Abstract
MicroRNAs (miRNAs) interact with target sites located in 3' untranslated regions (3'UTR) of mRNAs to down-regulate their expression when the appropriate miRNA is bound to target mRNA. To establish the functional importance of target localization in the 3' UTR, we modified the stop codon to extend the coding region of the transgene reporter through the miRNA target sequence. As a result, the miRNAs lost their ability to inhibit translation but retained their ability to function as siRNAs in mammalian cells in culture and in vivo. The addition of rare but not optimal codons upstream of the extended opening reading frame (ORF) made the miRNA target more accessible and restored miRNA-induced translational knockdown. Taken together, these results suggest that active translation impedes miRNA-programmed RISC association with target mRNAs, and support a mechanistic explanation for the localization of most miRNA target sites in noncoding regions of mRNAs in mammals.
MiRNAs are a class of short, 20- to 22-nt-long regulatory RNAs expressed in plants and animals1,2. Up to 4% of the human genome is predicted to code for over 400 miRNAs, which are estimated to regulate at least 30% of all human genes3-5. Although the specific functions of very few have been well established, a growing body of evidence indicates miRNAs play important regulatory roles in a vast range of biological processes 6-8. In plants, the majority of miRNAs hybridize to target mRNA with a near-perfect complementarity, and mediate an endonucleolytic cleavage through a similar, if not identical, mechanism used by the siRNA pathway 9. In animals, with few exceptions, most of the known miRNAs form an imperfect duplex/hybrid, with sequences located solely in the 3'UTR region of target mRNA (base-pairing of a minimum 7 nucleotide seed sequence is required) 10-12. The central mismatch between miRNA-mRNA hybridization may be responsible for the lack of RNAi-mediated mRNA cleavage events13,14. The association between miRNA-programmed RISC and target mRNA induces translational repression through a poorly understood mechanism. There is evidence supporting models in which translation repression occurs at the initiation stage or later steps, including elongation (reviewed in 15,16). Repressed mRNA and associated Ago proteins are enriched in Processing bodies (P-bodies), where endogenous cellular mRNAs are kept for storage and degradation17,18, which may partially explain why miRNA-mediated translational inhibition is often coupled with some RISC-independent target mRNA degradation 19.
In contrast to an siRNA which can target almost any part of an mRNA and be fully functional, almost all identified target sites for endogenous miRNA are located in the 3'UTR of target mRNAs in animals. This has been established by extensive bioinformatic sequence analyses as well as by experimental approaches 2. To further define the molecular events involved in miRNA-induced silencing, we cloned both the human mir-30 and Drosophila bantam miRNA target sites into the 3'UTR of the luciferase and GFP reporter genes so that by deleting one nucleotide in the stop codon, we were able to extend the ORF into the target site while maintaining the bioactivity of the protein. Using these reporter constructs as a starting point, in combination with the corresponding shRNA/miRNA expression cassettes, we provide experimental proof that there is a functional basis for the observed distribution of miRNA target sites in mammalian systems.
RESULTS
MiRNA-mediated repression is abolished in extended ORFs
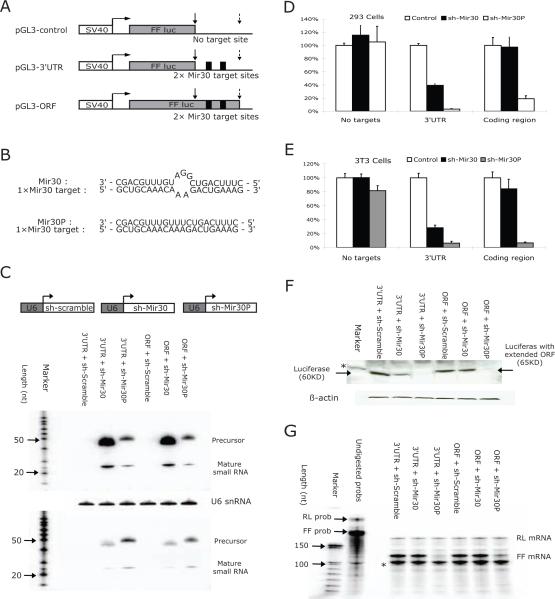
To establish if miRNAs can retain their negative regulatory activity if their targets remain in the 3' UTR of an mRNA but become embedded within the coding sequence, we constructed luciferase expression plasmids that contained no miRNA target sites; or tandem mir-30 target sites in the 3'UTR; or mir-30 target sites with an additional single base-insertion abolishing the stop-codon and extending the ORF through the mir-30 sites (Figure 1A). Each plasmid was tested for miRNA-induced silencing in mammalian cells. Specifically, luciferase plasmids were co-transfected with plasmids that can direct the expression of miRNAs, such as sh-mir-30 (mismatch); sh-mir-30P (perfect complementarity); or sh-Scramble (scrambled control) (Figure 1B,C). We first established that the mir-30 and mir-30P expressed from U6-driven cassettes were processed correctly and resulted in similar levels of the mature miRNA transcripts between transfection experiments (Figure 1C). As expected, co-transfection of plasmids expressing sh-mir-30, sh-mir-30P, or a scrambled shRNA with a Firefly luciferase (FF-luciferase) reporter construct without mir-30 target sites in HEK293 cells and NIH3T3 cells did not