Abstract
To pursue an earlier observation that the protein encoded by the UL34 gene binds to intermediate chain of dynein, we constructed a series of mutants from which sequences encoding the entire protein (ΔUL34) or amino-terminal [UL34Δ(3–119)] or carboxyl-terminal [UL34Δ(245–275)] domains were deleted. The mutant lacking the sequence encoding the carboxyl-terminal domain grew in all cell lines tested. The two other mutants replicated only in cell type-dependent manner and poorly. Rescue of ΔUL34 mutant with a fragment that does not encompass the UL31 ORF restored wild-type phenotype. UL34 protein interacts physically with UL31, and the UL31 deletion mutant appears to have a phenotype similar to that of UL34 deletion mutant. Experiments designed to determine whether the phenotypes of the deletion mutants have a common base revealed that cells infected with the ΔUL34 mutant accumulate UL31 RNA but not the corresponding protein. The UL31 protein accumulated, however, to near wild-type virus-infected cell levels in cells infected with ΔUL34 mutant and treated with the MG132 proteosomal inhibitor at 6 h after infection. This is evidence that the stability of an essential viral protein requires the presence of another protein. The observation raises the bar for identification of gene function on the basis of analyses of the phenotype of mutants in which the gene has been deleted or rendered inoperative.
Herpes simplex virus 1 and 2 (HSV-1 and HSV-2) multiply at the portals of entry into the body. From these sites the viruses are transported by retrograde flow to either the central nervous system in which they can cause encephalitis or to neurons in dorsal root ganglia where they can establish latent infections (1, 2). Much of the misery associated with HSV infections stems from encephalitis caused by viral replication in the central nervous system and reactivation of latent virus in dorsal root ganglia (reviewed in ref. 3). The mechanisms by which these viruses are transport retrograde from peripheral sites to the neuronal nucleus is poorly understood. What is known is that a structure containing the capsid surrounded by at least one (VP1–2) but probably more tegument proteins is transported along the microtubular network to the nuclear pore (4–6). The DNA then is released from the capsid into the nucleus. An earlier report from this and associated laboratories has shown that the intermediate chain of cytoplasmic neuronal dynein interacted physically in vitro with three viral proteins encoded by UL31, UL19, and UL34 (7). The interaction of UL34 with the intermediate chain of dynein was verified in reciprocal pull-down experiments and in mapping studies showing that it interacts with the amino-terminal domain of the intermediate chain of dynein. Preliminary studies have shown that, in turn, the intermediate chain of dynein interacts with the amino-terminal domain of UL34 protein. UL31 protein, however, appears to interact strongly with UL34; its interaction with dynein is secondary and caused by its affinity for UL34 protein. Virion protein no. 5, the major capsid protein and product of the UL19 ORF, tends to interact nonspecifically, and the significance of its presence in the complex pulled down by the intermediate chain of dynein remains to be established (7).
To define the role of UL34 protein in the intracellular virus transport, it was necessary to construct mutants in which the UL34 ORF was deleted or silenced. In this paper we report on the construction of three UL34 deletion mutants. During the construction of the mutants, Roller et al. (8) reported the construction of one UL34 deletion mutant and reported that this mutant does not replicate in the cell lines in which it was tested. This report describes the construction and properties of mutants lacking the entire UL34 ORF (ΔUL34), the amino terminal [UL34Δ(3–119)], or the carboxyl-terminal domain [UL34Δ(245–275)]. We report that ΔUL34 and UL34Δ(3–119) are phenotypically similar and replicate poorly in a cell line-dependent manner. Because the phenotypes of ΔUL34 and ΔUL31 mutants were similar, we examined the status of UL31 protein in cells infected with the ΔUL34 mutant. We report that in cells infected with ΔUL34 mutant the UL31 protein is unstable and does not accumulate. Relevant to this report are the following.
The nucleotide sequence of the UL31 ORF is predicted to encode a basic protein with a hydrophilic amino terminus and a nuclear localization signal (9, 10). In infected cells, UL31 protein is phosphorylated and predominantly dispersed throughout the nucleus. In nuclear fractionation studies, UL31 protein partitions with the nuclear matrix (11). A mutant lacking UL31 grew well in rabbit skin cells expressing the UL31, but poorly in cells lacking the gene. Electron microscopic studies indicated the presence of a small numbers of full capsids but few enveloped virions. The defect could be attributed to decreased yields of viral DNA and reduced cleavage of viral DNA for packaging (12). These results suggested that UL31 protein formed a network to enable the anchorage of viral products for the synthesis and/or packaging of viral DNA into virions.
