Abstract
Earlier reports have shown that cdc2 kinase is activated in cells infected with herpes simplex virus 1 and that the activation is mediated principally by two viral proteins, the infected cell protein 22 (ICP22) and the protein kinase encoded by UL13. The same proteins are required for optimal expression of a subset of late (γ2) genes exemplified by US11. In this study, we used a dominant-negative cdc2 protein to determine the role of cdc2 in viral gene expression. We report the following. (i) The cdc2 dominant-negative protein had no effect in the synthesis and accumulation of at least two α-regulatory proteins (ICP4 and ICP0), two β-proteins (ribonucleotide reductase major subunit and single-stranded DNA-binding protein), and two γ1-proteins (glycoprotein D and viral protease). US11, a γ2-protein, accumulated only in cells in which cdc2 dominant-negative protein could not be detected or was made in very small amounts. (ii) The sequence of amino acids predicted to be phosphorylated by cdc2 is present in at least 27 viral proteins inclusive of the regulatory proteins ICP4, ICP0, and ICP22. In in vitro assays, we demonstrated that cdc2 specifically phosphorylated a polypeptide consisting of the second exon of ICP0 but not a polypeptide containing the sequence of the third exon as would be predicted from the sequence analysis. We conclude that cdc2 is required for optimal expression of a subset of γ2-proteins whose expression is also regulated by the viral proteins (ICP22 and UL13) that mediate the activation of cdc2 kinase.
In recent years, studies from this and other laboratories have shown that, in cells infected with herpes simplex viruses (HSV), cellular proteins involved in regulation of the cell cycle are extensively altered with respect to function or stability. These changes reflect the function of specific viral genes and come about in several different ways. Thus, HSV-1 stabilizes cyclin D3 and maintains the function of cdk4 kinase but not of cdk2 kinase (refs. 1–4; S.J.A. and B.R., unpublished data). This activity is determined by the infected cell protein 0 (ICP0), an α-protein made immediately after infection. The net effect of this change is that, even though cyclin D3 and its partner cdk4 are activated, E2F proteins are not activated, and concurrently no evidence has been presented that the virus activates the expression of cellular S phase genes (4–6). The function of the stabilized cyclin D3 is not known. However, the substitution in ICP0 of aspartic acid 199 with alanine abolished the stabilization of the D cyclins. The mutant virus carrying this mutation replicated less well and is less pathogenic than the wild-type virus, suggesting that certain D type cyclins contribute to viral replication (3).
At the other end of the cell cycle, HSV infection activates cdc2 cell-cycle kinase (7, 8). To activate the kinase, at least one inhibitor of cdc2 (wee1) is down-regulated, whereas the cdc25C phosphatase is activated. Cdc2 (cdk1) is a family member of the cyclin-dependent serine/threonine kinases involved in the G2/M transition of the cell cycle (7). Although protein levels of cdc2 remain relatively stable throughout the cell cycle, its activity is regulated by both the presence of cyclin partners (cyclins A and B) and its phosphorylation status. Proteins involved in the phosphorylation of cdc2 are the kinases wee1 and myt1 (negative regulators), cdk activating kinase (positive regulator), and cdc25C phosphatase (positive regulator). Studies with a dominant-negative form of cdc2 have shown that such cells are unable to complete mitosis and accumulate in the G2/M phase of the cell cycle (9). Cellular proteins that have been identified as targets for cdc2 phosphorylation include proteins involved in transcription (RNA polymerase II and POU transcription factors) and translation (EF-1; refs. 10 and 11). Other substrates include casein kinase II and lamins (12, 13). The change in cdc2 is of interest from two points of view. First, infected cells do not go through mitosis. This conclusion is reinforced by the observation that, although cdc2 is activated, its natural partners, cyclins A and B, are degraded. Second, the function of at least two viral genes is required for activation of cdc2. These are ICP22 and the viral protein kinase specified by UL13 (8). Importantly, ICP22 and the UL13 viral protein kinase play a role in determining the synthesis of a subset of late (γ2) genes that includes US11, UL41, and UL38 (14–16). The question arises as to what role cdc2 kinase plays in the viral replicative cycle.
