Abstract
A broad region of chromosome 10 (chr10) has engendered continued interest in the etiology of late-onset Alzheimer Disease (LOAD) from both linkage and candidate gene studies. However, there is a very extensive heterogeneity on chr10. We converged linkage analysis and gene expression data using the concept of genomic convergence that suggests that genes showing positive results across multiple different data types are more likely to be involved in AD. We identified and examined 28 genes on chr10 for association with AD in a Caucasian case-control dataset of 506 cases and 558 controls with substantial clinical information. The cases were all LOAD (minimum age at onset ≥ 60 years). Both single marker and haplotypic associations were tested in the overall dataset and 8 subsets defined by age, gender, ApoE and clinical status. PTPLA showed allelic, genotypic and haplotypic association in the overall dataset. SORCS1 was significant in the overall data sets (p=0.0025) and most significant in the female subset (allelic association p=0.00002, a 3-locus haplotype had p=0.0005). Odds Ratio of SORCS1 in the female subset was 1.7 (p<0.0001). SORCS1 is an interesting candidate gene involved in the Aβ pathway. Therefore, genetic variations in PTPLA and SORCS1 may be associated and have modest effect to the risk of AD by affecting Aβ pathway. The replication of the effect of these genes in different study populations and search for susceptible variants and functional studies of these genes are necessary to get a better understanding of the roles of the genes in Alzheimer disease.
Keywords: Alzheimer disease, late-onset Alzheimer Diseasev, LOAD, genomic convergence, association, candidate genes, PTPLA, SORCS1
Introduction
Alzheimer disease (AD; MIM# 104300) is the leading cause of dementia in the elderly and the most common form of dementia occurring after the age of 40 (Rocca et al., 1991; Schoenberg et al., 1987). It is a devastating neurodegenerative disorder of later life with a largely unknown but complex etiology that includes a strong genetic component (Growden, 1995; Khachaturian, 1985). Understanding the genetics of AD is critical to developing new treatments.
Genetic studies have identified three genes underlying early onset AD. This form can be caused by over 120 mutations in genes encoding β-amyloid precursor protein (APP; MIM# 104760), presenilin 1 (PS1; MIM# 104311), and presenilin 2 (PS2; MIM# 600759) (Goate et al., 1991; Levy-Lahad et al., 1995; Rocchi et al., 2003; Sherrington et al., 1995). To date, five late-onset AD genome-wide linkage screens have been reported (Blacker et al., 2003; Kehoe et al., 1999; Myers et al., 2002; Pericak-Vance et al., 1997; Pericak-Vance et al., 1998; Pericak-Vance et al., 2000). In addition, numerous studies have tested hundreds of candidate genes for involvement in AD (Bertram et al., 2007). However, no single method can give us an answer in dissecting complex disease traits. A promising solution to the growing data flood from the latest genomic technologies (e.g. genome-wide association) is genomic convergence. Genomic convergence is a multifactor approach used in genetic research that combines different data analysis results from independent types to identify and prioritize susceptibility genes for a complex disease (Hauser et al., 2003). Examples include genetic linkage data, association data, gene expression data, and biological function.
Our previous linkage and candidate gene studies suggested a more extensive heterogeneous genetic background on chromosome 10 for AD than we previously appreciated (Liang et al., 2007). Rare and common polymorphisms in multiple genes may be involved together with non-genetic factors in this complex disease. Therefore, combining different sources of data is important to selecting and prioritizing candidate genes.
In a recent study (Xu et al., 2007) gene expression levels were compared in the brain tissue from AD patients and controls using the Serial Analysis of Gene Expression (SAGE) method. SAGE is a powerful method for profiling transcripts expressed in a given tissue (Velculescu et al., 1995) and provides expression data for thousands of gene products without a priori knowledge of their function. Brain samples from AD cases (ApoE 4/4, ApoE 3/4 and ApoE 3/3 genotypes) and controls (ApoE 3/3 genotypes) were chosen to establish SAGE libraries to analyze the differential gene expression between AD cases and controls as described elsewhere (Xu et al., 2007). We converged the previous genetic linkage screen results with the SAGE gene expression data to identify a set of 28 genes that reside under the linkage peak and are differentially expressed between patients and controls. Numerous SNPs in these genes were genotyped and examined for association with AD.
Common diseases are likely to have complex etiology. Multiple genes with multifaceted interactions among genes and other risk factors undoubtedly play roles in the Alzheimer disease process. Multifactor Dimensionality Reduction (MDR) is a computational data reduction method that can identify gene-gene interactions without requiring very large sample sizes. Since MDR is a nonparametric, genetic model-free method, no hypothesis concerning any statistical parameter and no genetic inheritance model is assumed (Ritchie et al., 2001). In the present study, we applied MDR to the 28 candidate genes to test for potential gene-gene interactions involved in Alzheimer disease.
Materials and methods
Study populations
The case-control data set consisted of 1064 individuals (506 cases and 558 controls, Table 1) with substantial clinical information. The samples were collected by the Center for Human Genetics Research (CHGR) at Vanderbilt University and the Miami Institute for Human Genomics (MIHG) at the University of Miami. AD was diagnosed according to the NINCDS-ADRDA criteria (McKhann et al., 1984). The cases were all late onset Alzheimer disease (minimum age at onset (AAO) ≥ 60 years). There were no significant differences in age at exam or gender between cases and controls (Table 1). All controls were ascertained in the same catchment area as cases, and had results within the normal range in the Mini-Mental State Exam (MMSE) or Modified Mini-Mental State Exam (3MS). To test the potential for heterogeneity effects in the data set, the overall data set was subsetted according to four variables: Gender, ApoE status, Age-of-Onset and Clinical status. The distribution is shown in the Supp. Table S1. Written consent was obtained from all participants in agreement with protocols approved by the institutional review board at each contributing center.
Table 1.
