Abstract
Purpose
Ample evidence supports an important role of tumor metastasis suppressor genes in cancer metastatic processes. We evaluated the association of genetic polymorphisms of tumor metastasis suppressor gene NME1 with breast cancer prognosis in a follow-up study of patients with primary breast cancer and further investigated the functions of these polymorphisms.
Experimental Design
NME1 genotypes were analyzed in a cohort of 1134 breast cancer patients recruited as part of the Shanghai Breast Cancer Study who were followed for a median of 7.1 years. In vitro biochemical analyses were carried out to examine the function of NME1 gene polymorphisms.
Results
Single nucleotide polymorphisms (SNPs) in the promoter region of the NME1 gene were found to be associated with breast cancer prognosis. Patients carrying the C allele in rs16949649 were associated with higher breast cancer-specific mortality (HR =1.4, 95% CI =1.1–1.9) as compared to those carrying the wild-type allele, and the association was more evident in patients with an early stage cancer (HR=1.7, 95% CI =1.2–2.5). SNP rs2302254 was also associated with breast cancer prognosis, and the association was statistically significant for the risk of breast cancer relapse, metastasis, and death (HR=1.3, 95% CI, 1.0–1.6). In vitro biochemical analyses showed that minor alleles in rs2302254 and rs3760468, which is in strong linkage disequilibrium with rs16949646, altered nuclear proteins binding capacity and reduced NME1 promoter activity, supporting the results from an association study of these SNPs with breast cancer survival.
Conclusion
Promoter polymorphisms in the NME1 gene may alter its expression and influence breast cancer survival.
Introduction
Breast cancer is the most common cancer and second leading cause of cancer deaths among women in the United States and many countries of the world. The principal factor contributing to cancer-related deaths in cancer patients is metastasis, a process by which the primary tumor spreads to distant locations (1). These metastatic tumors can remain dormant and undetectable for years before developing into a life-threatening disease for which few effective treatment options exist. Better management of metastasis will undoubtedly lead to an improvement in overall cancer survival.
Metastasis is a complex process involving many genes that regulate diverse and important signal transduction pathways. The mechanisms by which metastases arise from primary tumors and the factors that regulate cancer progression are not well understood. A novel class of genes, tumor metastasis suppressor genes, have been identified and implicated in playing an important role in metastatic processes (2;3). Metastasis-suppressor genes, usually transcriptionally silenced rather than mutated in aggressive tumors, affect many important signal transduction pathways (4).
The NME1 gene, also named Nm23, is the first of thirteen identified tumor metastasis suppressor genes (2;5). NME1 is a member of the nucleoside diphosphate (NDP) kinase family of proteins. It was isolated from highly metastatic melanoma cell lines by differential colony hybridization (2). Its metastatic suppressive potential has been confirmed, in vivo, in more than ten metastatically competent cell lines, including breast cancer. NME1 possesses multiple biochemical functions, including interactions with numerous proteins with NDP kinase activity and histidine protein kinase activity. NME1 over-expression results in reduced anchorage-independent colonization, reduced invasion and motility in response to multiple factors, and increased differentiation in in vitro assays (6–8). Reduced NME1 protein and mRNA expression in tumor specimens are correlated with characteristics of aggressive cancer, such as poor clinical prognosis and survival, lymph node infiltration, as well as invasiveness and metastasis in a variety of tumor types, including breast, melanoma, gastric, ovarian, cervical, and hepatocellular carcinomas (3). Furthermore, data from the NME1 gene knockout animals show that lack of NME1 expression promotes metastasis in the SV40 animal model of liver carcinogenesis (9). We evaluated whether NME1 gene polymorphisms may be associated with breast cancer progression in a cohort of 1134 breast cancer patients who participated in the Shanghai Breast Cancer Study (10). Furthermore, we investigated the functional significance of NME1 gene variants in a series of in vitro functional analyses.
Materials and Methods
Patient population
Included in this study are breast cancer patients who participated in the Shanghai Breast Cancer Study (SBCS) between August 1996 and March 1998 (10). Breast cancer patients were identified through a rapid case ascertainment system, supplemented by the Shanghai Cancer Registry, a population-based tumor registry. The study was approved by the relevant institutional review boards for human research. It is estimated that the Shanghai Cancer Registry has an overall coverage of over 85% of incident cancer case diagnoses in urban Shanghai. Of the 1602 potentially eligible patients, 1459 were recruited into the study with a response rate of 91%. Medical charts were reviewed to obtain information on cancer diagnosis, tumor-node-metastasis (TNM) stage, estrogen receptor (ER) and progesterone receptor (PR) status, and cancer treatments. Pathology slides for all cases were reviewed independently by two senior pathologists to confirm cancer diagnosis. Patients were followed through July 2005 for cancer recurrence and mortality using a combination of two in-person surveys and multiple record linkages to death certificates that are kept by the Vital Statistics Registry of the Shanghai Center for Disease Control and Prevention. Through interviews with patients, or next of kin for deceased patients, we obtained information on disease progression, recurrence, quality of life, and cause of death (if deceased). Of the 1455 eligible patients, 1378 were followed via in-person contact or by phone (active follow-up) at least once during the two follow-up periods. Among these, 266 deaths were identified, 237 from breast cancer, 26 from other diseases, and three from unknown causes. Survival status for the 77 participants who were not actively followed up with was established by linkage to the Shanghai Vital Statistic Registry, and 47 deaths were identified, all but one due to breast cancer. The remaining 30 patients had no match in the death registry and were assumed to be living on December 31, 2004, six months before our last search of the vital statistics registry, to allow for a possible delay of entry of death certificates into the registry. Breast cancer relapse information was not available for these 30 patients and thus they were excluded from the disease-free survival analysis. Four of the 1459 breast cancer patients were excluded from the survival study because of a lack of adequate follow-up information. The median follow-up time for the cohort was 7.1 years. A peripheral blood sample was obtained from 1193 patients. Patients who provided a blood sample had a similar distribution of traditional breast cancer risk factors and similar overall survival and disease-free survival rates compared to all study participants (11;12).
