Abstract
Cis-natural antisense transcripts (cis-NATs) have been speculated to be substrates for endogenous RNA interference (RNAi), but little experimental evidence for such a pathway in animals has been reported. Analysis of massive Drosophila melanogaster small RNA data sets now reveals two mechanisms that yield endogenous small interfering RNAs (siRNAs) via bidirectional transcription. First, >100 cis-NATs with overlapping 3′ exons generate 21-nt, Dicer-2 (Dcr-2)–dependent, 3′-end modified siRNAs. The processing of cis-NATs by RNA interference (RNAi) seems to be actively restricted, and the selected loci are enriched for nucleic acid–based functions and include Argonaute-2 (AGO2) itself. Second, we report that extended intervals of the thickveins and klarsicht genes generate exceptionally abundant siRNAs from both strands. These siRNA clusters derive from atypical cis-NAT arrangements involving introns and 5′ or internal exons, but their biogenesis is similarly Dcr-2– and AGO2-dependent. These newly recognized siRNA pathways broaden the scope of regulatory networks mediated by small RNAs.
cis-NAT pairs, for which the same genomic segment is transcribed on both strands, are common in the genomes of most higher eukaryotes1–3. Such transcript arrangements have been proposed as a potentially substantial source of endogenous double-stranded RNA (dsRNA)4, which might conceivably be processed by RNAi into siRNAs. In plants, select cis-NAT pairs generate siRNAs that function in stress response5,6. However, there has been no direct evidence for this mechanism in animals; indeed, the available evidence argues against the generation of RNAi triggers by cis-NATs7.
This is not to say that cis-NAT arrangements are not of regulatory consequence. For example, the steric hindrance of colliding polymerase complexes might disfavor simultaneous transcription of both strands8, or influence alternative splicing or polyadenylation9. On the other hand, if cis-NATs readily formed dsRNA that entered the RNAi pathway, one might expect that forced expression of antisense transcripts would efficiently trigger gene knockdown. This technique is not actually widely applicable10,11, probably because transcripts are likely to be segregated in vivo as ribonucleoprotein complexes at most stages of their life cycle. Therefore, complementary transcripts, even if coexpressed in the same cell, are not necessarily expected to have the opportunity to meet and anneal as dsRNA in vivo.
In this study, we examined large collections of D. melanogaster small RNA sequences and confidently identified two classes of endogenous siRNAs that derive from bidirectional transcription. These include > 100 cis-NAT pairs, most of which overlap on their 3′ ends. siRNA cloning patterns suggest that processed mRNAs engage in dsRNA interactions to yield 3′-modified, 21-nt RNAs. This does not seem to be a default pathway, as we identified hundreds of coexpressed cis-NATs that do not generate siRNAs. We also identified two exceptional regions of the genome whose siRNAs seem to define a distinct mechanism, on the basis of their largely intronic origin, their ability to generate unusually large numbers of siRNAs across extended genomic intervals and their involvement with candidate antisense noncoding transcripts. Contrary to current understanding of how dsRNA is processed, the biogenesis of these siRNAs is, unexpectedly, Loquacious (Loqs)-dependent, rather than r2d2-dependent. These observations collectively reveal new pathways for endogenous siRNA-mediated regulation in D. melanogaster.
RESULTS
Distinct sizes of various classes of 21–22-nt regulatory RNAs
We accumulated > 16 million mapped D. melanogaster small RNA reads from ten 454 samples12, a female head Solexa library13, a Solexa library from mass-isolated imaginal discs and brains14, nine Solexa libraries from early embryos and adult heads and bodies15, and three new Solexa libraries prepared from S2 cells and Kc cells (this study). In searching for new classes of small RNAs in these data, we sought to distinguish genuine regulatory RNAs from functionally irrelevant degradation products. Genuine regulatory RNAs have distinct sizes reflecting their particular biogenesis history, whereas random degraded RNAs are not expected to have particular size tendencies. For example, the reads mapped to ribosomal protein genes spanned the 18–26-nt window selected for library cloning (Fig. 1a), consistent with their probable identity as degradation fragments. In contrast, microRNAs (miRNAs) and miRNA* species, which are processed via consecutive cleavages of a precursor hairpin by Drosha and Dcr-1, show obvious size preference. Analysis of the first 382 miRNA clones suggested that 22-nt miRNA lengths were actually the most common in D. melanogaster16, and this preference held true across > 3.7 million miRNA reads, which collectively had a mean length of 21.91 nt (Fig. 1b).
Figure 1.
Distinct size distributions of different small RNA–generating loci. Each graph plots the number of small RNA reads on the y axis and the length of small RNAs on the x axis. (a) 93 ribosomal protein (RpL) genes. (b) 131 canonical miRNA genes; miRNA reads are blue and miRNA* reads are red. (c) The two most highly expressed hpRNA genes, hp-CG18854 and hp-CG4068 (ref. 14). (d) white siRNAs cloned from GMR-whiteIR heads20; sense reads are blue and antisense reads are red. (e) GFP siRNAs cloned from S2 cells carrying a GFP transgene and transfected with GFP dsRNA20; sense reads are blue and antisense reads are red. (f) All 5′ cis-NAT loci except the tkv/CG14033 cis-NAT. The combined overlap reads from both strands are blue, and the combined non-overlap exonic reads from both strands are red. (g) The non-overlap regions of all non-siRNA, 3′ cis-NAT loci, except the hp-CG18854/IP3K cis-NAT. In this and subsequent graphs, top-strand reads are blue and bottom-strand reads are red. (h) The overlap regions of 117 3′ cis-NAT siRNA loci. (i) The tkv-RC/CG14033 siRNA cluster. (j) The klar siRNA clusters.
A new class of endogenous small RNA derives from long inverted repeats (IRs) termed hairpin RNAs (hpRNAs), including CG18854 and a repeat cluster overlapping CG4068 (ref. 14). These produce a diverse set of reads that show preference for a length of 21 nt (Fig. 1c). This size distinction suggested that, despite their common derivation from hairpin transcripts, hpRNA biogenesis differs from that of miRNAs. Indeed, we found that hpRNA biogenesis is mediated not by Dcr-1, but instead by Dcr-2 (ref. 14), and generates functional siRNAs. Thus, endogenous Dcr-2 products seem to be shorter than Dcr-1 products.
We proposed that the internal bulges in hpRNA stems might push their average length above 21 nt and that processing of perfectly double-stranded substrates might yield products closer to 21 nt in length. Previous small-scale cloning of Dcr-2 products could not distinguish a preference for 21 nt or 22 nt17, and indirect analysis using tiling microarrays suggested that a perfectly double-stranded white IR RNA trigger18 is processed in transgenic animals with 22-nt periodicity19. We re-examined this using small RNA cloning data from white-IR heads reported by Zamore and colleagues20 and observed that white siRNAs actually show a strong preference for 21 nt (Fig. 1d). That study also described RNAs cloned from cultured S2 cells transfected with perfectly double-stranded GFP RNA, and we found that GFP siRNAs were similarly 21 nt (Fig. 1e). Taken together, these observations of miRNAs, hpRNAs and siRNAs show how functional differences in small RNA biogenesis can be inferred from subtle patterns in RNA size distributions.
