Abstract
Mesenchymal stem cells (MSC) derived from bone marrow stem cells (BMSC) and adipose tissue stem cells (ASC) of humans and rhesus macaques were evaluated for their cell cycle properties during protracted culture in vitro. Human ASCs (hASC) and rhesus BMSCs (rBMSC) underwent significantly more total population doublings than human BMSCs (hBMSC) and rhesus ASCs (rASC). The cell cycle profile of all MSCs was altered as cultures aged. hMSCs underwent an increase in the frequency of cells in the S phase at P20 and P30. However, rhesus MSCs from both sources developed a distinct polyploid population of cells at P20, which progressed to aneuploidy by P30. Karyotype analysis of MSCs revealed the development of tetraploid or aneuploid karyotypes in the rhesus cells at P20 or P30. Analysis of the transcriptome of the MSCs from early and late passages revealed significant alterations in the patterns of gene expression (8.8% of the genes were differentially expressed in hBMSCs versus hASCs, and 5.5% in rBMSCs versus rASCs). Gene expression changes were much less evident within the same cell type as aging occurred (0.7% in hMSCs and 0.9% in rMSC). Gene ontology analysis showed that functions involved in protein catabolism and regulation of pol II transcription were overrepresented in rASCs, whereas the regulation of IκB/nuclear factor-κB cascade were overrepresented in hBMSCs. Functional analysis of genes that were differentially expressed in rASCs and hBMSCs revealed that pathways involved in cell cycle, cell cycle checkpoints, protein-ubiquitination, and apoptosis were altered.
Introduction
Postnatal bone marrow and adipose tissue contain mesenchymal stem cells (MSCs). As with all mitotic stem cells, MSCs are faced with a decision for self-renewal or differentiation. MSCs have the ability to produce large numbers of differentiated progeny such as osteocytes, adipocytes, chondrocytes, and myelosupportive stroma (1–5).
The cell cycle is a highly ordered process that results in the faithful duplication and transmission of genetic information from one cell generation to the next (6). Primary mammalian somatic cells can replicate in vitro an estimated 50 cumulative population doublings, after which the cultures stop dividing (7). This phenomenon is termed Hayflick’s limit and is more readily known as replicative senescence. Although it has been shown that MSCs and other stem cell populations continuously grow in vitro for 10 to 20 passages, it seems that MSCs, similar to all other primary cells, are subject to the Hayflick limit (4, 8). It has been suggested that human MSCs derived from the marrow may become senescent during protracted culture, as indicated by their decreased differentiation potential, shortening of the mean telomere length, and morphologic alterations (9). An important factor involved in cell senescence is the maintenance of mean telomere length as a result of decreased telomerase activity. It has previously been reported by our group that cultures of MSCs derived from the bone marrow and adipose tissue underwent morphologic alteration, a decline in multilineage differentiation potential, and a marked decrease in telomerase activity in progressively increasing passages of MSCs (4).
It is unknown whether the MSCs, or a subpopulation of MSCs, are able to escape cellular senescence in a manner similar to immortalized or transformed cells. There is an increasing body of evidence that MSCs, and other stem cells, can undergo spontaneous transformation to malignant cells (10). The transformation of MSCs seems to be the direct result of spontaneous genetic alterations that accumulate during extended culture. Murine MSCs have been shown to undergo malignant transformation upon extended culture and form fibrosarcomas upon in vivo transplantation (11). The mechanisms of transformation observed in the murine MSCs were associated with chromosomal abnormalities, increased telomerase activity, and elevated c-myc expression levels. Human adipose tissue stem cells (hASCs) undergo immortalization and spontaneous transformation after protracted periods of expansion in vitro (10). Karyotype analysis of these transformed hASC cultures revealed numerous chromosomal alterations and rearrangements.
As MSCs from bone marrow adipose tissue and cord blood are being investigated for their potential as therapeutic interventions for numerous diseases, it is essential to fully understand the biological properties of these distinct MSC populations. It is also critical to characterize the biological limitations of these cells, particularly as they age. The data presented herein show that bone marrow– and adipose tissue–derived MSCs continuously cultured for protracted periods have altered cell cycle progression, resulting in both cellular crisis and senescence. However, extended culture of human MSCs failed to reveal any chromosomal alterations, whereas a high frequency of chromosomal aneuploidy was detected in all rhesus MSCs. Genome-wide transcriptome comparison of all four stem cell types at early and late passages indicates that the expression of genes involved in cell cycle, protein-ubiquitination, and apoptosis was altered. Despite the chromosomal alterations described, MSCs failed to generate tumors upon transplantation into immune deficient mice.
Materials and Methods
Cell Culture and Differentiation
Human and rhesus bone marrow stem cells (BMSCs) and ASCs were obtained and processed as previously described (4). Briefly, all MSC types were cultured in α-MEM (Invitrogen) supplemented with 20% fetal bovine serum (Atlanta Biological), 1% l-glutamine (Invitrogen), and 1% penicillin/streptomycin (Invitrogen) at 37°C in 5% CO2. Cultures were passaged when they reached 75% to 80% confluence using 0.5% trypsin plus 0.2% EDTA.
The multilineage potential of MSCs was examined using lineage-specific conditional medium for mesenchymal lineages as described before (4, 12–14). Adipogenic differentiation of MSCs was determined by staining the monolayers with 0.5% Oil Red-O solution. Osteogenic differentiation was assessed by staining the mineralization of differentiated cells with 40 mmol/L Alizarin red (pH 4.1; Sigma). For chondrogenic differentiation, MSC cell pellets were cultured in chondrogenic differentiation medium, which consisted of high-glucose DMEM supplemented with 500 ng/mL BMP-6 (R&D system); 10 ng/mL transforming growth factor β3; 10−7 mol/L dexamethasone; 50 µg/mL ascorbate 2-phosphate; 40 µg/mL proline; 100 µg/mL pyruvate; and 50 mg/mL insulin, transferrin, and selenium +premix (Becton Dickinson; 6.25 µg/mL insulin, 6.25 µg/mL transferrin, 6.25 ng/mL selenous acid, 1.25 mg/mL bovine serum albumin, and 5.35 mg/mL linoleic acid). The medium was replaced every 2 to 3 d for 21 d. Pellets then fixed in formalin, embedded in paraffin, and sectioned. The sections were stained with Toluidine Blue (15).
