Abstract
Background
Multi-drug resistant (MDR) bacteria have become a major concern in hospitals worldwide and urgently require the development of new antibacterial molecules. Peptide deformylase is an intracellular target now well-recognized for the design of new antibiotics. The bacterial susceptibility to such a cytoplasmic target primarily depends on the capacity of the compound to reach and accumulate in the cytosol.
Methodology/Principal Findings
To determine the respective involvement of penetration (influx) and pumping out (efflux) mechanisms to peptide deformylase inhibitors (PDF-I) activity, the potency of various series was determined using various genetic contexts (efflux overproducers or efflux-deleted strains) and membrane permeabilizers. Depending on the structure of the tested molecules, two behaviors could be observed: (i) for actinonin the first PDF-I characterized, the AcrAB efflux system was the main parameter involved in the bacterial susceptibility, and (ii), for the lastest PDF-Is such as the derivatives of 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide, the penetration through the membrane was a important limiting step.
Conclusions/Significance
Our results clearly show that the bacterial membrane plays a key role in modulating the antibacterial activity of PDF-Is. The bacterial susceptibility for these new antibacterial molecules can be improved by two unrelated ways in MDR strains: by collapsing the Acr efflux activity or by increasing the uptake rate through the bacterial membrane. The efficiency of the second method is associated with the nature of the compound.
Introduction
Multi-drug resistance (MDR) in Gram-negative bacteria is frequently reported in clinical isolates [1], [2]. This strongly limits therapeutic options and is a major cause of mortality in hospital acquired infections [1]–[4]. MDR is prevalent in important Gram-negative clinical pathogens such as Escherichia coli, Salmonella spp., Klebsiella spp., Enterobacter spp., and Pseudomonas spp. In these major pathogens, three major bacterial strategies are involved in the development of drug resistance: 1) the membrane barrier (acting to limit the required intracellular dose of an antibiotic), 2) the enzymatic barrier (producing detoxifying enzymes that degrade or modify the antibiotic), 3) the target protection barrier (mutation or expression of a molecule that impairs target recognition and thus antimicrobial activity) [5]. These Gram-negative bacteria, responsible for a large portion of antibiotic-resistant bacterial diseases, display a complex cell envelope comprising an outer membrane and an inner membrane delimiting the periplasm [6]. The outer membrane contains various protein channels which are involved in the transport of various compounds including several classes of antibiotics [6], [7]. Bacterial adaptation to reduce the outer membrane permeability is an increasing problem worldwide, which contributes, along with efflux systems, to the emergence and dissemination of antibiotic resistance. Consequently, it is important to explore the activity of existing and new antibiotic compounds by using different bacterial strains harbouring various resistance backgrounds and in the presence of diverse chemicals recently described as potent inhibitors of resistance mechanism or facilitator of antibiotic activity [8]–[10].
Face to this continuous emerging threat, several novel bacterial targets have been described as an alternate therapeutic solution to the emergence and dissemination of MDR bacterial isolates [11], [12]. Peptide deformylase (PDF) is involved in the cleavage of the N-formyl group of nascent polypeptide. This process is essential to the growth of bacteria [13] and represents a novel attractive target for new antibacterial molecules [14]–[16]. Peptide deformylase inhibitors (PDF-Is) have been characterized and described to be active in various bacterial pathogens. Most of them such as actinonin, the main characterized inhibitor, are pseudopeptidic molecule [17]. Recently we described a new series of heterocyclic compounds, with an indol scaffold, that inhibit efficiently bacterial PDF in the nM range [18], [19]. Two main mechanisms mediating resistance to peptide deformylase inhibitors in bacteria have been previously described. The first is amino acid substitutions within the target protein (Def) , and the second is the formylation bypass which results from mutational loss of methionyl tRNA formyltransferase (Fmt) or of the folD gene [20]–[22]. However a discrepancy of activity was observed with the efflux system that seriously compromised the PDF-Is action in some efflux producing strains [23], [24]. The role of AcrB and TolC component of efflux pump has been reported in the susceptibility of E. coli and Haemophilus influenzae [18], [23].
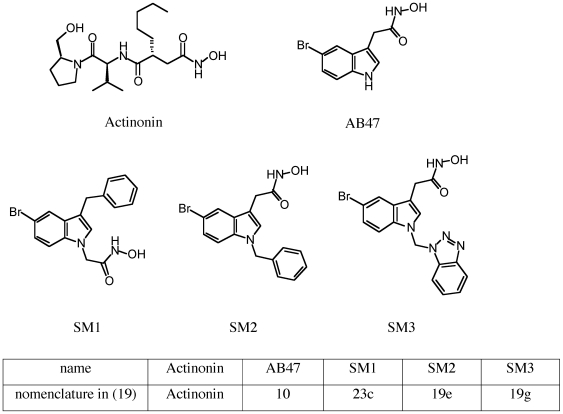
The MDR phenotype is often linked to with a general alteration of membrane properties including a decrease in membrane permeability associated with an overexpression of efflux pumps [5]. We decided to investigate the involvement of the membrane barrier (the “in and the out” transport) in the activity of some indolic PDF-Is structurally unrelated to actinonin group (Figure 1) and recently produced [18], [19] in various strains and clinical MDR isolates.
