relatives of lung cancer cases compared with smoking relatives of controls and showed that the familial aggregation of lung cancer in case relatives compared to control relatives occurred irrespective of the relative’s smoking history. Subsequent epidemiological case-control analyses have consistently provided evidence for a two- to threefold increased lung cancer risk in relatives of cases compared with those of controls2.
Direct evidence for a genetic predisposition to lung cancer is provided by the increased risk associated with constitutional TP53 (tumor protein p53)4 and RB1 (retinoblastoma)5,6 gene mutations, rare mendelian cancer syndromes such as Bloom’s7 and Werner’s syndromes8, and strongly familial lung cancer9. The genetic basis of inherited susceptibility to lung cancer outside the context of these disorders is at present undefined, but a model in which high-risk alleles account for all of the excess familial risk seems unlikely. Alternatively, part of the inherited genetic risk may be caused by low-penetrance alleles. This hypothesis implies that testing for allelic association should be a powerful strategy for identifying alleles that predispose to lung cancer.
We conducted a genome-wide association study (GWAS) of histologically confirmed non–small cell lung cancer (NSCLC) to identify common low-penetrance alleles influencing lung cancer risk. To minimize confounding effects from cigarette smoking and increase the power to detect genetic effects, we frequency matched controls to cases according to smoking behavior. We also matched controls to cases by age (within 5 year categories) and sex, and we further matched former smokers by years of cessation (Table 1). To minimize confounding by ethnic variation, we restricted our study population to individuals of self-reported European descent.
Table 1.
Characteristics of study populations
| Texas discovery set | Texas replication set | UK replication set | ||||
|---|---|---|---|---|---|---|
| Characteristic | Cases (n = 1,154) | Controls (n = 1,137) | Cases (n = 711) | Controls (n = 632) | Cases (n = 2,013) | Controls (n = 3,062) |
| Age (s.d.) | 62.1 (10.8) | 61.1 (8.9)c | 64.6 (9.9) | 57.1 (13.2)d | 64.5 (9.9) | 60.7 (10.6)d |
| Sex (% male) | 57.0 | 56.6 | 56.1 | 54.9 | 49.9 | 35.3 |
| Never smokers (%) | 0 | 0 | 0 | 0 | 6.25 | 36.9 |
| Former smokers (%) | 52.3 | 57.8 | 56.1 | 44.8 | 65.8 | 40.1 |
| Current smokers (%) | 47.8 | 42.2 | 43.9 | 55.2 | 27.9 | 23.0 |
| Cigarettes per day (s.d.)a | 28.0 (13.6) | 26.6 (14.3)c | 28.1 (39.0) | 24.7 (14.6)c | 22.6 (12.8) | 18.2 (11.5)d |
| Pack years (s.d.)a | ||||||
| Current smokers | 57.3 (30.6) | 47.1 (29.1) | 57.2 (33.3) | 38.5 (27.4)d | 51.0 (28.2) | 35.5 (19.6)d |
| Former smokers | 46.2 (31.2) | 42.8 (30.9)c | 48.1 (56.8) | 39.9 (31.9)c | 43.6 (28.3) | 27.7 (22.8)d |
| Years smoked (s.d.)a | ||||||
| Current smokers | 40.2 (11.0) | 39.0 (11.0)b | 41.8 (10.7) | 32.7 (13.4)d | 45.5 (10.1) | 41.1 (9.8)d |
| Former smokers | 31.9 (12.8) | 28.2 (11.9)d | 32.5 (12.3) | 26.5 (13.1)d | 38.8 (11.9) | 28.8 (13.7)d |
| Adenocarcinoma (%) | 54.6 | 49.9 | 22.6 | |||
| Squamous (%) | 26.6 | 26.9 | 33.6 | |||
| Other carcinoma (%) | 18.8 | 23.1 | 40.8 | |||
Excludes cases and controls who are never smokers.
P > 0.05 comparing case and control means.
0.01 < P < 0.05 comparing case and control means.
P < 0.0001 comparing case and control means.
