Abstract
Purpose
The present study used a new 16S rRNA-based microarray with probes for over 300 bacterial species better define the bacterial profiles of healthy root surfaces and root caries (RC) in the elderly.
Materials
Supragingival plaque was collected from 20 healthy subjects (Controls) and from healthy and carious roots and carious dentin from 21 RC subjects (Patients).
Results
Collectively, 179 bacterial species and species groups were detected. A higher bacterial diversity was observed in the Controls as compared to Patients. Lactobacillus casei/paracasei/rhamnosus and Pseudoramibacter alactolyticus were notably associated with most root caries samples. Streptococcus mutans was detected more frequently in the infected dentin than in the other samples, but the difference was not significant. Actinomyces were found more frequently in Controls.
Conclusion
Actinomyces and S. mutans may play a limited role as pathogens of RC. The results from this study were in agreement with those of our previous study based on 16S rRNA gene sequencing with 72% of the species being detected with both methods.
Keywords: Root caries, bacteria, microbiology, aged 80 and over, microarray analysis, 16S rRNA
Introduction
Root caries (RC) has become a significant oral health issue. Several studies agree that the RC prevalence is much higher in the dentate elderly than in other adult groups (6, 8, 17). Two factors are leading to an increasing number of exposed root surfaces in the elderly: the increased duration of life (14)] and the higher number of teeth being maintained in this population (5)]. Since RC is clearly a bacterial infection, it is therefore important to identify those bacterial species that are associated with this disease.
Based on culture-dependent studies, Streptococcus mutans, lactobacilli and some species of Actinomyces have been considered important in the etiology of RC (16, 21). In contrast, van Houte et al. (19) described the bacterial flora of RC as more diverse, and found that the above mentioned species played less of a role. More recently, culture-independent studies have focused either on single species such as S. mutans (11) and Actinomyces naeslundii (2), or on the analysis of microbial-derived organic acids (18). Several other studies have investigated advanced dentinal lesions from coronal caries with molecular methods (1, 3, 4, 9, 10), but they were not designed to study RC. Overall, the present understanding of the microflora of RC is limited compared to other infectious oral diseases (19).
Recently, we characterized the bacterial diversity of healthy root surfaces and root surface caries in the elderly based on sequencing 16S rRNA genes amplified from isolated DNA (15). We demonstrated that other species in addition to S. mutans, lactobacilli and Actinomyces may be also involved in the RC process (15).
Since only a limited number of subjects were analyzed, our findings only began to assess the bacterial associations with RC. The Human Oral Microbe Identification Microarray (HOMIM) allows for the simultaneous detection of about 300 of the most prevalent oral bacterial species, including those that cannot yet be grown in vitro (7). Overall, there are about 600 oral bacterial species, of which about 50% have not yet been cultivated (12; www.homd.org). The arrays are designed specifically for the characterization of bacterial profiles in oral clinical samples. More than 550 bacterial species were first identified by sequence analysis of approximately 20,000 16S rRNA clones derived from human oral samples (12, 13). In the present study, we used a new 16S rRNA-based microarray method to test the hypothesis that bacteria other than S. mutans, lactobacilli and Actinomyces are detected from root caries (RC) lesions in the elderly. Since HOMIM allowed the analysis of a more statistically significant number of subjects, we also tested for the significance of the bacterial association with healthy and carious roots. Moreover, as approximately half the samples had been analysed previously by 16S rRNA sequence analysis (15), we also compared the two methods for bacterial detection.
Materials and Methods
Subjects
Forty-one elderly subjects aged 70–101 years (36 females and 5 males) were analyzed. Twenty-one of these subjects were used in our previous study (15). Subjects were selected as previously described. The RC definition and diagnosis were according to the criteria of the World Health Organization (20). The control subjects (referred to as Controls) (n=20) had no RC and the RC group (referred to as Patients) (n=21) had at least one RC lesion at the time of examination. Mean age of the Controls was 84.3 years (range 72–92), mean number of teeth 23 (range 16–28), and number of males 3. Mean age in the Patients was 88.7 years (range 70–101), mean number of teeth 15 (range 5–26), and number of males 2.
Samples
The samples (n=83) were collected and processed as previously described (15). Briefly, plaque samples were obtained from the sound roots of Controls. From the Patients, the following samples were analyzed: plaque from one healthy root (plaque healthy), plaque from one carious lesion (plaque carious), and the underlying carious dentin from the same carious lesion (dentinal). All the samples were collected by one examiner (D. P.).
DNA extraction
Bacterial DNA was extracted using the QIAamp® DNA Mini Kit (Qiagen, GmbH, Hilden, Germany) according to the instructions of the manufacturer. The extracts were stored at −20°C until use.