Figure 1. Structures of PDF-Is used.
Results
Activity of new PDF-Is and membrane permeability
Actinonin was used as reference molecule to characterize the antibacterial activities of the newly produced PDF-Is for which an in vitro activity has been previously reported [19]. In Table 1 were presented the results obtained on isogenic strains in the absence or in the presence of various sub-inhibitory concentrations of the cyclic peptide antibiotic polymyxin B (Pol B) or its derivative the polymyxin nonapeptide (PMBN) known to increase membrane permeability [25], [26].
Table 1. Determination of antibacterial activity of various PDF-Is on E. coli strains.
| Strains | Actinonin MIC (µg/ml) | AB47 MIC (µg/ml) | SM1 MIC (µg/ml) | SM2 MIC (µg/ml) | Norfloxacin MIC (µg/ml) | Chloramphenicol MIC (µg/ml) | ||||||||||||
| Permeaa | 0 | PMBNb | Pol Bc | 0 | PMBNa | Pol Bb | 0 | PMBNa | Pol Bb | 0 | PMBNa | Pol Bb | 0 | PMBNa | Pol Bb | 0 | PMBNa | Pol Bb |
| AG100 | 128 | 2 (4) | 16 (128) | 512 | 8 (32) | 64 (128) | >512 | 4 (8) | >128 (>128) | >512 | 8 (16) | 128 (>128) | 0.25 | 0.125 | 0.125 | 8 | 2 | 4 |
| AG102 | 128 | 16 | 128 | 512 | 32 | 128 | >512 | 16 | >128 | >512 | 16 | 128 | 1 | 0.5 | 1 | 16 | 16 | 16 |
| AG100A | 1 | 0.125 (0.25) | 0.25 (0.25) | 64 | 16 (16) | 32 (32) | 128 | 4 (4) | 16 (32) | 64 | 8 (8) | 32 (32) | 0.06 | 0.06 | 0.03 | 1 | 1 | 0.5 |
| AG100Atet | 512 | 16 | >128 | 256 | 16 | 16 | >512 | 16 | >128 | >512 | 8 | >128 | 1 | 0.5 | 1 | 64 | 32 | 64 |
Values are means of four independent assays.
permeabilizer used.
PMBN was used at 1/5 MIC, in bracket the results with 1/10 MIC are indicated.
Pol B was used at 1/5 MIC, in bracket the results with 1/10 MIC are indicated.
In the absence of membrane permeabilizers, we observed a 128 fold decrease of actinonin MIC in the acrAB deleted strain compared to the parental ones. This suggests that AcrB pump is directly involved in the resistance observed in the parental strain towards this molecule. Regarding the other PDF-Is, we did not observed a susceptibility level similar to that obtained with actinonin. These results indicate that SM1, SM2, and AB47 compounds are not recognized as specific substrate for AcrAB efflux transporter or that another rate-limiting step is involved. Concerning SM3, whatever the tested strains or the conditions used, no antibacterial activity was detected (data not shown).
The effect of membrane permeabilizer, e.g. Pol B and PMBN, was assayed on the PDF-Is activities. The MICs for Pol B and PMBN were determined for each bacterial strain. From the respective MICs, a sub-inhibitory amount (MIC/5 and MIC/10) was added in the presence of each PDF-I. For actinonin, the presence of PMBN induced a serious decrease of MIC to the susceptible level whatever the strain tested. In the acrAB deleted strain, a small increase of susceptibility was noted (MIC of 0.25 µg/ml). Concerning the other molecules, the addition of PMBN, and Pol B at a lesser extent, induced a noticeable increase of susceptibility (Table 1). It is interesting to note that for SM1, SM2 and AB-47 whatever the strain background tested an important MIC decrease was induced in the presence of PMBN. Concerning SM3, no increase in the susceptibility was observed in the acrAB deleted strain in the absence or in presence of PMBN (data not shown). In addition, in the same conditions, presence of Pol B or PMBN, only a very limited effect was noted on the activity of usual antibiotics such as norfloxacin and chloramphenicol (Table 1).
The activity of the different PDF-Is was tested on other Gram-negative bacteria involved in human infectious diseases such as Pseudomonas aeruginosa, Enterobacter aerogenes and Klebsiella pneumoniae (Table 2). SM1 and actinonin exhibited no antibacterial activity on the P. aeruginosa strains including reference strain PA01 and clinical isolate 124. The addition of PMBN during the incubation noticeably increased the activity of actinonin, AB-47, SM1 (Table 2). A less efficient effect was obtained with SM2 (data not shown). It is interesting to note that the level of resistance for the P. aeruginosa clinical isolate (124 in table 2) which is a MDR strain with several mechanisms including efflux pump overexpression, was more significant for usual antibiotics (norfloxacin, tetracycline, etc).