Alzheimer disease case control data set
| Case | Control | ||
|---|---|---|---|
| Sample size (N) | 506 | 558 | |
| Gender | Female | 311 (61.5%) | 337 (60.4%) |
| Male | 195 (38.5%) | 221 (39.6%) | |
| Age-of-Onset | All | 72.5±6.3 | N/A |
| Female | 72.8±6.6 | N/A | |
| Male | 72.1±5.9 | N/A | |
| Age-of-Exam | All | 75.9±12.1 | 74.3±5.9 |
| Female | 75.8±14.1 | 73.5±5.7 | |
| Male | 76.2±8.1 | 75.5±6.0 | |
We also used independent family-based data sets as the validation sets to confirm any significant association identified in the case-control data set (Table 2). All individuals included in this study were Caucasian late-onset AD (LOAD) patients. AD diagnosis criteria were same as those for the case-control data set. Samples were ascertained by the following centers: the NCRAD repository at Indiana University (NCRAD); the Collaborative Alzheimer Project (CAP), including the University of Miami and Vanderbilt University and University of California at Los Angeles; and the National Institute of Mental Health repository (NIMH).
Table 2.
Family-based data set for Alzheimer disease
| Family | Overall | NIMH | NCRAD | CAP |
|---|---|---|---|---|
| Total pedigrees | 730 | 352 | 154 | 224 |
| Affected individuals | 1,521 | 807 | 315 | 390 |
| Unaffected individuals | 974 | 331 | 162 | 481 |
| Discordant Sib Pairs (DSP) | 1,337 | 629 | 269 | 439 |
| Independent Discordant Sib Pairs | 674 | 283 | 129 | 262 |
| Pedigrees with at least one DSP | 406 | 165 | 75 | 166 |
| Affected Relative Pairs (ARP) | 188 | 66 | 26 | 96 |
| Pedigrees with at least one ARP | 64 | 31 | 11 | 22 |
Following informed consent, blood samples were collected from each individual. Genomic DNA was extracted from whole blood by use of the Puregene system (Gentra Systems, Minneapolis, MN). All samples were coded and stored at 4°C until used.
Gene selection
The Serial Analysis of Gene Expression (SAGE) method was used to compare the gene expression levels in the brain tissue from AD patients and controls as described elsewhere (Velculescu et al., 1995; Li et al., 2006). Briefly, Six SAGE libraries were generated using hippocampus collected in the Kathleen Price Bryan Brain Bank, at the Duke University Alzheimer Disease Research Center, and the Brain Bank of the Center for Human Genetics (CHG), Duke University Medical Center, following a rapid autopsy protocol (Hulette et al., 1997). Four libraries were based on the short tag (14 bp, AD ApoE 4/4, ApoE 3/4, ApoE 3/3 and control ApoE 3/3). Two libraries were based on the long tag (21–22 bp, AD ApoE 3/3 and control ApoE 3/3). The number of times a tag observed in tissue was extracted from each library and compared between the AD and control samples using eSAGE software to form a compared ShortSAGE database. Chi-square and Fisher exact tests, as previously described (Hauser et al., 2003), were used to test differences in expression levels between AD and control for each tag in each compared SAGE data set. The SAGE tags were compared to the UniGene to interpret the SAGE results. The false discovery rate (FDR) was applied to the SAGE data to correct for multiple comparisons. p<10−10 was used as the cutoff to select genes whose expression levels were highly significantly different between AD cases and controls. Then the previous linkage study results for chromosome 10, shown in Table 3, were converged with gene expression data so that only differentially expressed genes that were also under linkage peaks in previous linkage screens were selected for the next step association analysis.
Table 3.
Summary of linkage study results on chromosome 10
| Chr. Location | Linkage region (Mb) | Peak LOD score | Dataset | Group |
|---|---|---|---|---|
| 10p | 5–25 | 3.00 | 50ca/50co | Zubenko et.al |
| 10q | 45–75 | 4.10 | 451 ASPs | Myers, et. al |
| 10q | 40–60 | 2.69 | 922 ASPs | Liang, et. al |
| 10q | 80–100 | 1.70 | 922 ASPs | Liang, et. al |
| 10q | 90–120 | 3.80 | 435 ASPs | Bertram et.al |
SNP selection and genotyping
Several criteria were applied to select SNPs as genetic markers. We included 10kb of flanking sequence on each side to include potential regulatory regions. Only tagSNPs were selected with the restriction that the linkage disequilibrium, r2, between selected SNPs should be less than 0.7. The web-based program, SNPselector (Xu et al., 2005), was used to automatically search available SNPs through the Ensembl and UCSC databases. It prioritizes these SNPs on their tagging for linkage disequilibrium based on the LD bin algorithm in LD select (Carlson et al., 2004) using HapMap data, SNP allele frequencies and source, function, regulatory potential and repeat status. Because we used the Illumina Goldengate genotyping method, we only selected SNPs with Illumina SNPscore greater than 0.6 to ensure the genotyping quality. A MAF greater than 20% was used as the cutoff for selecting the intronic SNPs. SNPs in conserved regions of eight-genome alignment (Human/chimp/mouse/rat/dog/chicken/fugu/zebra_fish, based on UCSC database), conserved transcription factor binding sites (TFBSs) in the human/mouse/rat alignment, CpG island, and microRNA genes were selected if the MAFs were greater than 10%. Coding SNPs were selected when the MAF was greater than 1%. To further reduce the SNPs to a manageable number, there were selected at the 1 SNP/10 kb spacing. Because TNFRSF6 showed significant results in linkage, candidate gene studies and SAGE gene expression analysis (Bertram et al., 2000; Feuk et al., 2000), more weight was given to it by genotyping one SNP per five kilobases.
The final set of 667 SNPs on chr10 was genotyped using the Illumina GoldenGate Oligo Pool Assay (OPA) on an Illumina BeadStation 500 GX (Illumina Inc., San Diego, CA). DNA samples from cases and controls were randomly sorted, and duplicated samples were implemented across plates for genotyping quality control.
We tested all SNPs for consistency with HWE in Haploview program (Barrett et al., 2005). SNPs were excluded if HWE p<0.001. TagSNPs were selected at different linkage disequilibrium levels using the Tagger option incorporated in Haploview for the future haplotypic association tests and gene-gene interaction tests.