Amplification of 16S rRNA genes
The 16S rRNA genes were amplified as previously described (12). Briefly, two separate PCR reactions were set up with, respectively, forward primer 5´-CCA GAG TTT GAT YMT GGC-3´ with reverse primer 5´-GAA GGA GGT GWT CCA RCC GCA -3´, and forward primer 5´-GAC TAG AGT TTG ATY MTG GC-3´ with reverse primer 5´-GYT ACC TTG TTA CGA CTT-3´. Two µl of genomic DNA template were added to the reaction mixture (final volume 25 µl) containing 20 pmol of each primer, 40 nmol of deoxytriphosphates, 1.5 mM Mg2+ and 1 U of Platinum High Fidelity Taq polymerase (Invitrogen, San Diego, CA). The samples were preheated at 94°C for 2 min, followed by 32 cycles of amplification under the following conditions: 94°C for 30 s, 55°C for 30 s, and 68°C for 1.5 min, with an additional 1 s for each cycle. A final 10 min elongation step at 68°C was added. The results of the PCR amplification were examined by electrophoresis in a 1% agarose gel. The two parallel PCR reactions were combined and the products purified with QIAquick PCR Purification Kit (Qiagen).
Labeling of PCR products
The labeled nucleotide Cy3-dCTP (GE Healthcare Biosciences, Pittsburgh, PA) was incorporated during a second, nested PCR reaction using the forward primer 5´-GAG TTT GAT YMT GGC TCA G-3´ and the reverse primer 5´-GYT ACC TTG TTA CGA CTT-3´. Seven µl of DNA template were added to a reaction mixture (final volume 25 µl) and amplified using the same cycling program (but with 40 cycles) as described above. Cy3-labeled amplicons were purified using a supplementary protocol with the QIAquick PCR Purification Kit (Qiagen). In this protocol, an extra wash was performed using a 35% guanidine hydrochloride aqueous solution, which aids in the removal of excess Cy-dye in the reaction.
Steps prior to hybridization
Printing of arrays
16S rRNA-based oligonucleotide reverse capture probes were custom synthesized with a 5´-(C6)-amine modified base, eight spacer thymidines and 18 to 20 nucleotides of target sequence, and printed (Michigan State University Research Technology Support Facility) on 25 × 76 mm aldehyde-coated glass slides. Sixty µM oligos were plated in v-bottom 384 well plates in a 2:1 solution with 2X spotting buffer giving a final spot concentration of 30 µM. By printing an OmniGrid Arrayer (GeneMachines) was used at 55% humidity. Post-processing of printed slides included immobilization by baking. Probes targeting more than two closely related species appeared as clusters (37 altogether). About 30% of the targeted species were identified using two probe sets. The array was divided in five sections allowing the processing of five samples simultaneously. Each section had two identical probe sets. Array locations were labeled by etching with a diamond pencil, and slides were stored in a slide box under dry conditions.
Blocking
Immediately before use, the slides were blocked to reduce non-reactive primary alcohols, to remove unreacted aldehyde groups and to minimize fluorescent background after hybridization. This was carried out using the Little Dipper Microarray Processor (SciGene, Sunnyvale, CA). First, the slides were extensively washed to remove un-bound DNA molecules and buffer substances by a series of washing steps using 10% SDS and ddH20. Then they were placed in a solution of sodium borohydride, 1 × PBS and 99% EtOH for 15 minutes at room temperature. Once complete, another set of washing step was performed, and slides were spun dry for 5 minutes and stored once again in a dessicator cabinet. The blocked slides and the 10 × 22 mm glass Lifterslips (Thermo Scientific, Hudson, NH) were first cleaned of any residual dust using an air line.
Hybridization
A hybridization solution was prepared using the purified labeled DNA, 20X SSC (2X final conc.), yeast tRNA (10 µg/µl), H20 and 0.25 µl 10% SDS (0.1% final). The solution was mixed and spun briefly in a centrifuge, then denatured at 100°C for 5 minutes. One ml of H20 was added to the Whatman paper inside the hybridization chamber to help prevent the slide from drying out during the incubation. Ten µl of denatured hybridization cocktail was then carefully injected under its respective cover slip using a pipette. The hybridization chamber was sealed, and hybridization was incubated overnight at 55°C. After hybridization the arrays were removed from the hybridization oven and washed at room temperature using the Little Dipper Microarray Processor. The slides were then spun dry in the attached centrifuge on the Little Dipper Microarray Processor and stored in a dark container until scanned as below. Each sample was hybridized in replicate and evaluated after one single hybridization.