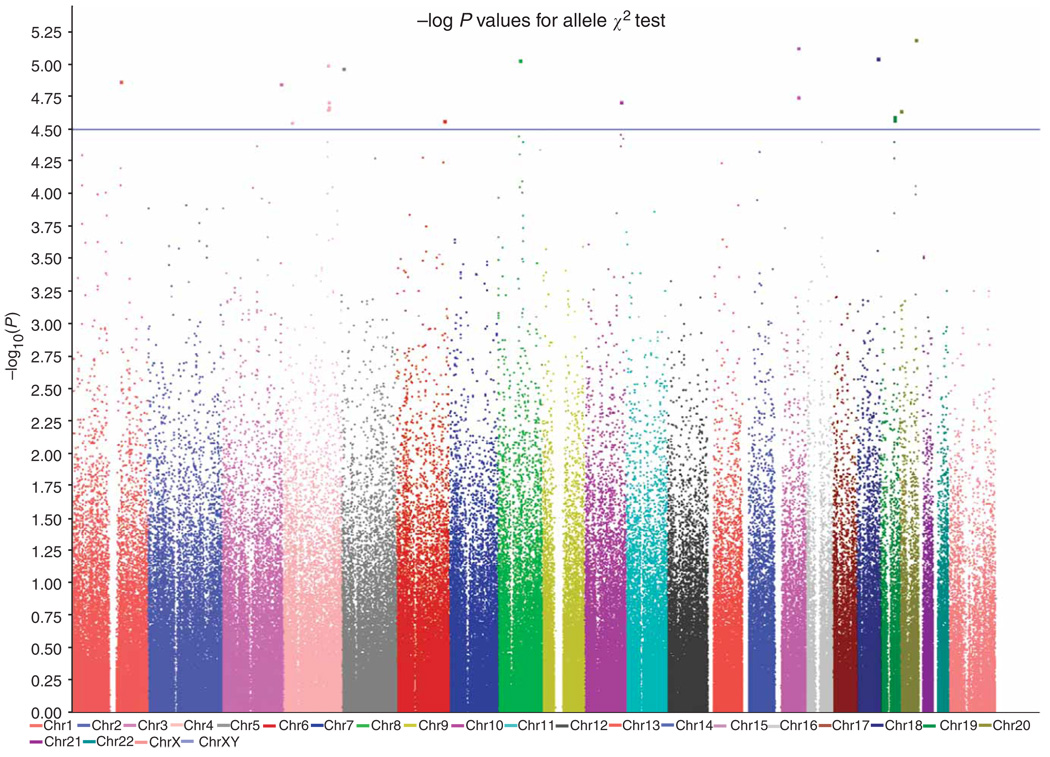
Using Illumina HumanHap300 v1.1 BeadChips, we genotyped 317,498 tagging SNPs in a series of 1,154 ever-smoking lung cancer cases and 1,137 ever-smoking controls (Texas discovery series; Table 2). There was no evidence of genome-wide inflation of χ2 tests, which can occur in the presence of population substructure. The GWAS identified several genomic locations as potentially associated with lung cancer risk (Fig. 1). We further verified that these findings were robust to potential substructure by conditioning on marker similarity either by using Cochran-Mantel-Haenszel tests (Supplementary Fig. 1 online) or by conditioning on eigenvectors (Supplementary Table 1 online).
Table 2.
Summary of ten fast-track SNPs analyzed in discovery and replication studies
| Discovery | Texas replication | UK replication | Combined | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. allelea | Chromosome | Position (bp) | Nearest gene or RNAb | OR (95% CI) |
P valuec (trend P) |
OR (95% CI) |
P valuec (trend P) |
OR (95% CI) |
P valuec (trend P) |
OR (95% CI) |
P value (ind.)d (P value (rep.)e) |
|
| rs2808630 | G | 1 | 156493941 | CRP | 0.76 | 2.05 × 10−5 | 0.93 | 0.426 | Not done | 0.82 | 7.40 × 10−6 | |
| (0.67–0.86) | (1.60 × 10−5) | (0.79–1.10) | (0.421) | (0.74–0.91) | (0.426) | |||||||
| rs7626795 | G | 3 | 191833163 | ILlRAP | 1.46 | 2.12 × 10−5 | 1.05 | 0.709 | 1.05 | 0.512 | 1.16 | 7.80 × 10−6 |
| (1.23–1.74) | (1.94 × 10−5) | (0.83–1.32) | (0.708) | (0.891–1.20) | (0.512) | (1.05–1.28) | (0.451) | |||||
| rs2202507 | C | 4 | 145615286 | GYPA | 1.30 | 8.49 × 10−6 | 0.92 | 0.267 | 1.00 | 0.970 | 1.06 | >1 × 10−5 |
| (1.16–1.46) | (8.67 × 10−6) | (0.79–1.06) | (0.262) | (0.91–1.09) | (0.970) | (1.00–1.14) | (0.556) | |||||
| rs11099666 | G | 4 | 148991033 | ARHGAP10 | 0.65 | 4.90 × 10−5 | 0.82 | 0.143 | 1.03 | 0.667 | 0.86 | >1 × 10−5 |
| (0.53–0.80) | (4.45 × 10−5) | (0.64–1.07) | (0.144) | (0.89–1.21) | (0.664) | (0.78–0.97) | (0.704) | |||||
| rs1481847f | A | 8 | 72944049 | MSC | 1.30 | 1.04 × 10−5 | 1.12 | 0.144 | 0.98 | 0.613 | 1.09 | 1.25 × 10−5 |
| (1.16–1.47) | (1.21 × 10−5) | (0.96–1.31) | (0.142) | (0.89–1.07) | (0.620) | (1.02–1.16) | (0.770) | |||||
| rs855974 | C | 10 | 119436858 | EMX2 | 0.74 | 1.42 × 10−5 | 1.02 | 0.849 | 1.00 | 0.948 | 0.92 | >1 × 10−5 |
| (0.65–0.85) | (1.37 × 10−5) | (0.85–1.21) | (0.848) | (0.91–1.11) | (0.948) | (0.85–0.99) | (0.880) | |||||
| rs8034191 | A | 15 | 76593078 | L0C123688 | 1.30 | 1.76 × 10−5 | 1.34 | 0.00036 | 1.33 | 1.94 × 10−11 | 1.32 | 3.15 × 10−18 |
| (1.15–1.47) | (1.94 × 10−5) | (1.14–1.57) | (0.00047) | (1.22–1.44) | (3.61 × 10−11) | (1.24–1.41) | (2.88 × 10−14) | |||||
| rs1051730 | G | 15 | 76681394 | CHRNA3 | 1.31 | 9.84 × 10−6 | 1.33 | 0.00042 | 1.32 | 2.33 × 10−10 | 1.32 | 7.00 ×10−18 |
| (1.16–1.48) | (1.14 × 10−5) | (1.13–1.55) | (0.00052) | (1.20–1.43) | (3.53 × 10−10) | (1.23–1.39) | (3.91 × 10−13) | |||||
| rs12956651 | G | 18 | 68265435 | CBLN2 | 0.59 | 1.09 × 10−5 | 0.84 | 0.232 | 1.04 | 0.633 | 0.85 | >1 × 10−5 |
| (0.47–0.75) | (1.12 × 10−5) | (0.62–1.12) | (0.226) | (0.88–1.23) | (0.636) | (0.76–0.96) | (0.859) | |||||
| rs6069045g | C | 20 | 52949270 | DOK5 | 0.75 | 6.24 × 10−6 | 0.91 | 0.243 | 1.05 | 0.266 | 0.93 | >1 × 10−5 |
| (0.66–0.85) | (8.60 × 10−6) | (0.77–1.07) | (0.240) | (0.96–1.16) | (0.264) | (0.87–0.99) | (0.704) | |||||