In this report, we show that cdc2 protein kinase is essential for the expression of US11, a representative γ2-gene whose expression is determined by ICP22 and the UL13 protein kinase. Cdc2 does not seem to be required for the accumulation of viral proteins expressed earlier in the course of the viral replicative cycle. We also report that at least one regulatory viral protein containing a cdc2 cognate consensus phosphorylation site is phosphorylated in vitro by the cdc2 kinase.
Materials and Methods
Cells and Viruses.
HeLa and HEp-2 cells were obtained initially from the American Type Culture Collection and maintained in DMEM with 10% (vol/vol) newborn calf serum. HSV-1(F) is the prototype wild-type HSV-1 strain used in this laboratory (17).
Cell Lysis and Nocodazole Treatment.
Cells grown in 25-cm2 flasks were harvested as follows. The medium was removed, and the cells were rinsed with PBS, scraped into 5 ml of PBS, pelleted by centrifugation, rinsed twice with PBS, lysed by the addition of high-salt lysis buffer [20 mM Tris, pH 8.0/1 mM EDTA/0.5% Nonidet P-40/400 mM NaCl/0.1 mM sodium orthovanadate/10 mM NaF/2 mM DTT/100 μg each of PMSF and tolylsulfonyl phenylalanyl chloromethyl ketone per ml/2 μg each of aprotinin and leupeptin per ml]. Cells were maintained in high-salt lysis buffer on ice for 1 h, and the insoluble material was pelleted by centrifugation. The soluble fraction (supernatant fluid) was collected, and the protein concentration was determined by Bradford assay (Bio-Rad). In experiments with nocodazole (Sigma), the drug was diluted to a concentration of 5 mg per ml of DMSO and used at 5 μg/ml. Asynchronous cell cultures were treated with nocodazole for 20 h. Untreated cells were exposed to an equivalent concentration (0.1%) of DMSO.
Production and Purification of Glutathione S-Transferase (GST) Fusion Proteins.
pGEX4T-1 encoding HSV-1 ICP0 codons 20–241 (pRB4994), ICP0 codons 543–768 (pRB4995), or UL34 codons 1–240 (pRB5704) have been described (18, 19). Escherichia coli BL21 cells were transformed with plasmids encoding the above GST fusion proteins or GST alone (pGEX4T-1), grown at 30°C until the optical density at 600 nm reached a value of 0.6 to 0.8, and induced with 100 μM isopropyl β-d-thiogalactoside for 2 h. Bacteria were collected by centrifugation, resuspended in PBS, and lysed by sonication, and finally Triton X-100 was added (1% final concentration). After the cell debris was clarified by centrifugation, GST fusion proteins were adsorbed to glutathione-agarose beads (Sigma). The beads were collected and rinsed in PBS, and fusion proteins were eluted with 10 mM glutathione in 50 mM Tris (pH 8.0). The eluted protein solution was dialyzed against PBS. Protein production was assessed by PAGE followed by Coomassie brilliant blue staining, and protein concentrations were measured by Bradford assay.
Transient Transfection/HSV-1 Infection.
Plasmid encoding a dominant-negative protein for cdc2 with C-terminal hemagglutinin (HA) tags was transiently transfected into HeLa or HEp-2 cells with Lipofectamine-Plus (GIBCO/BRL; ref. 20). Briefly, 2 μg of plasmid was diluted in serum and antibiotic-free DMEM and was complexed with Plus reagent followed by Lipofectamine. Cells were incubated in serum and antibiotic-free DMEM with plasmid DNA for 4 h, and then an equal volume of DMEM with 20% (vol/vol) serum and 2× antibiotics was added to the cell culture. Cells were either mock infected or infected with HSV-1(F) 36 h later as follows. The growth medium was replaced with the inoculum containing HSV-1(F) diluted in 199V (mixture 199 with 1% calf serum). After 2 h, the viral inoculum was aspirated and replaced with DMEM supplemented with 10% (vol/vol) newborn calf serum. The cells were harvested at indicated time points as described above.
Immunoblotting.