alter FF-luciferase expression, as measured by enzymatic activity in a dual-luciferase assay (Figure 1D, E). Moreover, this established there were no off-target effects using this reporter system from any of the U6-shRNA expressing constructs. Consistent with previous studies20, sh-mir-30 effectively down-regulated FF-luciferase expression by more than 60%, while sh-mir-30P inhibited FF-luciferase expression by >90% when tandem mir-30 target sites were present in the 3'UTR region (Figure 1D, E). Interestingly, when the same target sites were embedded within the extended coding region in both HEK293 cells and NIH3T3 cells (Figure 1D,E), sh-mir-30 9but not sh-mir30P)-induced repression was abolished (<3% for HEKs293 cells, and <15% for NIH3T3 cells).
Figure 1. MiRNA-mediated repression is abolished in extended ORFs.
The structure of the reporter constructs used in this study. pGL3-control containing no miRNA target sites; pGL3-3'UTR with two tandem mir-30 targets sites located in 3' untranslated region; and pGL3-ORF with upstream stop-codon abolished and mir-30 target sites covered by extended ORF. Grey box represents the ORF of FF-luciferase gene. Dark box represents the tandem mir-30 target sites with six base-pairs in between. Positions of upstream (original) stop-codon and downstream stop-codon are indicated by solid and dotted arrows, respectively.
(A) Schematic illustration of the interactions between mir-30 target sequence and guiding strand sequence of sh-mir-30 and sh-mir-30P, respectively. (C) NIH3T3 cells were co-transfected with plasmids, as described above. Sh-RNA expressed from U6-driven cassette was detected by Northern blot using either a probe against mir-30 (Up) or a probe against mir-30P (Down). Due to sequence similarity, cross-hybridization was observed. Endogenous U6 snRNA was also detected as an internal control.
(D)HEK293 cells and (E) NIH3T3 cells were co-transfected with different combinations of plasmids, and dual-luciferase assays were performed 36hr post-transfection. FF-luciferase activities were normalized with RL-luciferase, and the percentage of relative enzyme activity compared to the negative control (treated with sh-scramble) was plotted. Error bars represent the standard deviation from three independent experiments, each performed in triplicate.
(F) Protein analysis by Western blot was performed in transfected 3T3 cells. A protein band of β-actin was used as an internal control. Positions of the bands representing wild-type or mutant FF-luciferase were indicated by arrows. A non-specific band was indicated by an asterisk. (G) RNA levels of either FF-luciferase or RL-luciferase from transected 3T3 cells were detected by ribonuclease protection assay (RPA). Full-length probes and protected bands were indicated in the figure. A band labeled with an asterisk is possibly due to a truncated RL probe and, therefore, corresponding to RL-luciferase mRNA level.
The construct containing the tandem mir-30 target sites in the extended ORF was predicted to produce a FF-luciferase with extra amino acids at the C-terminal end. Although the luciferase activity from the extended ORF was in the linear range of the enzyme assay, it was about 100 times less active than the wild-type FF-luciferase (data not shown). A Western blot showed that, as expected, the ORF-extended protein migrated with a higher molecular weight and a signal intensity that was similar to the wild-type protein (Figure 1F). Importantly, relative changes in the protein band intensity for both the wild-type and extended ORF paralleled changes in luciferase activity measurements under all conditions when they were directly compared.
To confirm that both miRNA- and RNAi-mediated mechanisms were active, luciferase mRNA levels in transfected NIH3T3 cells were measured by ribonuclease protection assay. Co-expression of the sh-mir-30 and reporter containing the miRNA target in the 3'UTR resulted in 70% down-regulation of enzymatic activity and no detectable variation in mRNA, indicating that the reduction in protein level was primarily the result of translational repression, suggesting miRNA-mediated inhibition (Figure 1G). In contrast, the FF-luciferase-extended ORF mRNA concentration did not change in the presence of mir-30 expression, but was greatly reduced when mir-30P was co-expressed (Figure 1G). These results show that while miRNA-mediated translational inhibition was limited to targets in the untranslated region, RNAi-mediated activity directed against the same sequence remained functional, whether or not the site was within a coding sequence. This is consistent with a previous report 21 where only minor reductions of siRNA-mediated cleavage efficiency were observed when target sites were switched from an untranslated to translated region.