UL34 protein initially was identified as a substrate for the viral protein kinase encoded by US3 ORF (13). The protein has a hydrophilic amino-terminal domain but associates with membranes during viral replication (14).
Materials and Methods
Cells and Viruses.
The limited in vitro passage HSV-1 strain F [HSV-1(F)] is the prototype HSV-1 strain used in this laboratory (15). The sources and procedures for the cultivation of Vero, HEp-2, HeLa, 143TK−, and rabbit skin cells have been described (16).
Antibodies.
The production and properties of the polyclonal antibodies to UL31, UL34, and UL38 proteins have been described (11, 14, 17). The UL34 polyclonal antibody used in this study was purified by AffinityPak Immobilized Protein A column (Pierce).
Construction of UL34 Mutant Viruses.
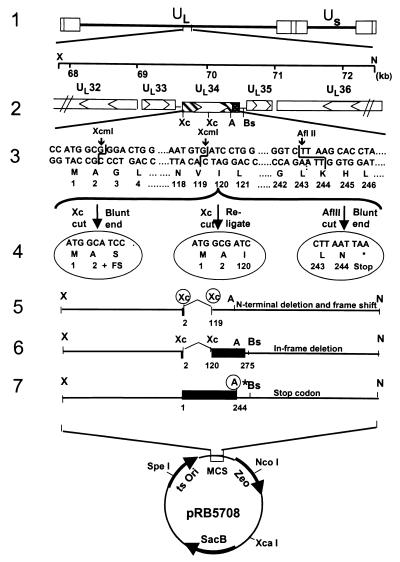
Recombinant viruses R5601 (ΔUL34 mutant), R5602 (UL34 lacking amino acids 3–119), and R5603 [UL34Δ(245–275), a mutant lacking the UL34 transmembrane and carboxyl-terminal domains] were constructed with the aid of a transfer plasmid derived from pKO5Y (pRB5708), a transfer vector regenerated from pKO5.1 (18, 19), and pKO3 (20). To construct the transfer plasmids pRB5713, pRB5714, and pRB5715, pRB260, which contains the BglII D fragment of HSV-1(F), was digested with XhoI/NotI, and a 4,664-bp fragment containing UL33, UL34, UL35 and portions of UL32 and UL36 (nucleotides 67,762–72,426) was inserted into pBluescript II KS(+) at XhoI/NotI sites to yield pRB5709 (line 2 in Fig. 1). pRB5709 was cleaved with XcmI to remove a fragment of 350 bp, which encodes codons 3–119 of UL34, blunt-ended, and religated. The resultant recombinant plasmid pRB5710 (lines 4 and 5 in Fig. 1) contained a frame shift after the first two codons of UL34. To make pRB5711 (lines 4 and 6 in Fig. 1), pRB5709 was cleaved with XcmI and religated. In pRB5711, codons 3–119 were deleted, but the remaining codons of UL34 remained in-frame. To construct pRB5712 (lines 4 and 7 in Fig. 1) pRB5709 was cleaved with AflII, blunt-ended, and religated. In pRB5712, a stop codon was introduced immediately after codon 244 of UL34 to preclude the translation of the carboxyl-terminal domain of UL34.
Figure 1.
Schematic representation of viral and plasmid DNA sequence arrangements. Line 1, A linear representation of the HSV-1 genome. The rectangles represent the inverted repeats flanking the unique sequences (UL and US represented by thin lines). Line 2, pRB5709, a 4.5-kb XhoI/NotI fragment of HSV-1(F) containing the UL33, UL34, UL35, and portion of UL32 and UL36 genes. The arrowheads indicate the direction of the transcription; solid dotted box indicates the putative transmembrane domain of the UL34 gene product. Line 3, DNA sequences of UL31 coding domains showing relevant restriction endonucleases sites used for construction of deletion mutants. Line 4, From left to right, pRB5710, pRB5711, and pRB5712. pRB5710 was derived by excision of a XcmI fragment in UL34, blunt-ending, and religation to produce a frame shift after the first two codons of UL34. pRB5711 was made by direct religation of the plasmid pRB5709 after removal of a XcmI fragment. This caused an in-frame deletion of codons 3–119. In pRB5712 the pRB5709 plasmid was cleaved with AflII, blunt-ended, and religated. In addition, a stop codon, and a unique restriction site (PacI), was introduced immediately after codon 244 of UL34. Lines 5–7, The transfer plasmids pRB5713, pRB5714, and pRB5715 were constructed by cloning the XhoI/NotI fragment from pRB5710, pRB5711, and pRB5712, respectively, into the transfer vector (pRB5708) at XhoI/NotI sites.