Genome-wide survey of cis-NAT–derived siRNAs
About 1,000 Drosophila loci are arranged as cis-NAT pairs1. However, there is currently little data to support the coexpression of cis-NAT pairs in individual cells, which would presumably be necessary for them to be relevant to an RNAi-type mechanism. We reasoned that such evidence might be revealed by analyzing the distribution and size of small RNAs emanating from the non-overlap versus the overlap regions of cis-NAT pairs.
The two major classes of cis-NAT mRNA pairs in D. melanogaster are divergently transcribed loci that overlap on their initiating 5′ exons (100 loci) and convergently transcribed loci that overlap on their terminal 3′ exons (793 loci). A plausible explanation for the fact that there is more 3′ cis-NAT pairs than 5′ cis-NAT pairs is that transcriptional exclusion mechanisms are preferentially inhibitory to transcriptional initiation8. If there are indeed general mechanistic differences between 5′ and 3′ cis-NAT arrangements, then that might be reflected in the types of RNAs that emanate from these loci. We therefore examined the length, number and density of unique small RNA reads that mapped to the exons of top- and bottom-strand mRNA annotations of cis-NAT overlap regions. We compared these data to reads that mapped to the non-overlap regions of the same cis-NATs. The full data set is presented in Supplementary Table 1 online.
These analyses yielded four conclusions. First, there were extremely few small RNAs emanating from either strand of 5′ cis-NAT overlap regions (Fig. 1f, blue data). The corresponding non-overlap regions of 5′ cis-NATs generated RNAs with broad distribution across lengths of 18–26 nt (Fig. 1f, red data). We conclude that 5′ cis-NATs make, at most, only minor contributions to siRNA pathways. The small RNAs derived from the exons of 3′ cis-NAT genes outside of annotated overlaps showed detectable 21-nt bias but were otherwise spread over a broad size range (Fig. 1g). In contrast, the overlap regions of 3′ cis-NAT pairs generated RNAs with a strong 21-nt bias (Fig. 1h), similar to other known Dcr-2 products. These RNAs came from the top and bottom strands in approximately equal numbers. Finally, the overlap regions of cis-NAT pairs that yielded 21-mers showed much greater small RNA density than did the non-overlap regions. This is evident in the gene schematics shown in Figure 2.
Figure 2.
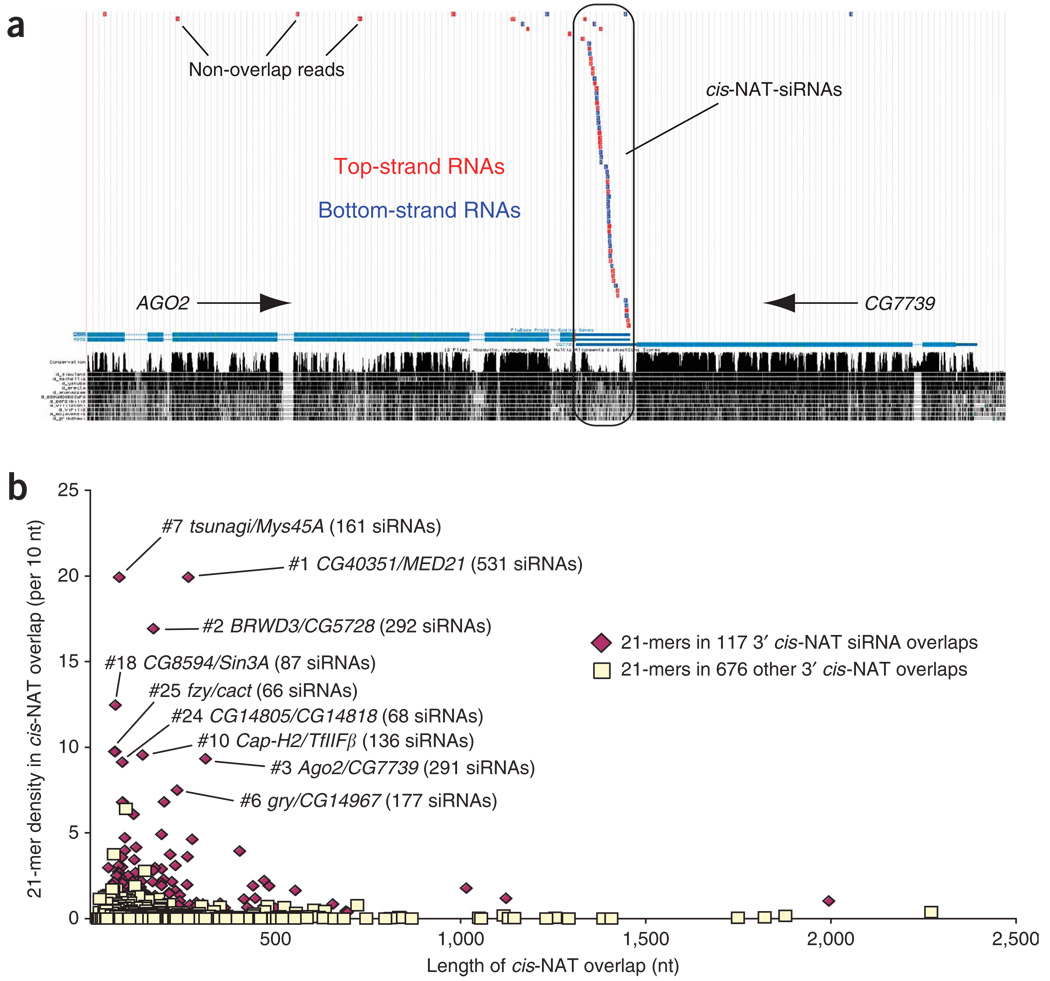
The overlap regions of a subset of 3′ cis-NAT loci preferentially generate siRNAs. (a) Small RNAs mapped to the AGO2/CG7739 cis-NAT via the UCSC genome browser42. Small RNAs from both strands mapped nearly exclusively to the mRNA-overlap region, and the vast majority (> 75%) of these are 21 nt in length (Supplementary Table 1). (b) A subset of 3′ cis-NAT overlaps are associated with particularly elevated siRNA density.
We propose from these data that 3′ cis-NATs are a substantial source of endogenous siRNAs in D. melanogaster. We estimated the minimum number of functional cis-NAT siRNA loci as follows. First, we selected cis-NAT overlap regions that produce a preponderance (> 67%) of 21-nt RNAs relative to all other sizes, which should reflect their preferred processing by an RNAi pathway. Second, we asked that there be at least 11 independent 21-nt reads generated by a given overlap. These criteria resulted in a set of 117 cis-NATs, with 40 pairs producing ≥40 siRNA reads (Supplementary Fig. 1a online). By comparison, we previously annotated 16 confident miRNAs on the basis of <40 reads12. In fact, this was a conservative estimate, as the annotated extent of many cis-NAT overlaps was incorrect (Supplementary Fig. 2a online and Methods). This contributed to the mild enrichment for 21-mers in annotated non-overlap regions of cis-NATs (Fig. 1f,g).