Analysis of Cell Cycle Status of MSCs
Single cell suspensions of each MSC type were obtained from cultures at various passages—P1, P10, P20, and P30. For DNA content analysis, cells were fixed in 70% ethanol, rehydrated in PBS, treated for 30 min with RNase A (1 mg/mL), and stained with 1 µg/mL of propidium iodide (PI) for 5 min. The intensity of fluorescence was determined by analysis on a fluorescent-activated cell sorter (FACS), which was a Becton Dickinson FACscan equipped with a 488-nm argon laser. Data acquisition was performed with CellQuest (Becton Dickinson) software, and percentages of G1, S, and G2 phase cells were calculated with MODFIT-LT software program (Verity Software House, Inc.).
Senescence-Associated β-Galactosidase Staining
Cell suspension and cultured cells on coverslips were washed in PBS, fixed in 4% formaldehyde in PBS for 4 min at room temperature, then briefly washed in PBS and incubated with β-Galactosidase solution (pH 6.0; Chemicon) for 8 to 16 h at 37°C (16).
Karyotype Analysis of MSCs
Metaphase chromosome spreads were prepared from cultures at the designated passages during the exponential phase of growth (65–75% confluence). For rhesus MSC cultures, 10 µL/mL colcemid (Boehringer Mannheim GMBH) was added directly to the cultures and incubated for two hours at 37°C, and human MSCs were incubated for 45 min at 37°C. Cells were removed from the flask using trypsinization, washed with PBS, and incubated in 0.075 mol/L KCl at 37°C for 15 min. Cells were fixed with methanol/acetic acid (3:1), G-banded, and analyzed. Slides were analyzed under a light microscope at ×10 and ×100 magnifications. Images of the individual metaphase spreads were captured and karyotyped using an automated imaging system for cytogenetics (CytoVision; Applied Imaging Corporation).
Microarray Hybridization and Data Analysis
Microarray-based interrogation of the transcriptome profiles exhibited by all four cell types was performed using Affymetrix human GeneChip (U133A 2.0).7 RNA was isolated from the four cell types at passage 20 and 30, and hybridized to the GeneChips using standard protocols. Total RNA was used to synthesize double-stranded cDNA (Superscript Choice System; Life Technologies Bethesda Research Laboratories). The resulting cDNA was purified and used for in vitro transcription to produce biotin-labeled cRNA (BioArray HighYield RNA Transcription Labeling kit; Enzo Diagnostics). The biotinylated cRNA was cleaned (RNAeasy Mini kit; Qiagen), fragmented, and hybridized on GeneChips containing 54,675 probes sets. After individual GeneChips had been washed, they were stained with streptavidinphycoerythrin (Molecular Probes), amplified by use of biotinylated antistreptavidin (Vector Laboratories), and scanned for fluorescence (GeneArray Scanner; Hewlett Packard) using Microarray Suite 5.0 software (MAS 5.0; Affymetrix).
For analyzing the data, the Affymetrix CEL files (containing scanned images, together with absolute calls for each gene) were transferred to the dChip program (version 1.3+)8 (17). Chips were normalized using Quantiles method, to stabilize MvA plots. This step was essential to eliminate any intensity-specific bias in probe-level data and to produce a matrix comprising of normally distributed data. Expression indices used the PM only (as opposed to PM-MM) method and were reported as log (base 2). Further-more, all fold change values reported in this article and the accompanying images are on a log (base 2) scale.
Probe sets whose targets were not detected were removed from the data matrix. A Student’s t test was then performed to identify genes expressed in a statistically significant manner (P < 0.05). A fold change cutoff of log2 1.2 (linear scale, 2.297) was then applied, so as to only consider genes whose expression was perturbed in magnitude as well as in a statistically significant manner.
Justification for use of human GeneChips for Rhesus samples
At the time these experiments were performed, a rhesus GeneChip was not available (it has since become available). As a result, both rhesus and human transcripts were profiled using human GeneChips. Given the high similarity between nonhuman primates (NHP) and human genomes (e.g., 98.77% similarity between NHP and human genomes; ref. 18), it has been hypothesized that human GeneChips could be used for gene expression profiling of NHPs. Several studies that have successfully used Affymetrix human GeneChips for gene expression profiling of NHPs have been published (19–26). These studies have used rhesus, chimpanzee, gorilla, orangutan, or African green monkey samples.
Terminal Deoxynucleotidyl-Transferase–Mediated dUTP Nick-End Assay for Apoptosis
Apoptotic MSCs were detected and quantified based on labeling of DNA strand breaks using terminal deoxynucleotidyl-transferase–mediated dUTP nick-end (TUNEL; 27) technology kit (Roche Diagnostics) according to manufacturer’s instructions. Briefly, MSC suspensions were fixed in 1% paraformaldehyde for 10 min. The DNA of the fixed cells was then labeled through the addition of fluorescein dUTP at strand break by terminal transferase by incubating for 1 h at 37°C. FACS analysis was then performed on the cell suspension. Cells grown on the coverslips were analyzed by fluorescent microscopy.
Western Blot Analysis
MSC cultures were washed twice with ice-cold PBS and then lysed in 40 AµL of lysis buffer (Promega) containing 1 µL of proteinase inhibitor cocktail (Sigma). The total protein concentration was measured using a Bradford assay containing Coomassie Plus protein reagent (Bio-Rad Laboratories) according to the manufacturer’s specifications. Equivalent amounts of total cell lysate were subjected to SDS-PAGE using 10% polyacrylamide gels. Proteins were electroblotted to polyvinylidene difluoride membrane (Millipore). The membranes were then blocked and incubated in anti-p53 (mouse monoclonal, 1:100; Abcam) and anti-glyceraldehyde-3-phosphate dehydrogenase antibody (rabbit polyclonal, 1:1,000; Abcam) overnight at 4°C. Alkaline phosphatase–conjugated anti-mouse or anti-rabbit IgGs (1:1,000) were used as secondary antibodies (Bio-Rad) for detection. The membranes were incubated with Western Blotting Detection Reagents (Bio-Rad) according to the manufacturer’s instructions and detected using the Versa Doc imaging system (Bio-Rad).
In vivo Tumorigenicity
Immune-deficient NIHIII 6-wk-old mice were inoculated s.c. with 1 × 106 cells obtained at specific passages (diploid and polyploid cells) in 200 µL of PBS (3 mice per MSC type per passage). Two hundred microliters of plain PBS were injected to one group of animal as negative control. One group of animal was injected with 1 × 106 A-549 cells, which is a transformed lung carcinoma cell line as a positive control. All animals were maintained in specific pathogen-free conditions, and all experiments conformed to the requirements of the Animal Welfare Act and the Institutional Animal Care and Use Committee.