To establish that miRNA-mediated repression between target sites in the 3'UTR and coding region was not limited to a single reporter system or cell line, we placed the same miRNA targets into an EGFP reporter gene and co-expressed with the various miRNAs (Figure 1) in both NIH3T3 (Supplementary Fig. 1) and HEK 293 cells (not shown) and obtained similar results.
As a final test for fidelity, we replaced the mir-30 sequences with a bantam miRNA target and corresponding miRNA originally identified in Drosophila melanogaster and not believed to have a direct mammalian counterpart22. Co-transfection studies using U6-bantam expression plasmids in mammalian cells (Supplementary Fig 2 A, B) gave virtually identical results as observed for the mir-30 constructs in both HEK293 cells and NIH3T3 cells. The ability of bantam miRNA to repress translation was lost when the target was part of the extended ORF, while the RNAi activity induced by bantam-P was equally robust, whether or not the target was embedded into a coding region (Supplementary Fig 2 C,D). Moreover, to establish that accessibility and functionality of the miRNA target were functions of its placement in the 3'UTR rather than its specific position in the 3'UTR, we found that varying its location relative to the stop codon and poly-A signal with the insertion of an irrelevant ~700 bp fragment had little effect on miRNA-induced silencing (Supplemental Fig 3).
ORFs are refractory to miRNA-mediated regulation in vivo
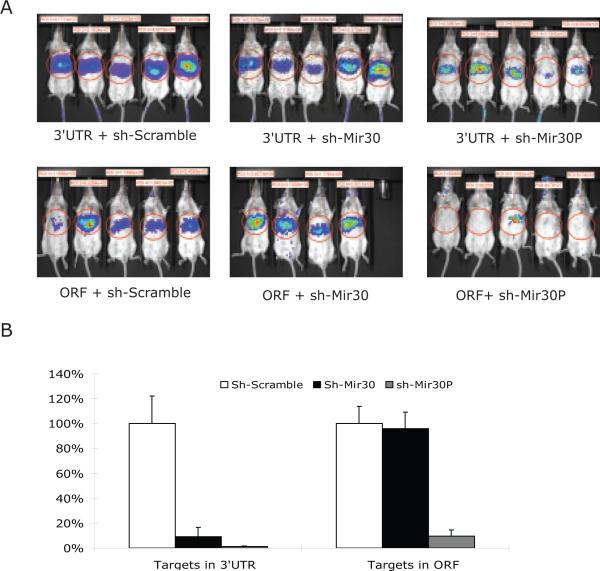
To establish that the regulatory miRNA circuit is biologically operative under physiological conditions in whole mammals, we examined the efficiency of the mir-30-Luciferase system (Figure 1) in mouse liver. We selected mir-30 because it is not believed to be highly expressed in mouse liver 23,24. Luciferase expression plasmids (Figure 1) were co-transfected into mouse liver via a hydrodynamic tail vein infusion, a method known to transfect up to 30% of mouse hepatocytes in vivo25. Four days later, luciferase expression was determined (Figure 2A). To control for variation in transfection efficiencies between individual animals, the FF-luciferase expression data was normalized (Figure 2B) to an added control plasmid expressing a third unrelated transgene product (see Methods).
Figure 2. MiRNA-mediated repression studies were concordant in mouse liver in vivo.
(A) The plasmids described in Figure 1 were transfected into mice by hydrodynamic tail injection (N = 5 per group, except group 4, N = 4, one animal died after injection). Real-time transgene expression was determined four days after injection. Levels of luciferase reporter activities were quantified as shown in each image.
(B) A control plasmid, RSV-hAAT was co-transfected within each sample as an internal control for transfection efficiency. The FF-luciferase activities were normalized to serum hAAT levels measured by ELISA. Percentage of relative luciferase activity compared to negative controls (treated with sh-scramble) was plotted. Error bars represent the standard deviation.
Data obtained from mouse liver was concordant with data from tissue culture cells. While RNAi-mediated knockdown activity was robust whether the target was in the 3'UTR or part of the extended coding region, miRNA-induced silencing was severely compromised when the target was included within the extended ORF.