Statistical methods
Association analysis in case-control data set
In the case-control data set, allelic association for single SNPs was tested using the χ2 test in Haploview (Barrett et al., 2005). Genotypic association was assessed by a 2 × 3 contingency table likelihood ratio test in SAS program (SAS Institute, 2003). Haplotypic association was analyzed using haplo.stats (Schaid et al., 2002). To save calculation time and satisfy computer memory limits, small genes (with less than 8 genotyped SNPs in the gene) were analyzed using individuals with less than 50% missing genotypes. Genes of medium size (with 8 to 26 genotyped SNPs) were analyzed using only individuals who had complete genotypes on all analyzed loci in that gene. There are seven large genes that were genotyped for 38 to 124 SNPs. They were analyzed on individuals without any missing genotypes and only tagSNPs with r2<0.5 were used. In addition, the tagSNPs were separated into groups of 10 with 2 SNPs overlapping across the groups. We estimated haplotype frequencies and tested association of each haplotype with a frequency of at least 1% in our case-control data set with age and gender adjusted score statistics. Haplotype logistic regression was modeled using a GLM algorithm including age and gender as covariates. The most frequent haplotype occurring in the similar percentage of cases and controls was selected as the referent haplotype. To evaluate the association of subsets of alleles from the full haplotype, a sliding window of three SNPs was used and the global score statistics were calculated.
Conditional analyses were conducted by dividing the case-control data set by known risk factors including age, gender and ApoE status. The difference in allele frequency between cases and controls was tested by a χ2 test or Fisher’s exact test when applicable (e.g. cell size <5). We also applied logistic regression to estimate the effects of the SNPs showing the most significant results across different analyses after controlling for age, gender and ApoE status. Nominal significance was declared at p=0.05 and FDR was used to correct multiple testing. Haploview and SAS (SAS institute, Cary, NC) programs were used for the analyses.
Association analysis in family data set
Family based association analysis was used to follow up the SNP with the strongest association in the female subset. The allelic association analyses were conducted using the association in the presence of linkage (APL) analysis (Martin et al., 2003b) and the pedigree disequilibrium test (PDT) (Martin et al., 2000). APL and PDT each have distinct advantages. APL can correctly infer missing parental genotypes in regions of linkage by estimating identity-by-descent (IBD) parameters and is a more powerful test under those conditions than PDT. However, APL only uses nuclear families for the association test. PDT can use data from extended pedigrees and gains power substantially when extended pedigree data are available. Genotype-PDT (GenoPDT) is an extension of PDT and was used to assess association between genotypes and risk of AD in the family data (Martin et al., 2003a).
Test for gene-gene interaction
TagSNPs were selected using r2>0.5 as a threshold to exclude SNPs from the interaction analysis that were in high LD by the Tagger option in Haploview. Potential gene-gene interactions were identified using Multiple Dimensionality Reduction. MDR performs an exhaustive search of all possible single-locus through k-locus interactions to evaluate all possible high/low risk models of disease (Ritchie and Motsinger, 2005). The statistical significance of the final best model was determined through permutation testing with 1000 permuted data sets. The significance of the final model was determined by comparing the prediction error and cross validation consistency of the final model with the distribution. A p value was extracted for the model by its theoretical location in the permutation distribution (Motsinger et al., 2006). In considering the possible unbalanced ratio of cases to controls in the data set, a balanced accuracy approach was used to evaluate the performance of the models (Velez et al., 2007).
In the present study, 1, 2, and 3 locus combinations were run and logistic regression (SAS program) was used to verify the interaction between genes in the identified model.
Results
Genomic convergence identified genes and genotyped SNPs
SAGE analysis revealed 41 genes on chromosome 10 that were differentially expressed between AD cases and controls with p<10−10. 28 of those genes were also under previously identified linkage peaks (Bertram et al., 2000; Myers et al., 2002; Pericak-Vance et al., 2000; Zubenko et al., 1998), (Liang et al., 2007) and thus considered for detailed analysis. The genomic size of the genes ranged from 10 kb to 1.3 Mb. The number of exons ranged from 2 to 38. Depending on the size of the gene, we genotyped 2–124 SNPs in the genes to for an average spacing of 1 SNP/10kb. The gene description, size and number of genotyped SNPs were summarized in the Supp. Table S2.
Of the 667 genotyped SNPs 70 SNPs were dropped from the analysis because of monomorphism, being out of HWE, or assay problems. Specifically, 58 SNPs failed Illumina quality control criteria, 10 SNPs were monomorphic and 2 SNPs were out of HWE in controls at p<10−5 (Bonferroni correction for 667 markers at p=0.05). In total, there were 597 SNPs that remained in the final analyses. 80% of those were intronic SNPs, 17% were functional SNPs (exon, promoter, UTR, exon/intron boundary, et al.) and the other 3% were from the 5’-upstream and 3’-downstream regions (Supp.Figure S1).
Association analysis
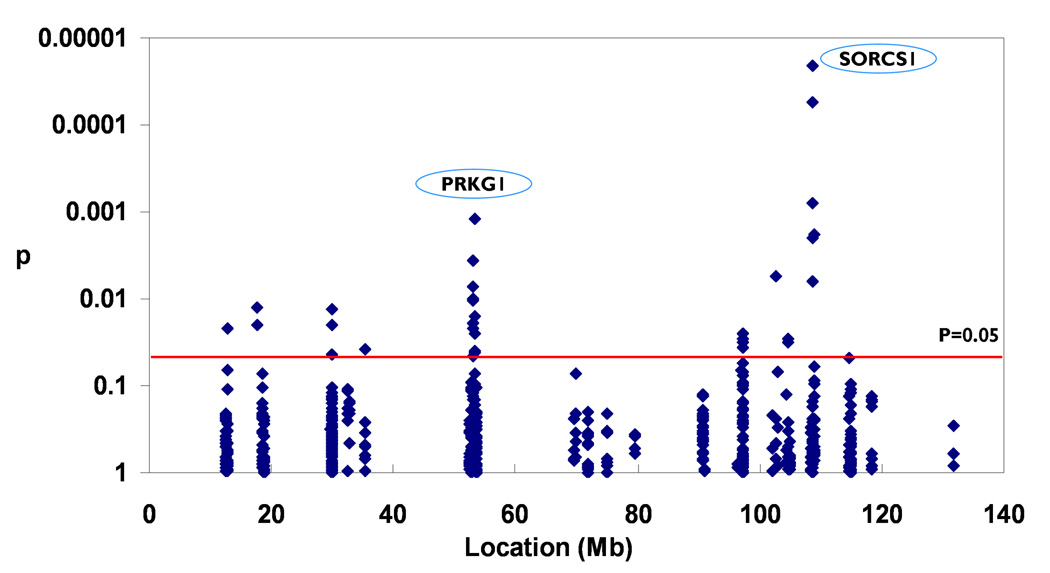
The allelic association of 597 genotyped SNPs was analyzed on the overall data set and eight subsets. Twenty-four SNPs in eight genes (PTPLA; MIM# 610467, SORCS1; MIM# 606283, PRKG1; MIM# 176894, SVIL; MIM# 604126, ACTR1A; MIM# 605143, BA108L7.2, CAMK1D; MIM# 607957, and PDZK7 (approved symbol PDZD7)) showed association with AD at nominal p<0.05 (Figure 1). After we applied FDR (q=20%) based on the number of genotyped SNPs in each gene to correct for multiple testing, two SNPs in two genes remained significant. The first SNP was rs10508533 in PTPLA with p=0.0022; the second is rs17277986 in SORCS1 gene with p=0.0025 (Figure 1). There were additional SNPs showing significant results in each subset defined by age, gender, ApoE status and clinical status. Five out of eight associated genes in the overall data set showed association in three or more subsets (Supp. Table S3). These five genes were PTPLA, SORCS1, SVIL, PRKG1 and PDZK7 (approved symbol PDZD7). The female subset had the most significant result (Supp. Table S3): p=0.00002 for a SNP (rs172777986) in SORCS1 (Figure 2). Close to rs17277986, four tag SNPs (rs2900717, rs10884399, rs11193170, rs4918280) were also significant at p<0.05. rs17277986 remained significant after FDR correction for multiple testing. The odds ratio (OR) of having the risk allele vs. not having risk allele was 1.34 (95% CI=1.11–1.63, p=0.003) in the overall data set. In the female subset, the OR was 1.70 with 95% CI of 1.33–2.19, and the p value was very significant (p<0.0001). The OR was also significant after the adjustment for age, gender and ApoE status (data not shown). SNP rs6480499 in PRKG1 was also significant in the female subset after FDR correction (Figure 2).