Results
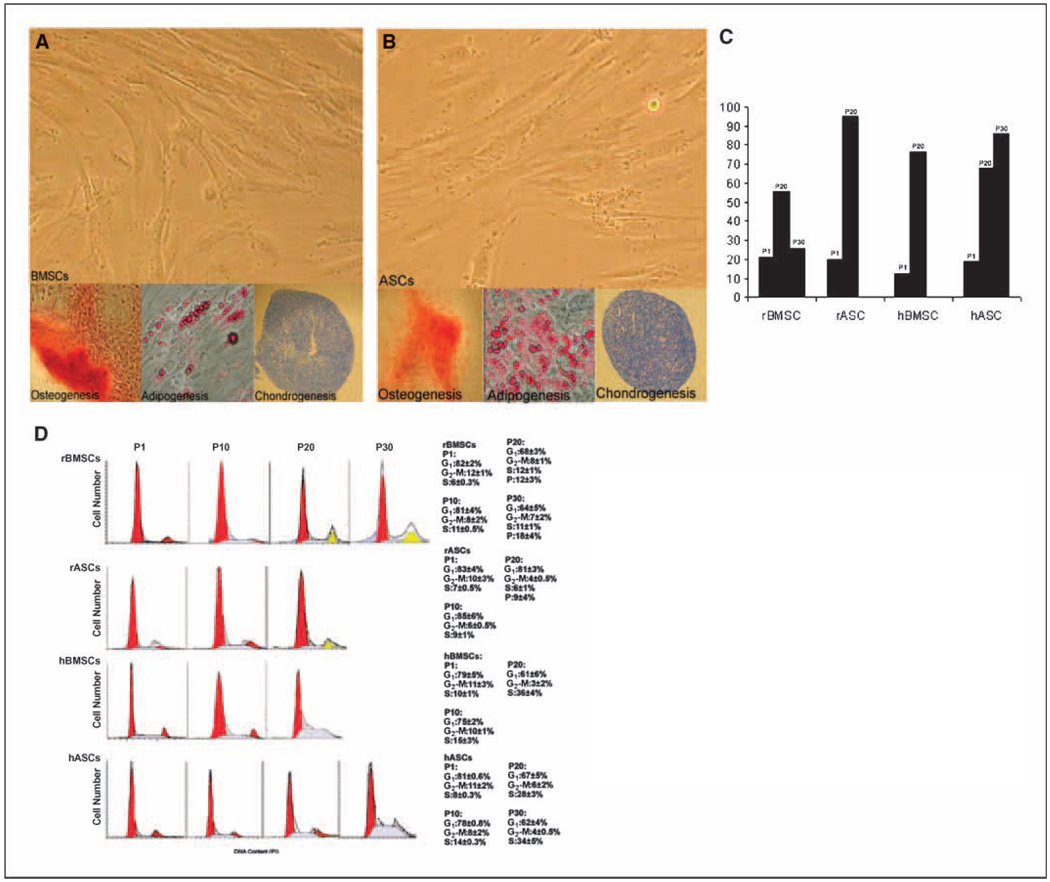
To characterize the effect of in vitro expansion on the biology of MSCs, four distinct groups of MSCs derived from human marrow (hBMSC) and hASC and rhesus monkey bone marrow and adipose tissue stem cells (rBMSC and rASC) were cultured for extended periods of time. We have reported that rASC and hBMSC can routinely be cultured for up to 20 passages, whereas rBMSCs and hACSs were expanded up to 30 passages (4). All MSC types had equivalent colony formation potential; furthermore, these lines displayed multiple in vitro differentiation capabilities to differentiate into osteogenic, adipogenic, and chondrogenic lineages under related inductive conditions at P1. All four MSC types retained their multilineage differentiation potential at least out to P10 (Fig. 1A and B ). Each MSC line showed a cell proliferation rate of ~ 48 hours in the early passages. Cells were allowed to proliferate and were replated (1,000 cells/cm2) when they reached no >80% confluence. After passage 10, there was a steady decline in the efficiency of proliferation in all cell populations. rASCs and hBMSCs showed a marked increase in the time required for cell doubling and showed an enlarged, flattened cellular morphology at P15, after which they ceased to undergo cell division but remained viable in culture. The culture doubling times ultimately exceeded 120 hours. At P20, rASCs and hBMSCs underwent cycle arrest, and the frequency of cells reactive for senescence-associated β-galactosidase were markedly increased to 95% and 76.4%, respectively (Fig. 1C ). Similar results were observed in hACSs at P25. Proliferation of hASCs ceased at P30; however, the cells remained viable in culture. Neither the proliferation rate nor the morphology of rBMSCs was significantly changed up to 30 passages. Human and rhesus BMSCs and ASCs maintain high levels of telomerase activity and long telomere length up to P10. However, telomerase levels significantly decreased in rASC and hBMSCs P20, and hASC P30 cultures. The reduction of telomerase activity in rBMSC P30 cultures was minimal, which may permit the maintenance of the rapid proliferation rate in these cells.
Figure 1.
Morphology and multilineage differentiation potential of MSCs. Cultures of rhesus MSCs (A) and human MSCs (B) capable of proliferating for several passages and exhibiting fibroblastic morphology in P1. Osteogenic differentiation of MSCs was confirmed by staining with Alizarin red. Adipogenic differentiation was confirmed by staining with Oil Red O. Chondrogenic differentiation was detected with Toluidine Blue. C, senescence was assessed by counting the number of reactive cells to senescence-associated β-galactosidase in cultures of four MSC types at low and higher passages. D, mitotic indeces of MSCs in early and late passage cultures. Single-variable histograms of DNA content indicate the percentages of cells in G0-G1, S, and G2-M phases of the cell cycle. The data are obtained from staining the DNA of P1, P10, P20, and P30 cultures of MSC types. rBMSC and rASCs showed a polyploid fraction in passage 20; a greater fraction of polyploidy (~18%) was seen in rBMSCs P30. hBMSC and hASC showed increasing fraction of cells arrested in S phase in P10 to P30.