Rare codons upstream of the ORF miRNA target rescues knockdown
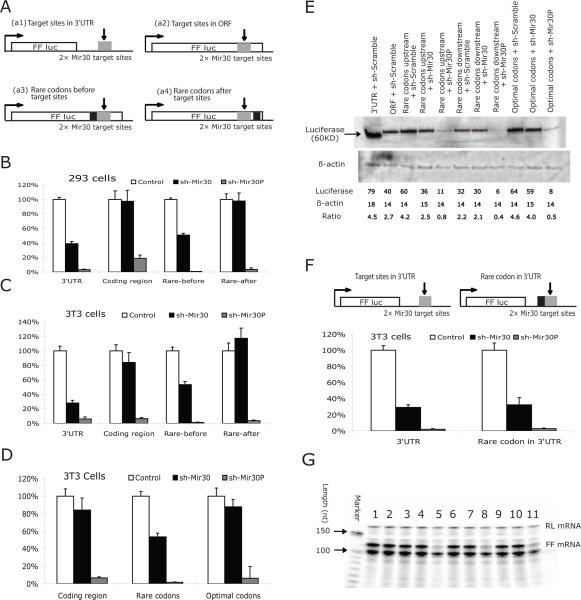
Since our results suggested that active translation of mRNAs precludes miRNA-induced knockdown, we predicted that ribosome hindrance would interfere with the ability of miRNA and its associated machinery to attach to its target site. To test this, we introduced a cluster (9 amino acids) of rare codons upstream of miRNA target sites located in extended luciferase ORF (Figure 3A), an approach used to cause ribosome pausing in eukaryotes26,27. Since we could not measure ribosome translocation directly, we constructed a number of different control sequences for direct comparisons. We inserted the same 9 amino acids in the identical location using an optimized set of codons, or placed the rare codons downstream of the miRNA target. When the rare codons were upstream of the target, miRNA-induced silencing from sh-mir-30 was restored to a level close to what was observed (rescue >80% and 70% in 293 and NIH 3T3 cells, respectively), while replacing the rare with optimal codons or placing the rare codons downstream of the miRNA target was unable to rescue miRNA-induced silencing (Figures 3B-E). This confirmed that the additional nucleotides or the extra amino acids were not responsible for the differential activity of the miRNA target. To eliminate the possibility that adding the 27 nucleotides altered the local RNA folding structure -- and, hence, the accessibility and efficacy of miRNA target sites -- we inserted these sequences upstream of mir-30 target sites, which remain in the 3'UTR in the FF-luciferase reporter construct. MiRNA-mediated repression was not changed (Figure 3F). RNA analyses (Figure 3G) confirmed that the rare/optimal codon clusters had no substantial effect on the steady-state mRNA levels.
Figure 3. Insertion of rare-codons upstream of the extended miRNA ORF rescues miRNA-mediated knockdown.
(A) The maps of the reporter constructs used in this study. Plasmids containing tandem mir-30 target sequence in either 3'UTR (a1) or ORF (a2) are the same as those described in Figure 1. A cluster of rare codons (represented as a dark box) were inserted either upstream (a3) or downstream (a4) of mir-30 target sequences. In another construct, the upstream rare codons (a3) were replaced with optimal-codon sequences that code for the same peptide sequence. The arrows and grey box represent the position of the miRNA target sequences.
(B) HEK293 cells and (C), (D) NIH3T3 cells were transfected with the reporter constructs illustrated in (A). Dual-luciferase assays were performed 36hrs post-transfection. FF-luciferase activities were normalized with RL-luciferase, and the percentage of relative enzyme activity compared to the negative control (treated with sh-scramble) was plotted. Error bars represent standard deviation from three independent experiments, each performed in triplicate.
(E) Protein levels of reporter genes were analyzed by Western blot in transfected 3T3 cells.
(F) NIH3T3 cells were transfected with constructs as indicated in the figure. Insertion of rare-codon cluster (dark box) upstream of mir-30 targets sites in the 3'UTR did not substantially change the miRNA-induced repression.
(G) RNA levels of reporter genes were analyzed by Ribonuclease Protection Assay. The loading sequence of line 1 to 11 is same as noted in (E).