pRB5710, pRB5711, and pRB5712 were cleaved with XhoI/NotI to recover the fragments with the mutated allele and inserted into pRB5708 at XhoI/NotI sites to yield the transfer plasmids pRB5713, pRB5714, and pRB5715 (lines 5–7 in Fig. 1). RR1 competent cells that harbored bacterial artificial chromosome (BAC)-HSV plasmids were transformed with the transfer plasmids DNA (pRB5713, pRB5714, or pRB5715) by electroporation. After incubation for 1 h at 30°C in LB broth, the bacteria were plated on prewarmed Zeocine (Zeo)/chloramphenicol (Cm) (20 μg/ml of each) plates and incubated overnight at 43°C for integration. The next day, six colonies were picked and diluted serially in LB, plated on Cm/10% sucrose (Suc) LB plates, and incubated at 30°C overnight. To further confirm the loss of the replacement vector, 24 Cm/Suc-resistant colonies (four colonies from each plate) were restreaked in duplicate on Cm/Suc LB and Zeo LB plates, respectively, and incubated at 30°C overnight. The Sucr/Cmr/Zeos colonies were further screened by PCR (95°C 4 min, then 35 cycles of 94°C, 1 min; 60°C, 1 min; 72°C 1 min). The primers used for this purpose were: PCR34F, CCCCCGCCGAGCTGGAGGTTGTCT (from nucleotides 69342 to 69365) and PCR34R, CGCAAGGCTCGTGCGTTGGTGTG (from nucleotides 70970 to 70948). PCR-confirmed colonies were grown in 200 ml of LB/Cm medium, and the recombinant BAC-HSV DNA was prepared with the aid of a Qiagen (Chatsworth, CA) plasmid purification kit. The purified recombinant viral DNAs also were analyzed by PCR for additional verification.
Transfection of Cell Lines with Recombinant BAC-HSV DNA.
Subconfluent cultures of Vero, HEp-2, 143TK−, or rabbit skin cells seeded in 25-cm2 culture flasks at 1 × 106 cells per dish were transfected with R5601, R5602, or R5603 recombinant BAC-HSV viral DNA by Lipofectamine reagent (Life Technologies, Grand Island, NY). For UL34 repair virus R5604, Vero cells were cotransfected with R5601 viral DNA and pRB5709 by using the same method.
Replication of UL34 Mutant Viruses.
Cultures of Vero, HEp-2, 143TK−, or rabbit skin cells were infected with 0.1 plaque-forming unit (pfu) of R5601(ΔUL34), R5603 [UL34Δ(245–275)], R5604 (UL34 repaired), or wild-type virus per cell. After 2 h of incubation at 37°C, the inoculum was removed, and the cells were rinsed three times with fresh medium and reincubated at 37°C. At 18 h after infection, the cells were harvested and titrated on rabbit skin cells by plaque assay.
Cell Fractionation.
Rabbit skin cells were mock-infected or exposed to 5 pfu of HSV-1(F) or R5601 (ΔUL34) per cell. At time points indicated in Results, the cells were harvested and rinsed once with PBS(A), and then resuspended into 120 μl of hypotonic lysis buffer [10 mM Hepes, pH 7.4/10 mM KCl/3 mM MgCl2/10 mM NaF/0.1% NP-40/0.1 mM EDTA/1 mM DTT/0.5 mM PMSF/0.2 mM Nα-p-tosyl-l-lysine-chloromethyl ketone (TLCK)] by gently pipetting up and down 10 times and stored on ice for 30 min. The cytoplasmic fraction was obtained by centrifugation at 2,500 rpm (500 g) for 5 min at 4° C. The nuclei were gently rinsed once with 200 μl of the hypotonic buffer and recentrifuged as above. To further fractionate the nuclear content, the nuclei were resuspended in 120 μl of lysis buffer (50 mM Hepes, pH 7.4/250 mM KCl/10 mM NaF/0.1% NP-40/0.1 mM EDTA/1 mM DTT/5% glycerol/0.5 mM PMSF/0.2 mM TLCK) and stored on wet ice for 30 min. The soluble and insoluble fractions were separated by centrifugation (13,000 rpm, 10 min).
Immunoblotting.