The enrichment of 21-nt RNAs in 3′ cis-NAT overlap versus non-overlap regions was especially evident in these top 117 loci. Their aggregate overlap length comprised 27,668 bp and generated 7,486 unique reads (271 reads per kilobase), of which 5,953 (80%) were 21 nt in length. In contrast, the corresponding non-overlap exonic regions of these 117 loci spanned 488,508 bp but generated only 8,848 unique reads (18 reads per kilobase), only 1,942 of which (22%) were 21 nt in length. This represents > 50-fold greater density of 21-nt RNAs in 3′ cis-NAT overlap regions compared to their annotated non-overlap regions. Comparison of the 117 cis-NAT siRNA loci and the remaining 676 3′ cis-NATs further highlighted that siRNA loci comprise a specialized subset of cis-NATs with great propensity to generate 21-nt RNAs from cis-NAT overlap regions (Supplementary Fig. 1b). This is clearly visualized in the siRNA density chart shown in Figure 2b.
Biogenesis of 3′ cis-NAT siRNAs
The relatively sharp demarcation of 21-mer mappings to 3′ cis-NATs suggested that the substrates were processed mRNAs with defined 3′ ends and precise complementary limits, instead of the heterogenous ends of primary transcripts before 3′ end cleavage. In support of this, we found rare examples in which a 3′ untranslated region (UTR) spanned several spliced exons on the other strand, in which case 21-nt RNAs were generated from exonic but not intronic sequence (for example, fbl/CG33969; Supplementary Fig. 2b). We also identified examples in which the genomic extent of siRNA-generating cis-NAT overlap differed between cell types. This was most evident when comparing cis-NAT siRNA patterns between the homogenous S2 and Kc cell populations (for example, for BRWD3/CG5728, CG5919/CG3308 and gry/CG14967 3′ cis-NATs; Supplementary Fig. 2c). Such variable siRNA patterns were probably due to alternative polyadenylation yielding distinct 3′ UTR limits in different cell types. We noticed that S2 cells yielded extended siRNA overlaps much more frequently than did Kc cells, and these uncertainties in particular were the basis of apparent ‘non-overlap’ 21-mer enrichments observed earlier (Fig. 1f,g).
To obtain further insight into the biogenesis of cis-NAT–derived siRNAs, we analyzed published small RNA data from Dcr-2 heterozygous and homozygous fly heads21. As reported, the overall miRNA content of these libraries was similar (Table 1). However, we observed a notable difference in the content of cis-NAT siRNAs generated from overlaps. When normalized for total library reads, we observed a > 12-fold decrease in cis-NAT siRNAs in Dcr-2 homozygous tissue. As a control, we analyzed the number of non–21-mer reads from cis-NAT regions outside of the overlap regions and observed no change in this population. These data indicate that, as suggested by their specific 21-nt size, the production of cis-NAT siRNAs is mediated by Dcr-2.
Table 1.
5′ and 3′ features of cis-NAT siRNAs
| (a) | Read counts normalized per million reads | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 21-mers in cis-NAT overlaps | Non–21-mers outside overlaps | ||||||||
| Library | Reads | miRNAs | % miRNA |
+ strand | − strand | Total | + strand |
− strand | Total |
| Dcr-2/CyO heads | 908,403 | 833,686 | 91.8 | 50.6 | 53.9 | 104.6 | 12.1 | 7.7 | 19.8 |
| Dcr-2/Dcr-2 heads | 734,243 | 671,471 | 91.5 | 4.1 | 4.1 | 8.2 | 5.4 | 15.0 | 20.1 |
| (b) | miRNA/hpRNA/siRNA counts normalized per 100,000 reads | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Library | Total Reads | miRNAs | β↓ | hpRNAs | β↑ | Exo-siRNAs | β↑ | Cis-NAT siRNAs | β↑ |
| Head-β | 42,354 | 16,128 | 1,354 | 20.0× | 20,034 (white) | 13.6× | 92.0 | 7.0× | |
| Head | 105,976 | 35,746 | 2.2× | 68 | 1,473 (white) | 13.2 | |||
| S2-β | 113,709 | 3,920 | 811 | 5.8× | 1,736 (GFP) | 5.4× | 137.1 | 4.9× | |
| S2 | 99,003 | 31,785 | 8.1× | 139 | 320 (GFP) | 2,803 | |||
(a) cis-NAT siRNAs are dependent on Dcr-2. Previously published small RNA data21 was analyzed for miRNAs and cis-NAT RNAs. The miRNA content of Dcr-2 heterozygous and homozygous heads is similar, as is the content of non–siRNA-sized RNAs derived from non-overlap regions of cis-NAT pairs. However, there is a strong depletion of 21-mers derived from the overlap regions of 3′ cis-NAT pairs in Dcr-2 homozygous tissue. (b) Behavior of various small RNA classes following β-elimination. Relevant reads were extracted from adult head and S2 cell data, cloned before and after β-elimination20, and normalized to counts per 100K. miRNAs are strongly depleted following β-elimination (β↓), whereas hpRNAs, white/GFP siRNAs and cis-NAT siRNAs are all strongly enriched (β↑).
Exogenous D. melanogaster siRNAs are processed by Dcr-2 and loaded into AGO2, where they are modified at their 3′ end by the Hen1 methyltransferase22,23. This modification can be inferred by their resistance to β-elimination, which affects small RNAs with free 3′ hydroxyl groups such as AGO1-loaded miRNAs20. On this basis, the enrichment of 21–22-nt RNAs following cloning from β-eliminated RNA has been interpreted as evidence for residence in AGO2 (refs. 20,22,24). We calculated a > two-fold depletion and > eight-fold depletion of mature miRNAs when cloning from β-eliminated head RNA and S2 RNA20, respectively (Table 1). Conversely, there was strong enrichment of both white (13.6-fold) and GFP (14.6-fold) siRNAs (derived from RNAi of white in adult heads and of GFP in S2 cells) following cloning from β-eliminated RNA (Table 1). Thus, miRNAs were depleted following β-elimination, whereas exogenous siRNAs were correspondingly enriched. Similar results were recently reported21.
Analysis of cis-NAT overlap clones also clearly showed their enrichment in the appropriate libraries. There was a five-fold enrichment of cis-NAT overlap clones following β-elimination of S2 RNAs and a seven-fold enrichment of such clones following β-elimination of head RNAs (Table 1). The number of distinct cis-NAT loci that generated cloned RNAs was also strongly increased following β-elimination of head and S2 RNAs. In summary, the specific generation of 21-nt RNAs from mRNA overlaps, their selective loss in Dcr-2 homozygous tissue and their enrichment following β-elimination experiments, lead us to conclude that 3′ cis-NATs produce siRNAs via a Dcr-2 and AGO2 pathway. However, our observations do not exclude the possibility that some cis-NAT–derived small RNAs might be loaded into AGO1.
cis-NAT siRNAs may only subtly regulate host gene activity
We selected the AGO2/CG7739 and tsunagi/Mys45A cis-NAT siRNA loci (Fig. 2b) for functional tests, as they generated the third- and seventh-highest number of overlap siRNAs among all libraries and were similarly ranked when considering only S2 reads. We tested the effect of loqs, Dcr-2 and AGO2 knockdown on the levels of these transcripts in S2 cells using quantitative PCR (qPCR) and normalization to transcript levels following treatment with GFP dsRNA. Certain transcripts appeared mildly upregulated in certain conditions, but we did not observe convincing evidence from consistent derepression under both Dcr-2 and AGO2 knockdown conditions (Supplementary Fig. 3a online).