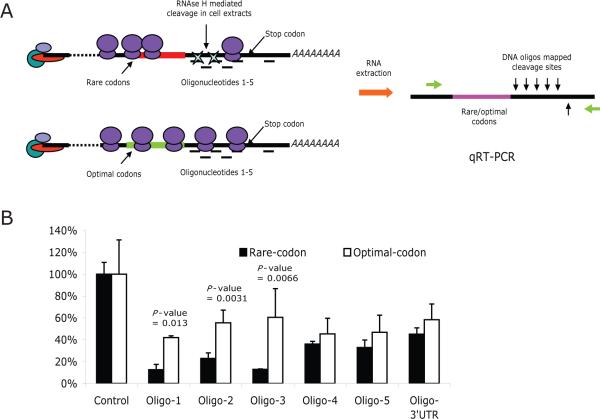
To further validate that the rescue of the miRNA repression was due to the brief translational pause mediated by rare codons26,27, we mapped the accessibility of sequences downstream of the rare and optimal codons using a DNA-oligo-RNase H approach 28 (Figure 4A). The sequences immediately downstream (~70 nucleotides - Oligos 1-3 in Figure 4) of the rare codons were more accessible to RNase H-mediated cleavage compared to the same sequences in the mRNAs containing the optimal codons (Figure 4B). In contrast, sequences farther downstream of the rare/optimal codons in the 3'UTR were similar in their accessibility to RNase H cleavage (Oligos 4 and 5, Figure 4A), indicating that the difference in accessibility is specific to the region just downstream of the rare codon track (Figure 4B). In addition, and consistent with our prediction (Figure 4A), RNase H-mediated cleavage was equal or modestly less robust in sequences contained upstream of the rare vs. optimal codon mRNA sequences, suggesting a slight backup of ribosomes upstream of the rare-codon insertion (Supplementary Fig 4). Since the steady-state production of protein (Fig 3E) and the average density of ribosomes along the mRNA as determined by polysome gradient fractionation (supplementary Fig 5) was not substantially altered by the rare-codon insertion, the ribosomal pause during active translation over the specific region covered by oligos 1-3 was likely quite brief.
Figure 4. Insertion of rare codons increases the accessibility of downstream sequences to RNase H-mediated cleavage.
(A) Experimental strategy. Cells were transfected with the luciferase reporter constructs containing the cluster of rare or optimal codons (Figure 3A). After fixing the ribosomes on the mRNA by the addition of cycloheximide, one of six oligos corresponding to the region between rare/optimal codons and target 3'UTR was added into the cell extracts. The hybridization of DNA oligos at the target site within the mRNA results in cleavage mediated by the endogenous RNase H activity in the cell extracts. The extent of the cleavage represents the relative RNA accessibility, which was quantified by real-time RT-PCR using two primers flanking the cleavage sites. (B) Quantification of RNase H-mediated cleavage. The values are presented as the relative PCR signal compared to control samples treated with a scrambled oligonucleotide and normalized for a GFP mRNA obtained from a co-transfected control plasmid.
Repressed reporter mRNAs are associated with polyribosomes
Our data was consistent with the requirement of a stable association between miRNA-RISC and target mRNA for miRNA-induced translation repression. We next investigated whether this association results in exclusion of the target mRNA from the translational machinery by analyzing the polysome profiles of repressed target mRNAs. Whole cell extracts were prepared from NIH3T3 cells transfected with either a luciferase or an EGFP reporter gene containing tandem mir-30 target sequence in either 3'UTR or ORF, as well as plasmids expressing sh-mir-30 or sh-scramble. Polysome sedimentation profiles of luciferase reporter mRNA and control RL-luciferase mRNA were measured by RPA (Supplementary Fig 6), while EGFP mRNA and actively translated β-actin mRNA were determined by Northern blot (Supplementary Fig 7). Importantly, reporter mRNAs containing target sequences in their 3'UTRs or in the extended ORF and co-expressed with sh-scramble or sh-mir-30, displayed distribution profiles similar to actively translated mRNA (RL-luciferase or actin) (Supplementary Fig 6B and Supplementary Fig 7 A-E). To establish that these mRNAs were actually associated with polyribosomes, we performed polysome gradient analyses after treatment with puromycin or EDTA, both of which release polysomes. As shown in Supplementary Figs. 6 and 7, the miRNA-repressed mRNAs shifted to the slow-sedimenting part of the gradient to the same degree as actively translated mRNA after puromycin (Supplementary Fig 6C and Supplementary Fig 7F and H) or EDTA treatment (Supplementary Fig 6D and Supplementary Fig 7G and I). These results strongly favor a model where miRNA-targeted mRNAs remain associated with the polyribosome. At first consideration, these results appear concordant with some studies 29-33 but contrast with other studies where miRNA-repressed mRNAs were found in the fast-sedimenting34,35 or puromycin-resistant, slow-sedimenting pseudopolysomal fractions 36.