Cell lysates harvested as above were solubilized in 4× gel loading buffer [1× buffer is 2% (vol/vol) SDS/50 mM Tris, pH 6.8/2.75% (vol/vol) sucrose/5% (vol/vol) 2-mercaptoethanol/0.1% bromophenol blue] and boiled for 5 min. Equivalent amounts of total protein were subjected to electrophoresis on 10% bisacrylamide gels, transferred to nitrocellulose membranes, blocked for 2 h with 5% (vol/vol) nonfat dry milk in PBS, and reacted with the appropriate antibodies. Antibody to HA tag (Santa Cruz Biotechnology) was diluted 1:500 in PBS with 1% BSA and 0.05% Tween-20 and reacted with the membrane for 2 h at room temperature. Secondary antibody (Bio-Rad) conjugated to alkaline phosphatase was diluted 1:3,000 in PBS with 1% BSA and 0.05% Tween-20 and was reacted to the membrane for 1 h at room temperature; the blot was rinsed in AP buffer (100 mM Tris, pH 9.5/100 mM NaCl/5 mM MgCl2) and developed by the addition of AP buffer containing 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. The reaction was stopped by the addition of stop buffer (100 mM Tris, pH 7.6/10 mM EDTA). All rinses were done in PBS containing 0.05% Tween-20.
Cdc2 Kinase Assay.
Cells were harvested as described above and lysed in high-salt lysis buffer. Equivalent amounts of protein were brought up to a total volume of 300 μl in high-salt lysis buffer. Lysates were precleared with preimmune serum [5% (vol/vol)] for 2 h, mixed with 50 μl of 50% (vol/vol) protein A (Sigma) slurry for 2 h, and centrifuged to pellet the protein A slurry. The supernatant fluids were transferred to new tubes; cdc2 was immunoprecipitated by the addition of cdc2 antibody (1:100; Santa-Cruz Biotechnology, sc-54) and recovered by the addition of 20 μl of 50% (vol/vol) protein A slurry. The beads were rinsed twice with high-salt lysis buffer, twice with low-salt lysis buffer (20 mM Tris, pH 8.0/1 mM EDTA/0.5% Nonidet P-40/1 mM NaCl/2 mM DTT), and twice with incomplete kinase buffer (50 mM Tris, pH 7.4/10 mM MgCl2/5 mM DTT). Kinase assays were done to determine whether certain proteins could serve as substrates for cdc2 kinase. Complete kinase buffer (40 μl) was added to each sample {10 μM ATP, 20 μCi [γ-32P]ATP (1 Ci = 37 GBq), and 2 μg of histone H1 (Roche Molecular Biochemicals) or GST fusion proteins}. Samples were reacted for 20 min at 30°C, and the reaction was terminated by the addition of 17 μl of 4× gel loading buffer. The samples were heated to 95°C for 5 min, subjected to electrophoresis in 10% bisacrylamide gels, transferred to nitrocellulose membrane, and analyzed by autoradiography and PhosphorImager (Storm 860, Molecular Dynamics).
Roscovitine (Calbiochem) was used in certain in vitro kinase studies as follows. Cdc2 immune complexes were collected as above, and 20 μl of incomplete kinase buffer containing 0, 2, 10, or 40 μM of roscovitine was added to the cdc2-containing beads for 5 min at 30°C. Then, 20 μl of 2× complete kinase buffer was added to the beads, and the reaction was allowed to proceed for 20 min at 30°C, after which the samples were processed as above.
Immunofluorescence Staining.
Cells were grown on microscope cover slips in six-well plates and transfected with pCMVcdc2-dn as described above. At 36 h after transfection, cells were infected with HSV-1(F) as above. At indicated times, the wells were rinsed with 199V and fixed in methanol for 20 min at −20°C. Cells were incubated for 1 h in 20% (vol/vol) human serum in PBS, rinsed in PBS, and incubated with antibodies to HA (1:100; Santa Cruz Biotechnology) as well as ICP0, ICP4, ICP6, gD, or US11 (all diluted at 1:500) in 10% (vol/vol) human serum in PBS. Cells were rinsed with PBS, incubated with secondary antibodies conjugated to FITC (for cdc2-dn), or Texas red (viral proteins) for 1 h in 10% (vol/vol) human serum in PBS, and rinsed with PBS. Cells were mounted with 90% (vol/vol) glycerol in PBS and analyzed with the aid of a Zeiss confocal microscope.
Results
Transfection of cdc2 Dominant Plasmid Results in Attenuation of cdc2 Kinase Activity in HSV-1(F)-Infected Cells.