Microarray validation
Reproducibility
One sample was hybridized on two different sections on the same slide to test section-to-section reproducibility. Further, one sample was hybridized on two different slides to test the slide-to-slide reproducibility. Duplicate probe sets for each clinical sample under the same cover slip tested the constant validation of reproducibility. Specifity Pools of genomic DNA from 10 target bacterial species or phylotypes (e.g., from cloned inserts) were processed on one array to confirm the specificity of the bacterial probes and to identify any possible cross-reacting probes. After these experiments, those probes that were determined to cross-react or not specifically detect the target bacterial species, were not considered in the analysis or deleted from the probe panel.
Controls
Negative controls (buffer only and blank spots, reverse complement to the universal probe, probes that detected non-human DNA) were included in both the original and duplicate probe set. They were used to identify high background- and non-specific binding.
Scanning and analysis
An Axon 4000B microarray scanner (Molecular Devices, Sunnyvale, CA) was used for scanning the slides. They were scanned using a wavelength of 532 nm to visualize probes hybridized with Cy3-labeled amplicons. The PMT value (photomultiplier tube) was adjusted accordingly, with Power at 100%. Median pixel intensities for each individual spot could be calculated using the analysis function of the GenePix Pro (Molecular Devices) accessory software. The median background intensity for each individual feature was subtracted from the median feature intensity yielding a normalized “median intensity score” for each individual feature. The generated gpr files were exported to the HOMIM tool website for further analysis (http://bioinformatics.forsyth.org/homim/). This analysis allowed the determination of the presence or absence of a particular microorganism based on specific criteria set for that individual spot, and thus microbial profile maps for each sample, and subsequent cluster analyses. The cluster method used was UPGMA with a correlation distance function. The similarity measurement was calculated with the Person’s Correlation Coefficient with value from −1 to +1, when identical: 1, and completely opposition: −1. The signal intensities were categorized from 0 to 5.
Statistical analysis
When comparing the prevalence of subjects with a specific species in the two subject groups, a chi-square test was used to calculate p-values. When comparing p-values of bacterial species in two different samples in one group, a sign test was used. H0 was defined as “the true prevalence in the two groups is identical”.
Results
General findings
In the present study, bacterial profiles of healthy and diseased roots from 41 elderly subjects were investigated. Overall, nine bacterial phyla comprising 164 bacterial species and 15 clusters (species groups detected by probes targeting more than two closely related species (Table 1, Table S1). The number of bacterial species per sample ranged from 5 to 45 (Table 2). Some species (such as Fusobacterium nucleatum subsp. polymorphum oral taxon 202, Capnocytophaga sputigena oral taxon 775 and Lactobacillus casei/paracasei/rhamnosus oral taxon 749/716/568) were dominant while others appeared to be present in low concentration. A higher bacterial diversity was observed in the Controls as compared to the Patients; while the number of species within the various samples from each patient was reasonably stable (Table 2).
Table 1.
Number of species belonging to various bacterial phyla identified in the different sample categories, and comparison of data from microarray and sequencinga
| Phylogenetic group |
Total | Control samples |
Patient samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plaque healthy root |
Plaque carious root |
Dentinal | |||||||||||||
| MT | MS | SS | MT | MS | SS | MT | MS | SS | MT | MS | SS | MT | MS | SS | |
| Firmicutes | 76 | 63 | 102 | 49 | 38 | 49 | 63 | 39 | 67 | 50 | 38 | 73 | 55 | 44 | 55 |
| Bacteroidetes | 25 | 19 | 50 | 19 | 11 | 24 | 15 | 14 | 24 | 12 | 7 | 27 | 15 | 12 | 17 |
| Actinobacteria | 22 | 15 | 44 | 7 | 3 | 16 | 8 | 5 | 13 | 16 | 5 | 16 | 17 | 14 | 30 |
| Proteobacteria | 20 | 16 | 19 | 14 | 9 | 13 | 15 | 13 | 10 | 14 | 12 | 10 | 13 | 9 | 3 |
| Fusobacteria | 6 | 5 | 21 | 6 | 5 | 15 | 4 | 3 | 11 | 3 | 3 | 7 | 6 | 4 | 6 |
| Spirochetes | 7 | 6 | 4 | 5 | 4 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 3 | 3 | |
| TM7 | 4 | 4 | 3 | 2 | 2 | 2 | 4 | 1 | 1 | 2 | 2 | 1 | |||
| Synergistes | 4 | 3 | 2 | 1 | 1 | 1 | 4 | 3 | 1 | 1 | 1 | 3 | 1 | 2 | |
| OP11 | 1 | 1 | 1 | ||||||||||||
| Total | 165 | 131 | 245 | 101 | 71 | 123 | 114 | 80 | 129 | 101 | 67 | 137 | 113 | 89 | 117 |
MT = Microarray total (includes samples from 20 Controls and 21 root caries subjects). MS and SS= microarray and sequencing subset, respectively (includes analysis of the same samples from 10 Controls and 11 root caries subjects), with sequencing data from Preza et al. (15).