Allele nomenclature was set according to definitions for top SNPs provided for Illumina Hap300 v1.1 platform.
Nearest known transcribed sequence, as reported by Illumina.
Top line is P value from χ2 test for alleles, bottom line is P value from Armitage-Doll trend test.
P values from joint analysis of data from all stages. P values Indicated as > 1 × 10−5 are not accurately computed because the replication P values were much less significant than the discovery P values.
P values from replication data only.
SNP rs1481848 was genotyped for rs1481847 by Taqman.
Previous name for this SNP was rs7353629.
Figure 1.
Results from genome-wide association analysis of directly tested SNPs in the Texas discovery set using Illumina 300K HumanHap v1.1 Beadchips.
We performed a fast-track replication of the ten most significant associations from the GWAS in two additional case-control datasets (Table 1). One replication set was drawn from the same case-control population in Texas (711 cases and 632 controls) as the discovery phase, following the same criteria for matching. The other replication set was from the UK (2,013 cases and 3,062 controls). Table 1 shows adequate frequency matching in the discovery phase for smoking behavior, age and sex, cigarette smoking intensity and years of smoking exposure, but currently smoking cases reported heavier packyears (cigarettes per day × years smoked) than currently smoking controls. The Texas replication set included more recently recruited participants for whom matching was incomplete. The UK replication set was not matched, and included some small-cell lung cancers and some lifetime never smokers. We could not assess potential effects of substructure in the replication sets, but the Texas replication used the same study population and control selection procedures as the discovery set, and previous studies from the same UK controls showed that population substructure did not influence risk estimation for colorectal cancer10.
We replicated the elevated risks associated with two of the ten SNPs selected for validation in these additional case-control series, rs10151730 and rs8034191, both mapping to an 88-kb region of chromosome 15 (Table 2 and Fig. 2). Through joint analysis of genotype data for cases and controls from the three series (Table 3 and Supplementary Table 2 online), we found unequivocal evidence for an association between these two SNPs and lung cancer risk. For rs8034191 and rs1051730, the combined P values were 3.15 × 10−18 and 7.00 × 10−18, respectively (Table 2). P values from the replication data were < 10−12 (Table 2), and a similar level of significance was obtained when the joint tests were Bonferroni adjusted for 315,450 tests (results not shown). No other SNP showed significant evidence for association. Using Cochran-Mantel-Haenszel analysis, we did not observe any heterogeneity in the odds ratios (ORs) among the series (P > 0.9) for these two SNPs. Combined adjusted ORs for lung cancer associated with rs8034191 and rs1051730 were 1.32 (95% CI: 1.24–1.41) and 1.32 (95% CI: 1.23–1.39), respectively. Combined adjusted ORs among all ever-smokers from the three studies were 1.28 for heterozygotes for both SNPs, and 1.81 and 1.80 for homozygotes with minor alleles of rs8034191 and rs1051730, respectively (Table 3).