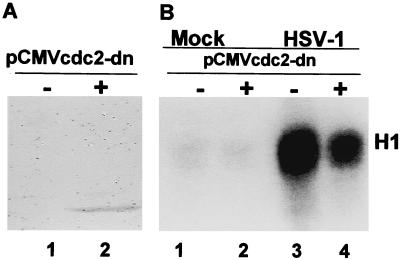
In an earlier report, we showed that HeLa or HEp-2 cells infected with HSV-1 exhibit a striking increase in the activity of cdc2 kinase (8). The objective of the experiments described in this section was to determine whether cdc2-activity would be diminished in cells transfected with a plasmid encoding a cdc2 dominant-negative protein and then infected with HSV-1(F). cdc2 was readily detected in cells 36 h after transfection with an expression plasmid encoding a dominant-negative cdc2 tagged with an HA amino acid sequence (pCMVcdc2-dn) in immunoblots with the aid of HA-specific antibody (HeLa cells, Fig. 1A) or by immunofluorescence (HEp-2 cells, Fig. 2). On the basis of this observation, replicate 25-cm2 flask cultures were transfected with vector alone or plasmid expressing pCMVcdc2-dn. At 36 h after transfection, the cells were mock infected or infected with HSV-1(F) and incubated at 37°C. The lysates (100 μg) of cells harvested 14 h after infection were tested for cdc2 kinase activity with histone H1 as the substrate. The results (Fig. 1B) showed that the cdc2 kinase activity expressed in lysates of infected cells transfected with the plasmid encoding cdc2 dominant-negative protein was reduced compared with that of infected cells transfected with the vector alone.
Figure 1.
(A) Immunoblot of electrophoretically separated lysates of HeLa cells reacted with antibody to HA tag. HeLa cells were mock transfected or transfected with cytomegalovirus expression plasmids carrying HA-tagged dominant-negative cdc2 (pCMVcdc2-dn). The cells were harvested 36 h after transfection, solubilized, electrophoretically separated on 10% bisacrylamide gels, and probed with antibody to HA. (B) Autoradiographic image of histone H1 phosphorylated in vitro by immunoprecipitated cdc2. HeLa cells were infected with HSV-1(F) 36 h after transfection with pCMVcdc2-dn and incubated for an additional 14 h at 37°C. Histone H1 was mixed with immune-precipitated Cdc2 from harvested, lysed cells. The reaction mixtures were separated on denaturing gels as described in Materials and Methods.
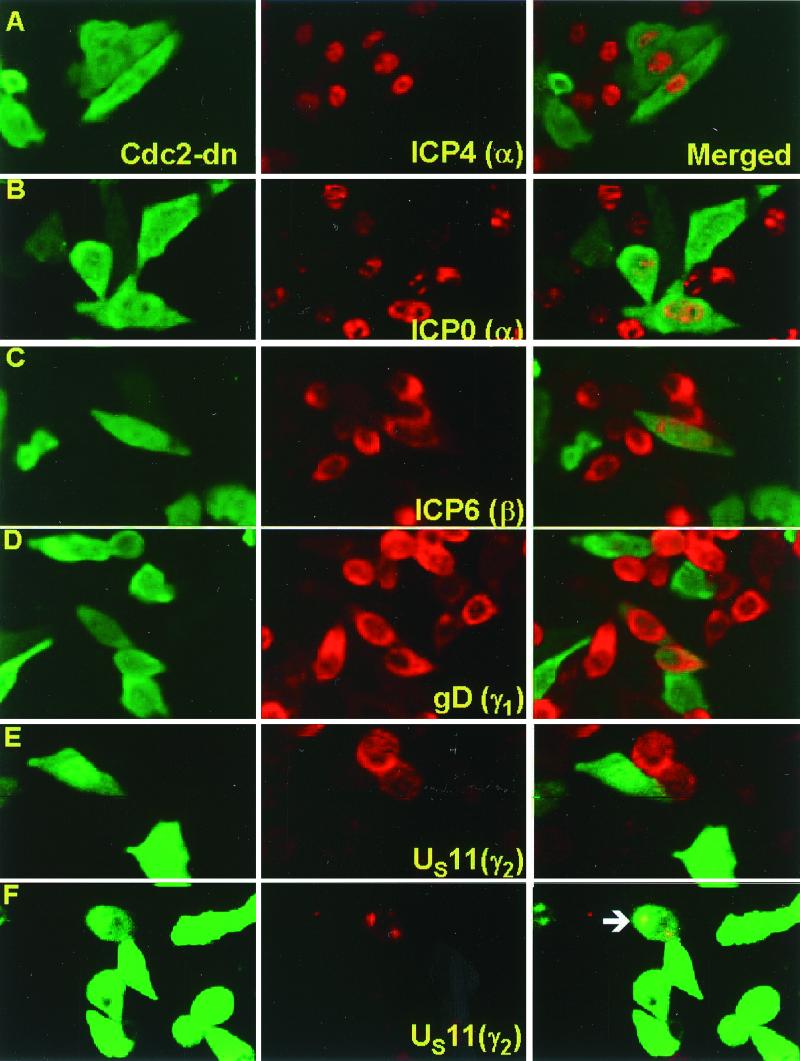
Figure 2.