Whole-cell lysates or cytoplasmic and nuclear fractions obtained as described above were resuspended in disruption buffer (50 mM Tris⋅HCl, pH 6.8/100 mM DTT/2% SDS/10% glycerol/0.1% Bromophenol blue, subjected to electrophoresis on a 12% denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and reacted with antibodies as indicated in Results.
Detection of UL31 RNA.
Replicate cultures of rabbit skin cells were mock-infected or exposed to 10 pfu of R5601(ΔUL34), R5604 (ΔUL34 repaired), or wild-type HSV-1(F) per cell and incubated at 37°C. At 6, 18, or 35 h after infection replicate cultures were lysed directly in the culture dish with TRIzol reagent (Life Technologies). Five micrograms of the total RNA extracted from each sample according to manufacturer's instruction was separated on a 1.2% agarose/formaldehyde gel, transferred onto a nitrocellulose membrane, and probed with a UL31-specific DNA fragment, which had been 32P-labeled by nick-translation.
Electron Microscopy.
Cells were mock-infected or infected with 5 pfu R5601 (ΔUL34), R5602 [UL34Δ(3–119)], R5603 [UL34Δ(245–275)], or wild-type HSV-1(F) virus per cell and incubated at 37°C. The cultures were fixed at 18 h after infection, sectioned, processed for electron microscopy, and examined in a Siemens 102 microscope as described (4).
Results
Construction of UL34 Null and Partial Deletion Mutants.
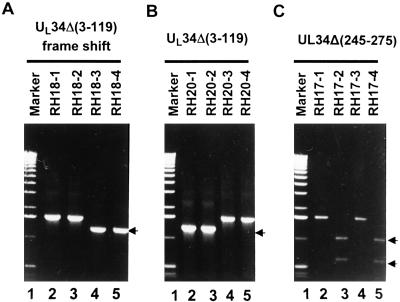
As detailed in Materials and Methods and illustrated in Fig. 1, we constructed three mutants lacking or silencing all or portions of the UL34 ORF with the aid of a BAC-HSV system (18). The construction of the transfer plasmids pRB5713, pRB5714, and pRB5715 is described in Materials and Methods and illustrated in Fig. 1. In brief, RR1 cells harboring BAC-HSV were transformed with pRB5713, pRB5714, or pRB5715. After a series of selections described in Materials and Methods, the clones meeting selection criteria were further analyzed by PCR. The PCR-positive clones yielded an amplified product of expected sizes (Fig. 2 A and B) or were cleaved with PacI (Fig. 2C), the marker restriction site created in place of the AflII site (line 4 in Fig. 1). All of the negative clones, on the other hand, yielded a 1.6-kb wild-type DNA fragment. A positive clone of each virus was transfected into a series of cell lines, but the ΔUL34 and the UL34(Δ3–119) mutants formed plaques and replicated only in rabbit skin cells. Stocks were made in rabbit skin cells, infected, and maintained at 34°C.
Figure 2.
Verification of Camr/Zeos/Sucr colonies for the presence of UL34 recombinant HSV-BAC clones by PCR. (A and B) Photographs of electrophoretically separated PCR products amplified directly from positive colonies. (C) Restriction digests of the PCR products amplified from the potential positive colonies for UL34Δ(245–275). The PCR product was clarified by Gene-clean (Biol01) and digested with PacI, a restriction site expected if the integration of pRB5715 had taken place. RH18 1–4, RH20 1–4, and RH17 1–4 represent individual colonies selected for analysis. Marker, 1-kb ladder of molecular weight marker. The arrows identify the PCR-positive clones, which gave amplified DNA band of expected size or were cleaved by the appropriate restriction endonuclease.
Growth Properties of the Null Mutants.
Table 1 summarizes the results of an experiment in which the cells were exposed to 0.1 pfu of virus per cell on the basis of virus titers determined in rabbit skin cells. The cells were harvested at 18 h after infection and titrated in rabbit skin cells. In essence, only UL34Δ(245–275) formed plaques and replicated in all cell lines at levels approximately 10-fold lower than those of the wild-type or repaired viruses. All other mutants failed to form plaques or replicate in Vero, HEp-2, or 143TK− cells at levels significantly above background. In rabbit skin cells, the yields of UL34Δ(3–119) and ΔUL34 were approximately 10-fold higher than background and almost 1,000-fold lower than the wild-type of mutant viruses. Interestingly, in rabbit skin cells the yields were at least 2-fold higher at 34°C than at 37°C.
Table 1.