Table 2. Activity of PDF-Is on MDR Gram-negative isolates.
| P. aeruginosa | E. aerogenes | K. pneumoniae | |||||||||
| PA01 | 124 | ATCC 13048 | EA5 | EA27 | EA289 | EA298 | EA294 | ATCC 11296 | KP55 | KP63 | |
| Actinonin | 128 | 128 | >128 | 128 | 128 | >128 | 32 | 128 | 128 | 128 | 128/64 |
| Actinonin + PMBN a | 4 | 16 | 8 | 64 | 64 | 32 | 0.5 | 2 | 32 | 32 | 32 |
| SM1 | 128 | 128 | >128 | >128 | >128 | >128 | 64 | 128 | >128 | >128 | 128 |
| SM1 + PMBN a | 2 | 8 | 8 | 32 | 32 | 32 | 8 | 16 | 64 | 64 | 128–64 |
| AB47 | 128 | >128 | >128 | >128 | >128 | 256 | 128 | 128 | 128 | 128 | 128 |
| AB47 + PMBN a | 4 | 8 | 32 | 128 | 128 | 128 | 16 | 32 | 64 | 64 | 64–32 |
| CM | 256 | 256 | 8 | >128 | >128 | >128 | 32 (16) b | 32 | 16 (4) b | 256 | >512 |
| NFX | 2 | 64 | 0,5 | 128 | 128 | 128 | 16 (32) b | 32 | 1 (0.5) b | 16 | 8 |
| CAZ | 8 | 32 | 1 | >512 | >512 | >512 | >512 | >512 | 1 | >512 | >512 |
| ERY | 1024 | 1024 | 512 | 512 | 512 | 512 | 32 | 128 | 128 | 512 | 512 |
| TC | 16 | 64 | 4 | 4 | 16 | 8 | 0.5(0.25) b | 1 | 2 (2) b | >128 | 8 |
Values are means of four independent assays listed in µg/ml.
CM, chloramphenicol; NFX, norfloxacin; CAZ, ceftazidime; ERY, erythromycin; TC, tetracycline.
PMBN was used at 1/5 MIC.
in bracket the results obtained with 1/5 PMBN are indicated.
We observed similar profiles with other Gram-negative bacterial pathogens and the activity of actinonin was noticeably increased in the presence of membrane permeabilizer. The tested clinical MDR isolates (E. aerogenes EA5 and EA27; K. pneumoniae KP55 and KP63) exhibited a lower susceptibity towards other PDF-Is assayed under the same conditions. Moreover, in the derivative strains of MDR E. aerogenes clinical isolate EA27, we observed a significant role of the efflux pump: when the tolC or acrAB genes were knocked out, the presence of PMBN induced a noticeable increase of actinonin and SM1 activity, e.g. compare EA289 and EA298 strains in Table 2. Under these conditions, it is worthy of note that the tolC deletion was more efficient compare to acrA for actinonin (MICs of 0.5 and 2 respectively). A similar level was reached in the two acrA and tolC deleted strains for SM1 and AB47 (8 and 16, 16 and 32 respectively). It has been previously demonstrated that at least two distinct efflux pumps are active, AcrAB-TolC and another unidentified pump, in the EA27 clinical isolate and its EA289 derivative strain [27]. In these MDR isolates, the results indicated that the AcrAB-TolC could be the main transporter involved for actinonin efflux. By contrast, regarding SM1 and AB-47, additional pump may also modulate the activity of these molecules since in EA298 with PMBN the MICs was 8 and 16 compare to actinonin MIC (0.5). It is interesting to note that, in general,the new antibacterial molecules tested here seemed to be more efficient against clinical isolates compare to several hospital-used antibiotics (e.g., erythromycin, ceftazidime).
Effect of efflux pump inhibitors on PDF-Is activity
The impact of efflux pumps in antibiotic susceptibility has been largely demonstrated [28]–[30]. The results shown in Table 1 suggested an involvement of AcrAB pump in the actinonin efflux and at lesser extent for the other tested PDF-Is. In this context, to get more insight in the process we determine the effect of a well-described efflux pump inhibitor, PAβN which is able to block the activity of various efflux pumps in addition to AcrAB-TolC [8], [9]. In Table 3, the results indicated that the PAβN induced only a limited increase of PDF-Is activities compared to PMBN. Moreover, when the two molecules, PMBN and PAβN, were conjointly added no cumulative effect was observed (data not shown). Since AcrAB was involved in modulating actinonin activity, these results suggest that PAβN was not an effective competitive substrate for efflux of PDF-Is. In other words, the affinity constant of efflux pump for PDF-Is may be stronger compare to PAβN.