Accumulation of viral proteins belonging to different kinetic classes in cells expressing cdc2-dn protein. HEp-2 cells were infected with HSV-1(F) 36 h after transfection with plasmid pCMVcdc2-dn. The cells were fixed 12 h after infection and reacted with anti-HA tag (FITC) or an antibody against a viral protein (Texas red). The slides were examined in a Zeiss confocal microscope, and images were captured with software provided by the manufacturer. (A) Monoclonal antibody against ICP4. (B) Monoclonal antibody against ICP0. (C) Monoclonal antibody against ICP6, the major subunit of ribonucleotide reductase. (D) Monoclonal antibody against glycoprotein D. (E) Monoclonal antibody against US11 protein. (F) Monoclonal antibody against US11 protein. In this image, the signal for green fluorescence was amplified to make apparent a very weak signal for cdc2-dn protein in the cells identified by the arrow and showing the presence of US11 protein.
Cdc2 Activity Is Required for the Synthesis of US11, a Late (γ2) Protein.
In this series of experiments, HEp-2 cells grown in coverslip cultures were infected with HSV-1(F) 36 h after transfection with pCMVcdc2-dn. The cells were fixed 12 h after infection with HSV-1(F) and reacted with antibody to HA tag and a viral protein as indicated in the legend to Fig. 2. The results were as follows. There was an abundance of cells expressing cdc2-dn and α-proteins ICP4 (Fig. 2A) or ICP0 (Fig. 2B), β-proteins ICP6 (Fig. 2C) or ICP8 (data not shown), and γ1-proteins glycoprotein D (Fig. 2D) or UL26 protein (data not shown). The striking feature of the data was the absence of cells expressing cdc2-dn and US11, a γ2-protein (Fig. 2E). A small number of cells expressing very small amounts of cdc2-dn protein exhibited also small amounts of US11 protein. In Fig. 2F, the fluorescein isothiocyanate immunofluorescence was amplified artificially to make apparent the barely visible fluorescence caused by cdc2-dn.
We conclude that the cdc2 is required for the expression of at least one γ2-gene and that the genes expressed earlier in infection do not seem to require the cell-cycle kinase.
HSV-Encoded Proteins with Putative cdc2 Phosphorylation Sites.
The experiments described above indicate that cdc2 activity is required for the synthesis of late (γ2) proteins. Elsewhere, this laboratory reported that cdc2 activation requires the function of ICP22 and of UL13 protein kinase, because deletion of either one of these genes abolished the activation of cdc2 kinase (8). Cdc2 is a proline-directed kinase known to phosphorylate the first serine or threonine in the consensus substrate site S/T-P-X-R/K/H in which the fourth amino acid must be basic (20, 21). On the basis of this motif, we have identified 27 HSV proteins as potential substrates of cdc2 kinase. The list includes ICP4 (four sites), ICP0 (three sites), ICP22, glycoproteins C, D, E, G, H, I, K, and L, and the proteins encoded by UL2, 12, 19, 21, 29, 30, 34, 36, 37, 38, 39, 46, 47, 50, 52, and US2 (reviewed in ref. 22).
HSV-1 ICP0 Exon II Is a Substrate for cdc2 Kinase.