As dsRNA affords only incomplete suppression of gene activity, we also tested null alleles of animal mutants. We performed qPCR of RNA isolated from the heads of Dcr-2 and AGO2 heterozygous and homozygous adult flies. We again did not observe significant changes in cis-NAT transcript levels (Supplementary Fig. 3b,c). It is possible that these animal tests were obscured because a proportion of tissue may not have coexpressed these cis-NATs on a cell-by-cell basis. However, as these tests were directed at some of the more highly expressed cis-NAT siRNA loci, a conservative conclusion is that cis-NAT siRNAs may have only subtle effects on the expression of their parental loci.
Entry of cis-NAT pairs into the siRNA pathway is restricted
Despite the lack of regulatory evidence for qPCR tests, we sought evidence for the active restriction of cis-NATs into the siRNA pathway. It was reported that certain classes of D. melanogaster genes actively avoid the acquisition of miRNA binding sites25. Even though the endogenous function of individual miRNA binding sites often cannot be assessed in laboratory tests, their exclusion from certain types of genes can be inferred to reflect their regulatory capacity. Inspired by this line of reasoning, we sought to determine whether any co-expressed cis-NATs are denied entry into the RNAi pathway. By analogy with the phenomenon of miRNA target-site avoidance, the existence of coexpressed cis-NATs that do not generate siRNAs would provide evidence for the functional impact of cis-NAT siRNAs.
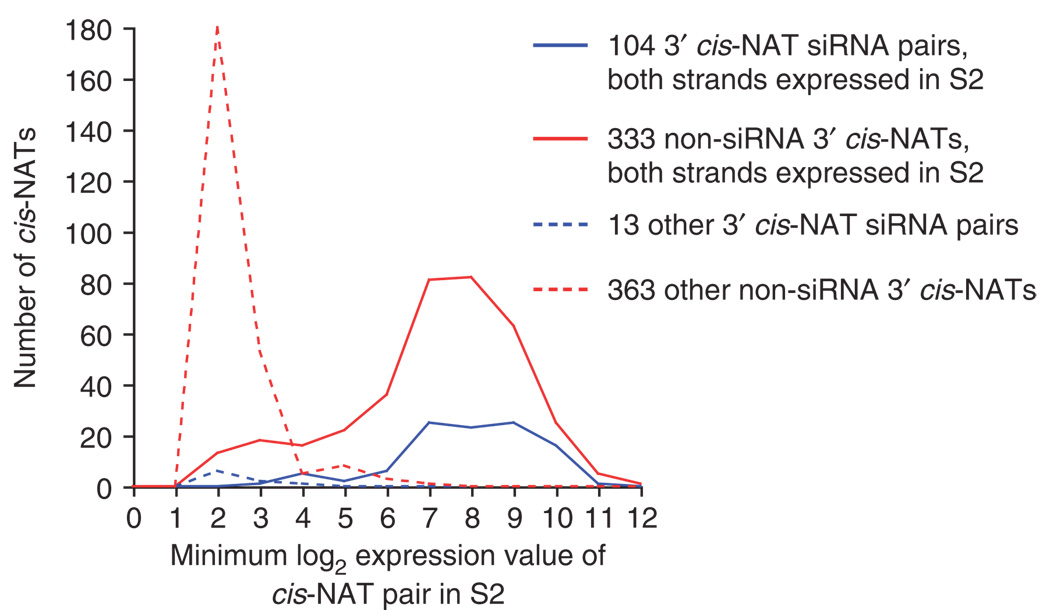
We tested this by analyzing S2 cell gene expression profiles using Affymetrix microarrays. Although the assessment of coexpression on a cell-by-cell basis is not trivial, it can be approximated by examining gene expression in a homogenous cell population. Of 13,632 annotated genes interrogated by the Affymetrix platform, 7,274 (53%) were reproducibly called present in S2 cells. Of the 117 annotated cis-NAT siRNA pairs, we observed 104 cases in which both top- and bottom-strand genes of a cis-NAT pair were called present in S2 cells. This enrichment is consistent with the idea that the genes involved in cis-NAT siRNA arrangements are more broadly expressed than average genes. However, of the 676 non-siRNA cis-NATs, 333 top- and bottom-strand gene pairs were also called present at a minimum level that captured almost all of the genuine cis-NAT siRNA loci (Fig. 3). In other words, there are three times as many coexpressed cis-NATs in S2 cells than there are cis-NATs that generate siRNAs. Indeed, many non-siRNA cis-NATs consisted of gene pairs that were highly expressed. These data provide compelling evidence that only a minority (∼25%) of coexpressed cis-NAT pairs are competent to enter the siRNA pathway.
Figure 3.
Most coexpressed 3′ cis-NAT pairs do not generate siRNAs. Gene expression in duplicate isolates of S2 cell RNA was assessed using Affymetrix arrays. For each cis-NAT pair, we recorded the lower gene expression value; thus, the transcripts in a given pair have expression that is equal to or higher than the value plotted on the x-axis. The vast majority of cis-NAT siRNA loci include pairs in which both genes are called ‘present’ in S2 cells. However, many more non-siRNA cis-NATs involve gene pairs for which both members are called ‘present’ in S2 cells.
cis-NAT siRNA loci enriched for nucleic acid–related genes
Because of the apparently active selection of cis-NATs by the RNAi pathway, we wondered whether there was an underlying molecular logic to the genes regulated by this network. To test this, we used GOToolbox26 to ask whether there was a functional signature of Gene Ontology (GO) terms among cis-NAT siRNA loci. We were particularly interested to identify GO terms that distinguished the 117 cis-NAT siRNA loci from the 676 non-siRNA cis-NATs, which would provide evidence for the active selection of specific molecular functions by the siRNA pathway.
We found that terms such as ‘nucleus’ and ‘transferase’ were highly enriched among both cis-NAT siRNA genes and non-siRNA cis-NAT genes, relative to all other D. melanogaster genes (Table 2), consistent with the idea that some gene functions are preferentially encoded by cis-NAT gene arrangements1. However, the other most highly significant enrichments concerned intriguing sets of genes with nucleic acid–based functions, including nuclease activity (both DNase and RNase), transcription factor complexes and chromosome pericentric regions (Table 2 and Supplementary Table 2 online); lipid metabolism genes were also overrepresented. Therefore, genes with nucleic acid–based functions are preferentially incorporated into cis-NAT siRNA systems. Moreover, this selection is not a simple consequence of the types of gene functions that are generally arranged as cis-NATs in the genome. Taken together, these observations implicate the endogenous regulatory impact of cis-NAT siRNAs in D. melanogaster.
Table 2.