Table 3. Effect of efflux pump inhibitors and membrane permeabilizers.
| Bacteria | Actinonin | AB47 | ||||
| E. coli | 0 | PMBNa | PAβN | 0 | PMBNa | PAβN |
| AG100 | 128 | 2 | 32 | 512 | 8 | 128 |
| AG102 | 128 | 16 | 64 | 512 | 32 | 128 |
| AG100A | 1 | 0.125 | 0.5 | 64 | 16 | 16 |
| AG100Atet | 512 | 8 | 32 | 256 | 16 | 128 |
| P. aeruginosa | ||||||
| 124 | 128 | 16 | 32 | >128 | 8 | 128 |
Values are means of four independent assays.
PMBN was used at 1/5 MIC.
PAβN was used at 20 µg/ml.
Same results were obtained when the assays were performed on P. aeruginosa strain (Table 3). We have similarly determined the effect of other inhibitors such as reserpine, verapamil and omeprazol. No significant change in the MICs of PDF-Is was obtained in the various E. coli strains (data not shown).
Activity of PDF-Is on lipopolysaccharide (LPS) deep rough mutants, effect of detergents and chelators
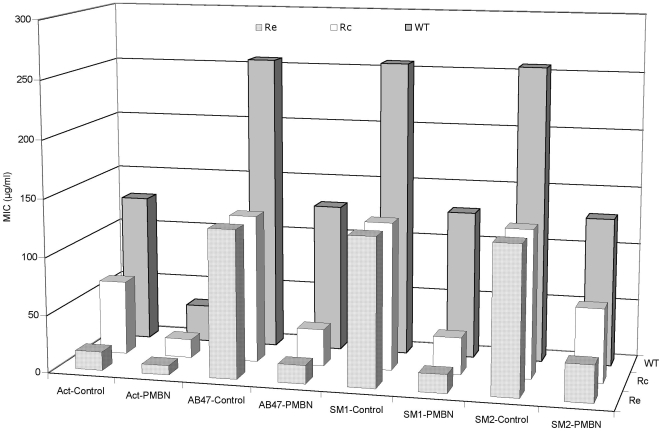
In Gram negative bacteria, the LPS constitutes the outer leaflet of outer membrane and may impair the penetration of antibacterial agents [6]. Since we have observed that the addition of membrane permeabilizer such as Pol B or PMBN, are capable to induce a serious decrease of MICs, we tested a series of Salmonella typhimurium LT2 mutants producing truncated LPS [31] to assess a putative role of LPS structure in the PDF-Is activity. These strains have been previously used as standard strains to assess the role of LPS on the level of diffusion through the outer membrane [31]. As shown in Figure 2, the actinonin activity was increased by 8 fold in LPS truncated mutants while only 2.5 fold activity increased was observed with other compounds. By contrast, in the presence of PMBN, we observed an effective restoration of susceptibility and a quite similar MIC was obtained for actinonin, AB47 and SM1. This suggests that the truncated LPS increased the bacterial susceptibility to actinonin probably by facilitating the diffusion through the LPS layer of outer membrane. Regarding the activity of the other molecules, the presence of LPS barrier is not the limiting step since PMBN addition was required to reach the same level of antibacterial activity.
Figure 2. Activity of PDF-Is on Salmonella wild-type strain SL696 and its LPS-deficient mutants.
Various isogenic strains, WT (intact LPS), Rc (truncated LPS: lipidA-KDO-hep-hep-Glc) and Re (truncated LPS: lipidA-KDO) previously described [31] were used. Act-Control, actinonin alone; Act-PMBN, actinonin + PMBN; AB47-Control, AB47 alone; AB47-PMBN, AB47 + PMBN; SM1-Control, SM1 alone; SM1-PMBN, SM1 + PMBN; SM2-Control, SM2 alone; SM2-PMBN, SM2 + PMBN. PMBN was used at 1/5 MIC. MIC values are in µg/ml.
An alternate way to bypass the membrane barrier is to use chaotropic agents or detergents [6]. The assays with actinonin and SM1 were carried out on E. coli AG100 in the presence of a chelator agent such as EDTA or in the presence of various detergents including SDS, DOC or Tx-100. Interestingly, no significant effect was observed except in the case of actinonin and EDTA for which the activity is noticeably increased: a decrease of 64 fold was obtained for actinonin MIC (Table 4). These results fitted well with the results of Figure 2 that indicated a role for LPS in the actinonin activity.
Table 4. Effect of EDTA and detergents on PDF-Is activity.
| E. coli strain | Actinonin | SM1 | ||||||||
| 0 | SDS | Tx-100 | DOC | EDTA | 0 | SDS | Tx-100 | DOC | EDTA | |
| AG100 | 128 | 128 | 128 | 64 | 2 | >128 | >128 | >128 | >128 | >128 |
Values are means of four independent assays.
The different compounds were used at concentration for which no direct antibacterial effect was observed. EDTA was used at 1 mM ; SDS at 100 µg/ml; Tx-100 at 1000 µg/ml; DOC at 1000 µg/ml.