Table 2.
Species diversity in the different sample categories, and a comparison between microarray and sequencing resultsa
| Samples | Comparison | |||||
|---|---|---|---|---|---|---|
| MT | MS | SS | ||||
| Range | Mean | Range | Mean | Range | Mean | |
| Controls | ||||||
| Control | 15–45 | 26 | 21–33 | 26 | 16–41 | 28 |
| Patients | ||||||
| Plaque healthy root | 8–43 | 22 | 12–33 | 20 | 9–35 | 25 |
| Plaque carious root | 8–35 | 19 | 8–30 | 16 | 8–31 | 24 |
| Dentinal | 5–38 | 22 | 15–38 | 22 | 10–34 | 21 |
MT = Microarray total (includes samples from 20 Controls and 21 root caries subjects). MS and SS = microarray and sequencing subset, respectively (includes analysis of the same samples from 10 controls and 11 root caries subjects), with sequencing data from Preza et al. (15).
Species distribution within the four sample categories
Several bacterial species were commonly found in all categories, including: species of Streptococcus (present in 95% of the samples), Campylobacter (90%), Veillonella and Selenomonas (83%), Capnocytophaga (65%), Prevotella (63%) and Eubacterium and Actinomyces (59%). The specific species present in the highest number of samples were as follows: Campylobacter gracilis oral taxon 623 (84%), F. nucelatum subsp. polymorphum oral taxon 202 (70%), Veillonella atypica oral taxon 524 (53%), Selenomonas infelix oral taxon 126 and Parvimonas micra oral taxon 111 (52%), Dialister invisus oral taxon 118 (47%), Eubacterium saburreum oral taxon 494 and Synergistes sp. oral taxon 363, clone W090 (46%) (Table S1). Twenty-two species were found only in the plaque, dentin, or plaque and dentin of the carious roots; however, their prevalence were low (e.g., present in only one or two Patients) (Table S1).
For bacterial species previously associated with RC, such as Streptococcus, Lactobacillus and Actinomyces, the species with the highest prevalence were Streptococcus parasanguinis oral taxon 721/411 (45%), L. casei/paracasei/rhamnosus oral taxon 749/716/568 (47%) and Actinomyces gerencseriae oral taxon 618 (45%). S. mutans oral taxon 686 was found in only 19% (e.g., in three control samples, three plaque samples from healthy and carious roots, and seven dentinal samples). In the Control group, S. mutans oral taxon 686 was found together with L. casei/paracasei/rhamnosus oral taxon 749/716/568 (one subject) or Actinomyces (two subjects). In the Patient group, the combination of S. mutans oral taxon 686, lactobacilli and Actinomyces was as follows: S. mutans oral taxon 686, lactobacilli and Actinomyces were found together in one plaque sample from carious roots and two plaque samples from healthy roots and dentinal samples. S. mutans oral taxon 686 and lactobacilli (L. casei/paracasei/rhamnosus oral taxon 749/716/568) were detected together in only three dentinal samples. S. mutans and Actinomyces were found together in one plaque sample from carious roots and one dentinal sample. Lactobacilli and Actinomyces together were similarly prevalent in the three Patient sample categories (20–25% of the samples). S. mutans oral taxon 686 alone was found in only one plaque from carious roots.
Cluster analysis
A cluster analysis was performed in order to group samples with similar bacterial profiles. The cluster analysis was based on both the presence of species and their relative quantity measured as spot intensities (level 1–5). No specific clustering was observed, and no distinct patterns of bacterial species characterized the different sample categories.
Statistical analysis
Although the species detected did not form distinct clusters, when looking at the four sample categories, there were significant differences between these categories (Table 3). The prevalences of species in the four sample categories are shown in Table 3 and Fig. 1. A total of 182 bacterial species, clusters and species groups (genus level) were included. Each sample category was compared pairwise with the three other categories. Significant differences of prevalence in at least one of the comparisons were observed for 22 target species and clusters (Table 3).
Table 3.