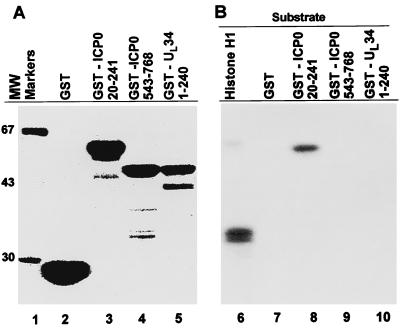
On the basis of the above sequence analysis, we next determined whether either ICP0 or UL34 fusion proteins could serve as substrates for cdc2 kinase. The potential cdc2 phosphorylation sites in ICP0 are located within exon II at codons 89–92 (S-P-P-R) and 224–227 (S-P-T-H). A third site lies in exon III at codons 371–374 (S-P-H-R). UL34 has one potential site at codons 112–115 (T-P-E-R). To assess their ability to serve as substrates, we generated chimeric proteins consisting of glutathione S-synthetase fused to ICP0 amino acids 20–241 encoded by exon II (GST-ICP0 20–241) and containing two potential sites, to ICP0 amino acids 543–768 (GST-ICP0 543–768) encoding a portion of exon III and containing no consensus phosphorylation sites, or to the first 240 amino acids of UL34 (GST-UL34 1–240) predicted to contain one consensus phosphorylation site. GST does not contain consensus sites and serves as an additional negative control. The synthesis and accumulation of fusion proteins were assessed by PAGE followed by Coomassie brilliant blue staining (Fig. 3A). The concentrations of fusion proteins were determined as described in Materials and Methods.
Figure 3.
(A) Coomassie blue staining of GST fusion proteins. GST alone, GST-ICP0 20–241, GST-ICP0 543–768, and UL34 1–240 were purified with the aid of glutathione-agarose beads, electrophoretically separated on 10% bisacrylamide gels, and stained with Coomassie blue. (B) Autoradiographic image of cdc2-mediated phosphorylation of histone H1 or indicated GST fusion proteins. Cdc2 was immunoprecipitated from uninfected HeLa cells with cdc2 antibody, and histone H1 or GST fusion proteins were tested as substrates for cdc2. Reactions were electrophoretically separated on 10% bisacrylamide gels, transferred to nitrocellulose membrane, and exposed to film.
In the first series of experiments, cdc2 was immunoprecipitated from 200 μg of HeLa cell lysate and reacted with histone H1 or GST fusion proteins as substrates for in vitro kinase activity (Fig. 3B). The results showed that both histone H1 and GST-ICP0 20–241 were substrates for cdc2 kinase, whereas GST alone and GST-ICP0 543–768 were not phosphorylated by cdc2. The GST-UL34–240 protein showed minimal phosphorylation by cdc2 and probably does not serve as a substrate for cdc2.
Because asynchronous cell cultures typically have between 5 and 10% of cells in G2/M where cdc2 is active, the objective of the second series of experiments was to increase cdc2 activity in uninfected cells. In these studies, histone H1 or the GST-ICP0 20–241 fusion protein was reacted with cdc2 immunoprecipitated from 100 μg of HeLa cell lysate from replicate asynchronous cell cultures that were either untreated or treated with nocodazole for 20 h. As expected (Fig. 4A), treatment with nocodazole resulted in an increased phosphorylation of both histone H1 and the GST-ICP0 20–241 fusion protein. GST-ICP0 543–768 fusion protein served as a negative control and was not phosphorylated by the nocodazole-induced kinase activity.
Figure 4.
(A) Autoradiographic image of nocodazole-induced cdc2-mediated phosphorylation of histone H1 or indicated GST fusion proteins. Replicate HeLa cell cultures were mock-treated or treated with nocodazole for 20 h. cdc2 was immunoprecipitated from lysates and reacted with indicated substrates. Reaction mixtures were electrophoretically separated on 10% bisacrylamide gels, transferred to nitrocellulose membrane, and exposed to film. (B) Autoradiographic image of electrophoretically separated histone H1 or GST-ICP0 20–241 polypeptide reacted with immune-precipitated cdc2 in the presence of various concentrations of roscovitine. The cdc2 kinase was immune-precipitated from HEp-2 cells treated with nocodazole for 20 h and reacted with roscovitine before the addition of complete kinase buffer and substrates as described in Materials and Methods. Reactions were electrophoretically separated on 10% bisacrylamide gels, transferred to nitrocellulose membrane, and exposed to film. (C) Quantification of the amount of radioactivity in substrates phosphorylated by cdc2 in the presence or absence of roscovitine. Radioactivity in each band shown in B was quantified with the aid of Storm 860 PhosphorImager. The radioactivity in each band was normalized with respect to that of untreated (no roscovitine) reaction mixtures (100%). The dashed line represents 50% reduction in phosphorylation.