Discussion
The worldwide dissemination of « pandrug » and « multidrug» resistant pathogens have severely compromised the efficacy of our antibiotics and dramatically increased the occurence of therapeutic failure [3], [4], [32]. To efficiently combat multi-resistant pathogens, it is urgently require to define new targets and to produce novel classes of antibiotics. Several new potent antibacterial molecules have been recently tested and among them, the PDF-I group presents attractive properties including a novel and specific bacterial target and mode of action [13], [33].
In this antibacterial group, new derivatives of 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide have been recently described [18], [19]. It is important to define the activity of these compounds taking into account the bacterial adaptation to antibacterial agents. The special architecture of Gram-negative envelope screens the enter of molecules and strongly control the diffusion of toxic molecules including antibiotics, detergents, etc [6], [7], [31]. The outer membrane comprising a lipid bilayer that is impermeable to large and charged molecules, is the first barrier against toxic compounds [5], [6]. Moreover, efflux mechanisms also control the intracellular concentration of compounds [5], [30]. Consequently, the bacterial susceptibility to PDF-Is is assayed in various genetic and culture conditions modifying the membrane permeability.
When the molecules are assayed on isogenic strains expressing various level of AcrAB efflux pump, it is observed that actinonin is a substrate for AcrB efflux transporter: the acrAB deleted strain exhibits a noticeable susceptibility to actinonin in contrast to other PDF-Is. However, actinonin is also recognized by other RND efflux system including AcrD, AcrF, etc. These pumps are overexpressed in the tetracycline induced acrAB deleted strain [34] and the MIC for actinonin is strongly increased. Interestingly, the efflux pump inhibitor used, PAβN, which is able to restore susceptibility to several usual antibiotic family in various enterobacteriaceae [8], [9], has no strong effect on the level of PDF-Is susceptibility. The MICs are only reduced in the strain overproducing efflux pumps different to AcrAB suggesting that these efflux transporters are involved, partially, in the PDF-Is resistance. These data suggest that actinonin and PAβN do not share the same affinity sites in the efflux pump cavities as previously mentioned for certain pump substrates [35]–[37]. Regarding the respective susceptibility of the various strains which produce different levels and types of documented efflux pumps [34], [35], the tested efflux mechanisms seems to be not the main and sole process involved in the lack of activity of PDF-Is, with the exception of the involvement of AcrAB in actinonin activity. The difference in the chemical structure of the various molecules (Figure 1) may support these discrepancies in the recognition and transport by efflux pumps observed here. It is interesting to mention that actinonin seems to be an effective substrate for AcrAB-TolC compare to SM1, SM2 and AB47. These results obtained with the tolC deleted MDR isolate (EA298) confirm the involvement of a TolC-dependent channel which has been previously noted [18], associated with the expression of the AcrB family (e.g. AcrD, AcrF, etc) in the bacterial susceptibility to actinonin.
Influx or penetration represents the other aspect of membrane role in controlling molecule uptake [5]. Recent studies have evidenced that PMBN, a cationic peptide that perturbs the outer membrane, is able to increase the rifampin susceptibility in resistant strains [38]. Studies of PMBN mode of action against Gram-negative bacterial cells have shown that bacterial membranes are the primary target : PMBN exhibits the ability of polymyxins to disrupt or disorganise the cell envelope but fails to kill the cells at low concentrations [25], [38]. The PMBN treatment shows an increase in the permeability of the E. coli membrane. At sub-inhibitory concentration, PMBN promotes an increase of PDF-Is susceptibility whatever the compound (actinonin, AB47, SM1, SM2) or the bacteria tested (E. coli, P. aeruginosa, etc). This improvement of PDF-Is activity is also observed when the bacteria overproduce efflux pumps (AcrB or other pumps) demonstrating that the influx/diffusion through the membrane is a limiting step for the AB47, SM1 and SM2 antibacterial activity. It is worthy of note that AB47, SM1 and SM2 exhibited a same core structure with aromatic rings, a structure different from that of actinonin (Figure 1). In addition, we observed a noticeable decrease of MICs for actinonin, SM1 and SM2, and at less extent with AB47 in the acrAB deleted strain treated by PMBN. The comparison with usual antibiotics tested under the same conditions indicates that penetration is the key step for AB47, SM1 and SM2. A similar result was obtained with other Gram-negative bacteria including P. aeruginosa, K. pneumoniae and E. aerogenes.