Species with significant differences in their association with healthy and diseased roots
| Bacteria | Samples | Sample comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control vs Patient | Within Patients | |||||||||
| Lactobacillus spp.a | 5 | 10 | 10 | 15 | ns | ns | .005 | ns | ns | ns |
| Lactobacillus casei/paracasei/rhamnosusa | 5 | 10 | 10 | 14 | ns | ns | .012 | ns | ns | ns |
| Pseudoramibacter alactolyticusa | 2 | 8 | 5 | 9 | ns | ns | .032 | ns | ns | ns |
| Streptococcus oralis | 9 | 5 | 14 | 11 | ns | ns | ns | .012 | ns | ns |
| Streptococcus parasanguinis | 9 | 4 | 13 | 11 | ns | ns | ns | .022 | ns | ns |
| Dialister pneumosintes | 0 | 5 | 0 | 0 | .048 | ns | ns | ns | ns | ns |
| TM7 sp. clone AH040/BS003 | 0 | 5 | 2 | 0 | .048 | ns | ns | ns | ns | ns |
| H. parainfluenzae/A. aphrophilus | 8 | 2 | 5 | 6 | .032 | ns | ns | ns | ns | ns |
| Actinomyces clusterb | 7 | 1 | 5 | 5 | .02 | ns | ns | ns | ns | ns |
| Campylobacter spp. | 20 | 20 | 16 | 19 | ns | .048 | ns | ns | ns | ns |
| Campylobacter showae | 11 | 6 | 4 | 6 | ns | .025 | ns | ns | ns | ns |
| Propionobacterium spp. | 4 | 3 | 1 | 0 | ns | ns | .048 | ns | ns | ns |
| Streptococcus cristatus/ S. sp. clone BM053e | 4 | 0 | 0 | 3 | .048 | .048 | ns | ns | ns | ns |
| Cardiobacterium hominise | 9 | 1 | 3 | 4 | .004 | .043 | ns | ns | ns | ns |
| Capnocytophaga cluster ce | 17 | 11 | 10 | 13 | .043 | .02 | ns | ns | ns | ns |
| Eubacterium saburreume | 15 | 7 | 9 | 7 | .012 | ns | .012 | ns | ns | ns |
| Eubacterium spp.e | 17 | 10 | 12 | 10 | .02 | ns | .02 | ns | ns | ns |
| Leptotrichia cluster de | 17 | 14 | 10 | 9 | ns | .02 | .009 | ns | ns | ns |
| Capnocytophaga sputigenae | 12 | 2 | 4 | 5 | .001 | .011 | .028 | ns | ns | ns |
| Campylobacter concisuse | 13 | 4 | 4 | 5 | .004 | .004 | .012 | ns | ns | ns |
| Gemella morbillorume | 14 | 7 | 5 | 7 | .029 | .005 | .029 | ns | ns | ns |
| Fusobacterium nucleatum ss polymorphume | 20 | 15 | 12 | 11 | .021 | .001 | 0.00 | ns | ns | ns |
| Total no. of samples | ||||||||||
| Total no. of samples | 20 | 21 | 21 | 21 | ||||||
Species associated with RC subjects.
Actinomyces cluster= All species.
Capnocytophaga cluster= C. gingivalis oral taxon 337, C. granulosa oral taxon 325, C. sp. clone AH105, C. sp. clone S3, C. sp. oral taxon 325, clone BB167, C. sp. clone TFI Cap08
Leptotrichia cluster= L. buccalis oral taxon 563, L. sp. oral taxon 498/462/463/417, clone IK040/GT018/GT020/ C3MK102
Species associated with control subjects.
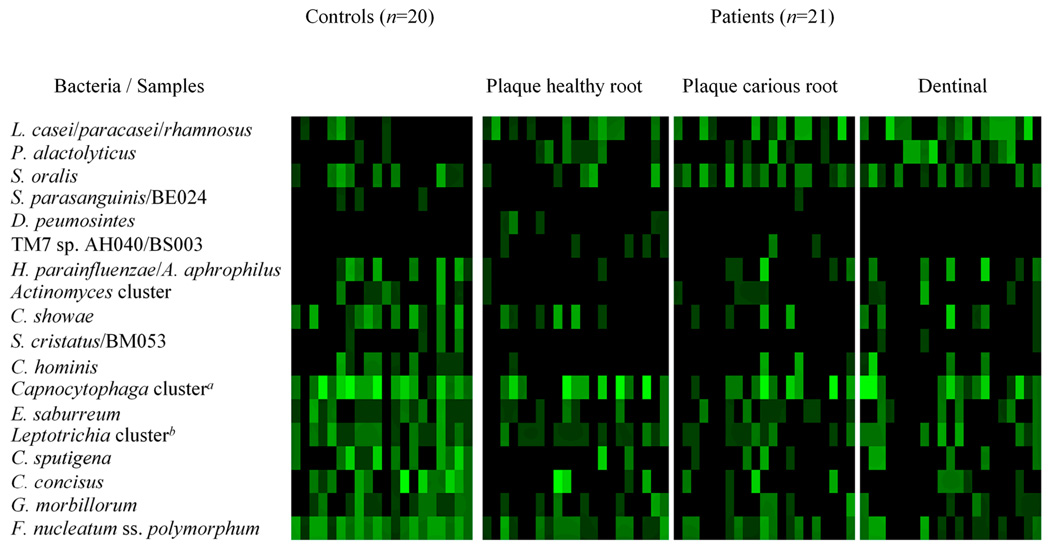
Fig. 1.