Yield of UL34 mutants in different cell lines
| Virus | RSC | Vero | HEp-2 | 143TK− |
|---|---|---|---|---|
| ΔUL34 | 5.2 × 105 | 4.8 × 104 | 5.1 × 104 | 4.5 × 104 |
| UL34D(245–275) | 4.0 × 107 | 6.2 × 107 | 3.5 × 107 | 2.6 × 107 |
| UL34 repair | 5.6 × 108 | 8.6 × 108 | 4.0 × 108 | ND |
| HSV-1(F) | 6.0 × 108 | 8.2 × 108 | 5.6 × 108 | ND |
Calculated as total pfu of viruses recovered from the infected cell culture. Titration of the progeny viruses was done on rabbit skin cells (RSC). ND, not done.
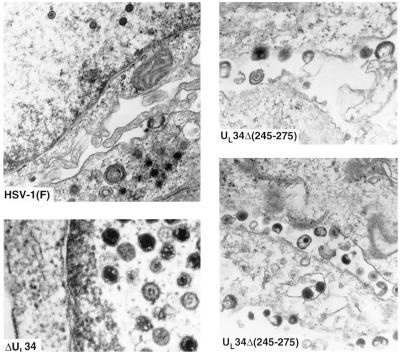
Electron microscopic studies (Fig. 3) revealed that rabbit skin cells infected with ΔUL34 mutant accumulated largely empty capsids in the nucleus. Enveloped particles were found in only a few cells either in the cytoplasm or in extracellular space. In contrast, cells infected with UL34Δ(245–275) exhibited significant numbers of virus particles in extracellular space.
Figure 3.
Electron micrographs of cells infected with R5601(ΔUL34) or R5603 [UL34Δ(245–275)]. Rabbit skin cells exposed to 5 pfu of R5601, R5603, or the wild-type HSV-1(F) per cell and incubated at 37°C. At 18 h after infection, the cells were fixed and processed for electron microscopy as described (4).
Accumulation of UL34 and UL31 Protein in Cells Infected with Wild-Type, Repaired, or Deletion Mutants.
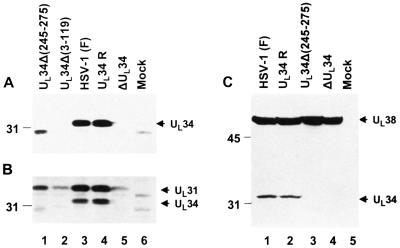
We report two series of experiments. In the first, rabbit skin cells were tested for the accumulation of UL34 protein in cells infected with wild-type parent, repaired virus (R5604), R5603 [UL34Δ(245–275)], R5602 [UL34Δ(3–119)], or R5601 (ΔUL34) mutants. The immunoblot shown in Fig. 4A indicates that the UL34 protein was not detected in cells infected with the ΔUL34 or UL34Δ(3–119) mutant. Cells infected with the UL34Δ(245–275) mutant accumulated reduced amount of a more rapidly migrating protein consistent with deletion of the carboxyl-terminal domains. In Fig. 4C the capsid protein 19C encoded by UL38 ORF served as a loading control and also demonstrated that it was synthesized in cells infected with each of the mutants or wild-type viruses. Because of the small amounts of the UL34 protein shown in cells infected with the wild-type virus, the smaller amounts of the truncated protein accumulating in cells infected with the UL34Δ(245–275) mutants were not detected.
Figure 4.
Photograph of immunoblots of electrophoretically separated whole-cell extracts of rabbit skin cells mock-infected or infected with wild-type or recombinant viruses. Rabbit skin cells (1 × 106 cells) were mock-infected or infected with R5601 (ΔUL34), 5602 [UL34Δ(3–119)], R5603 [UL34Δ(245–275)], R5604 (UL34 repaired virus), or wild-type HSV-1(F) viruses at 5 pfu per cell and incubated at 37°C. Cells were harvested 18 h after infection and resuspended in 1× SDS disruption buffer, boiled for 5 min, and centrifuged, and the supernatants were subjected to electrophoresis on a denatured polyacrylamide gel (12%). After transfer to a nitrocellulose membrane, the transferred proteins then were reacted with different primary antibodies. (A) Photograph of an immunoblot reacted with UL34 polyclonal antibody. Arrow indicates the infected cell protein specifically reacted with the UL34 antibody. A truncated protein band of expected size in R5603 [UL34Δ(245–275)]-infected cells (lane 1) also is detected by the antibody. (B) The same blot from A was reprobed with polyclonal antibody to UL31; the detected band of UL31 is indicated by an arrow. (C) A duplicated experiment (same as A and B) probed first with the antibody to UL34 and subsequently with antibody to UL38. Arrows indicate the infected cell proteins specifically reacted with UL34 and UL38 antibodies.