Selection of single nucleotide polymorphisms (SNPs) and genotyping
We focused this study on the evaluation of genetic variants in the promoter region since metastasis-suppressor genes are usually transcriptionally silenced, rather than mutated, in aggressive tumors. Twelve SNPs in the 5’ promoter region of NME1 gene have been documented in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) and the information for minor allele frequencies were provided for five of them. In order to have adequate power to evaluate the potential association, we selected four SNPs for our study, including rs16949649 (-1465 T/C), rs3760468 (-1242 A/T), rs3760469 (-1181 T/G), and rs2302254 (-873 C/T), with a minor allele frequency (MAF) ≥5%. Since rs16949649 is in strong linkage disequilibrium (LD) with both rs3760468 (R2 of 1 and D’ of 1) and rs3760469 (R2 of 0.967 and D’ of 0.985), based on Chinese HapMap data, rs16949649 was selected to tag the remaining two SNPs. Therefore, SNPs rs16949649 and rs2302254 were included in the present study.
Genomic DNA was extracted from buffy coats (WBC) using a Puregene (Gentra Systems, Minneapolis, MN) or Qiagen (Valencia, CA) DNA purification kit following the manufacturer's protocol. The allelic discrimination of the NME1 polymorphisms were assessed with the ABI PRISM 7900 Sequence Detection Systems (Applied Biosystems, Foster City, CA) using the Taqman assay (assay IDs: C_34107066_10 for rs16949649; C_2646888_1_ for rs2302254). The final volume for each reaction was 5 µL, consisting of 2.5 µL TaqMan Universal PCR Master Mix, 0.25 µL primers/TaqMan probes mix, and 5.0 ng genomic DNA. The PCR profile consisted of an initial denaturation step at 95°C for 10 minutes and 40 cycles of 92°C for 15 seconds and 60°C for 1 minute. The fluorescence level was measured with the ABI PRISM 7900HT sequence detector (Applied Biosystems). Allele frequencies were determined by ABI SDS software.
The laboratory staff was blind to the identity of study patients. Quality control samples were included in genotyping assays. Each 384-well plate contained two water, eight CEPH 1347-02 DNA, eight blinded quality control DNA, and eight unblinded quality control DNA samples. The concordance rate for the quality control samples were 98.6% for both SNPs. In addition, 45 DNA samples of the Chinese participants used in HapMap and 24 DNA samples used in Perlegen were genotyped as an additional quality control. The genotypes of the same samples generated from our study were compared to data downloaded from HapMap (http://www.hapmap.org) and Perlegen (http://genome.perlegen.com). The concordance rates between data generated in our lab and those from HapMap (http://www.hapmap.org) and Perlegen (http://genome.perlegen.com) on these samples were 98.4% for rs16949649 and 96.8% for rs2302254.
NME1 promoter/reporter constructs and luciferase assays
A 2.0 kb promoter fragment of NME1 gene was PCR amplified by using forward primer CCCTGTATAGATTGGTCTTTTGGTGTCGTCTAAAAAC and backward primer CATCTCTTGGCCCCATCTTCCTGTCCTTTAGTTAAC. This fragment was cloned into pGL4.20 promoter-less luciferase reporter vector (Promega, WI) to generate pGL-NME1, which contains the major allele sequence and was the template for generating other haplotype constructs. The minor allele sequences of rs16949649, rs3760468, rs3760469, and rs2302254 were generated by using QuikChange Site-Directed Mutagenesis Kit (Strategene, La Jolla, CA) with the following pair of oligonucleotides, CATCTCACAACATAAGATTCGGCTCTATTCTAACGGCAT and ATGCCGTTAGAATAGAGCCGAATCTTATGTTGTGAGATG (for rs16949649), CTATTACATATTAATTTGCTTTTTTGCTTCTTGTTTTCCTA and TAGGAAAACAAGAAGCAAAAAAGCAAATTAATATGTAATAG (for rs3760468), CTCCAAGAGAGCATACTGTTGTATCCCTAGGGCCTAG and CTAGGCCCTAGGGATACAACAGTATGCTCTCTTGGAG (for rs3760469), GTGTTCTGCAAAATGGGTTCTCCGGCAGCGGATC and GATCCGCTGCCGGAGAACCCATTTTGCAGAACAC (for rs2302254). Similarly, the other haplotype constructs were generated by using a combination of different pairs of oligonucleotides. All DNA constructs were verified by sequencing analysis. Transfection was performed with the use of FuGene 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN) in triplicate for each of the constructs. Briefly, 2×105 cells of MDA231 and MCF7 were seeded in 24 well plates and co-transfected with pGL4.73, a Renilla expressing vector which served as a reference for transfection efficiency, and different allelic containing luciferase reporter constructs, respectively. Thirty-six to 48 hours later the cells were lysed with Passive Lysis Buffer and luminescence (relative light units) was measured using the Dual-Luciferase Assay System (Promega, WI). NME1 promoter activity was measured as a ratio of firefly luciferase activity to renilla luciferase activity, and the mean from five independent experiments are presented.