Distribution of species with significant differences within the sample categories Each column represents one sample. Five different intensities of green correspond to signal intensities of the arrays.
a Capnocytophaga cluster= C. gingivalis oral taxon 337, C. granulosa oral taxon 325, C. sp. clone AH105, C. sp. clone S3, C. sp. oral taxon 325, clone BB167, C. sp. clone TFI Cap08
b Leptotrichia cluster= L. buccalis oral taxon 563, L. sp. oral taxon 498/462/463/417, clone IK040/GT018/GT020/ C3MK102
Altogether, 546 significance tests were performed. Based on random distribution, 5%, i.e., 27 of the comparisons would be expected to be significantly different at a p=0.05 level. The results showed, however, 34 significant results; suggesting that 7 out of the 34 significant results (approximately 21%) listed in Table 3 are true positives. Lactobacilli were present in a significantly higher number of dentinal samples. The most dominant species of this group was L. casei/paracasei/rhamnosus oral taxon 749/716/568. Besides lactobacilli, Pseudoramibacter alactolyticus oral taxon 538 was significantly associated with the dentinal samples. The putative cariogenic pathogens, S. mutans oral taxon 686 and Actinomyces, showed no significant differences. The Capnocytophaga cluster (C. gingivalis oral taxon 337, C. granulosa oral taxon 325, C. sp. clone AH105, C. sp. clone S3, C. sp. oral taxon 325, clone BB167, C. sp. clone TFI Cap08) showed a significant association with the Control samples (Table S1). C. sputigena oral taxon 775, Campylobacter concisus oral taxon 575, Gemella morbillorum oral taxon 046 and F. nucleatum subsp. polymorphum oral taxon 202 were significantly associated with Controls in all three comparisons. The probability of getting three significant results for one species, if the true prevalence of the two groups was identical, is approximately 0.0001. Thus the association of these species with healthy subjects is reasonably substantiated.
Comparison of microarray analysis with results from sequencing
The microarray results correlated well with the results of our previous study on bacterial diversity based on cloning and sequencing of 16S rRNA genes (Table 4) (15). Of the species determined by clonal analysis, 72% were also detected by HOMIM. More specifically, of the 164 species detected by microarray, 89 were detected by both methods. The microarray and gene sequencing data were directly compared as to species associated with either healthy or carious roots according to the previous study (Table 4). Some of the disease-associated species, e.g., Propionibacterium sp. oral taxon 191, strain FMA5 and Actinomyces sp. oral taxon 448, clone IP073, were either not, or only rarely, detected by the microarrays. Probes to these species are therefore being reevaluated. Actinomyces species were previously described as increased in their prevalence from the Controls to the dentinal samples, while the present study did not show such a correlation. Both S. mutans oral taxon 686 and L. casei/paracasei/rhamnosus oral taxon 749/716/568 were associated with the dentinal samples with both methods. S. mutans oral taxon 686 was moderately prevalent, and lactobacilli, especially L. casei/paracasei/rhamnosus oral taxon 749/716/568, showed a clear association with the dentinal samples in both studies.
Table 4.
Comparison between microarray and previously published 16S rRNA gene sequencing data [15].