The second experiment was based on the observations reported earlier that UL34 protein interacts with UL31 in vitro (7). It was of interest to determine whether UL31 protein accumulates in these cells. In this experiment the immunoblot shown in Fig. 4A was reacted with antibody to UL31 described elsewhere. The results shown in Fig. 4B were that the levels of UL31 accumulating in cells infected with the UL34 mutants were very much reduced relative to those of cells infected with wild type or R5604 (UL34-repaired virus). Because the DNA fragment used to repair the ΔUL34 mutant did not encompass the sequences encoding the promoter or coding sequences of the UL31 gene, it was of interest to pursue the interaction between UL34 and UL31 further.
The Time Course of Accumulation of UL31 Protein.
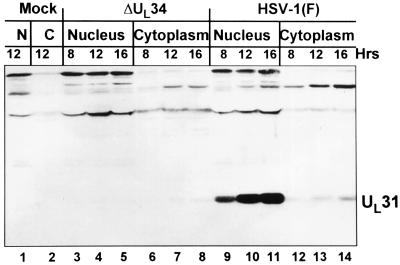
As shown in Fig. 5, we examined the time course of accumulation of UL31 protein in rabbit skin cells infected with wild-type and mutant viruses. In these experiments rabbit skin cells were infected with wild-type parent or ΔUL34 mutant, harvested at 8, 12, or 16 h after infection, and processed as described in Materials and Methods and the Fig. 5 legend. The results shown in Fig. 5 indicate that in wild-type virus-infected cells UL31 protein accumulated mainly in the nucleus but small amounts were also present in the cytoplasm. In mutant-infected cells trace amounts of UL31 were present in the cytoplasm and none were detected in the nucleus.
Figure 5.
Immunoblot of electrophoretically separated cell fractionates of rabbit skin cells infected with R5601(ΔUL34) or wild-type HSV-1 (F). Rabbit skin cells (1 × 106) were exposed to R5601 or HSV-1(F) at 5 pfu per cell and incubated at 37°C. Cells were harvested at time points of 8, 12, and 16 h after infection, and cell fractionation was done as described in Materials and Methods. The cytoplasmic and nuclear fractions obtained were resuspended in 1× SDS disruption buffer and subjected to electrophoresis on a denatured polyacrylamide gel (12%). After transfer to a nitrocellulose membrane, the separated infected cell proteins were reacted with the polyclonal antibody to UL31. Lanes 1 and 2, nuclear and cytoplasmic fractions of mock-infected cells. Lanes 3–8, nuclear fractions (lanes 3–5) and cytoplasmic fractions (lanes 6–8) of cells infected with R5601 and harvested at 8 h (lanes 3 and 6), 12 h (lanes 4 and 7), and 16 h (lanes 5 and 8) after infection. Lanes 9–14, nuclear (lanes 9–11) and cytoplasmic (lanes 12–14) fractions of cells infected with HSV-1(F) virus, in the same arrangement as that in lanes 3–8.
UL31 Protein Is Degraded in the Absence of UL34.
The failure of UL31 to accumulate in cells infected with ΔUL34 mutant raised the question of whether UL34 protein is required for the synthesis of the protein or its stability. To answer this question two series of experiments were done.
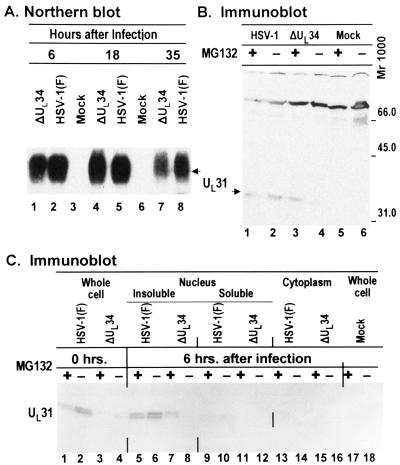
In the first, we examined the presence of UL31 ORF-specific RNA in rabbit skin cells mock-infected or infected with ΔUL34 or wild-type parent virus. In the experiment shown in Fig. 6A, equal amounts of total RNA extracted from cells harvested at 6, 18, or 35 h after infection were electrophoretically separated in denaturing gels, transferred to a nitrocellulose membrane, and hybridized to a 32P-labeled probe derived from the coding domain of UL31. The transcript of UL31, 1.3 kb is size, accumulated in both wild-type and mutant virus-infected cells at all three time points tested.