To verify that GST-ICP0 20–241 polypeptide is specifically phosphorylated by cdc2 kinase in the immunoprecipitation mixture, the proteins immunoprecipitated by cdc2 antibody from nocodazole-treated cells were treated with roscovitine, a specific inhibitor of cdk2, cdc2, and cdk5, followed by addition of substrates for cdc2 kinase (23). Against a panel of purified enzymes, roscovitine selectively inhibits cdc2, cdk2, and cdk5 at an IC50 dose of <0.7 μM. The drug inhibits other kinases at much higher concentrations. Immune complexes precipitated with cdc2 antibody from nocodazole-treated HEp-2 cells were exposed to final concentrations of roscovitine of 1, 5, or 20 μM in the presence of histone H1 and GST-ICP0 20–241 polypeptide (Fig. 4B). At doses as low as 1 μM roscovitine, there was a 65% reduction in the phosphorylation of histone H1 and a 55% reduction in the phosphorylation of GST-ICP0 20–241 fusion proteins compared with phosphorylation with no roscovitine (Fig. 4C). At increasing concentrations of roscovitine, there was a further decrease in the phosphorylation of both histone H1 and GST-ICP0 20–241 fusion protein.
Discussion
One puzzling aspect of the studies reported earlier was the apparent activation of cdc2 at times late in infection, when cyclins A and B, the natural partners of cdc2, were degraded and could no longer be detected in the infected cells (8). The notion that the virus simply ignored cdc2 as it dealt with cyclins A and B was dispelled by two observations. Foremost, there was an actual increase in the activity of cdc2 kinase. Moreover, two different viral proteins mediated both the activation of cdc2 and the disappearance of cyclins A and B, because in their absence, these effects were abolished (8). ICP22 and the UL13 protein kinase have been of particular interest, because they regulated the accumulation of a subset of late or γ2-proteins exemplified by US11 (15, 16). At least some aspects of the puzzle have become less opaque on the basis of this report. Specifically, we show that in the presence of a cdc2 dominant-negative protein, US11 protein did not accumulate or accumulated only in cells in which cdc2 dominant-negative protein was not expressed at a high level. This observation is based on the expression of cdc2 dominant-negative mutants and contrasts with the use of drugs that block both cdc2 and cdk2 as well as possibly other cellular kinases (24, 25). On the basis of this observation, we can construct the hypothesis that cdc2 plays a role in late gene expression. Because cdc2 levels may be very low in nondividing or resting cells, one function of ICP22 and of UL13 protein kinase is to activate cdc2 to ensure optimal expression of the US11 protein.
One may wonder why HSV goes about this circuitous way to ensure optimal expression of the γ2-proteins exemplified by US11 protein when it could acquire active homologs of either the substrate or the cdc2 kinase responsible for its accumulation. For example, whereas HSV-1 stabilizes cyclin D3, other herpesviruses have acquired a functional homolog of at least one of the D type cyclins (3, 26–29). Scavenging the cell for cellular factors to achieve its goals is the modus operandi of HSV as opposed to that of other herpesviruses. One possible explanation is that HSV proteins, like ICP0, which stabilizes cyclin D3, perform multiple functions. The cyclin D homologs acquired by other herpesviruses, on the other hand, seem to perform a single function. If coding capacity is at a premium, HSV could encode more functions per unit length of its DNA than viruses encoding homologs of cellular regulatory proteins.
The substrate of cdc2 relevant to the expression of γ2-genes is not known, and the reason why a subset of γ2-genes are regulated differently from other viral genes remains an interesting and important question. Cdc2 kinase could act on a viral regulatory protein. In this report, we show that cdc2 phosphorylated a polypeptide encoding exon II but not exon III of ICP0, consistent with their amino acid sequences. Experiments designed to identify viral regulatory proteins phosphorylated in vivo will help to clarify this issue. As indicated above, multiple HSV-1 glycoproteins contain phosphorylation motifs for cdc2, and gI of VZV has been shown to be phosphorylated by cdc2 (29). It should be pointed out, however, that recently published studies indicate that cdc2 may also alter the function or intracellular location of transcriptional factors (30–34). The trail initiated by the discovery of the role of ICP22 in the expression of the γ2-gene expression has led to cdc2 kinase and ultimately may lead to the identification of the factors that directly affect the accumulation of γ2-gene products.