Top Gene Ontology (GO) term enrichments among cis-NAT-siRNA loci
| GO ID | Term | Number in genome |
Number in 676 non- siRNA 3′ cis-NATs |
P-value enriched in non-siRNA cis-NATs |
Number in 117 siRNA 3′ cis-NATs |
P-value enriched in siRNA cis-NATs |
3′ cis-NAT siRNA genes |
|---|---|---|---|---|---|---|---|
| GO:0004518 | Nuclease activity |
68 | 9 | 0.298 | 10 | 2.15E-08 |
CG8038, CG10214, CG4853, CG8862, CG7741, AGO2, mus205, CG3308, Ercc1, Rrp45, CG9666, CPTI, IP3K1, α4GT1 |
| GO:0016740 | Transferase activity |
855 | 99 | 2.27E-07 | 27 | 1.63E-05 |
CG11337, Gcn2, βggt-I, lok, fbl, Rpb7, CG5508, CG40351, Mes-4, mus205, CG5537, CG18854, CG5038, Csk, Pitslre, l(1)G0020, Pi4KIIα, skf, Fpps, CdsA, Magi, r-l, Fps85D, mad2, CG5728, aop, aft, CG10214, CG17328, TfIIFβ, Ets97D, tsu, Mys45A, CG31365, wuho, lok, Rpb7, bigmax, gro |
| GO:0005634 | Nucleus | 1424 | 147 | 5.36E-08 | 37 | 2.65E-05 |
Sin3A, MP1, cav, dmt, CG40351, Mes-4, Patj, CG16903, Su(var)3–7, CG11419, Jra, MED21, mago, hyd, Pitslre, ref(2)P, l(1)G0020, Rrp45, CG11508, CdsA, Pc |
| GO:0006629 | Lipid metabolic process |
125 | 9 | 0.135 | 9 | 4.77E-05 |
α4GT1, CG5508, Hmgcr, Pi4KIIα, Lsd-1, Fpps, CG16935, CdsA, CG8199 |
| GO:0004540 | RNase activity | 31 | 5 | 0.458 | 5 | 5.26E-05 | CG8038, CG10214, CG7741, AGO2, Rrp45 |
| GO:0004536 | DNase activity | 32 | 4 | 0.117 | 5 | 6.15E-05 | CG10214, CG4853, CG8862, CG3308, Ercc1 |
| GO:0008134 | Transcription factor binding |
82 | 9 | 0.063 | 7 | 1.06E-04 | gro, Sin3A, Mes-4, CG4756, Jra, MED21, cact |
| GO:0044255 | Cellular lipid metabolic process |
113 | 9 | 0.121 | 8 | 1.41E-04 |
α4GT1, CG5508, Hmgcr, Pi4KIIα, Fpps, CG16935, CdsA, CG8199 |
| GO:0000775 | Chromosome, pericentric region |
27 | 3 | 0.177 | 4 | 4.64E-04 | mad2, cav, Su(var)3–7, fzy |
The 117 cis-NAT siRNA loci (234 genes) and 676 non-siRNA cis-NAT loci (1,352 genes) were compared with the background data set of all D. melanogaster genes using GOToolbox26. The table lists the most highly enriched GO terms in the cis-NAT siRNA gene set, except for general GO terms comprising > 10% of protein-encoding genes; that is, terms such as intracellular part, intracellular, cell part, cell, catalytic activity, intracellular organelle and organelle. Some terms are highly enriched in both classes of cis-NATs, but many nucleic acid and lipid metabolic gene functions are enriched only among the cis-NAT siRNA loci (bold). The entire GO analysis is available in Supplementary Table 2.
An unexpected class of bidirectionally transcribed exonic-intronic siRNAs
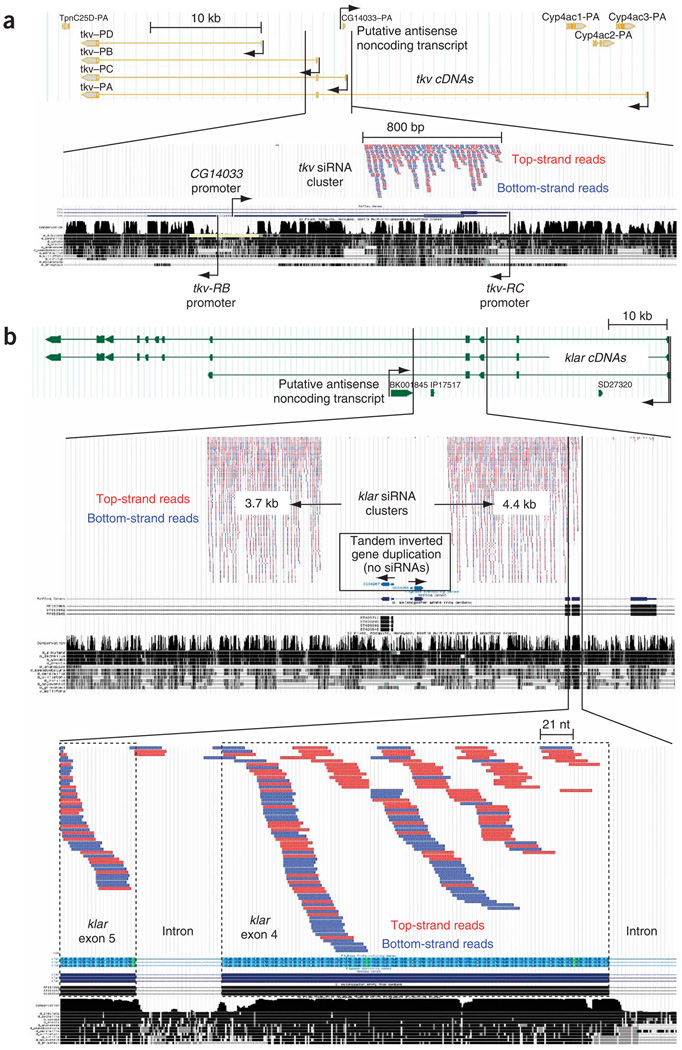
In our computational screen, we recovered an abundantly expressed cis-NAT siRNA locus in the thickveins (tkv) region (Fig. 4a). This locus was cloned specifically from male bodies but not from male heads or female bodies, suggesting that its expression in the adult is restricted to the male sexual apparatus or germ line. It was also abundant in a library constructed from mass-isolated imaginal discs and brains. This locus showed several unusual features. First, whereas the vast majority of cis-NAT siRNA loci overlap on their 3′ ends, the tkv/CG14033 cis-NAT pair overlap on their 5′ exons, a configuration that is otherwise highly unfavorable for entry into an siRNA pathway (Fig. 1f). Second, this locus produced bidirectional 21-nt RNAs (Fig. 1i) not only from annotated exons, but also from the annotated tkv-RC intron (Fig. 4a). The structure of this intron-exon junction was verified by the existence of multiple cDNA clones, none of which was unspliced27. The extent of the tkv/CG14033 cluster was also not coincident with the CG14033 transcript, and thus seemed unlikely to be an mRNA phenomenon as deduced for cis-NAT siRNA loci. Unexpectedly, CG14033 was a candidate for a noncoding locus, because its longest open reading frame was only 74 amino acids (aa) long and was poorly conserved in other Drosophilids27.
Figure 4.