Figure 6.
(A) Autoradiographic image of Northern blot of formaldehyde agarose gel-separated RNAs isolated from rabbit skin cells infected with ΔUL34 virus or wild-type HSV-1(F) virus. Infected cells were harvested at time points indicated, and total RNAs were isolated. An equal amount (4 μg) of total RNAs was loaded onto each lane. The UL31 mRNA was probed with a 32P-labeled, 520-bp BfaI DNA fragment within the UL31 coding sequences. (B) Immunoblot of electrophoretically separated cell extracts of rabbit skin cells infected with R5601(ΔUL34) or HSV-1(F). Replicate 25 cm2 cultures of rabbit skin cells were exposed to 10 pfu of ΔUL34 or HSV-1(F) per cell and incubated at 37°C. Proteasome inhibitor MG132 (final concentration, 5 μM) was added 6 h after infection. The cells were harvested at 14 h after infection, solubilized, electrophoretically separated on denaturing poilyacrylamide, transferred to a nitrocellulose sheet, and reacted with the UL31 polyclonal antibody. (C) Replicate cultures of rabbit skin cells were infected as above but were either mock-treated or treated to MG132 at time of infection (0 h) or at 6 h after infection. The cells were harvested, processed as described in Materials and Methods, and reacted with the polyclonal antibody against UL31.
In the second series of experiments replicate cultures of rabbit skin cells were infected with wild-type or mutant virus and incubated in medium containing proteosome inhibitor MG132 6 h after infection. The cells were harvested at 14 h after infection and processed as described in Materials and Methods and the Fig. 6B legend. In this series of experiments the anti-UL31 polyclonal antibody also reacted with a cellular protein with a Mr of approximately 70,000. The reactivity of this protein with the UL31 antibody served as a useful lane loading control. The results were as follows: UL31 protein was readily detected in lysates of cells infected with wild-type virus (Fig. 6B, lanes 1 and 2). The amounts of UL31 protein present in lysates of cells treated with MG132 proteasome inhibitor were equivalent to or slightly less than those accumulating in untreated infected cells (Fig. 6B, compare lanes 1 and 2). In contrast, in cells infected with ΔUL34-infected cells, UL31 protein accumulated in larger amounts in cells exposed to MG132 as compared with untreated infected cells (Fig. 6B, lanes 3 and 4). The results indicate that in the absence of UL34 the UL31 protein is made but degraded.
In the third series of experiments replicate cultures of rabbit skin cells were mock-treated or treated with MG132 at the time of infection (0 h) or at 6 h after infection with either ΔUL34 or wild-type parent virus. The cultures treated at 6 h after infection were fractionated into nuclear soluble, insoluble, and cytoplasmic fractions on the basis of earlier studies showing that UL31 associates largely with an insoluble fraction. The results were as follows: (i) Wild-type virus-infected cells exposed to MG132 at the time of infection or at 6 h after infection produced slightly decreased levels of UL31 protein (Fig. 6C, lanes 1 and 5) whereas cells either untreated or exposed to MG132 at the time of infection with ΔUL34 accumulated very little UL31 protein (Fig. 6C, lanes 3 and 4). (ii) In cells infected with ΔUL34 and treated with MG132 at 6 h after infection, UL31 protein accumulated predominantly in the insoluble nuclear fraction (Fig. 6C, lane 7), that is, in the same compartment as the UL31 protein made in wild-type virus-infected cells (Fig. 6C, lanes 5 and 6). (iii) Again, no appreciable UL31 protein accumulated in untreated ΔUL34 infected cells (Fig. 6C, lanes 4, 8, 12, and 16). The results of this experiment suggest that in the absence of UL34 a key event associated with proteasomal function must take place some time between 0 and 6 h after infection to enable the accumulation of UL31 protein. This event is not required in the presence of UL34 protein. In cells infected with ΔUL34 and exposed to MG132, the UL31 protein accumulated in the insoluble fraction of the nucleus.
Discussion
The appropriate test of the capacity of a HSV mutant to be transported retrograde from a peripheral site to the nucleus of a dorsal root neuron is in a suitable experimental animal system. Although transport requires that the virus infect nerve endings at the peripheral site but not actually viral replication at that site, in practice, replication at the peripheral site of inoculation insures that the dorsal root ganglia is seeded with sufficient virus to be easily detected and quantified. To determine the role of ΔUL34 in retrograde transport of virus in neurons of dorsal root ganglia, we made several deletion mutants. Our results summarized in the initial sections of this report indicated that the mutants of interest, ΔUL34 and UL34Δ(3–119), replicated in cell type-dependent manner but poorly nevertheless.