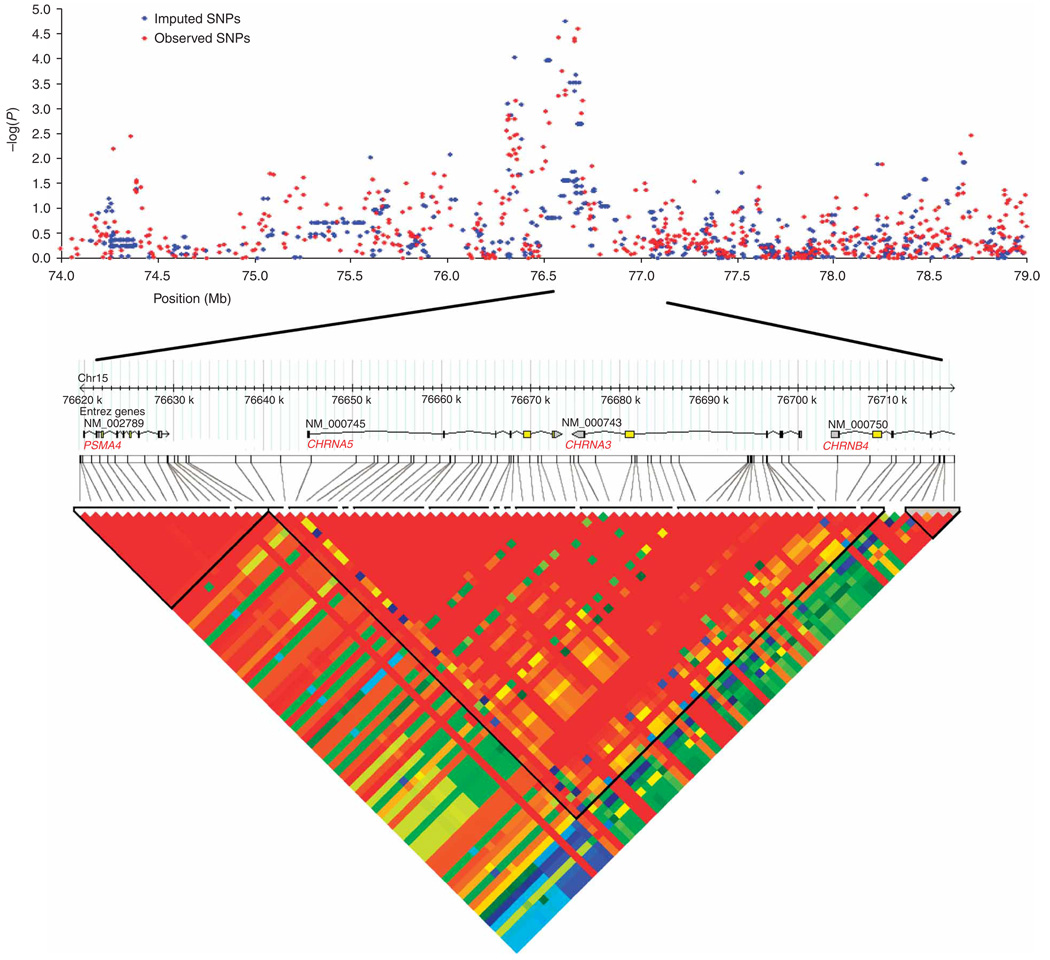
Figure 2.
The 15q25.1 locus. The top panel shows SNP single marker association results. Results in blue depict genotyped SNPs, and results in red are for imputed SNPs. All known genes and predicted transcripts in the local area are shown. Positions are that of University of California Santa Cruz Genome Browser March 2006 assembly; NCBI Build 36.1. The bottom panel shows the LD structure at 15q21.4. Boxes are shaded according to the standardized disequilibrium coefficient, D, derived from Phase 1 genotypes in Haploview (v3.2).
Table 3.
Association of rs8034191 and rs1051730 genotypes with lung cancer risk among ever smokers before and after adjustment for age, sex and packyears of cigarette exposure
| SNP | Allele | Case | Control | OR (95%CI) | P value | AdjORa (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| Texas discoveryb | |||||||
| rs8034191 | AA | 426 | 493 | ||||
| AG | 536 | 522 | 1.19 (1.00–1.42) | 5.60 × 10−2 | 1.18 (0.99–1.42) | 0.063 | |
| GG | 191 | 122 | 1.81 (1.39–2.35) | 8.45 × 10−6 | 1.79 (1.37–2.33) | 1.72 × 10−5 | |
| rs1051730 | GG | 424 | 501 | ||||
| AG | 541 | 511 | 1.25 (1.05–1.49) | 1.32 × 10−2 | 1.25 (1.04–1.49) | 1.50 × 10−2 | |
| AA | 188 | 125 | 1.78 (1.37–2.31) | 1.52 × 10−5 | 1.76 (1.35–2.28) | 2.85 × 10−5 | |
| Texas eplicationb | |||||||
| rs8034191 | AA | 259 | 269 | ||||
| AG | 328 | 253 | 1.35 (1.06–1.71) | 1.37 × 10−2 | 1.33 (1.04–1.7) | 2.50 × 10−2 | |
| GG | 111 | 69 | 1.67 (1.18–2.36) | 3.60 × 10−3 | 1.68 (1.17–2.42) | 5.20 × 10−3 | |
| rs1051730 | GG | 259 | 266 | ||||
| AG | 330 | 260 | 1.30 (1.03–1.65) | 2.77 × 10−2 | 1.31 (1.03–1.69) | 3.11 × 10−2 | |
| AA | 113 | 68 | 1.71 (1.21–2.41) | 2.47 × 10−3 | 1.75 (1.22–2.53) | 2.50 ×10−3 | |
| UK replicationb | |||||||
| rs8034191 | AA | 670 | 448 | ||||
| AG | 858 | 415 | 1.38 (1.17–1.63) | 1.50 × 10−4 | 1.31 (1.10–1.56) | 2.35 × 10−3 | |
| GG | 303 | 97 | 2.09 (1.61–2.70) | 2.25 × 10−8 | 1.89 (1.45–2.47) | 3.14 × 10−6 | |
| rs1051730 | GG | 687 | 445 | ||||
| AG | 848 | 418 | 1.31 (1.11–1.55) | 1.36 × 10−3 | 1.26 (1.06–1.50) | 8.87 ×10−3 | |
| AA | 295 | 93 | 2.05 (1.58–2.67) | 7.02 × 10−8 | 1.85 (1.41–2.43) | 9.10 × 10−6 | |
| Pooledc | |||||||
| rs8034191 | AA | 1,355 | 1,210 | ||||
| AG | 1,722 | 1,190 | 1.29 (1.16–1.44) | 2.71 × 10−6 | 1.28 (1.14–1.43) | 1.40 × 10−5 | |
| GG | 605 | 288 | 1.88 (1.6–2.20) | 1.46 × 10−14 | 1.81 (1.53–2.13) | 2.54 × 10−12 | |
| rs1051730 | GG | 1,370 | 1,212 | ||||
| AG | 1,719 | 1,189 | 1.28 (1.15–1.42) | 6.49 × 10−6 | 1.28 (1.15–1.43) | 1.10 × 10−5 | |
| AA | 596 | 286 | 1.84 (1.57–2.17) | 8.90 × 10−14 | 1.80 (1.52–2.13) | 4.59 × 10−12 |
Subjects missing packyear information were deleted from these analyses.