Figure 1. Allelic association test in the overall data set.
PTPLA and SORCS1 were associated with AD after FDR correction for multiple testing
Figure 2. Allelic association test in the female subset.
SORCS1 and PRKG1 showed association after FDR correction for multiple testing
Twenty-seven SNPs in twelve genes (CACNB2; MIM# 600003, PTPLA, SORCS1, SVIL, SORBS1; MIM# 605264, PRKG1, CAMK1D, CNNM2; MIM# 607803, PDZK7 (approved symbol PDZD7), TCF7L2; MIM# 602228, TNFRSF6; MIM# 134637, and NT5C2; MIM# 600417) showed association with AD at nominal p<0.05 in the genotypic analysis (Supp. Figure S2). SNPs in CACNB2 and PTPLA were significant after FDR correction (q=0.2) for multiple testing (p value of 0.0030 and 0.0036 for rs1277738 in CACNB2 and rs10508533 in PTPLA, respectively). Supp. Table S4 shows genotypic association in subsets for SNPs having genotypic association with AD at p<0.05 in the overall data set. Similar to allelic association, there were additional SNPs showing significant results in each subset. The female subset had the most significant results. The SNP rs17277986 in SORCS1 showed genotypic association after FDR correction (Supp. Figure S3) in this subset (p=0.0002).
Haplotypic association analysis for each gene was done on the overall data set. Global p values for score statistics are shown in Supp. Table S5. The haplotype in PTPLA had the most significant p value of 0.009. A haplotype containing ten tagSNPs in SVIL gene was marginally associated with AD (p=0.049). Since a SNP in SORCS1 was strongly associated with AD in the female subset, it was also analyzed for haplotypic association in the female subset only. The global p value was 0.15, which was not significant (Supp. Table S5). However, the three 3-SNP sliding window haplotypes were highly significant (smallest p=0.0005, Figure 3). All these three haplotypes contained rs17277986, which showed the most significant single SNP allelic association in the same data set.
Figure 3. Haplotype analysis in SORCS1 in the female subset.
20 tagSNPs were analyzed in SORCS1. Bars show the haplotypes in the 3-SNP sliding window; triangles show the single marker allelic association –log p values. The single marker allelic association results for three tagSNPs around the SNP rs17277986 with r2<0.8 are also shown in the figure. The gene structure is shown at the bottom.
Analytical convergence of the genetic association tests
Across all the association analyses including allelic, genotypic and haplotypic association tests, several genes showed association. After we applied FDR to correct for multiple testing for allelic and genotypic association tests, only five genes showed association in at least one of the analyses. SNPs in PTPLA were significant in the overall data set across all three association tests. SNPs in SORCS1 had a significant allelic effect in the overall data set, and it was highly significant in the female subset across all three association tests. Although SNPs in CACNB2 did not show allelic association in the overall data set, SNP rs1277738 in CACNB2 had the strongest genotypic effect in overall data set, and showed genotypic association at nominal p<0.05 in four subsets (Age-of-onset between 60 and 75, ApoE-negative, female and possible AD). SNPs in SVIL were marginally significant in the haplotypic association test in the overall data set.
Follow up of SNPs in SORCS1 and PTPLA genes
To confirm the effect of SORCS1, we genotyped the most significant SNP (rs17277986) in our validation data sets. When we analyzed female and male individuals together, none of the datasets showed significant results at p<0.05. When we analyzed females only, the overall data set was not significant (APL p=0.06) (Table 4). The results in family data sets were similar to the population based case-controls data set, but not significant after correction for multiple comparisons.
Table 4.
Family-based association tests for rs17277986 in SORCS1 in validation data sets
| Female & Male |
Female only |
|||||||
|---|---|---|---|---|---|---|---|---|
| All | CAP | NIMH | NCRAD | All | CAP | NIMH | NCRAD | |
| APL | 0.189 | 0.696 | 0.301 | 0.189 | 0.057 | 0.526 | 0.309 | 0.010 |
| Sum_PDT | 0.862 | 0.085 | 0.099 | 0.742 | 0.904 | 0.030 | 0.254 | 0.668 |
| Geno_PDT | 0.606 | 0.269 | 0.052 | 0.402 | 0.798 | 0.103 | 0.182 | 0.562 |
All: combined data set; CAP: the Collaborative Alzheimer Project; NCRAD: the NCRAD repository at Indiana University; NIMH: the National Institute of Mental Health repository
The two SNPs genotyped in PTPLA were significant across all the previous association analyses (rs10508533 had OR=1.34 with p=0.003; rs4453117 had OR=1.31 with p=0.003). This gene is small, spanning only 27 kb. We followed up the results by genotyping seven more SNPs in the case-control data set to give this gene a better coverage. The linkage disequilibrium between all genotyped SNPs in cases is shown in the Supp. Figure S4. Most of the SNPs were significant in allelic and genotypic association test (Table 5). The overall haplotypic association test considering all SNPs in the gene was significant with global score statistic p value = 0.03. The 3-SNP sliding window haplotypic association test was significant for all haplotypes (Supp. Figure S5).