The thickveins (tkv) and klarsicht (klar) loci define a distinct class of exceptionally abundant siRNAs derived from bidirectional transcription. (a) The tkv locus covers > 50 kb and comprises several alternatively spliced transcripts. The CG14033 transcript is generated from the strand opposite to tkv, and its promoter resides between tkv-RB and tkv-RC. A dense cluster of siRNAs maps to the second half of CG14033 and overlaps the short tkv-RC coding exon on the other strand. (b) The klar locus extends over 100 kb and comprises several alternatively spliced transcripts. Two massive clusters of siRNAs derive from both strands of an intron; the 4.4-kb cluster overlaps two short klar coding exons. Note klar primary transcription across the CG32467/CG32468 inverted gene pair, which theoretically adopts an extended hairpin structure (Supplementary Fig. 4), does not generate siRNAs. At the bottom, a magnified view of klar exons that overlap the 4.4-kb cluster shows that siRNA production in this region is clearly related to klar exons.
A search for other intronic-exonic, bidirectionally transcribed, siRNA loci revealed two massive clusters of small RNAs that map to the klarsicht (klar) locus (Fig. 4b). We recovered ∼180,000 unphased 21-nt RNAs from our libraries (Fig. 1j), and these tiled densely across both strands of neighboring 3.7-kb and 4.4-kb regions (Fig. 4b). The sheer length of these siRNA clusters rendered them unlikely to be produced by the same system that generates cis-NAT siRNAs (Fig. 2b), which are almost exclusively < 500 bp. Similarly to the tkv locus, the klar siRNA clusters were mostly intronic, but overlapped short klar coding exons at the 5′ end of the cluster (Fig. 4b). The klar small RNAs were cloned most abundantly from S2 cells and Kc cells; however, both clusters were also expressed in 0–1-hour-old and 2–6-hour-old embryos. Thus, the klar siRNA system is endogenously active in the animal. Unlike tkv, there was not an annotated antisense transcript that included the siRNA clusters. However, an uncharacterized, possibly noncoding, EST mapped (BK001845) close to the 5′ end of the cluster on the klar antisense strand (Fig. 4b).
Notably, the genomic region between the two klar siRNA clusters contains two transcription units, CG34267 and CG34268, that comprise an inverted gene duplication (Fig. 4b). Although the predicted proteins are short (77 aa and 83 aa), they are associated with spliced cDNA evidence and encode highly related polypeptides. In theory, the primary klar transcript that traverses the siRNA clusters would produce a long inverted repeat across CG34267/CG34268 (Supplementary Fig. 4 online) that resembles artificial RNAi-inducing transcripts as well as endogenous hpRNAs14. Nevertheless, no siRNAs were produced by this dsRNA region. This makes the strategy by which klar siRNA clusters are selected for processing even more mysterious.
Overall, the tkv and klar siRNA loci show many coherent features. First, these loci generate large numbers of 21-nt small RNAs that tile with nearly single-nucleotide resolution across long genomic intervals up to several kilobases in length. Second, the 5′ ends of the siRNA clusters overlap short coding exons of protein-encoding genes, but are contiguous with substantial lengths of intronic sequence. Third, the transcript pairs consist of a protein-encoding mRNA and a possibly noncoding RNA. Altogether, these characteristics define a second class of siRNAs derived from bidirectional transcription.
klar siRNAs are generated by a Dcr-2/Loqs/AGO2 pathway
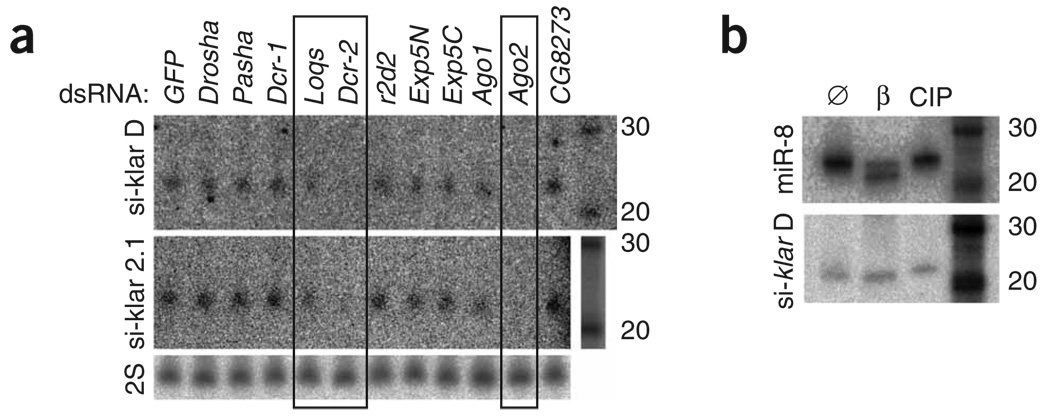
Although most klar siRNAs were individually rare, two out of six probes tested detected endogenous 21-nt RNAs in S2 cells. This allowed us to analyze their biogenesis using knockdowns of candidate RNAi and miRNA factors. To do so, we collected RNA from cells treated with dsRNA against GFP, Drosha, Pasha, Dcr-1, Loqs, Dcr-2, r2d2, Exportin-5, AGO1, AGO2 and CG8273 (encoding a double-stranded RNA binding domain (dsRBD) protein of unknown function) and analyzed them by northern blot. Consistent with their classification as siRNAs, we found that their accumulation was most dependent on Dcr-2 and AGO2 (Fig. 5a). In addition, we observed that klar siRNAs are blocked at their 3′ termini (Fig. 5b), providing further evidence for their loading into AGO2. Notably, their accumulation showed a substantial requirement for Loqs, which was previously known as a miRNA-specific Dcr-1 cofactor28–30. Equally unexpected was the finding that r2d2, a Dcr-2 cofactor that loads siRNAs into AGO2 (refs. 31,32), was not apparently required for klar siRNA accumulation. qPCR tests verified comparable knockdown of loqs and r2d2 in these experiments (Supplementary Fig. 5 online). These biogenesis data provide direct evidence for the production of klar siRNAs by a canonical Dcr-2 and AGO2 pathway, but introduce unexpected complexity in their requirement for dsRBD partner proteins.
Figure 5.
Biogenesis and structural features of klar siRNAs. (a) We carried out northern analysis on RNA isolated from S2 cells treated with the indicated dsRNAs. Quantification with the hybridized 2S signal across three northern experiments showed 27% klar siRNAs in AGO2 knockdown cells, 20% in Dcr-2 knockdown cells and 58% in loqs knockdown cells. (b) 5′ end and 3′ end analysis. S2 cell RNA was β-eliminated to probe 3′ ends and treated with phosphatase to probe 5′ ends. Compared to untreated RNA (Ø), most miR-8 shifts to a faster-migrating species following β-elimination (β), but klar siRNAs are resistant, indicating 3′ blockage. The slight reduction in mobility of miR-8 and klar siRNAs following phosphatase (CIP) treatment indicates 5′ phosphates.
DISCUSSION
Endogenous siRNAs from 3′ cis-NAT loci in Drosophila
There has been substantial controversy over whether complementary transcripts produced by bidirectional transcriptional can anneal in vivo to yield dsRNA that is recognized by the RNAi machinery. In plants, there is strong evidence that select cis-NAT loci engage an siRNA-mediated autoregulatory loop, especially under stress conditions5,6. However, evidence has also been presented against the broad engagement of RNAi pathways by plant cis-NATs, in favor of perhaps influencing alternative splicing or polyadenylation9. Similarly, experimental studies in mammalian cells have argued against the involvement of RNAi by cis-NATs7 in favor of indirect modes of cis-NAT regulation8.