AdjOdds Ratio column was adjusted for age, sex and packyears.
AdjOdds Ratio column was adjusted for age, sex, packyears and centers.
rs1051730 and rs8034191 map to a 100-kb region of strong linkage disequilibrium (LD) on chromosome 15 extending from 76,593,078 bp to 76,681,394 bp (Fig. 2). Three genes map to this region: CHRNA3 and CHRNA5 (nicotinic acetylcholine receptor alpha subunits 3 and 5) and PSMA4 (proteasome alpha 4 subunit isoform 1), as well as the hypothetical gene LOC123688 isoform 1. Although rs1051730 and rs8034191 are separated by 88 kb, the genotypes are highly correlated (r2 = 0.88 in the discovery set and 0.81 in HapMap for the population of European ancestry (CEU)). Intervening genotyped markers in the region showed weaker associations with lung cancer in the discovery set (Fig. 2), but the imputed SNP rs931794 at position 76,613,235 in LOC123688 showed the most significant association with lung cancer risk (P = 1.8 × 10−6).
We determined the haplotype block structure across the entire region (Fig. 2). To further study genetic effects in the candidate region, we estimated haplotypes from nine SNPs genotyped on the Illumina panel spanning the haplotype block that includes rs1051730 and rs8034191. A single extended haplotype was significantly associated with lung cancer risk (P = 7.0 × 10−5), but this did not improve the prediction of case status over that provided by the individual SNPs rs1051730 or rs8034191 (Supplementary Table 3 online). This result provides evidence against multiple alleles or loci in the region contributing to disease susceptibility.
There is a growing body of evidence implicating the nicotinic acetylcholine receptor pathway in both the etiology and the progression of lung cancer11–13. Specifically, nicotine has been reported to promote cancer cell proliferation, survival, migration, invasion and tumor angiogenesis through the acetylcholine receptor pathway. The nicotinic acetylcholine receptor may also be a key player in nicotine-mediated suppression of apoptosis in lung cancer cells12. Furthermore, it has been demonstrated that stimulation of nicotinic cholinergic receptors by nicotine promotes growth of human mesothelial cells14. CHRNA3 is perhaps the more attractive candidate susceptibility gene for lung cancer. A previous study has shown15 that the nicotinic acid receptor could increase risk of lung cancer through a mechanism in which the CHRNA3 subunit binds NNK and subsequently upregulates nuclear factor kappa B to induce cell proliferation. PSMA4 is a component of the ATP- and ubiquitin-dependent nonlysosomal pathway, and although it is involved in the processing of class I major histocompatibility complex (MHC) peptides, there is little evidence to date for a role in lung cancer.