Table 5.
Allelic and genotypic association of all genotyped SNPs in PTPLA in the overall data set
| SNP | Allele variation | Nucleotide position a | function | MAF | Allelic Asso. | Genotypic Asso. | |
|---|---|---|---|---|---|---|---|
| 1 | rs4453117 | A>C | 17,666,100 | 3’UTR | 0.42 | 0.0011 | 0.0039 |
| 2 | rs2357285 | C>T | 17,672,568 | Intron | 0.06 | 0.4504 | 0.7087 |
| 3 | rs1053926 | C>T | 17,676,315 | Exon 6, ns | 0.30 | 0.0141 | 0.0432 |
| 4 | rs7921987 | C>G | 17,680,752 | Intron | 0.32 | 0.0688 | 0.1316 |
| 5 | rs7073625 | A>G | 17,682,131 | Intron | 0.37 | 0.0223 | 0.0857 |
| 6 | rs2252808 | C>T | 17,685,820 | boundary | 0.44 | 0.0017 | 0.0035 |
| 7 | rs10508533 | G>T | 17,689,895 | Intron | 0.29 | 0.0029 | 0.0082 |
| 8 | rs2461891 | C>T | 17,695,987 | Intron | 0.25 | 0.0035 | 0.0102 |
| 9 | rs7918263 | A>T | 17,697,158 | Intron | 0.38 | 0.0007 | 0.0014 |
The map location is from NCBI dbSNP, build 35.
Gene-gene interaction among genes on chromosome 10
Overall data set
To reduce the computational burden and to eliminate highly redundant data, 300 tagSNPs were selected to represent the 597 SNPs at an r2>0.5 threshold to test for gene-gene interaction in MDR (Table 6). The single best model was found between two SNPs in CACNB2. This 2-locus model involves rs1277738 and rs10741083. The average prediction accuracy was 51.43% and the cross-validation consistency was 40%, using 5-fold cross-validation. Because other models outperformed this model in other cross validation intervals, the p value was not significant in the permutation test (p=0.37). However, this model is in the top 10 models in 3 out 5 of the cross-validation intervals, being the top model 67% of the time.
Table 6.
Summary of MDR results for genes on chromosome 10
| # locus | Best model | Accuracy (%) | Cross validation | P |
|---|---|---|---|---|
| 1-locus | CACNB2(rs1277738) | 50.35 | 40% | 0.636 |
| 2-locus | CACNB2(rs1277738) | 51.43 | 40% | 0.374 |
| CACNB2(rs10741083) | ||||
| 3-locus | CACNB2(rs1277738) | 49.96 | 20% | 0.710 |
| CACNB2(rs2489214) | ||||
| CAMK1D(rs4750255) |
When we eliminate noise by using the “FORCELOCI” function in MDR and let the program do the permutation only on the two SNPs in the best 2-locus model, the permutation p value was significant (p=0.003). The result was confirmed in logistic regression when including the interaction term. The interaction was significant with p=0.004 in the model. Table 6 shows that SNP rs1277738 in CACNB2 was consistently identified across 1-, 2-, 3-locus models in MDR. It showed a main effect (best 1-locus model) in the MDR analysis. This SNP also had strongest genotypic effect in single marker association test (p=0.003).
Because several models may perform almost equally in different cross validation intervals, looking at only the top model might not be sufficient to identify important genes in this complex disease. Therefore, we considered the frequency of each gene in the top ten MDR models ranked by prediction accuracy. Table 7 shows the three most frequently identified genes in the top 10 MDR models. CACNB2 was in the top 10 models among all cross-validation intervals in the 1-, 3-locus models, and in 80% of cross-validations for 2-locus model. In more than 60% of cross validation intervals, PTPLA was in the top 10 MDR models. SORCS1 was in top 10 models among 40% of cross-validation intervals.
Table 7.
The frequency of 3 genes in the top 10 MDR models from 5 cross-validations
| 1-locus model | 2-locus model | 3-locus model | |
|---|---|---|---|
| Overall | |||
| CACNB2 | 100% | 80% | 100% |
| PTPLA | 80% | 60% | 60% |
| SORCS1 | 40% | 40% | 40% |
| Female | |||
| CACNB2 | 100% | 40% | 40% |
| PTPLA | 20% | 40% | 40% |
| SORCS1 | 100% | 100% | 80% |
Female subset
Our previous analysis showed a strong effect of SORCS1 in the female subset, so we performed MDR in this subset as well (Table 8). The single SNP rs17277986 in SORCS1 was identified as the single best model with prediction accuracy of 54.74% and 80% cross-validation consistency (p=0.10). This SNP was the one showing the strongest single marker allelic effect in the female data set. No 2-locus model had a prediction accuracy greater than 50%. The 3-locus model contains rs17277986 in SORCS1 and two SNPs (rs1409207 and rs2482100) in CACNB2 with 52.49% prediction accuracy and 40% cross-validation consistency. Although the permutation p was not significant at p<0.05, the SORCS1 gene was always in the top 10 MDR models among all cross-validation intervals for 1-, 2- locus models. CACNB2 was in the top 10 1-locus models among all cross-validation intervals and 40% of the cross-validation intervals for 2-, 3-locus models (Table 7).
Table 8.
Summary of MDR results in the female subset
| # locus | Best model | Accuracy (%) | Cross validation | P |
|---|---|---|---|---|
| 1-locus | SORCS1(rs17277986) | 54.74 | 80% | 0.098 |
| 2-locus | SORCS1(rs11193054) | 48.67 | 20% | 0.884 |
| CACNB2(rs1277738) | ||||
| 3-locus | SORCS1(RS17277986) | 52.49 | 40% | 0.297 |
| CACNB2(rs1409207) | ||||
| CACNB2(rs2482100) |
Discussion
While both linkage and expression analyses are powerful methods when applied individually, the number of possible genes they present as candidates for Alzheimer disease or any complex disorder remains extremely large. Thus, focusing and prioritizing effort on jointly identified candidate genes is a key to success using these techniques (Hauser et al., 2003). Our previous study (Liang et al., 2007) suggested more extensive locus heterogeneity in AD than was previously suspected. We designed the current study to converge all available data (linkage, candidate gene, and gene expression data) to identify susceptibility genes altering AD risk.