The frequent failure of forced antisense transcripts to engage the RNAi pathway makes it clear that complementary mRNAs probably do not efficiently associate as dsRNA in vivo. However, it has been difficult to assess whether any dsRNA at all is generated from cis-NATs. A major problem for their study lies in the challenge of determining confidently that a given genomic locus is transcribed on both strands in the same cell. Even if one can detect coexpression of top and bottom strands of a cis-NAT in RNA extracted from a seemingly homogenous cell line, it is hard to rule out that expression of the two strands might not be inversely correlated on a cell-by-cell basis.
The study of large-scale small RNA sequence data allowed us to bypass the difficulty of detecting cis-NAT dsRNA by visualizing downstream siRNA production directly. On the basis of genomic patterns of small RNA enrichment, 21-nt specificity, dependence on Dcr-2 and 3′ end modifications, we were able to confidently infer that many 3′ cis-NATs transit a bona fide siRNA pathway. Although the RNAi pathway accepts at least 100 3′ cis-NAT pairs, gene expression analysis of S2 cells indicated that 75% of coexpressed cis-NAT pairs are rejected from the siRNA pathway. In addition, the highly significant enrichment for various nucleic acid–based (DNA and RNA) functions among cis-NAT siRNA genes indicated that this pathway is not a fortuitous consequence of frequent cis-NAT arrangements in the D. melanogaster genome. Rather, we infer that cis-NAT siRNA loci are actively selected from among bulk cis-NAT gene pairs. Notably, a concurrent study identified 17 cis-NAT siRNA loci expressed in mouse oocytes33, suggesting that this regulatory strategy is conserved in animals.
Intriguing cis-NAT siRNA loci with RNA-based functions
Our report of genuine endogenous siRNA loci now provides direction to efforts to understand the role of endogenous RNAi in D. melanogaster. We describe here some particularly compelling cis-NAT siRNA loci that are highly expressed and have RNA-based regulatory functions.
First, the AGO2/CG7739 cis-NAT (AGO2 being the very protein that binds siRNAs and effects siRNA-mediated gene regulation) generated the third-highest number of cis-NAT siRNAs. Although our experimental tests were mostly inconclusive, this potential for self-regulation is reminiscent of autoregulation of ARGONAUTE-1 and DICER-LIKE-1 by miRNAs34,35. Perhaps this regulatory arrangement has a specialized use in the fly.
Second, with regard to the CG18854/IP3K cis-NAT, we recently showed that the spliced mRNA of the CG18854 locus is processed into functional siRNAs by a Dcr-2– and AGO2-dependent mechanism termed the hpRNA pathway14. We noticed that the 3′ end of CG18854 overlaps extensively with the 3′ exon of inositol 1,4,5-triphosphate kinase 1 on the other strand, and generates the fifth-highest number of overlap siRNAs of all cis-NATs. Therefore, the CG18854 locus participates prominently in both cis-NAT siRNA and hpRNA pathways.
Third, the Tsunagi/Mys54A and mago/Magi cis-NATs generated the seventh- and twelfth-highest number of overlap siRNAs amongst all cis-NATs. Notably, tsunagi and mago encode physically interacting components of the exon junction complex36. It is commonly recognized that proteins that function as components of a common machinery are co-transcriptionally regulated. These cis-NATs provide a curious example for the common post-transcriptional regulation of functionally related proteins.
In general, the fact that cis-NAT siRNA loci are strongly enriched for nucleic acid–based functions suggests this as a major molecular axis for siRNA-mediated gene regulation in D. melanogaster. In this regard, it is perhaps relevant that Dcr-2 mutants were reported to have abnormal nucleolar morphology37, and AGO2 mutants were found to have defects in chromosome condensation, centromeric function and nuclear division38. The most highly significantly enriched GO-terms for confident cis-NAT siRNA include genes that function in these pathways (Table 2). Even though Dcr-2 and AGO2 null mutants are reasonably healthy and do not wildly deregulate cis-NAT siRNA transcripts, directed genetic interaction studies may prove useful in gaining further insight into the regulation of RNA and DNA pathways by cis-NAT siRNAs.
Distinct mechanisms for the production of siRNAs via bidirectional transcription
We showed that in addition to cis-NAT siRNA loci, there are at least two exceptional regions of the genome that generate siRNAs via bidirectional transcription and a Dcr-2– and AGO2-dependent pathway. These loci, located within the klar and tkv genes, probably define an independent mechanism of siRNA generation. Several salient features support this idea: (i) they generate 1–2 orders of magnitude more siRNA clones than do the most highly expressed cis-NAT siRNA loci; (ii) they are not 3′ cis-NATs; (iii) they originate from much larger genomic intervals than do cis-NAT siRNA overlaps; (iv) their antisense partners are potentially noncoding RNAs; (v) their predominant genomic annotations are introns, not exons; (vi) both siRNA clusters overlap coding exons at their 5′ ends. A functional understanding of their impact on klar and tkv activity awaits genetic knockouts of these siRNA clusters.
In summary, our studies reveal distinct mechanisms for the production of bona fide endogenous siRNAs via bidirectional transcription in D. melanogaster. Together with concurrent studies of endogenous siRNAs in D. melanogaster21 and mice33,39, this work opens windows onto studies of host-directed functions of RNAi and novel mechanisms for small regulatory RNA biogenesis.
METHODS
Small RNA cloning and sequencing
We used a published protocol40 to generate libraries from ∼18–26-nt RNAs isolated from (i) S2 cells, (ii) Kc cells and (iii) a mixture of Kc cells and S2 cells before and after 48-h treatment with 5 × 10−6 M 20-hydroxyecdysone (Sigma). These were analyzed using the Solexa/Illumina platform, respectively yielding 2,265,651, 2,151,714 and 699,873 reads for which 3′ linker sequences could be confidently identified and clipped. The entire data sets of clipped reads were deposited in NCBI-GEO under the platform GPL6573 as GSM272652 (S2), GSM272653 (KC) and GSM272651 (S2+KC).
Functional analysis of cis-NAT siRNA and klar siRNAs
We used previously described templates28,41 to generate dsRNAs for S2 cell knockdowns. We soaked 2 × 106 S2 R+ cells in six-well plates with 20 µg ml−1 dsRNA41. Quantitative reverse-transcription PCR (qPCR) was performed on AGO2, CG9937, tsunagi and Mys45A using SYBR green. These raw data were normalized to rp49 values, and then each of the knockdown samples was normalized to the value from GFP knockdown. We performed a total of eight qPCR reactions on two different knockdown samples for each condition.