| Bacteria | Control sample |
Patient samples |
||||||
|---|---|---|---|---|---|---|---|---|
| Plaque healthy root |
Plaque carious root |
Dentinal | ||||||
| MS | SS | MS | SS | MS | SS | MS | SS | |
| (n=10) | (n=11) | (n=11) | (n=11) | |||||
| Propionibacterium sp. strain FMA5 a | 7 | |||||||
| Pseudoramibacter alactolyticus a | 1 | 3 | 1 | 5 | 3 | |||
| Prevotella multisaccharivorax a | 1 | 1 | 1 | |||||
| Enterococcus faecalis a | 1 | 1 | 1 | |||||
| Lactobacillus spp.a | 2 | 5 | 3 | 4 | 6 | 7 | 5 | |
| Lactobacillus casei/paracasei/rhamnosus a | 2 | 5 | 1 | 4 | 2 | 6 | 5 | |
| Actinomyces spp. | 8 | 5 | 10 | 5 | 5 | 4 | 6 | 8 |
| Selenomonas sp. clone CS002 | 3 | 2 | 1 | 1 | 5 | 7 | 4 | 6 |
| Streptococcus mutans | 3 | 1 | 2 | 3 | 1 | 3 | 4 | 5 |
| Actinomyces sp. clone IP073 | 1 | 1 | 2 | 5 | 2 | 3 | 1 | 4 |
| Dialister invisus | 5 | 3 | 5 | 4 | 7 | 5 | 8 | 5 |
| Selenomonas sp. strain GAA14 | 3 | 1 | 3 | 3 | 6 | 5 | 7 | 4 |
| Prevotella denticola | 2 | 1 | 2 | 5 | 5 | 5 | 3 | 3 |
| Prevotella melaninogenica | 1 | 2 | 2 | 5 | 1 | 5 | 1 | 2 |
| Selenomonas sputigena | 5 | 4 | 6 | 10 | 5 | 7 | 3 | 4 |
| Corynebacterium matruchotii | 1 | 8 | 3 | 4 | 2 | 4 | ||
| Campylobacter gracilis | 9 | 6 | 10 | 9 | 9 | 6 | 10 | 3 |
| Prevotella nigrescens | 3 | 5 | 4 | 6 | 2 | 3 | 2 | 2 |
| Selenomonas infelix | 7 | 5 | 6 | 4 | 4 | 3 | 3 | 2 |
| Streptoccocus gordonii/anginosus | 5 | 6 | 3 | 3 | 3 | 5 | 6 | 3 |
| Leptotrichia spp. | 9 | 9 | 7 | 6 | 4 | 3 | 4 | 2 |
| Granulicatella adiacens | 1 | 4 | 2 | 3 | 1 | |||
| Streptococcus sanguinis | 1 | 5 | 1 | 2 | 2 | 1 | ||
| Selenomonas noxia b | 6 | 7 | 5 | 3 | 3 | 2 | 6 | 1 |
| Fusobacterium nucleatum ss polymorphum b | 10 | 10 | 6 | 3 | 6 | 4 | 6 | 1 |
| Streptococcus anginosus b | 1 | 3 | 2 | 3 | 1 | 3 | 1 | |
| Streptococcus mitis bv2 b | 2 | 5 | 1 | 3 | 1 | 2 | ||
| Kingella oralis b | 7 | 5 | 2 | 3 | 1 | 3 | ||
The numbers refer to the numbers of samples with that particular species present.
SS=Sequencing subset. Previously published sequencing results [15].
MS=Microarray subset. Species detected in the same sequenced samples [15].
Species associated with Patients.
Species associated with Controls.
Discussion
The present study investigated the bacterial flora of RC using HOMIM, a new rRNA-based microarray technique, which allows the detection of both cultivable and not yet-cultivated species. As we previously demonstrated by sequence analysis of 16S rRNA genes amplified from clinical samples (15), the bacterial diversity of root surfaces of healthy and diseased teeth was extensive. In the present study, 164 species and 15 clusters were detected in the 83 samples examined.
The dominance hierarchy of phyla in the different sample categories was reasonably similar, the more prominent exception being Actinobacteria. This phylum was the second most dominant in dentinal samples, but less common in the other categories. Several species were clearly associated with Control (healthy) subjects, including F. nucleatum subsp. polymorphum oral taxon 202, G. morbillorum oral taxon 046, C. sputigena oral taxon 775 and C. concisus oral taxon 575 (Table 3 and Fig 1). The differences between Patients and Controls were more distinct than the differences between samples from healthy or diseased sites within the Patients. Species such as F. nucleatum subsp. polymorphum oral taxon 202, P. alactolyticus oral taxon 538 and L. casei/paracasei/rhamnosus oral taxon 749/716/568 that showed significant results in the statistical comparison between the Patient and Control group, have also previously been notably associated with either healthy or carious roots (15).
No obvious patterns were found when cluster analyses were used in an attempt to distinguish the four categories of samples, suggesting that there are no distinct groups of bacterial species that are associated with either RC, in comparison with other samples from Patients, or with Patients as compared to Controls, i.e, no particular bacteria appear to be consistently involved in the pathogenesis of RC. Notably, we did not see an association between Actinomyces and RC, which is in contrast to conclusions from earlier studies [16]. Similarly, while certain reports have associated S. mutans oral taxon 686 with RC, our previous [15] and present results indicate that it may play only a limited role. Conversely, Lactobacillus spp. and P. alactolyticus oral taxon 538 were notably associated with most RC samples. Consequently, it is clear that there is subject-to-subject variation concerning the microbial etiology of RC— it is not just S. mutans oral taxon 686.