The cell cycle distribution of MSCs at various passages was determined by FACS analysis after the cellular DNA was stained with propidium iodide. Single-variable histograms of DNA provided data describing the percentages of cells in G0-G1, S, and G2-M. More than three fourths (81% ± 2%) of four MSC populations from P1 were in the quiescent G0-G1 phase of the cell cycle; whereas, the remaining cells were divided between the S (5.7%–10.1%) and G2-M (9.7%–12.1%) phase. Each type of MSC displayed a different mitotic index at higher passages. The distribution of cells in the various stages of the cell cycle were not altered up to P10, except a slight increase in the number of cells in S phase (Fig. 1D ). Intracellular PI staining indicated a unique polyploid population in the rMSCs at P20. The level of polyploidy increased further in rBMSCs at P30 and higher. Unlike rhesus MSCs, human MSCs never developed polyploidy throughout the prolonged culture. hBMSCs showed a progressive increase in the frequency of cells in S phase at P20 (36%); whereas, hASCs showed 28% and 34% of the cells in S phase at P20 and P30, respectively.
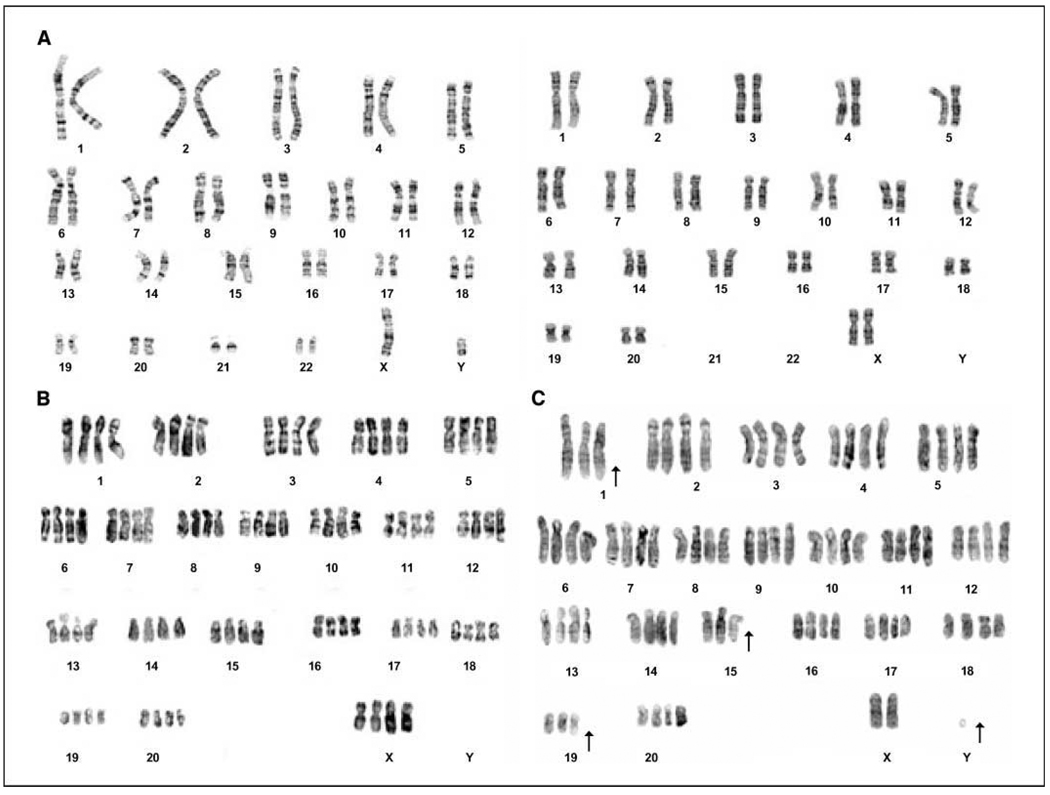
The four MSC types were karyotyped in early and late passages to determine whether the polyploidy seen in prolonged cell culture resulted in altered chromosome content. hBMSCs and hASCs had the normal diploid number of chromosomes (2n, n = 23; Fig. 2A , left) at all passages analyzed. The early passage rBMSCs and rASCs had the normal diploid chromosome number of 42 (2n, n = 21; Fig. 2A , right). However, at the later passages, the ploidy of the rhesus MSCs shifted from diploid to tetraploid in P20 (84 chromosomes, 4n; Fig. 2B ). About 40% of the P20 population of rhesus MSC had a tetraploid karyotype. By P30, ~70% of the rBMSCs contained a tetraploid karyotype.
Figure 2.
Karyotype analysis of MSCs. A, human MSCs maintained the diploid karyotype (2n, n = 23) throughout cell culture (left ). Rhesus MSCs showed stable karyotype in early passages (right ). B, rhesus MSC karyotypes were changed from the normal diploid (2n, n = 21) to tetraploid in P20. C, a subpopulation of P30 rBMSC cultures displayed an aneuploid karyotype.
Aneuploid cells (10%) were observed in the rBMSCs at P30 (Fig. 2C ). The chromosome content of rMSCs beyond P30 was variable, ranging from 55 to 81 (Table 1). This finding was even more evident in the higher passages (P90). The karyotype of rBMSCs from passage 90 showed that 20% of the metaphase chromosome spreads were diploid 42, XY; 40% had an 84, XXYY karyotype; and the remaining 40% of the cells were aneuploid. Aneuploid cells were not observed in rhesus ASCs or in any of the human cells. It seems that these aneuploid cells were derived from the tetraploid cells, by the random loss of chromosomes. Therefore, prolonged in vitro culture of rBMSCs resulted in the progression from a diploid to a tetraploid karyotype, and finally aneuploidy. Despite the fact that a high frequency of aneuploid rBMSCs was detected at later passages, no chromosomal rearrangements were detected in any of the samples analyzed. There were no chromosomal rearrangements detected in any of the human MSC samples analyzed at any passage.
Table 1.
Rhesus BMSCs showed 40% diploid metaphases (42, XY), whereas 20% metaphases had 84, XXYY karyotype at P90
| MSC type | Passage (3 donors) | Diploid cells (%) | Tetraploid cells (%) | Aneuploidy (%)* |
|---|---|---|---|---|
| rBMSC | P1 | 100% | 0% | — |
| P10 | 100% | 0% | — | |
| P20 | 60% | 40% | — | |
| P30 | 20% | 70% | 10% | |
| P90 | 20% | 40% | 40% | |
| rASC | P1 | 100% | — | — |
| P10 | 100% | — | — | |
| P20 | 90% | 10% | — | |
| hBMSC | P1 | 100% | — | — |
| P10 | 100% | — | — | |
| P20 | 100% | — | — | |
| hASC | P1 | 100% | — | — |
| P10 | 100% | — | — | |
| P20 | 100% | — | — | |
| P30 | 100% | — | — |
NOTE: The rest of the cells in that population were aneuploid.