Gene expression data were from Serial Analysis of Gene Expression (SAGE) analysis. The gene expression levels of forty-one genes were highly significantly different at p<10−10. Interestingly, when we compared the direction of gene expression between AD and controls, we found that all but one gene was up-regulated in AD the ApoE 4/4 and ApoE 3/4 samples compared to the ApoE 3/3 samples. It is possible that the ApoE ε4 allele elevates the expression of other genes. The only down-regulated gene was BA108L7.2 encoding sideroflexin 3. The difference for this gene was found when we compared the long tag SAGE library for AD to control ApoE 3/3 samples.
Given the assumption that genes showing positive association in multiple tests have higher likelihood of being important in the disease process, we performed an analytical convergence on the results from allelic, genotypic and haplotypic association tests. The most consistently associated SNPs across the three analyses are rs10508533 in PTPLA in the overall data set (allelic association p=0.0022) and rs17277986 in SORCS1 in the female subset (allelic association p=0.00002).
PTPLA is protein tyrosine phosphatase-like, member A. Little is known about its function, but it could be involved in the protein phosphorylation process that is important for the function of some proteins in the AD pathology. Among all nine genotyped SNPs in PTPLA, seven showed allelic association and six showed genotypic association. Three of those six SNPs are functional SNPs. This gene is not in the same LD block with any nearby genes based on HapMap data. Therefore, the three functional SNPs could have potential roles in the biological function of PTPLA gene or are in LD with a susceptible SNP affecting PTPLA function, but are unlikely to be in LD with susceptibility SNPs in another gene.
SORCS1 encodes sortilin-related VPS10p domain containing type 1 receptor. It contains a leucine-rich domain and mediates intracellular sorting and trafficking functions. It is highly expressed in the brain (Hermey et al., 1999) and neuronal activity can differentially affect its expression (Hermey et al., 2004). SORCS1 is a substrate of γ-secretase, and γ-secretase cuts amyloid precursor protein (APP) and generates amyloid β peptide (Aβ), one of the hallmarks of Alzheimer disease. rs17277986 in SORCS1 was significant in the overall, age-of-onset greater than 76 year old, ApoE 4-positive, and definite/probAD data sets, with p=0.0025, 0.0178, 0.0073, and 0.0034 respectively. It was most significant in the female subset (p=0.00002). It survived not only an FDR multiple testing correction, but also a more conservative Bonferroni correction. Four nearby tagSNPs (r2<0.8) were also significant in the female subset (p=0.0008, 0.002, 0.0064, 0.0018 for rs2900717, rs10884399, rs11193170 and rs4918280, respectively). Another SNP (rs12571141) had a p=0.00006 in this subset, but it was in high LD with rs17277986 (r2=0.97), suggesting it was measuring the same effect. Haplotypes containing the most significant SNP (rs17277986) were all significant with the smallest p value = 0.0005.
All the interesting SNPs are in intron 1 of SORCS1. This gene is 590kb and intron 1 itself is 207kb and the largest intron in the gene. Most of the regions in intron 1 are conserved (>70% identity) over the 100bp calculation window among mammalians (data not shown). Thus, this region may be important as a functional element in the gene.
Grupe et al. (Grupe et al., 2006) found an association between SNP rs600879 (164 bp away from rs17277986) in SORCS1 and Alzheimer disease with a p=0.0043 in their combined data set. SNP rs600879 (108,913,108 bp) is 608 bp away from the first intron/exon boundary. However, it was not consistently replicated in all of their individual datasets as it was significant in only two of them (p=0.017 and 0.040). However, the sample size of the homozygotes with the minor allele was small (between 4 and 9 samples for cases and controls) because the minor allele frequency of this SNP was 11.2% and 8.5% for cases and controls, respectively. The SNP was not significant in the other two data sets.
In our study, SNP rs17277986 in SORCS1 was significantly associated with AD in females in the case-control and independent family-based data sets. There are several possible hypotheses for the role of SORCS1 in AD.
First, it could be due to the estrogen effect. Studies on the estrogen-replacement therapy in Alzheimer disease suggested that estrogen may provide some protection against memory loss and lower the risk of developing AD (Burns and Murphy, 1996; Tang et al., 1996; Wickelgren, 2003). Second, this variant may be in or near intronic regulatory sequences that might govern cell type-specific or tissue-specific expression of SORCS1. Third, genomic imprinting may also play a role for the significant difference of the alleles between AD cases and controls, where differential gene expression depends upon whether the inheritance is through the mother or father (Hall, 1990). Methylation has been proposed as a mechanism of imprinting (Holliday, 1989) and is supported by the findings of the increased number of unmethylated sites in AD patients in comparison to controls (Payao et al., 1998). Fourth, there may be interaction between mitochondrial DNA mutations and this autosomal locus such that a particular mitochondrial genotype is required for individuals carrying this autosomal risk variant to express disease. It was suggested that mitochondria are involved in apoptosis and there is evidence of oxidative damage to mitochondrial DNA in AD cases (Mecocci et al., 1994; Green and Reed, 1998). Although there is not evidence that any of these possible explanations are in fact correct, they provide plausible areas for further research.
Epistasis, or gene-gene interaction, is crucial in detecting polymorphisms associated with an increased risk of disease (Moore, 2003; Thornton-Wells et al., 2004; Sing et al., 2004). In the present study, we applied Multifactor Dimensionality Reduction to investigate the potential associations between AD susceptibility and candidate genes coverged from linkage, gene expression and association studies.
MDR identified two SNPs (rs1277738 and rs10741083) in CACNB2 as having an interaction effect among genes on chromosome 10. These two SNPs were both intronic SNPs, one was in intron 2 and the other was in intron 5. They are 216 kb apart and not in linkage disequilibrium with each other (r2=0.002). One SNP, rs1277738, had a very strong genotypic association (p=0.003) with AD and it was identified as having a main effect in MDR analysis. Another SNP, rs10741083 did not show single marker association with AD. However, the interaction effect between this SNP and the main effect SNP had higher prediction accuracy (Table 6), suggesting that the interaction effect was not absolutely driven by rs1277738, which had the main effect.