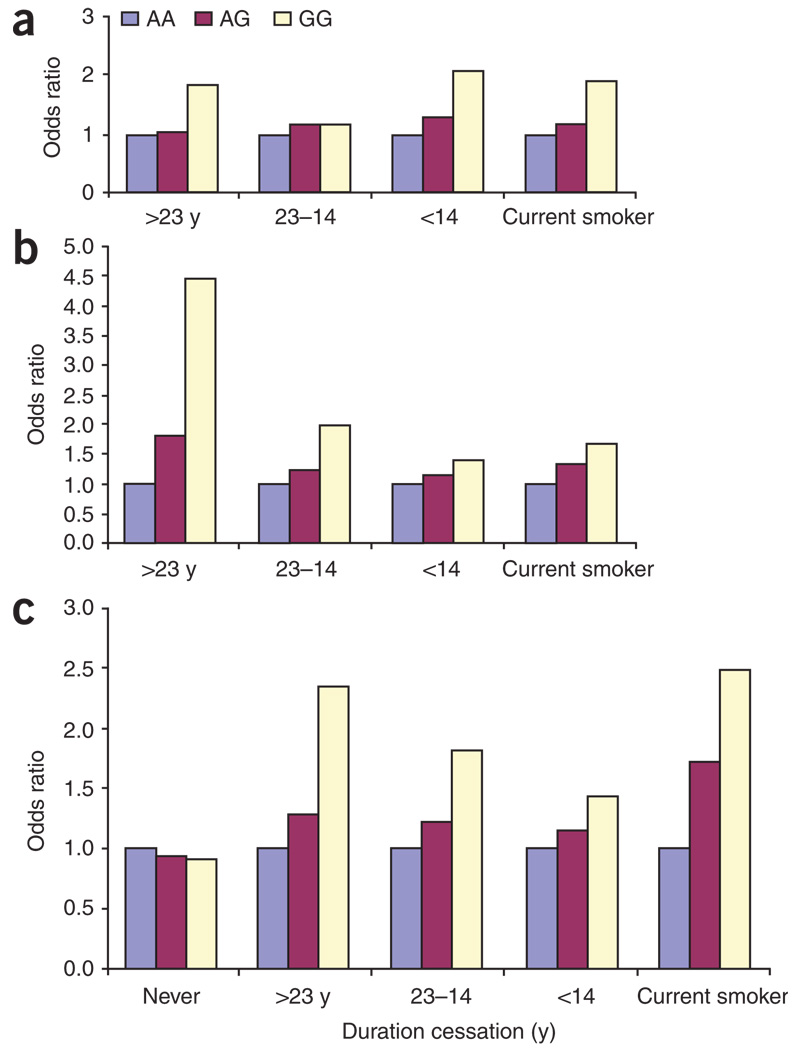
Because CHRNA3 and CHRNA5 may have a role in nicotine dependence16, we evaluated the relationship between the SNPs and lung cancer risk by smoking phenotype. Even though cases and controls from Texas were frequency matched on smoking behavior, lung cancer cases who smoked reported higher cumulative levels of exposure than controls who smoked (Table 1). Hence, it might be conjectured that the genetic associations we have identified relate to smoking behavior, which in turn modulates lung cancer risk, rather than a direct effect of a genetic susceptibility factor per se. There was, however, no consistent trend of genotypic risk associated with different strata of smoking behavior and years since smoking cessation among former smokers (Fig. 3 and Supplementary Table 4 online). We also did not observe any significant change in risk of lung cancer associated with rs8034191 or rs1051730 after adjusting for age, sex and packyears of smoking (Table 3) in the Texas populations. For the UK population, smoking adjustment decreased the ORs slightly. As shown in Figure 3, for the UK sample, the OR among participants who had never smoked was nearly 1 for both risk genotypes. These results, if subsequently confirmed with a larger sample of never-smoking cases and controls, would indicate that these SNPs play a role in determining lung cancer risk only among ever-smokers. We found similar risks associated with genotypes for heavier and lighter smokers (Supplementary Table 5 online), with marginally higher genotypic risks among lighter smokers. Adjusting for genotype of either candidate SNP did not affect the association between smoking and lung cancer risk, indicating that the candidate SNPs and smoking have independent effects on lung cancer risk in our study. (Supplementary Table 6 online).
Figure 3.
Effects of SNPs according to smoking behavior in current, former and never smokers adjusting for age, sex and packyears of tobacco smoke exposure. (a–c) The x axis indicates the extent of exposure, starting with never smokers (UK population, panel c only), followed by former smokers who quit 24 or more years ago, former smokers who quit 15–23 years ago, former smokers who quit less than 15 years ago and current smokers. Panel a presents data from the Texas discovery set, panel b presents data from the Texas replication set and panel c presents data from the UK replication set.
To characterize in further detail the relationships between genotypes and smoking, we carried out additional exploratory studies. We analyzed whether rs8034191 or rs1051730 were associated with selected measures of nicotine dependence, that is, number of cigarettes consumed per day and packyears of exposure (Supplementary Table 7 online). Results showed weak evidence that these SNPs influence smoking behavior; however, the effects seemed consistently significant across studies in only former but not in current smokers. Collectively, these data provide evidence that, although the nicotinic acetylcholine receptor may have a role in smoking behavior, variation at 15q5.4 defined by rs8034191 or rs1051730 directly contributes to lung cancer susceptibility. A previous study16 found an association with rs16969968, a marker in strong LD with rs1051730, with an index of nicotine dependence (Fagerstrom index) in nondiseased individuals. Our study shows a weak effect of rs8034191 or rs1051730 on smoking behaviors and an extremely significant effect on lung cancer risk, whether or not an adjustment for smoking behavior is made during the analysis.