Discussion
Taken together, our studies, using multiple expression systems, cells, and miRNA targets, are in good concordance. While our results suggest that location within the 3'UTR may not cause a large functional difference, there does appear to be a functional reason for the localization of miRNA targets in the 3'UTR. We propose that these functional constraints may be the primary explanation for the observed miRNA target distribution pattern found in mammalian cells. However, other studies have reported that artificially designed, mismatched siRNA or shRNA co-delivery studies can result in some translational repression when mRNAs contained target sequences in the coding regions 37,38. The source of these contradictions is not completely clear, but in several studies, the mismatched synthetic siRNAs were provided in very high concentrations. Other factors possibly contributing to the degree of repression between miRNAs and their corresponding targets may include sequence composition 4, number of target sites 39, local RNA structure40 and distance between target sites41. Adding or removing miRNA target sites in coding regions may not elucidate the true natural functional differences between a target site residing in the coding region and 3'UTR. In our study, we carefully designed our reporter constructs such that there was only one nucleotide difference between the mRNA sequences we directly compared. Therefore, the abolishment of miRNA-induced gene repression should be a direct result of changing the 3'UTR to ORF without making major alterations in the mRNA sequence.
Our data support a model whereby miRNA-programmed RISC is required to remain attached to the target mRNA to effectively silence translation in cis. Moreover, when target sites remain in the same site of the mRNA but become part of the coding region, ribosomal complexes override and inhibit the miRNA-programmed RISC from attaching to the target site. If the translational process is slowed, we speculate there is less physical constraint by the ribosomes, thus allowing miRNA-programmed RISC to attach to the target.
This process seems to be functionally distinct from RISC RNAi-mediated RNA degradation, since converting the miRNA to give it perfect complementarity to the target still resulted in loss of the mRNA, presumably through the RNAi pathway whether the miRNA target was part of the extended coding sequence or in the 3'UTR. This is consistent with the finding that, unlike in mammals, miRNA target sites in plants are widely distributed across coding regions, since nearly all of them have perfect complementarity with their target sequences and function through an RNAi-mediated degradation pathway. Curiously, the only known mammalian miRNA which targets the coding region in the mRNA has perfect complementarity with its targets and also functions through RISC-mediated cleavage42. Nonetheless, we cannot exclude the possibility that some functional miRNA targets exist in coding regions. If such sites are identified, it will be of great interest to determine if they are preceded by rare codons. In fact, Agami and colleagues provide evidence for a functional miRNA target in the coding region of an endogenous mammalian gene.43 The mRNA was active when it had an extensive 17bp but not the more classical 7bp 5' seed match with the mRNA sequence. This suggests that the down-regulation may have been mediated by RNAi cleavage rather than translational down-regulation44.
Also of interest in future studies is determining when functional miRISC-mRNA complexes can be assembled in the post-transcriptional life of an mRNA . Our results show that if translocation of ribosome is slow, miRISC complexes can still form after translational initiation begins. We favor a model where miRNA-RISC biding to actively translating mRNAs result in reduced translational elongation/termination concordant with a reduction in ribosomal initiation and possible nascent peptide destabilization.32,33
While we provide evidence for why endogenous miRNA target sites are found in non-coding regions, it is logical to ask why relatively few miRNA targets are localized in the 5'UTR. When the translation initiation complex forms around the cap structure, the 40S subunit of ribosome will scan the 5'UTR until identifying the first AUG where the 60S subunit joins to form an 80S ribosome. It is possible that the scanning process impairs the formation of miRNA-RISC complexes in some 5'UTRs depending on its structure, which can be quite complex. Our preliminary studies were consistent with this because we found a great degree of discordance between different miRNA target 5'UTR insertions and the degree of translational repression (Gu and Kay, unpublished data). Nonetheless, there are examples of miRNAs that do function with 5'UTR targets. While one study shows that the mir-122 target sites located in the 5'UTR region of HCV are important to maintain robust viral replication 45, another reports that the mRNA-bearing miRNA target sites in the 5'UTR can be repressed as effectively as those having miRNA target sites in the 3'UTR46. Further studies are needed to establish the extent to which functional miRNA targets are present in these non-coding regions.
METHODS
Plasmid constructions
Both strands of 2XMir30 target sites were chemically synthesized (sense strand: 5'-AATTCGCTGCAAACAAAGACTGAAAGAACTAGTGCGCTGCAAACAAAGAC TGAAAGCTGCA;antisense strand5'-GCTTTCAGTCTTTGTTTGCAGCGCACTA GTTCTTTCAGTCTTTGTTTGCAGCG:), annealed, purified, and inserted between EcoRI and PstI sites 67 bp downstream of FF-luciferase coding region in pGL3 construct with modified 3'UTR sequences. PCR-based point mutagenesis approaches were used to create a single-point insertion to disrupt stop codon of the FF-luciferase gene. A similar approach was used to generate the GFP reporter system and FF-luciferase reporter system with bantam target sequences. An ~700 bp long sequence in the middle of a kanamycin-resistant gene coding region was PCR amplified, then inserted into various cloning sites upstream or downstream of miRNA target sites to reposition the miRNA targets within different regions of the 3'UTR.