CACNB2 encodes a voltage-dependent calcium channel beta 2 subunit. The beta 2 subunit works as a complex with other subunit (e.g. alpha unit) and the protein complexes play pivotal roles in signal transduction and homeostasis processes (Opatowsky et al., 2003). The polymorphisms in the introns may affect the binding site that binds alpha unit. If the interaction between the alpha and beta subunit is affected, the function of the whole protein complex is subsequently affected.
Interpreting results is a challenge in exploring gene-gene interactions. MDR analysis on genes on chromosome 10 had low cross-validation consistency on the best model (two SNPs in CACNB2), although the interaction was significant in the logistic regression analysis when noise was eliminated from the data set. It may be the case that multiple models are all good predictors of disease risk. If a gene is always in the top MDR models, it also suggests that the gene may have an effect. We discovered three genes (CACNB2, PTPLA, and SORCS1) that showed consistent evidence of association with AD in our previous studies. MDR identified CACNB2 as the single best model, but not PTPLA or SORCS1. By considering the frequencies of specific models in the top ten MDR models, we were able to find that these three genes were in the top ten MDR models most of the time. This is another way to identify important effects in the disease pathology. The data suggest that each gene has a modest effect and that an extensive epistasis or gene-gene interaction is underlying the disease etiology.
Our genomic convergence study suggests that genetic variations in PTPLA, SORCS1 and CACNB2 genes may be associated and have modest effect to the risk of AD. In Alzheimer disease, the amyloid precursor protein (APP) is enzymatically cut to generate Amyloid-β. Amyloid-β accumulates and aggregates to form an oligomer and cause apoptosis which can cause AD (hardy and Higgins, 1992). The SORLA gene is associated with AD and it inhibits the generation of Amyloid-β (Rogaeva et al., 2007). SORCS1 is a homolog of SORLA and may have a similar function as SORLA to inhibit the generation of Aβ. CACNB2 is a calcium channel protein and affects the calcium level which could cause mitochondrial damage and then induce apoptosis (Finlin et al., 2006). PTPLA is a phosphatase, but relatively little is known about its biological function. Base on its phosphatase function, we can conjecture that it is involved in the phosphorylation of the Tau protein; phosphorylated Tau forms neurofibrillary tangles, which is one of the hallmarks of AD. PTPLA might also be involved in the phosphorylation of GSK-3β protein which binds β-catenin. β-catenin is associated with elevated Aβ 42 (Prager et al., 2007; Shim et al., 2007). β-catenin can also bind to transcription factors and induce unscheduled cell cycles, which is also hypothesized to be related to AD (Figure 4). In conclusion, this study suggests that genetic variations in PTPLA, SORCS1 and CACNB2 genes might alter the risk for Alzheimer disease by affecting multiple pathways. The replication of the effect of these genes in different study populations and search for susceptible variants and functional studies of these genes are necessary to get a better understanding of the roles of the genes in Alzheimer disease.
Figure 4. Hypothesized pathways involved in Alzheimer disease.
Supplementary Material
Acknowledgments
We would like to thank all of the individuals who participated in this study. This study was supported by NIH grants AG019757, AG021547, AG20135, and a grant from the Alzheimer Association (IIRG-00-2006). Funding for collection and distribution of NCRAD samples was provided, in past, by NIH grant AG21886. This work used the core resources of the GCRC (MO1 RR-00095) and the CHGR at VUMC, and the MIHG at University of Miami.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K, Selkoe DJ, Saunders AJ, Tanzi RE. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Burns A, Murphy D. Protection against Alzheimer's disease? Lancet. 1996;348:420–421. doi: 10.1016/S0140-6736(05)64533-3. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Prince JA, Breen G, Emahazion T, Carothers A, St Clair D, Brookes AJ. apolipoprotein-E dependent role for the FAS receptor in early onset Alzheimer's disease: finding of a positive association for a polymorphism in the TNFRSF6 gene. Hum Genet. 2000;107:391–396. doi: 10.1007/s004390000383. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Correll RN, Pang C, Crump SM, Satin J, Andres DA. Analysis of the complex between Ca2+ channel beta-subunit and the Rem GTPase. J Biol Chem. 2006;281:23557–23566. doi: 10.1074/jbc.M604867200. [DOI] [PubMed] [Google Scholar]
- Goate AM, Chartier-Harlin MC, Mullan MC, Brown J, Crawford F, Fidani L, Guiffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance MA, Roses AD, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;33:53–56. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Growden JH. Advances in the Diagnosis of Alzheimer's Disease. In: Iqbal K, Mortimer JA, Winblad B, Wisniewski H, editors. Research Advances in Alzheimer's Disease and Related Disorders. New York: John Wiley and Sons, Ltd; 1995. pp. 139–153. [Google Scholar]
- Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, Kauwe JS, Maxwell TJ, Cherny S, Doil L, Tacey K, van LR, Myers A, Wavrant-De VF, Kaleem M, Hollingworth P, Jehu L, Foy C, Archer N, Hamilton G, Holmans P, Morris CM, Catanese J, Sninsky J, White TJ, Powell J, Hardy J, O'donovan M, Lovestone S, Jones L, Morris JC, Thal L, Owen M, Williams J, Goate A. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JG. Genomic imprinting: review and relevance to human diseases. Am J Hum Genet. 1990;46:857–873. [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hauser MA, Li YJ, Takeuchi S, Walters R, Noureddine M, Maready M, Darden T, Hulette C, Martin E, Hauser E, Xu H, Schmechel D, Stenger JE, Dietrich F, Vance J. Genomic convergence: identifying candidate genes for Parkinson's disease by combining serial analysis of gene expression and genetic linkage. Hum Mol Genet. 2003;12:671–677. [PubMed] [Google Scholar]
- Hermey G, Plath N, Hubner CA, Kuhl D, Schaller HC, Hermans-Borgmeyer I. The three sorCS genes are differentially expressed and regulated by synaptic activity. J Neurochem. 2004;88:1470–1476. doi: 10.1046/j.1471-4159.2004.02286.x. [DOI] [PubMed] [Google Scholar]
- Hermey G, Riedel IB, Hampe W, Schaller HC, Hermans-Borgmeyer I. Identification and characterization of SorCS, a third member of a novel receptor family. Biochem Biophys Res Commun. 1999;266:347–351. doi: 10.1006/bbrc.1999.1822. [DOI] [PubMed] [Google Scholar]