In conclusion, we have identified and replicated a locus associated with lung cancer risk. Given that the carrier frequencies of rs8034191 and rs1051730 are ∼50% in populations of European ancestry, they may be of importance from a public health perspective. These data are the strongest evidence to date for common susceptibility alleles for lung cancer risk. CHRNA5 and CHRNA3 are promising candidate genes in this region of 15q25.1.
METHODS
Study populations
For detailed descriptions of the component studies, see Supplementary Methods online. The study protocols were approved by the Institutional Review Board of The University of Texas M.D. Anderson Cancer Center and by a review board at the Institute for Cancer Research Foundation. Informed consent was obtained from all participants.
Genotyping
Genotyping procedures and quality control approaches are described in Supplementary Methods. We retained data from 315,860 SNPs from Illumina analysis that had genotyping results in 90% or more subjects, but 410 were monomorphic for analysis in individuals of European descent (and hence not informative). Confirmatory genotyping in Houston was conducted on an independent sample of 711 cases and 632 controls using a Taqman genotyping platform for the ten most significant SNPs identified in the discovery phase. The Texas replication sample comprised independent cases and controls from the discovery set who were from the same study population source but who tended to be more recently enrolled participants with incomplete frequency matching. Genotyping of UK samples was conducted by competitive allele-specific PCR KASPar chemistry (KBiosciences).
Statistical analyses
We used similarity in genotypes as implemented in PLINK to identify individuals and clusters of individuals who deviated by more than 4 standard deviations from other study subjects, and we excluded these outliers. We identified genetically related subjects using PLINK software, which uses the similarity in identity by state of genotypes to estimate identity by descent values17, setting the clustering value at 0.0001 and excluding 639 markers that deviated from Hardy-Weinberg equilibrium in the controls (P < 0.0001) and 584 SNPs with minor allele frequency (MAF) < 0.01.
Association between SNP genotype and disease status was primarily assessed using the allelic 1 degree-of-freedom (d.f.) test or Fisher’s exact test where an expected cell count was <5. We also carried out association analysis using the Armitage-Doll trend test18. The ORs associated with each SNP and the 95% confidence intervals were estimated by allele and by genotype using unconditional logistic regression. None of the markers associated with lung cancer risk showed deviations from Hardy-Weinberg equilibrium (P > 0.05).
We evaluated the adequacy of the case-control matching and the possibility of differential genotyping of cases and controls using quantile-quantile plots of test statistics. A test inflation factor λ was calculated by dividing the median of the test statistics by the expected median from a χ2 distribution with 1 d.f.19. The mean and median of the χ2 tests in Figure 1 were 1.0196 and 0.4675, very close to the expected values of 1.00 and 0.456. Comparison of the median χ2 test with its expected value yielded a λ value of 1.025, very close to expected, indicating that population substructure, if present, did not have any substantial effect upon the discovery stage analyses presented here.
We used HelixTree for preliminary analyses and for initial data manipulation; we then transferred data to PLINK17 and EIGENSTRAT20. We evaluated the association of markers with lung cancer risk allowing for potential effects of population substructure by using a Cochran-Mantel-Haenszel test22 in PLINK. Strata were defined by a nearest neighbor cluster analysis of genetic similarity, which identified 44 clusters. We also carried out a second analysis to allow for substructure effects using EIGENSTRAT20. All genetic data from the discovery set were used to obtain correlation matrices among the subjects. Spectral analysis was done to extract those eigenvectors explaining the largest proportion of interindividual variation. A scree plot of the associated eigenvalues showed a point of inflection when three eigenvalues were included, and these three eigenvalues alone exceeded 2.0 (results not shown). Results from all analyses were very similar for significantly associated SNPs whether or not adjustments for population structure were made (Supplementary Table 1).
We used SAS Genetics v9.1 to conduct association tests for Hardy-Weinberg equilibrium and to perform haplotype analyses. Logistic regression, implemented in SAS version 9.1, was used to perform analyses adjusting for smoking and other covariates. We conducted joint analysis of data generated from multiple phases using standard methods for combining raw data based on the Mantel-Haenszel method. We used Cochran’s Q statistic to test for heterogeneity.
We used Haploview21 software (v3.2) to infer the LD structure of the genome in the regions containing loci associated with disease risk. To impute SNPs from multimarker tags, we used a procedure described previously22 based upon haplotype frequencies from HapMap release 21, build 35.