The addition of sub-inhibitory concentration of PMBN facilitates the penetration of these molecules and partially counterbalances the activity of efflux pumps. The use of polymyxins, alone or in combination with other antibiotic classes has been recently questionated due to the increase of infectious disease caused by MDR pathogens [11], [26], [39]. In this study, it is important to mention that low concentration of Pol B and PMBN (1/5 and 1/10 MICs) facilitates the activity of PDF-Is. This facilitator effect indicates that these new antibacterial agents probably not used the porin to enter the cell as reported for β-lactams and fluoroquinolones [6], [7], but a diffusion pathway through the lipid bilayer. The use of several isogenic strains previously selected for membrane permeability studies [31] indicate that LPS truncated mutants are more sensitive to actinonin than the wild type parental strain. This result fits well with the increase of actinonin susceptibility observed in the presence of EDTA, a potent cationic chelator which affects the LPS organization [6], [38]. Thus, the diffusion through LPS layer, in addition to efflux pump is important for actinonin activity. In contrast, for the other PDF-Is which exhibit different chemical structure, the modification of LPS (via mutation, or EDTA addition) did not improve the activity.
To conclude, two processes are able to modulate the antibacterial activity of the new molecules tested and their respective involvement seems to be related to their structure. The level of actinonin susceptibility is controlled mainly by the efflux pump expression, the AcrAB-TolC system playing a central role in pumping the molecule out of the bacterial cell. The LPS may be partially involved in the uptake pathway of actinonin through the outer membrane. In contrast, the other tested molecules are more dependent on the permeation rate and PMBN appears as an efficient adjuvant to ensure a correct internal concentration. Concerning the molecules tested here in the presence of membrane permeabilizer, SM1 exhibits a level of activity quite similar to that observed with actinonin.
It is interesting to mention that during this work, we have been able to enhance the antibacterial activity of these molecules by two independent ways, firstly by eliminating the Acr efflux transporters, secondly by increasing the uptake rate through bacterial membrane. In the case of actinonin, this indicates that we can balance the activity of efflux pump by increasing the penetration rate. In the case of other compounds, we can bypass the limited diffusion by increasing the membrane permeability.
Materials and Methods
Bacterial Strains
The various strains are presented in Table 5. Briefly, Escherichia coli K-12 AG100 strain (argE3 thi-1 rpsL xyl mtl delta (gal-uvrB) supE44), AG100A, AG102 and AG100Atet derivatives have been characterized [34], [35]. Salmonella enterica Typhimurium strains, SL696 WT, SL1069 Rc, SL1102 Re which produce different types of lipopolysaccharide (LPS) have been characterized [31]. Pseudomonas aeruginosa strains are PA01(ATCC) and the clinical isolate 124 [43] Enterobacter aerogenes ATCC 13048 and clinical resistant isolates EA5, EA27, KP55 and KP63 have been previously described [40], [42].
Table 5. Bacterial strains used in this study.
| Bacterial strains | Major characteristics | Origin |
| Escherichia coli | ||
| AG100 | argE3 thi-1 rpsL xyl mtldelta (gal-uvrB) supE44 | 34 |
| AG100A | AG100 Δ acrAB:: Kanr | 34 |
| AG102 | AG100 overproducing AcrAB | 35 |
| AG100Atet | AG100Atet, AG100A selected on tetracycline | 34 |
| Enterobacter aerogenes | ||
| ATCC13048 | ATCC strain | 40 |
| EA5 | MDR clinical isolate | 40 |
| EA27 | MDR clinical isolate | 40,41 |
| EA289 | Kans derivative of EA27 | 41 |
| EA298 | EAEP289 tolC:: Kanr | 41 |
| EA294 | EAEP289 acrA::Kanr | 41 |
| Klebsiella pneumoniae | ||
| ATCC11296 | ATCC strain | 42 |
| KP55 | MDR clinical isolate | 42 |
| KP63 | MDR clinical isolate | 42 |
| Pseudomonas aeruginosa | ||
| PA01 | ATCC strain | 43 |
| 124 | MDR clinical isolate | 43 |
| Salmonella enterica Typhimurium | (LT2) | |
| SL696 | WT, metA22, trpB2, strAi20 | 31 |
| SL1069 | SL696 Re derivative | 31 |
| SL1102 | SL696 Re derivative | 31 |
Chemicals
PDF-Is used were : Actinonin (Sigma), AB-47, SM1, SM2 and SM3 were previously described and characterized [18], [19] (Figure 1). Phenylalanine arginine β-naphthylamide (PAβN), reserpine, verapamil were assayed as efflux pump inhibitors [34], [44] and norfloxacin, tetracycline and chloramphenicol were used as reference antibiotics. Polymyxin B (Pol B) was used as membrane permeabilizer in addition to polymyxin B nonapeptide (PMBN), a cationic peptide that perturbs the outer membrane bilayer without the killing action of unmodified polymyxin [25], [26]. EDTA was used to produce divalent cation-poor growth conditions [38]. SDS, DOC and Triton X-100 were used as detergents.
Antibiotic susceptibility tests
Bacteria were grown in Mueller–Hinton (MH) broth at 37°C. Susceptibilities to antibiotic compounds (polymyxin B, PMBN, norfloxacin, ceftazidime, tetracycline and chloramphenicol) and efflux inhibitors (PAβN, reserpine, verapamil) were determined by broth dilution method, as previously described [40], [44] Minimal inhibitory concentrations (MICs) were determined with an inoculum of 106 CFU in 1 mL of MH broth containing two-fold serial dilutions of each antibiotic. Isolates were classified as susceptible, intermediately susceptible or resistant to the antibiotics tested according to the Antibiogram Committee of the French Society for Microbiology [45].