Aneuploidy includes the presence of a total chromosome number ranging from 55 to 81.
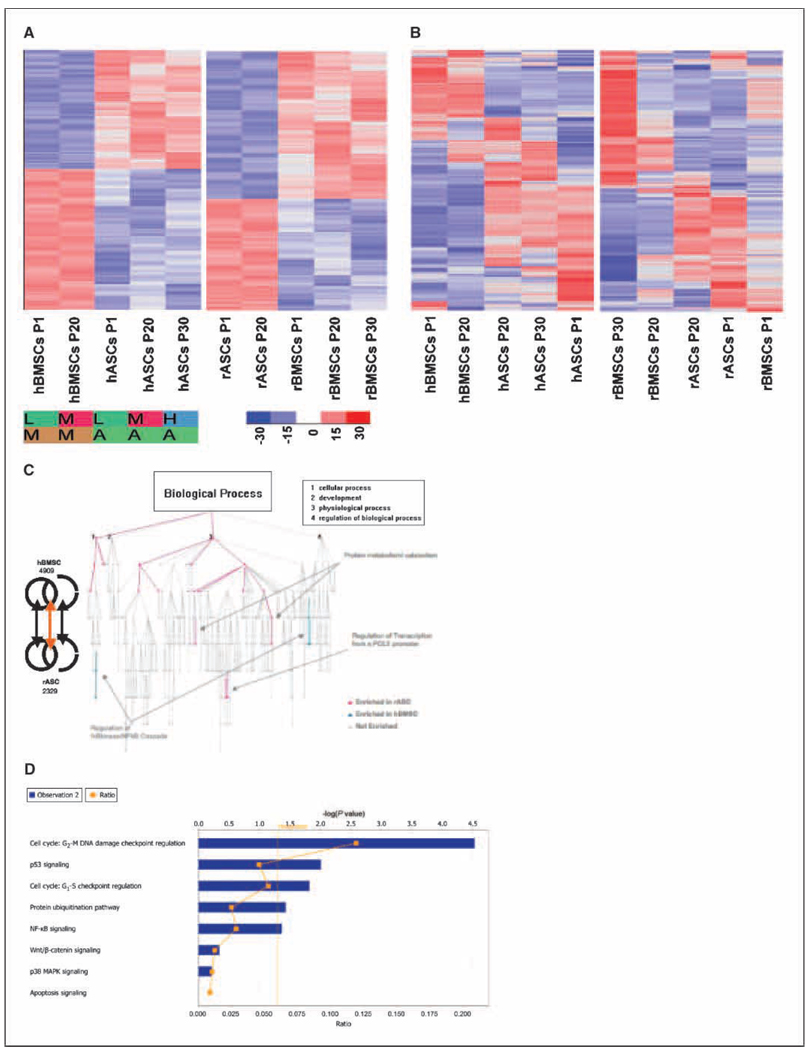
Genome-wide transcriptome comparison of MSCs was performed, to obtain a global overview of alterations in gene expression patterns that may play a role in the manifestation of the biological alterations. These experiments were performed using human Affymetrix GeneChip (U133 2.0), and the results are expressed either as a function of cell-lineage or extent of in vitro passaging. Transcriptome analysis was performed to compare the different cell types and passages (Supplementary Table S1–S8). Hierarchical clustering was used to display genes that are differentially expressed in each stem cell type (Fig. 3A ).
Figure 3.
Differential gene expression by stem cell type. A, hierarchical clustering was used to display genes differentially expressed in each stem cell type. The extent of blue (decreased fold change) or red (increased fold change) colors is directly proportional to the magnitude of differential expression of these genes. Hierarchical cluster for hASCs and hMSCs at different passages (from left to right: hBMSC passage 1, hBMSC passage 20, hASCs passage 1, hASCs passage 20, and hASCs passage 30). Hierarchical cluster for rASCs and rMSCs at different passages (from left to right: rASCs passage 1, rASCs passage 20, rBMSCs passage 1, rBMSC passage 20, and rBMSC passage 30). To perform these comparisons, probe sets whose target was not detected in any sample were eliminated from the data matrix. GeneChips were grouped by stem cell type and members of each group were pooled, before a Student’s t test was used to identify those genes that were expressed in a statistically significant manner (P = <0.05). Alog 2 fold change cut-off of 1.2 (numerical scale fold change, 2.297) was then applied to further select genes. These criteria ensure that only those genes whose expression was not only highly differential between experiments but which were also expressed in a statistically significant manner. Genes that exhibit highly inconsistent expression patterns, as well as genes that do not exhibit any change in expression between different stem cell types, are therefore excluded from the data matrix. B, hierarchical clustering displaying genes differentially expressed as a function of time (or passage). Genes that were found to be differentially expressed in human stem cell lines as a function of time did not fall into any particular gene ontology category, in a statistically significant manner, when compared with genes that were differentially expressed in rhesus stem cell lines. The extent of blue (decreased fold change) or red (increased fold change) colors is directly proportional to the magnitude of differential expression of these genes. C, Gene Ontology Tree. We carried out functional characterization of genes differentially expressed in human and rhesus cells at different passages, by comparing the number (and significance) of gene ontologies represented in these lists. This graph shows a dendrogram comparison of gene ontology (biological process) specifically differential between rASC and hBMSC. Red, ontologies enriched in rASC; blue, ontologies enriched in hBMSC. Inset, the top four biological processes hit. Of particular interest to us was the specific and significant enrichment of regulation of IκB kinase/NF-κB cascade, cell-cell adhesion, and G protein–coupled receptor signaling pathway in hBMSC but not rASC. Similarly, categories such as protein metabolism and catabolism, regulation of transcription from a POLII promoter, RNA processing, and regulation of cell cycle were enriched in rASCs compared with hBMSCs. D, comparison of the RNAisolated from rBMSC P1, P20, and P30 was performed on D-chip normalized data by SOM algorithm within the Spotfire DecisionSite application. Ingenuity pathways analysis of a cluster of >220 genes up-regulated only at the p30 revealed a statistically significant preponderance of genes involved in the cell cycle, p53, protein ubiquitination pathway, NF-κB signaling, Wnt/β-catenin signaling, and p38 MAPK signaling.