We tested the following locked nucleic acid (LNA) probes for their ability to detect klar siRNAs in S2 cell RNA. Of these, klar2.1 and klarD hybridized to endogenous 21-nt RNAs on northern blots and were subsequently used to analyze klar siRNA biogenesis across a panel of RNA samples treated with various dsRNAs against miRNA and RNAi factors: klar_2.1, 5′-TGGACACCCATTCGATAGAATCGGA-3′; klar_2.2, 5′-CAACCGAGGTGTAAGCACTCATGTT-3′; klarA_probe, 5′-AGAGACAGGCCCAACAAAAAGACGA-3′; klarB_probe, 5′-GCGACCTTTAATCAACACCTCAA-3′; klarC_probe, 5′-CTCATCATTAAGGCAAATCCGAAGA-3′; klarD_probe, 5′-AGAGGCACAGGAAGAAACGCTCGAA-3′.
cis-NAT bioinformatics
FlyBase release 5.5 annotations (January 2008) were used to identify cis-NATs in the D. melanogaster genome27. A set of overlaps between exons in the plus strand and those in the minus strand were first identified, and alternatively spliced transcript annotations were collapsed to single genes to avoid duplicate analyses. For each gene pair, those transcripts that contained overlapping exons were then identified. The length of the overlap region (lin) was the sum of the overlapping exon regions from both strands. The sum of the lengths of distinct exon regions that were outside the overlap region of the transcripts in the cis-NAT pair was the length outside the overlap (lout). Small RNAs of length greater than or equal to 18 nt that appear within and outside the overlap region of a cis-NAT were identified from the 454 and Solexa libraries. The normalized number of reads of the small RNAs were recorded as output; that is, if the clone count of a small RNA is c and its number of blast hits is b, then, the normalized clone count is (c/b).
Our cis-NAT read analysis made it evident that many annotations are either incorrect with truncated 3′ UTRs or are alternatively polyadenylated in an individual library resulting in cell-specific cis-NAT overlap regions. We manually corrected a small number of loci that gave rise to substantial numbers of siRNAs in corrected overlaps. The FlyBase annotations and revised overlaps are as follows: (i) CG12016/CG11526, chr3L:3322274–3322491 to chr3L: 3322000–3322491; (ii) CG31898/fy, chr2L:8401446–8401477 to chr2L:8,401, 438–8,401,647; (iii) dmt/hyd, chr3R:5540344–5540397 to chr3R:5,539,735–5,540,500; (iv) BRWD3/CG5728, chr3R:20154175–20154348 to chr3R: 20150000–20158919 only in S2 cells; (v) CG5919/CG3308, chr3R:17096644–17097129 to chr3R:17,096,119–17,098,531 only in S2 cells; (vi) gry/CG14967, chr3L:3211110–3211347 to chr3L:3209271–3218451 only in S2 cells; and (vii) CG8594/Sin3A, chr2R:8462743–8462813 to chr2R:8460808–8464448 only in S2 cells. We did not attempt to correct the many cis-NATs whose reannotation individually affected few reads; however, summed together, these were sufficient to account for the couple of hundred 21-nt non-overlap reads observed above background (Fig. 1f,g).
Another relevant cis-NAT revision concerned CG18854, a FlyBase-annotated protein-encoding gene that we have recently shown to be a hairpin RNA (hpRNA) transcript that generates endogenous siRNAs. CG18854 produces relatively abundant 3′ cis-NAT siRNAs with IP3K1. However, because the CG18854 non-overlap region generates thousands of 21-nt reads via the hpRNA pathway, it was necessary to exclude hpRNA-derived CG18854 siRNAs from the analysis.
Small RNA enrichments within and outside the cis-NAT overlap region were assessed on the basis of uniquely mapped small RNA, as certain mRNA degradation fragments map to many (sometimes hundreds) of locations. Small RNA enrichment (Esr) and the 21-mer enrichment (E21) of a cis-NAT pair were calculated as follows. Let nsr,in, nsr,out, n21,in and n21,out be the sum of clone count of unique small RNAs and 21-mers in and outside the overlap, respectively. Then, Esr is defined as the ratio of the number of small RNAs in overlap to the length of overlap over those outside overlap to the length outside the overlap; that is, Esr = (nsr,in/lin)/(nsr,out/lout). Similarly, E21 is defined as the ratio of the number of 21-mers in overlap to the length of overlap over those outside overlap to the length outside the overlap; that is, E21 = (n21,in/lin)/ (n21,out/lout). Note that enrichment values are inherently variable as a function of overlap and non-overlap lengths, which necessarily differ for each individual cis-NAT. Therefore, the enrichment value of any particular cis-NAT is slightly less meaningful than the overall distribution of enrichment values across cis-NAT sets.
To extract cis-NATs enriched with 21-mers in the overlap, we adopted a threshold for the ratio (n21,in/ nsr,in) > 0.67. Of the total 793 3′ cis-NATs found, 275 that satisfied the above threshold were then binned with respect to the number of 21-mers in the overlap. We empirically set cut-off values of > 10 overlap 21-mers, for which > 67% of all overlap reads were 21-nt in length. This resulted in 117 confident 3′ cis-NAT siRNA loci.
To assess enrichments of GO terms in cis-NAT siRNA genes, we used GOToolBox (http://burgundy.cmmt.ubc.ca/GOToolBox/)26. We used the hypergeometric test option of GO-Stat with the gene set and the reference set specified as follows: (i) genes from the 117 confident 3′ cis-NAT siRNA loci compared to all D. melanogaster genes and (ii) genes from the 676 non-siRNA cis-NATs compared to all D. melanogaster genes.
Affymetrix Drosophila Genome Arrays were interrogated using independent preparations of total RNA from S2 cells treated with GFP dsRNA28. The expression data were processed using the R software environment for statistical computing and graphics (http://www.r-project.org/) using GC-RMA normalization.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Hodges and G. Hannon for Solexa sequencing, A. Viale and the Sloan-Kettering Genomics Core for hybridizing Affymetrix microarrays, and N. Socci for assistance with microarray analysis. K.O. was supported by a grant from the Charles Revson Foundation. E.C.L. was supported by grants from the Burroughs Wellcome Foundation, the V Foundation for Cancer Research, the Sidney Kimmel Cancer Foundation and the US National Institutes of Health (R01-GM083300 and U01-HG004261).
Footnotes
Accession codes. NCBI-GEO. The microarray data were deposited under platform GPL1322, accession number GSM286644. The entire data sets of clipped reads were deposited in NCBI-GEO under the platform GPL6573 as GSM272652 (S2), GSM272653 (KC) and GSM272651 (S2+KC).
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 3.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 4.Lipman DJ. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 1997;25:3580–3583. doi: 10.1093/nar/25.18.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol. Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faghihi MA, Wahlestedt C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome Biol. 2006;7:R38. doi: 10.1186/gb-2006-7-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jen CH, Michalopoulos I, Westhead DR, Meyer P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 2005;6:R51. doi: 10.1186/gb-2005-6-6-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 11.Bunch TA, Goldstein LS. The conditional inhibition of gene expression in cultured Drosophila cells by antisense RNA. Nucleic Acids Res. 1989;17:9761–9782. doi: 10.1093/nar/17.23.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyler DM, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008 May 7; doi: 10.1038/nature07015. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. doi: 10.1016/j.cub.2008.05.006. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 19.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 20.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr. Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008 April 10; doi: 10.1126/science.1157396. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Martin D, et al. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang F, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in. Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, et al. r2d2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 32.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008 April 10; doi: 10.1038/nature06908. advance online publication. [DOI] [PubMed] [Google Scholar]
- 34.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deshpande G, Calhoun G, Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008 April 10; doi: 10.1038/nature06904. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau NC, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 41.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.