Acknowledgments
We thank Joanne Trgovich for invaluable discussions, Sander van den Heuvel for kindly providing pCMVcdc2-dn, and Nancy Markovitz and Pascal Lopez for assistance with confocal analyses. These studies were aided by National Cancer Institute Grants CA47451, CA71933, and CA78766 and by the United States Public Health Service.
Abbreviations
- HSV
herpes simplex virus
- GST
glutathione S-transferase
- HA
hemagglutinin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200375297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200375297
References
- 1.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 2.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehmann G L, McLean T I, Bachenheimer S L. Virology. 2000;267:335–349. doi: 10.1006/viro.1999.0147. [DOI] [PubMed] [Google Scholar]
- 5.Song B, Liu J J, Yeh K C, Knipe D M. Virology. 2000;267:326–334. doi: 10.1006/viro.1999.0146. [DOI] [PubMed] [Google Scholar]
- 6.Advani S J, Weichselbaum R R, Roizman B. J Virol. 2000;74:7842–7850. doi: 10.1128/jvi.74.17.7842-7850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King R W, Jackson P K, Kirschner M W. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 8.Advani S J, Brandimarti R, Weichselbaum R R, Roizman B. J Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- 9.Van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 10.Gebara M M, Sayre M H, Corden J L. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- 11.Belle R, Minella O, Cormier P, Morales J, Poulhe R, Mulner-Lorillon O. Prog Cell Cycle Res. 1995;1:265–270. doi: 10.1007/978-1-4615-1809-9_21. [DOI] [PubMed] [Google Scholar]
- 12.Bosc D G, Slominski E, Sichler C, Litchfield D W. J Biol Chem. 1995;270:25872–25878. doi: 10.1074/jbc.270.43.25872. [DOI] [PubMed] [Google Scholar]
- 13.Nikolakaki E, Meier J, Simos G, Georgatos S D, Giannakouros T. J Biol Chem. 1997;272:6208–6213. doi: 10.1074/jbc.272.10.6208. [DOI] [PubMed] [Google Scholar]
- 14.Purves F C, Ogle W O, Roizman B. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purves F C, Roizman B. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogle W O, Roizman B. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ejercito P, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Bruni R, Roizman B. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye G-J, Vaughan K T, Vallee R B, Roizman B. J Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin O, Meggio F, Draetta G, Pinna L A. FEBS Lett. 1992;301:111–114. doi: 10.1016/0014-5793(92)80221-2. [DOI] [PubMed] [Google Scholar]
- 21.Holmes J K, Solomon M J. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 22.Roizman B, Sears A E. In: Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Lippincott–Raven; 1996. pp. 2231–2295. [Google Scholar]
- 23.Meijer L, Borgne A, Mulner O, Chong J P J, Blow J J, Inagaki N, Inagaki M, Delcros J G, Moulinoux J P. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 24.Schang L M, Phillips J, Schaffer P A. J Virol. 1998;72:5626–5637. doi: 10.1128/jvi.72.7.5626-5637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schang L M, Rosenberg A, Schaffer P A. J Virol. 1999;73:2161–2172. doi: 10.1128/jvi.73.3.2161-2172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholas J, Cameron K R, Honess R W. Nature (London) 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 28.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Nature (London) 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 29.Ye M, Duus K M, Peng J, Price D H, Grose C. J Virol. 1999;73:1320–1330. doi: 10.1128/jvi.73.2.1320-1330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto N, Ogashiwa M, Okumura E, Endo T, Iwashita S, Kishimoto T. FEBS Lett. 1994;352:236–242. doi: 10.1016/0014-5793(94)00964-3. [DOI] [PubMed] [Google Scholar]
- 31.Caelles C, Hennemann H, Karin M. Mol Cell Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connolly T, Caligiuri M, Beach D. Mol Biol Cell. 1997;8:1105–1115. doi: 10.1091/mbc.8.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jans D A, Moll T, Nasmyth K, Jans P. J Biol Chem. 1995;270:17064–17067. doi: 10.1074/jbc.270.29.17064. [DOI] [PubMed] [Google Scholar]
- 34.El-Hodiri H M, Che S, Nelman-Gonzalez M, Kuang J, Etkin L D. J Biol Chem. 1997;272:20463–20470. doi: 10.1074/jbc.272.33.20463. [DOI] [PubMed] [Google Scholar]