Microarray data were initially analyzed in the context of genes that were differentially expressed in a statistically significant manner within a particular comparison (either as a function of species or passage). The gene sets were analyzed by a pathways network algorithm to identify those pathways that were significantly perturbed at the level of gene expression. The definition of significantly perturbed pathway is one that contains a select number of genes whose expression was altered (based on a Student’s t test) at a level significantly higher than just by chance. The probability of a select number of genes from a particular pathway being present in the data set that includes all perturbed genes can be calculated based on the total number of genes on the chip, the total number of genes perturbed, the total number of genes in a select pathway, and the total number of genes for that particular pathway that were part of the perturbed group. As an example, lets assume that all 30,000 rhesus genes are represented on our chip and we identified 3,000 genes as differentially expressed. Hence, there is a one in a hundred chance of a gene ending up as significant. If the pathway X were composed of 30 genes, then it would be expected that 3 of those genes would be part of the perturbed gene list. However, if six genes from that pathway end up in the list, it might be significant. The probability of 9 or 12 genes being part of the perturbed list will be even lower.
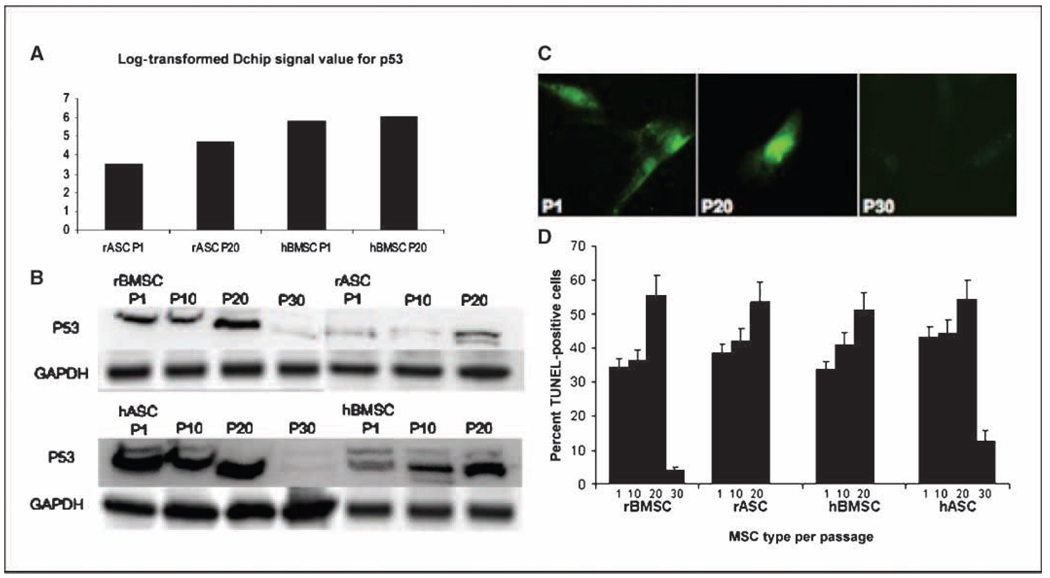
The initial analysis indicated that a great majority of rhesus transcripts were recognized by the probes on the human Gene-Chip, confirming previous studies (Supplementary Fig. S1A and B; refs. 19–25). Hierarchical clustering was used to display genes differentially expressed as a function of time (passage). Genes that were differentially expressed in human stem cell lines as a function of time did not decrease into any particular gene ontology category, in a statistically significant manner, when compared with genes that were differentially expressed in rhesus stem cell lines. Not surprisingly, significant differences were detected when the GeneChip data were compared on the basis of lineage, rather than on the basis of passage time (Fig. 3B ). Analysis of gene ontology indicated that genes involved in certain functions were overrepresented in a statistically significant manner between hBMSCs and rASCs. As an example, genes involved in biological processes regulation of cell cycle, RNA processing, regulation of IκB kinase, and nuclear factor-κB (NFκB) cascade were overrepresented in hBMSCs, whereas those involved in cell adhesion and G protein–coupled receptor-signaling, protein metabolism, protein catabolism, and regulation of transcription from a POLII promoter were overrepresented in rASCs (Fig. 3C ). The differences observed in other analyses, such as comparison between hBMSCs versus rBMSCs, and rASCs versus hASCs were minimal. Transcriptome analysis of rBMSCs from low and high passages by Self Organizing Maps (SOM) algorithm within the Spotfire DecisionSite application identified a cluster of >220 genes, which were specifically and significantly upregulated only at the rBMSC p30, compared with p1 and p20. Ingenuity Pathways analysis of these data revealed a statistically significant preponderance of genes involved in the cell cycle, p53, protein ubiquitination pathway, NF-κB signaling, Wnt/β-catenin signaling, and p38 mitogen-activated protein kinase (MAPK) signaling within this cluster (Fig. 3D ). Comparisons of genes differentially expressed in hBMSCs, hASCs, and rASCs at different time points was performed to identify canonical pathways that were perturbed in a statistically significant manner. The canonical pathways related to protein ubiquitination, nucleotide excision repair pathway, NF-κB signaling, cell cycle:G1-S checkpoint regulation, and cell cycle:G2-M DNA damage checkpoint regulation were significantly altered when rASC was compared with hBMSC cell types (Supplementary S2A–E). In contrast, no such changes were observed when rASC and rBMSC cell types were compared. These results underscore the importance of cell cycle regulation, as well as apoptosis in stem cell biology, and correlate with the observed differences in karyotype changes (Fig. 2). It is likely that dysregulation of critical checkpoint pathways and apoptosis mediators are involved in the polyploidy and aneuploidy in the rASCs, compared with hBMSCs (Fig. 2). In particular, Cyclin D (CCLD2) was expressed at a level >50-fold higher in rASCs compared with hBMSCs (log2 fold change, 5.654; Supplementary Fig. S2D). Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns that contribute to the temporal coordination of each mitotic event. CCLD2 forms a complex with and functions as a regulatory subunit of CDK4 or CDK6, whose activity is required for the G1-S transition. The expression level of the CDK4/6 complex was also elevated in the rASCs (Supplementary Fig. S2D). On the other hand, the expression of Smad3, a Tumor Growth factor-β responsive transcriptional regulator that directs the activity of a number of cyclin kinase pathway members, was reduced in rASCs compared with hBMSCs. Smad3 is known to regulate the expression of p15INK4, a cyclin-dependent kinase inhibitor, and this effect was probably reflected in the higher expression of CDK4/6. As shown in Fig. 4A , the levels of p53 were higher in hBMSCs compared with rASCs, although this difference was not statistically significant at the level of mRNA. Analysis of canonical pathways represented in generated gene lists indicated that several members of p53 signaling pathway were expressed in hBMSCs but not in rASCs (Fig. 4A ). Moreover, our Western blot results (Fig. 4B ) have shown that the level of p53 was lower in rASCs compared with hBMSCs. Several genes known to act downstream of p53 protein in cell cycle regulation were also expressed at a lower levels in rASCs, e.g., p27Kip1 (statistically significant reduction), p27Cip1 (reduced but not statistically significant), and cyclin-dependent kinase 2 (statistically significant reduction). Similarly, members of the NF-κB signaling pathway, including NF-κB itself, p65/RelA, Bcl-10, AKT, MAPKKK7, IRAK1/4, TRAF (2,3,5 and 5,6), and Nck-1/Nik-1 (an alternate pathway for recruitment of NF-κB) were all expressed at higher levels in hBMSCs compared with rASCs (Supplementary Fig. S2C). Conversely, mediators of NF-κB inhibition, such as IκB and IκBκB, showed higher levels of expression in rASCs compared with hBMSCs.