Statistical methods for obtaining P values
We obtained P values combining data from the discovery phase as well as the two replication phases following a procedure outlined previously23. Specifically, we set the critical value for the discovery phase to be the least significant result among the ten SNPs retained for follow up (P = 4.9 × 10−5). We obtained the joint test statistics by comparing allele frequencies in cases versus controls from all studies according to their sample sizes (results from the two replication phases were combined prior to joint analysis). We used the joint statistic value conditioning on the critical value P = 4.9 × 10−5 using the program CaTS to estimate the P value required to reach observed joint Z value. The pointwise P value so derived can be adjusted for multiple testing using a Bonferroni approach by multiplying the pointwise P value by the number of tests (results not shown). For several cases in which the replication P value was very much larger than the discovery P value, the CaTS software could not provide a result because of numerical overflow, and these results were indicated by > 1 × 10−5, which was the least significant P value obtained before the overflow. We also provided P values from the replication phase only by combining results from the Texas replication and UK studies, and adjusting for center effects using a Cochran-Mantel-Haenszel procedure implemented in SAS.
URLs
Haploview, http://www.broad.mit.edu/mpg/haploview/; Eigenstrat, http://genepath.med.harvard.edu/~reich/EIGENSTRAT.htm; CaTS, http://www.sph.umich.edu/csg/abecasis/CaTS/.
Supplementary Material
ACKNOWLEDGMENTS
Partial support for this study has been provided by US National Institutes of Health grants R01CA133996, R01CA55769, P50 CA70907 and R01CA121197, the Kleberg Center for Molecular Markers at M.D. Anderson Cancer Center, and by support from the Flight Attendants Medical Research Institute. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403. We thank the Kelsey Research Foundation for facilitating control selection in Texas. At the Institute for Cancer Research, work was undertaken with support primarily from Cancer Research UK. We are also grateful to the National Cancer Research Network, HEAL and Sanofi-Aventis. A. Matakidou was the recipient of a clinical research fellowship from the Allan J. Lerner Fund. We are also thankful for the unstinting efforts of the study coordinators and interviewers, including S. Honn, P. Porter, S. Ritter and J. Rogers. We also thank the study participants, who had the most critical role in this research.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Tokuhata GK, Lilienfeld AM. Familial aggregation of lung cancer in humans. J. Natl. Cancer Inst. 1963;30:289–312. [PubMed] [Google Scholar]
- 2.Amos CI, Xu W, Spitz MR. Is there a genetic basis for lung cancer susceptibility? Recent Results Cancer Res. 1999;151:3–12. doi: 10.1007/978-3-642-59945-3_1. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson S, et al. Familial risk of lung carcinoma in the Icelandic population. J. Am. Med. Assn. 2004;22:2977–2983. doi: 10.1001/jama.292.24.2977. [DOI] [PubMed] [Google Scholar]
- 4.Hwang SJ, et al. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum. Genet. 2003;113:238–243. doi: 10.1007/s00439-003-0968-7. [DOI] [PubMed] [Google Scholar]
- 5.Sanders BM, Jay M, Draper GJ, Roberts EM. Non-ocular cancer in relatives of retinoblastoma patients. Br. J. Cancer. 1989;60:358–365. doi: 10.1038/bjc.1989.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinerman RA, et al. Hereditary retinoblastoma and risk of lung cancer. J. Natl. Cancer Inst. 2000;92:2037–2039. doi: 10.1093/jnci/92.24.2037. [DOI] [PubMed] [Google Scholar]
- 7.Takemiya M, Shiraishi S, Teramoto T, Miki Y. Bloom’s syndrome with porokeratosis of Mibelli and multiple cancers of the skin, lung and colon. Clin. Genet. 1987;31:35–44. doi: 10.1111/j.1399-0004.1987.tb02764.x. [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka A, Hirai T, Ohtake Y, Kitagawa M. Lung cancer associated with Werner’s syndrome: a case report and review of the literature. Jpn. J. Clin. Oncol. 1997;27:415–418. doi: 10.1093/jjco/27.6.415. [DOI] [PubMed] [Google Scholar]
- 9.Bailey-Wilson JE, et al. A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am. J. Hum. Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb EL, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14,704 first-degree relatives. Hum. Mol. Genet. 2006;15:3263–3271. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, et al. Nicotine induces hypoxia-inducible factor-1alpha expression in human lung cancer cells via nicotinic acetylcholine receptor-mediated signaling pathways. Clin. Cancer Res. 2007;13:4686–4694. doi: 10.1158/1078-0432.CCR-06-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam DC, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638–4647. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- 13.Minna JD. Nicotine exposure and bronchial epithelial cell nicotinic acetylcholine receptor expression in the pathogenesis of lung cancer. J. Clin. Invest. 2003;111:31–33. doi: 10.1172/JCI17492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trombino S, et al. Alpha7-nicotinic acetylcholine receptors affect growth regulation of human mesothelioma cells: role of mitogen-activated protein kinase pathway. Cancer Res. 2004;64:135–145. doi: 10.1158/0008-5472.can-03-1672. [DOI] [PubMed] [Google Scholar]
- 15.Ho YS, et al. Tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) induces cell proliferation in normal human bronchial epithelial cells through NFkappaB activation and cyclin D1 up-regulation. Toxicol. Appl. Pharmacol. 2005;205:133–148. doi: 10.1016/j.taap.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balding DJ. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 19.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 20.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.de Bakker PI, et al. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 23.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.