Rare codon sequences (5'- GCG CCG GTA ACG GTA CCG GCG ACG GCG -3') or optimal codon sequences (5'- GCC CCC GTC ACC GTC CCC GCC ACC GCC-3') were inserted either 53bp upstream or immediately downstream of mir-30 target sites. Mir-30/mir-30P or bantam/bantam-P shRNAs were designed as a passenger strand, followed by mir-22 loop sequence (CCTGACCCA) followed by the guiding strand sequence. These were cloned downstream of U6 Pol III promoter.
Cell culture and Transfections
Adherent HEK293 and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) with L-glutamine and 10% heat-inactivated fetal bovine serum with antibiotics. All transfection assays were done using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. HEK293 and NIH3T3 cells at 90% confluency were transfected in 24-well plates with 50ng FF-luciferase or EGFP reporter DNA, 50ng shRNA expression DNA, and 5ng RL-luciferase DNA, unless specified otherwise. Unless indicated, cells were assayed 36hrs after transfection.
Dual-Luciferase Assay
FF-luciferase and RL-luciferase were measured using Promega's dual-luciferase kit (cat E1980) protocol and detected by a Modulus Microplate Luminometer (Turner BioSystems).
Western Blots
NIH3T3 cells (36h post-transfection) were lysed with mammalian protein extraction reagent from M-PER (PIERCE cat #78501) with protease Inhibitors (Roche cat #11836153001). The samples were denatured in Laemmli sample buffer (Bio-RAD #161-0737) for 5 minutes at 95°C and separated in 10% (w/v) SDS-PAGE gels. The denatured proteins were then electro-transferred onto a PVDF membrane blocked with 5% (w/v) fat-free milk powder in PBS and 0.5% (v/v) Tween 20 for 1hr. Either an anti-FF-luciferase antibody (diluted 1:5000, Abcam), anti-GFP antibody (diluted 1:1000, Abcam), or anti-β-actin antibody (diluted 1:8000, Sigma) was used. Following three washes in PBS for 5 minutes, a secondary antibody (HRP-anti-mouse IgG, Sigma, dilution 1:10000) was added for 1h at room temperature, followed by three 5-minute washes in PBS. Antibody-bound proteins were visualized using the ECL Western blotting analysis system (Amersham, RPN2109).
Northern Blots and RPA
Total RNA was isolated using Trizol (Invitrogen). The DNA-free kit (Ambion Cat #1906) was used to purify total RNA from contaminating DNAs. Ten-20 ug of total RNA was electrophoresed on 1% (w/v) agarose gel. After transfer onto Hybond-N1 membrane (Amersham Pharmacia Biotech), target mRNAs were detected using P32-labeled full-length cDNA probes.
RPA assays were carried out according to Ambion PRA III kit (cat # AM1414). P32-labled antisense RNA probes against either FF-luciferase or RL-luciferase were generated by in vitro transcription (Ambion MAXIscript Kit, Cat #AM1308). DNA templates were produced by PCR using primer sets (FF-luc: ATCCATCTTGCTCCAACACC and TTTTCCGTCATCGTCTTTCC; RL: GATAACTGGTCCGCAGTGGT and ATTTGCCTGATTTGCCCATA). Total RNA from NIH3T3 cells were isolated by Trizol (Invitrogen) 36hrs after transfection and purified using a DNA-free kit (Ambion Cat #1906). Hybridization reactions were at 55°C overnight and RNase digestion was at 37°C for 30 minutes using the RNase A/T1 cocktail provided in the RPA III kit.