- Holliday R. DNA methylation and epigenetic mechanisms. Cell Biophys. 1989;15:15–20. doi: 10.1007/BF02991575. [DOI] [PubMed] [Google Scholar]
- Hulette CM, Welsh-Bohmer KA, Crain B, Szymanski MH, Sinclaire NO, Roses AD. Rapid brain autopsy. The Joseph and Kathleen Bryan Alzheimer's Disease Research Center experience. Arch Pathol Lab Med. 1997;121:615–618. [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De VF, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Khachaturian Z. Diagnosis of Alzheimer's disease. Arch Neurol. 1985;42:1097–1104. doi: 10.1001/archneur.1985.04060100083029. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu Y-H, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Li YJ, Xu P, Qin X, Schmechel DE, Hulette CM, Haines JL, Pericak-Vance MA, Gilbert JR. A comparative analysis of the information content in long and short SAGE libraries. BMC Bioinformatics. 2006;7:504. doi: 10.1186/1471-2105-7-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Martin ER, schnetz-boutaud N, Bartlett J, Anderson B, Zuchner S, Gwirtsman H, Schmechel D, Carney R, Gilbert JR, Pericak-Vance MA, Haines JL. Effect of heterogeneity on the chromosome 10 risk in late-onset Alzheimer disease. Hum Mutat. 2007;28:1065–1073. doi: 10.1002/humu.20567. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003a;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Hauser ER, Kaplan NL. Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet. 2003b;73:1016–1026. doi: 10.1086/378779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman G, Folstein M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of the department of health and human services task force on Alzheimer's disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Ann Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Motsinger AA, Ritchie MD, Shafer RW, Robbins GK, Morse GD, Labbe L, Wilkinson GR, Clifford DB, D'Aquila RT, Johnson VA, Pollard RB, Merigan TC, Hirsch MS, Donahue JP, Kim RB, Haas DW. Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogenet Genomics. 2006;16:837–845. doi: 10.1097/01.fpc.0000230413.97596.fa. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Opatowsky Y, Chomsky-Hecht O, Kang MG, Campbell KP, Hirsch JA. The voltage-dependent calcium channel beta subunit contains two stable interacting domains. J Biol Chem. 2003;278:52323–52332. doi: 10.1074/jbc.M303564200. [DOI] [PubMed] [Google Scholar]
- Payao SL, Smith MD, Bertolucci PH. Differential chromosome sensitivity to 5-azacytidine in Alzheimer's disease. Gerontology. 1998;44:267–271. doi: 10.1159/000022023. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass ML, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer's disease. Neurobiol Aging. 1998;19:S39–S42. doi: 10.1016/s0197-4580(98)00037-2. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease: evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Prager K, Wang-Eckhardt L, Fluhrer R, Killick R, Barth E, Hampel H, Haass C, Walter J. A structural switch of presenilin 1 by GSK-3beta mediated phosphorylation regulates the interaction with beta-catenin and its nuclear signaling. J Biol Chem. 2007;282:14083–14093. doi: 10.1074/jbc.M608437200. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Motsinger AA. Multifactor dimensionality reduction for detecting gene-gene and gene-environment interactions in pharmacogenomics studies. Pharmacogenomics. 2005;6:823–834. doi: 10.2217/14622416.6.8.823. [DOI] [PubMed] [Google Scholar]
- Rocca WA, Hofman A, Brayne C, Breteler MM, Clarke M, Copeland JR, Dartigues JF, Engedal K, Hagnell O, Heeren TJ. Frequency and distribution of Alzheimer's disease in Europe: a collaborative study of 1980–1990 prevalence findings. The EURODEM-Prevalence Research Group. Ann Neurol. 1991;30:381–390. doi: 10.1002/ana.410300310. [DOI] [PubMed] [Google Scholar]
- Rocchi A, Pellegrini S, Siciliano G, Murri L. Causative and susceptibility genes for Alzheimer's disease: a review. Brain Res Bull. 2003;61:1–24. doi: 10.1016/s0361-9230(03)00067-4. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute. SAS, Version 9.1. 2003. [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg BS, Kokmen E, Okazaki H. Alzheimer's disease and other dementing illnesses in a defined United States population: incidence rates and clinical features. Ann Neurol. 1987;22:724–729. doi: 10.1002/ana.410220608. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev E, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, DaSilva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shim SB, Lim HJ, Chae KR, Kim CK, Hwang DY, Jee SW, Lee SH, Sin JS, Leem YH, Lee SH, Cho JS, Lee HH, Choi SY, Kim YK. Tau overexpression in transgenic mice induces glycogen synthase kinase 3beta and beta-catenin phosphorylation. Neuroscience. 2007;146:730–740. doi: 10.1016/j.neuroscience.2007.01.041. [DOI] [PubMed] [Google Scholar]
- Sing CF, Stengard JH, Kardia SL. Dynamic relationships between the genome and exposures to environments as causes of common human diseases. World Rev Nutr Diet. 2004;93:77–91. doi: 10.1159/000081252. [DOI] [PubMed] [Google Scholar]
- Tang MX, Jacobs D, Stern Y, Marder K, Schofield P, Gurland B, Andrews H, Mayeux R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer's disease. Lancet. 1996;348:429–432. doi: 10.1016/S0140-6736(96)03356-9. [DOI] [PubMed] [Google Scholar]
- Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20:640–647. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, Moore JH. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007 doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- Wickelgren I. Estrogen research. Brain researchers try to salvage estrogen treatments. Science. 2003;302:1138–1139. doi: 10.1126/science.302.5648.1138. [DOI] [PubMed] [Google Scholar]
- Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. BioInformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PT, Li YJ, Qin XJ, Kroner C, Green-Odlum A, Xu H, Wang TY, Schmechel DE, Hulette CM, Ervin J, Hauser M, Haines J, Pericak-Vance MA, Gilbert JR. A SAGE study of apolipoprotein E3/3, E3/4 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Mol Cell Neurosci. 2007;36:313–331. doi: 10.1016/j.mcn.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB, Stiffler JS, Hurtt MR, Kaplan BB. A genome survey for novel Alzheimer disease risk loci: results at 10-cM resolution. Genomics. 1998;50:121–128. doi: 10.1006/geno.1998.5306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.