Half of the samples were included in a comparison between the present results and those of previous analyses using cloning and sequencing (Table 4). Combining the two methods, a total of 320 species were detected in either RC or the overlying plaque. These species were divided between nine of the 12 phylogenetic groups observed in oral samples (Table 1). Several species observed in the sequencing study, and present on the microarrays, were not detected (Table S2). Most of these species were only rarely detected. The discrepancies may be partly ascribed to stochastic effects when it comes to finding bacteria with relatively low copy number. In some cases, such as Propionibacterium species oral taxon 191, strain FMA5, species observed in a large number of clones were not found by the arrays. These probes are presently being re-evaluated. Nevertheless, our data indicate that HOMIM represents a rapid and reliable means of identifying microbial profiles directly from oral clinical samples, such as plaques samples on root surfaces.
Supplementary Material
Acknowledgements
We thank the Cathinka Guldberg-Centre in Oslo, Norway, particularly Marianne Wenaasen and Sabah Tariq for patient management. We are very thankful to Professor Leiv Sandvik for his statistical advice. The study was supported by the Faculty of Dentistry, University of Oslo, Oslo, Norway and NIH grant DE11443 (bjp).
References
- 1.Banerjee A, Yasseri M, Munson M. A method for the detection and quantification of bacteria in human carious dentine using fluorescent in situ hybridisation. J Dent. 2002;30:359–363. doi: 10.1016/s0300-5712(02)00052-0. [DOI] [PubMed] [Google Scholar]
- 2.Bowden GH, Nolette N, Ryding H, Cleghorn BM. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. doi: 10.1177/00220345990780120601. [DOI] [PubMed] [Google Scholar]
- 3.Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol. 2004;42:3128–3136. doi: 10.1128/JCM.42.7.3128-3136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhour KL, Nadkarni MA, Byun R, Martin FE, Jacques NA, Hunter N. Molecular analysis of microbial diversity in advanced caries. J Clin Microbiol. 2005;43:843–849. doi: 10.1128/JCM.43.2.843-849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fejerskov O, Baelum V, Ostergaard ES. Root caries in Scandinavia in the 1980's and future trends to be expected in dental caries experience in adults. Adv Dent Res. 1993;7:4–14. doi: 10.1177/08959374930070010501. [DOI] [PubMed] [Google Scholar]
- 6.Fure S. Five-year incidence of caries, salivary and microbial conditions in 60-, 70-and 80-year-old Swedish individuals. Caries Res. 1998;32:166–174. doi: 10.1159/000016449. [DOI] [PubMed] [Google Scholar]
- 7.HOMIM. 2008 posting date. http://mim.forsyth.org/. [online]
- 8.Imazato S, Ikebe K, Nokubi T, Ebisu S, Walls AW. Prevalence of root caries in a selected population of older adults in Japan. J Oral Rehabil. 2006;33:137–143. doi: 10.1111/j.1365-2842.2006.01547.x. [DOI] [PubMed] [Google Scholar]
- 9.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadkarni MA, Caldon CE, Chhour KL, Fisher IP, Martin FE, Jacques NA, Hunter N. Carious dentine provides a habitat for a complex array of novel Prevotella-like bacteria. J Clin Microbiol. 2004;42:5238–5244. doi: 10.1128/JCM.42.11.5238-5244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nascimento MM, Hofling JF, Goncalves RB. Streptococcus mutans genotypes isolated from root and coronal caries. Caries Res. 2004;38:454–463. doi: 10.1159/000079627. [DOI] [PubMed] [Google Scholar]
- 12.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. 2000. [DOI] [PubMed] [Google Scholar]
- 14.Petersen PE, Yamamoto T. Improving the oral health of older people: the approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol. 2005;33:81–92. doi: 10.1111/j.1600-0528.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- 15.Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46:2015–2021. doi: 10.1128/JCM.02411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravald N. Root surface caries. Curr Opin Periodontol. 1994:78–86. [PubMed] [Google Scholar]
- 17.Saunders RH, Jr, Meyerowitz C. Dental caries in older adults. Dent Clin North Am. 2005;49:293–308. doi: 10.1016/j.cden.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Silwood CJ, Lynch EJ, Seddon S, Sheerin A, Claxson AW, Grootveld MC. 1H-NMR analysis of microbial-derived organic acids in primary root carious lesions and saliva. NMR Biomed. 1999;12:345–356. doi: 10.1002/(sici)1099-1492(199910)12:6<345::aid-nbm580>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73:1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 20.WHO. Geneva, Switzerland: World Health Organization; Oral health surveys: basic methods. (4th., ed) 1997
- 21.Zambon JJ, Kasprzak SA. The microbiology and histopathology of human root caries. Am J Dent. 1995;8:323–328. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.