Effect of membrane permeabilizers and efflux pump inhibitors
The efflux pump inhibitor (EPI) PAβN was used as previously reported: MICs of each antibiotic were determined in the presence of PAβN at a concentration which has no effect on bacterial cells [44].To evaluate a possible role of efflux or membrane barrier in the PDF-Is activity, we developed an in vitro assay using different conditions: PDF-Is alone, PDF-Is + polymyxin (at 1/10 or 1/5 MIC), PDF-Is + EPI (PAβN or other EPIs), and PDF-Is + polymyxin + EPI. Norfloxacin and chloramphenicol belonging to unrelated structural antibiotic groups were used as internal standard. The results were scored after 18 h at 37°C and were expressed as µg/ml (MIC).
Acknowledgments
We thank E. Goemaere for participation in determining certain antibiotic susceptibilities.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Centre National de la Recherche Scientifique (CNRS, France), a grant ANR-06-MIME-010 from Agence Nationale de la Recherche (ANR, France), the Service de Sante des Armees and the Universite de la Mediterranee. L. Mamelli was supported by a postdoctoral grant from the ANR-MIME, S. Petit was supported by a PhD fellowship from the Research Agency. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chopra I, Schofield C, Everett M, O'Neill A, Miller K, et al. Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect Dis. 2008;8:133–139. doi: 10.1016/S1473-3099(08)70018-5. [DOI] [PubMed] [Google Scholar]
- 2.Blot S, Depuydt P, Vandewoude K, De Bacquer D. Measuring the impact of multidrug resistance in nosocomial infection. Curr Opin Infect Dis. 2007;20:391–396. doi: 10.1097/QCO.0b013e32818be6f7. [DOI] [PubMed] [Google Scholar]
- 3.Rice LB. Emerging issues in the management of infections caused by multidrug-resistant gram-negative bacteria. Cleve Clin J Med. 2007;74(Suppl 4):S12–20. doi: 10.3949/ccjm.74.suppl_4.s12. [DOI] [PubMed] [Google Scholar]
- 4.Paterson DL. Impact of antibiotic resistance in gram-negative bacilli on empirical and definitive antibiotic therapy. Clin Infect Dis. 2008;47(Suppl 1):S14–20. doi: 10.1086/590062. [DOI] [PubMed] [Google Scholar]
- 5.Davin-Régli A, Bolla JM, James CE, Lavigne JP, Chevalier J, et al. Membrane permeability and regulation of drug "influx and efflux" in enterobacterial pathogens. Curr Drug Targets. 2008;9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 8.Lomovskaya O, Bostian KA. Pratical applications and feasibility of efflux pump inhibitors in the clinic–a vision for applied use. Biochem Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Mahamoud A, Chevalier J, Alibert-Franco S, Kern WV, Pagès JM. Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J Antimicrob Chemother. 2007;59:1223–1229. doi: 10.1093/jac/dkl493. [DOI] [PubMed] [Google Scholar]
- 10.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Rafailidis PI. Re-emergence of colistin in today's world of multidrug-resistant organisms: personal perspectives. Expert Opin Investig Drugs. 2008;17:973–981. doi: 10.1517/13543784.17.7.973. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill AJ. New antibacterial agents for treating infections caused by multi-drug resistant Gram-negative bacteria. Expert Opin Investig Drugs. 2008;17:297–302. doi: 10.1517/13543784.17.3.297. [DOI] [PubMed] [Google Scholar]
- 13.Boularot A, Giglione C, Artaud I, Meinnel T. Structure-activity relationship analysis and therapeutic potential of peptide deformylase inhibitors. Curr Opin Investig Drugs. 2004;5:809–822. [PubMed] [Google Scholar]
- 14.Jain R, Chen D, White RJ, Patel DV, Yuan Z. Bacterial Peptide deformylase inhibitors: a new class of antibacterial agents. Curr Med Chem. 2005;12:1607–1621. doi: 10.2174/0929867054367194. [DOI] [PubMed] [Google Scholar]
- 15.Johnson KW, Lofland D, Moser HE. PDF inhibitors: an emerging class of antibacterial drugs. Curr Drug Targets Infect Disord. 2005;5:39–52. doi: 10.2174/1568005053174618. [DOI] [PubMed] [Google Scholar]
- 16.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen DZ, Patel DV, Hackbarth CJ, Wang W, Dreyer G, et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 18.Boularot A, Giglione C, Petit S, Duroc Y, Alves de Sousa R, et al. Discovery and refinement of a new structural class of potent peptide deformylase inhibitors. J Med Chem. 2007;50:10–20. doi: 10.1021/jm060910c. [DOI] [PubMed] [Google Scholar]
- 19.Petit S, Duroc Y, Larue V, Giglione C, Leon C, et al. Structure-activity relationship analysis of the peptide deformylase inhibitor 5-Bromo-1H-indole-3-acetohydroxamic Acid. Chem Med Chem. 2009;4:261–275. doi: 10.1002/cmdc.200800251. [DOI] [PubMed] [Google Scholar]