Hydrodynamic tail injection and luciferase imaging
Animals studies were done in concordance with NIH guidelines and the Stanford Animal Care Committee. Female BALB/c mice, 6-8 weeks of age (Jackson Laboratory, Bar Harbor, Maine) were hydrodynamically infused with a mixture of 2 ug FF-luciferase DNA, 2 ug of the appropriate shRNA plasmid, 2ug of an RSV-hAAT expression cassette DNA , and 34 ug pBluescript plasmid DNA (Stratagene), and were then imaged for luciferase. As described 47, raw light values were reported as relative detected light photon per minute, and normalized serum hAAT expression
Polyribosome Fractionation
Polysomal mRNA was prepared based on the method described previously 48. Briefly, before being harvested, cells were incubated with 0.1 mg ml-1 cycloheximide for 3 minutes at 37°C. NIH3T3 cells were harvested directly on their culture dish in lysis buffer (15 mM Tris-Hcl, pH 7.4, 15 mM MgCl2, 0.3 M NaCl, 1% (v/v) Triton X-100, 0.1 mg ml-1 cycloheximide, 1 mg ml-1 heparin), and loaded onto 10-50% (w/v) sucrose gradients composed of the same extraction buffer lacking Triton X-100. The gradients were sedimented at 35,000 rpm for 180 minutes in a SW41 rotor at 4°C. Fractions of equal volumes were collected from the top using an ISCO fraction collector system. RNAs were extracted by phenol/chloroform followed by isopropernol precipitation, 75% (v/v) ethanol washes, and re-suspended in DNase I reaction buffer (Turbo DNase, Ambion).
Mapping accessibility
This approach is modified from a previous publication49. HEK293 cells were transfected with plasmids expressing FF-luciferase reporter gene embedded with the cluster of rare or optimal codons along with a GFP control plasmid. Thirty-six hours post-transfection, cells were harvested after incubation with 0.1 mg ml-1 cycloheximide for 3 minutes at 37°C. After three washes with PBS, approximately 2×107 cells were pelleted and re-suspended in 2x volume of the cell pellet in hypotonic swelling buffer (7 mM Tris-HCl pH7.5; 7 mM KCl; 1 mM MgCl2; 1 mM beta-mercaptoethanol). After a 10-minute incubation on ice, samples were Dounce homogenized (VWR, San Diego, CA) 40 times with a tight pestle B followed by addition of one tenth of the final volume of neutralizing buffer (21 mM Tris-HCl pH 7.5; 116 mM KCl; 3.6 mM MgCl2; 6 mM beta-mercaptoethanol). After centrifugation of the homogenates at 20,000g for 10 minutes at 4°C, supernatants were collected. The RNase H-mediated-cleavage experiments were carried out in a total volume of 300 uL, containing 280 uL cell extracts, 1 mM dithiothreitol (DTT), 20-40 units RNase-Inhibitor (Promega, Madison, WI) and 50 nM each of the defined sequence antisense deoxyribooligonucleotides (ODNs) (Supplementary Table 1). The ODNs were incubated in the extracts for 5 minutes at 37°C. Total RNA was extracted by phenol/chloroform extraction. After the RT reaction (Invitrogen RT kit, cat # 18080-051) with oligo dT primer, real time-PCR (Qiagen, QuantiTect SYBR green PCR kit) was performed with two primers flanking the cleavage sites. (upstream: AGGCCAAGAAGGGCGGAAAG or ACCGCGAAAAAGTTGCGCGG; downstream: TCACTGCATTCTAGTTGTGG). All results are obtained with R > 0.98). Each oligonucleotide was tested six times in two separate experiments. P values were calculated using the student's t-test.
Supplementary Material
Acknowledgements
This work was supported by NIH Grant DK 78424. We thank Benjamin Hu for helping prepared some of the samples; Dr. Randy Cevailos for technical assistance with the polyribosome fractionation experiments and Dr. Dirk Haussecker for critical reading of the manuscript.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 7.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Triboulet R, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 9.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–71. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 15.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–6. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 19.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–33. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 21.Gu S, Rossi JJ. Uncoupling of RNAi from active translation in mammalian cells. Rna. 2005;11:38–44. doi: 10.1261/rna.7158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 24.Takada S, et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yant SR, et al. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez J, et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell. 2005;17:405–16. doi: 10.1016/j.molcel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Lemm I, Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol. 2002;22:3959–69. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scherr M, et al. Detection of antisense and ribozyme accessible sites on native mRNAs: application to NCOA3 mRNA. Mol Ther. 2001;4:454–60. doi: 10.1006/mthe.2001.0481. [DOI] [PubMed] [Google Scholar]
- 29.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–25. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 30.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 31.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–7. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 32.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–14. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 33.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 36.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007 doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 37.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 38.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–91. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–42. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long D, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–94. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 41.Saetrom P, et al. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–42. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 43.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. Rna. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 45.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 46.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 48.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. Rna. 1998;4:1500–13. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu S, Ji J, Kim JD, Yee JK, Rossi JJ. Inhibition of infectious human immunodeficiency virus type 1 virions via lentiviral vector encoded short antisense RNAs. Oligonucleotides. 2006;16:287–95. doi: 10.1089/oli.2006.16.287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.