HSV-1 proteins studied in detail appear to perform several functions. The reported functions of UL34 protein in addition to those involving possible association with dynein motors reported here and elsewhere include failure of viral DNA packaging and failure of capsid envelopment but no interference with capsid assembly (8). Curiously, the phenotype of UL34 deletion mutants appears to be strikingly similar to that of UL31 protein although the two proteins appear to have a superficially different distribution in the wild-type virus-infected cell (8, 12). In the course of investigations reported here on the interaction of UL34 and UL31 proteins, we discovered that UL31 was absent or grossly diminished in cells infected with the ΔUL34 mutant. In this report we show that UL31-specific RNA accumulated in infected cells and that UL31 protein was readily detected in cells infected with ΔUL34 mutant and treated with proteasome inhibitor MG132. The conclusion is that UL31 protein is made and degraded in the absence of UL34 protein. This is evidence that a viral protein depends on another protein for its stability in infected cells. It raises obvious questions as to why it is degraded, whether its accumulation is inimical to the infected cell, and whether the functions of UL34 with respect to stabilization of UL31 protein could be differentiated from other putative functions of the UL34 protein. We also noted that a key event possibly associated with viral functions must take place between 0 and 6 h after infection for UL31 to accumulate in ΔUL34-infected cells but that this event is not required in wild-type virus-infected cells.
Another, potentially more critical, issue is whether the growth defect of ΔUL34 mutant reflects the absence of UL31 or whether among the multifaceted functions of these proteins the key function in viral morphogenesis requires the cooperation of both proteins. Preliminary studies support the latter hypothesis inasmuch as ΔUL34 mutant did not replicate better in cells expressing the UL31 protein reported by Chang et al. (12) than in cells lacking HSV sequences (data not shown). The significance of the studies reported here is that mutants in specific gene functions may have secondary effects on the accumulation of other gene products. Marker rescue tests, the standard method for attribution of a specific phenotype to a mutated gene, may not be in all instances sufficient to ascribe the deduced function to the gene.
Acknowledgments
We thank George Church for plasmid pKO3 and Escherichia coli strain EMG2, Brian Horsburgh and Frank Tufaro for pKO5.1 and HSV-BAC, Shu-Fen Chou for electron microscopy, and Sunil Advani for helpful discussion. These studies were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766), the United States Public Health Service.
Abbreviations
- HSV
herpes simplex virus
- BAC
bacterial artificial chromosome
- Cm
chloramphenicol
- Zeo
Zeocine
- Suc
sucrose
- pfu
plaque-forming unit
References
- 1.Roizman B, Sears A E. In: Fields' Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Lippincott–Raven; 1996. pp. 2231–2295. [Google Scholar]
- 2.Kristensson K, Lycke E, Roytta M, Svennerholm B, Vahlne A. J Gen Virol. 1986;67:2023–2028. doi: 10.1099/0022-1317-67-9-2023. [DOI] [PubMed] [Google Scholar]
- 3.Whitley R J. In: Fields' Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Lippincott–Raven; 1996. pp. 2297–2342. [Google Scholar]
- 4.Batterson W, Furlong D, Roizman B. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tognon M, Furlong D, Conley A J, Roizman B. J Virol. 1981;40:870–880. doi: 10.1128/jvi.40.3.870-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sodeik B, Ebersold M W, Helenius R. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye G J, Vaughan K T, Vallee R B, Roizman B. J Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roller R J, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 10.Kennelly P J, Krebs E G. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 11.Chang Y E, Roizman B. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y E, Van Sant C, Krug P W, Sears A E, Roizman B. J Virol. 1997;71:8307–8315. doi: 10.1128/jvi.71.11.8307-8315.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purves F C, Spector D, Roizman B. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purves F C, Spector D, Roizman B. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 16.Baines J D, Roizman B. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward P L, Ogle W O, Roizman B. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horsburgh B C, Hubinette M M, Qiang D, MacDonald M L E, Tufaro F. Gene Ther. 1999;6:922–930. doi: 10.1038/sj.gt.3300887. [DOI] [PubMed] [Google Scholar]
- 19.Horsburgh B C, Hubinette M M, Tufaro F. Methods Enzymol. 1999;306:337–352. doi: 10.1016/s0076-6879(99)06022-x. [DOI] [PubMed] [Google Scholar]
- 20.Link A J, Phillips D, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]