Figure 4.
The effect of prolonged in vitro culture on MSC apoptosis. A, the expression of the p53 gene in low and high passage cultures of rASCs and hBMSCs. B, Western blot analysis of the expression level of p53 protein in MSCs. C, MSCs were examined for apoptosis using a fluorescent DNAfragmentation assay (TUNEL stain). Microscopic fields of MSCs from P1, P20, and P30 were photographed individually to detect apoptosis cells. D, apoptosis level in MSC populations were performed by FACS using TUNEL stain.
The differences in gene expression among these mediators of cell cycle signaling (p53 and NF-κB) may account for some of the phenotypic differences between these stem cell lineages. Our data indicate an increase in p53 in protein levels in cultures of MSCs for at least 20 passages. However, p53 protein expression was markedly down-regulated in cultures of hASCs and rBMSCs P30.
The frequency of apoptotic MSCs at the various passages was also analyzed. The TUNEL assay was used to identify DNA fragmentation in cultures of early and late passages of MSC lines (Fig. 4C ). Approximately 36% of P1 MSCs were reactive to TUNEL labeling for all four cell lines. They showed an increased frequency of cells that underwent apoptosis out to P20 (50.9 to 55.4 among MSC types). The percentage of apoptotic rBMSCs and hASCs were markedly decreased at P30 (Fig. 4D ).
MSCs from early (P1) and late (P20 and P30) cultures were injected into the s.c. flank of immune deficient mice (n = 3 per MSC type) to examine their tumorigenic potential. Interestingly, none of the MSC types (even MSCs with aneuploid karyotypes) produced tumors in immune deficient mice, after 120 days.
Discussion
In this study, the cell cycle characteristics of adult human and rhesus monkey MSCs were analyzed, and MSCs obtained from bone marrow and adipose tiisue were compared. Our data suggest that long term in vitro expansion of MSCs can cause cell cycle dysregulation in both human- and rhesus-derived cells. In long-term cultures, human MSCs showed distinct cell cycle properties compared with the rhesus MSCs. Rhesus diploid MSCs developed abnormal mitotic division leading to tetraploidy. These populations of MSCs lost their ability to maintain chromosome stability during further cell divisions. The tetraploid population underwent the random loss of individual chromosomes resulting in aneuploidy. One possible mechanism of chromosome instability is the loss of telomeres (28). Telomeres shorten as cells age in most somatic cells due to loss of sufficient telomerase activity. Our group previously reported the significant loss of telomerase activity in hBMSCs and hASCs at P20 and P30, respectively. rASCs P20 also showed major reduction in telomerase activity, which may cause these MSCs to enter the crisis phase. rBMSCs showed minimal reduction of telomerase activity (4). It has been previously reported that tetraploid cells may produce aneuploid daughter cells with high efficiency (29). One cause for aneuploid progeny within a tetraploid population is the inheritance of twice the normal complement of centrosomes, which causes the formation of multipolar spindles and generates random chromosome segregation in subsequent division (30–32). Aneuploid rhesus cells did not show a consistent loss of any particular chromosome, indicating that the chromosome loss was random. The number of chromosomes in the aneuploid cells remained under the tetraploid number (84). Individual cells did not show more than four copies of a single chromosome. Thus, aneuploidy in rhesus cells was not the result of nondisjunction because nondisjunction would lead to five copies of some chromosomes. In many cases, the development of polyploidy is associated with malignancy and poor patient prognosis (33–35). An intrinsic capacity to eliminate tetraploid cells is a p53-dependent pathway (32, 36). The transcriptome analysis also suggested significant failure of apoptotic mechanisms leading to polyploidy. The level of apoptosis increased with passaging of MSCs up to P20. There was a loss of p53 expression in rBMSC and hASC at P30, which was accompanied by the loss of apoptosis in these cells. The considerable decrease in the frequency of apoptotic cells may result from the absence of p53 expression; this might be the reason for the uncontrolled proliferation of rBMSCs in passages above 30.
Human MSC cultures retained the normal diploid (2n) karyo-type up to passage 20 and 30 for hBMSCs and hASCs, respectively. A significant percentage of cycling human MSCs were arrested in S phase of the cell cycle at P20 and higher. S phase is responsible for protecting the integrity of the genome and preventing genetic instability (37). The data presented here show that the arrest in S phase of the cell cycle observed in the hMSCs coincided with markedly reduced p53 expression. It has been previously reported that S phase checkpoint mechanisms arrest the cell cycle in a p53-independent fashion (38, 39).
One of the key regulators of the anticancer pathways of the cell is the p53 protein. p53 is a transcription factor that plays a central role in controlling apoptosis, growth arrest, and senescence. It has the ability to halt progression through the cell cycle at the G1-S checkpoint in response to DNA damage. It also activates DNA repair proteins when extensive DNA damage is detected, and if the damage is irreparable, p53 will initiate apoptosis. Interestingly, the microarray analysis data indicated that the nucleotide excision repair pathway was one of the most significantly perturbed pathways in the rASCs compared with the hBMSCs. Almost all of the significant differences that mapped to this pathway (Supplementary Fig. S2B) were expressed at a higher level in hBMSCs, probably reflecting the preferential recruitment of p53 in these cell types, compared with rASCs. The mechanism for the formation of tetraploidy in somatic cells is unknown, but evidence suggests that it may result from endoreduplication, which has been associated with p53 inactivation. Furthermore, p53 inactivation can cause centrosome amplification, leading to multipolar spindles and subsequent missegregation, resulting in aneuploidy (33).