- 20.Dean CR, Narayan S, Richards J, Daigle DM, Esterow S, et al. Reduced susceptibility of Haemophilus influenzae to the peptide deformylase inhibitor LBM415 can result from target protein overexpression due to amplified chromosomal def gene copy number. Antimicrob Agents Chemother. 2007;51:1004–1010. doi: 10.1128/AAC.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolis PS, Hackbarth CJ, Young DC, Wang W, Chen D, et al. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duroc Y, Giglione C, Meinnel T. Mutations in three distinct loci cause resistance to peptide deformylase inhibitors in Bacillus subtilis. Antimicrob Agents Chemother. 2009;53:1673–1678. doi: 10.1128/AAC.01340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean CR, Narayan S, Daigle DM, Dzink-Fox JL, Puyang X, et al. Role of the AcrAB-TolC efflux pump in determining susceptibility of Haemophilus influenzae to the novel peptide deformylase inhibitor LBM415. Antimicrob Agents Chemother. 2005;49:3129–3135. doi: 10.1128/AAC.49.8.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritsche TR, Sader HS, Cleeland R, Jones RN. Comparative antimicrobial characterization of LBM415 (NVP PDF-713), a new peptide deformylase inhibitor of clinical importance. Antimicrob Agents Chemother. 2005;49:1468–1476. doi: 10.1128/AAC.49.4.1468-1476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahalan AZ, Dixon RA. Role of the cell envelope in the antibacterial activities of polymyxin B and polymyxin B nonapeptide against Escherichia coli. Int J Antimicrob Agents. 2008;31:224–227. doi: 10.1016/j.ijantimicag.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Landman D, Georgescu C, Martin DA, Quale J. Polymyxins revisited. Clin Microbiol Rev. 2008;21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cholet R, Chevalier J, Bryskier A, Pagès JM. The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob Agents Chemother. 2004;48:3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 29.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 30.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 31.Plesiat P, Nikaido H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol Microbiol. 1992;6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 32.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 33.Fieulaine S, Juillan-Binard C, Serero A, Dardel F, Giglione C, et al. The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms. J Biol Chem. 2005;280:42315–4224. doi: 10.1074/jbc.M507155200. [DOI] [PubMed] [Google Scholar]
- 34.Viveiros M, Jesus A, Brito M, Leandro C, Martins M, et al. Inducement and reversal of tetracycline resistance in Escherichia coli K-12 and expression of proton gradient-dependent multidrug efflux pump genes. Antimicrob Agents Chemother. 2005;49:3578–3582. doi: 10.1128/AAC.49.8.3578-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elkins CA, Mullis LB. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob Agents Chemother. 2007;51:923–929. doi: 10.1128/AAC.01048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su CC, Yu EW. Ligand-transporter interaction in the AcrB multidrug efflux pump determined by fluorescence polarization assay. FEBS Lett. 2007;581:4972–4976. doi: 10.1016/j.febslet.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Z, White RJ. The evolution of peptide deformylase as a target: contribution of biochemistry, genetics and genomics. Biochem Pharmacol. 2006;71:1042–1047. doi: 10.1016/j.bcp.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pankuch GA, Lin G, Seifert H, Appelbaum PC. Activity of meropenem with and without ciprofloxacin and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52:333–336. doi: 10.1128/AAC.00689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malléa M, Chevalier J, Bornet C, Eyraud A, Davin-Régli A, et al. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 41.Pradel E, Pagès JM. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother. 2002;46:2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevalier J, Pagès JM, Eyraud A, Malléa M. Membrane permeability modifications are involved in antibiotic resistance in Klebsiella pneumoniae. Biochem Biophys Res Commun. 2000;274:496–499. doi: 10.1006/bbrc.2000.3159. [DOI] [PubMed] [Google Scholar]
- 43.Salmi C, Loncle C, Vidal N, Letourneux Y, Fantini J, et al. Squalamine: an appropriate strategy against the emergence of multidrug resistant gram-negative bacteria? PLoS ONE. 2008;3:e2765. doi: 10.1371/journal.pone.0002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevalier J, Mulfinger C, Garnotel E, Nicolas P, Davin-Régli A, et al. Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS ONE. 2008;3:e3203. doi: 10.1371/journal.pone.0003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavallo JD, Chardon H, Chidiac C, Courvalin P, Dabernat H, et al. 2008. Comité de l'Antibiogramme de la Société Francaise de Microbiologie [ http://www.sfm.asso.fr/publi/general.php?pa=1]