Whereas all the MSCs studied showed altered cell cycle progression at higher passages, the rhesus MSCs displayed an aneuploidy karyotype after long-term culture. However, none of these abnormal MSCs were tumorigenic in immunodeficient mice. Tetraploidy and DNA anueploidy are implicated in the early phase of carcinogenesis. Recent studies have indicated that the inherent instability of an aneuploid karyotype drives a chain reaction of aneuploidizations that leads to chromosomal instability. It is indicated that cells need to accumulate specific genomic changes with tetraploidization before full transformation (40). Although rhesus MSCs showed ploidy, these changes did not satisfy the criteria of cancer cells. However, the extended continuous culture of these aneuploidy cells may result in full transformation and tumor formation upon transplantation in vivo. These results indicate that long-term culture of MSCs results in significant changes in cell cycle kinetics, suggesting the importance of biosafety studies of MSCs before their clinical application.
Acknowledgments
Grant support: National Center for Research Resources, NIH, grant number RR00164, and a grant from the State of Louisiana Millennium Health Excellence Fund and the Louisiana Gene Therapy Research Consortium.
We thank Dr. Darwin Prockop and the Center for Gene Therapy at Tulane for providing the human BMSCs, Dr. Xavier Alvarez, and the staff of the confocal microscopy and the flow cytometry core laboratories for their help.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Available at http://biosun1.harvard.edu/complab/dchip.
References
- 1.Zuk PA, Zhu M, Ashjian P, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodbury D, Reynolds K, Black IB. Adult bone marrow stromal stem cells express germline, ectodermal, endodermal, and mesodermal genes prior to neurogenesis. J Neurosci Res. 2002;69:908–917. doi: 10.1002/jnr.10365. [DOI] [PubMed] [Google Scholar]
- 3.Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 4.Izadpanah R, Trygg C, Patel B, et al. Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue. J Cell Biochem. 2006;99:1285–1297. doi: 10.1002/jcb.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern S, Eichler H, Stoeve J, Kluter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 6.Israels ED, Israels LG. The cell cycle. Stem Cells. 2001;19:88–91. doi: 10.1634/stemcells.19-1-88. [DOI] [PubMed] [Google Scholar]
- 7.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 9.Bonab MM, Alimoghaddam K, Talebian F, Ghaffari SH, Ghavamzadeh A, Nikbin B. Aging of mesenchymal stem cell in vitro. BMC Cell Biol. 2006;7:14. doi: 10.1186/1471-2121-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubio D, Garcia-Castro J, Martin MC, et al. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035–3039. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 11.Miura Y, Miura M, Gronthos S, et al. Defective steogenesis of the stromal stem cells predisposes CD18-null mice to osteoporosis. Proc Natl Acad Sci U S A. 2005;102:14022–14027. doi: 10.1073/pnas.0409397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 13.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 14.Izadpanah R, Joswig T, Tsien F, Dufour J, Kirijan JC, Bunnell BA. Characterization of multipotent mesenchymal stem cells from the bone marrow of rhesus macaques. Stem Cells Dev. 2005;14:440–451. doi: 10.1089/scd.2005.14.440. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya I, Colter DC, Prockop DJ. BMP-6 enhances chondrogenesis in a subpopulation of human marrow stromal cells. Biochem Biophys Res Commun. 2001;284:411–418. doi: 10.1006/bbrc.2001.4898. [DOI] [PubMed] [Google Scholar]
- 16.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and utlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiyama A, Watanabe H, Toyoda A, et al. Construction and analysis of a human-chimpanzee comparative lone map. Science. 2002;295:131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- 19.Ylostalo J, Randall AC, Myers TA, Metzger M, Krogstad DJ, Cogswell FB. Transcriptome profiles of host gene expression in a monkey model of human malaria. J Infect Dis. 2005;191:400–409. doi: 10.1086/426868. [DOI] [PubMed] [Google Scholar]
- 20.Chismar JD, Mondala T, Fox HS, et al. Analysis of result variability from high-density oligonucleotide arrays comparing same-species and cross-species hybridizations. Biotechniques. 2002;33:516–518. doi: 10.2144/02333st01. [DOI] [PubMed] [Google Scholar]
- 21.Enard W, Khaitovich P, Klose J, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 22.Kayo T, Allison DB, Weindruch R, Prolla TA. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci U S A. 2001;98:5093–5098. doi: 10.1073/pnas.081061898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uddin M, Wildman DE, Liu G, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci U S A. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vahey MT, Nau ME, Taubman M, Yalley-Ogunro J, Silvera P, Lewis MG. Patterns of gene expression in peripheral blood mononuclear cells of rhesus macaques infected with SIVmac251 and exhibiting differential rates of disease progression. AIDS Res Hum Retroviruses. 2003;19:369–387. doi: 10.1089/088922203765551728. [DOI] [PubMed] [Google Scholar]
- 25.Dillman JF, Phillips CS. Comparison of non-human primate and human whole blood tissue gene expression profiles. Toxicol Sci. 2005;87:306–314. doi: 10.1093/toxsci/kfi243. [DOI] [PubMed] [Google Scholar]
- 26.Caceres M, Lachuer J, Zapala MA, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortunel NO, Out HH, Ng HH, et al. Comment on “‘Stemness‘: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 28.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 29.Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- 30.Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci U S A. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 33.Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene. 2004;23:6845–6853. doi: 10.1038/sj.onc.1207860. [DOI] [PubMed] [Google Scholar]
- 34.Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003;2:202–210. [PubMed] [Google Scholar]
- 35.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 36.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 37.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 38.Bartek J, Lukas J. Pathways governing G1-S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 39.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 40.Inomata K, Oga A, Kawauchi S, Furuya T, Sasaki K. Global genomic changes induced by two-stage carcinogen exposure are precancerous alterations in non-transformed human liver epithelial THLE-3 cells. Int J Oncol. 2005;27:925–931. doi: 10.3892/ijo.27.4.925. [DOI] [PubMed] [Google Scholar]