Electrophoretic mobility shift assay
Biotin-labeled, double stranded oligonucleotide probes 5′-CATATTAATTTGCTTATTTGCTTCTTGTTT-3′ and 5′-CATATTAATTTGCTTTTTTGCTTCTTGTTT-3′, and 5’-GTTCTGCAAAATGGGCTCTCCGGCAGCGGA-3’ and 5’-GTTCTGCAAAATGGGTTCTCCGGCAGCGGA-3’ containing the minor and major allele sequence of rs3760468 and rs2302254, respectively, in the NME1 gene promoter were synthesized. The probes were incubated with nuclear protein extracts from MDA-MB-231 and MCF7 breast cancer cells, in the presence or absence of competitors, i.e. unlabelled probe. Protein-DNA complexes were resolved by polyacrylamide gel electrophoresis and detected using a LightShift Chemiluminescent EMSA kit (Pierce Biotechnology, Rockford, IL).
Statistical analysis
Included in this study were 1134 patients with both survival status and genotype information, approximately 95% of the subjects who provided a blood sample (n=1193). The primary outcomes were breast cancer specific mortality and risk of breast cancer relapse, metastasis, and death (disease-free survival). The endpoints included cancer recurrence/metastasis or death due to breast cancer for the analysis of disease-free survival. Death due to causes other than breast cancer was excluded from survival analyses. Survival time was calculated as the time from cancer diagnosis until the occurrence of the study endpoints, censoring at the date of last contact or non-cancer death. The Kaplan-Meier method was used to estimate the survival function, and differences in survival across genotype groups were examined using the log-rank test. The Cox proportional hazard model was employed to compute hazard ratios (HRs) after adjusting for potential confounders. The proportional hazard assumption of the Cox model was examined by graphic evaluation of Schoenfeld's residual plot. All statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, North Carolina, USA) and all tests were based on two-sided probability.
Results
Patient characteristics
Descriptive data on breast cancer patients included in this study are summarized in Table 1, along with five-year survival rates by demographic characteristics of breast cancer patients and clinico-pathologic features of cancer. During the average 7.1 years of follow-up, 245 deaths and 310 disease-related events (including 223 deaths from breast cancer and 87 relapses or metastasis) were identified among the 1134 patients.
Table 1.
Overall survival by demographics and known breast cancer prognosis factors, the Shanghai Breast Cancer Study
| Covariables | Levels | Number of cases | Number of deaths | 5-y survival, % | P |
|---|---|---|---|---|---|
| Age at diagnosis | <42 | 282 | 64 | 84.0 | 0.011 |
| 42–46 | 283 | 41 | 88.3 | ||
| 47–52 | 278 | 67 | 81.3 | ||
| >52 | 291 | 73 | 80.8 | ||
| Education | <Middle School | 139 | 37 | 79.1 | 0.278 |
| Middle School | 502 | 109 | 83.5 | ||
| > Middle School | 493 | 99 | 84.8 | ||
| TNM stage * | 0–I | 284 | 25 | 93.3 | <0.001 |
| IIa | 400 | 66 | 88.2 | ||
| IIb | 252 | 66 | 79.8 | ||
| III–IV | 125 | 61 | 60.0 | ||
| ER * | Positive | 504 | 107 | 83.9 | 0.456 |
| Negative | 286 | 54 | 85.0 | ||
| PR * | Positive | 505 | 107 | 83.8 | 0.450 |
| Negative | 276 | 52 | 84.8 | ||
| Chemotherapy * | Yes | 1,061 | 225 | 84.0 | 0.417 |
| No | 59 | 15 | 79.7 | ||
| Radiotherapy * | Yes | 435 | 140 | 75.2 | <0.01 |
| No | 538 | 79 | 89.4 | ||
| Tamoxifen use * | Yes | 729 | 127 | 88.5 | 0.510 |
| No | 211 | 40 | 86.3 | ||
| Total | 1,134 | 245 |
Patients with missing data were not included in the analyses
Associations of NME1 SNPs with breast cancer survival
The minor allele frequencies were 0.425 for rs16949649 and 0.26 for rs2302254. Genotype distributions of both SNPs were in agreement with Hardy-Weinberg equilibrium.
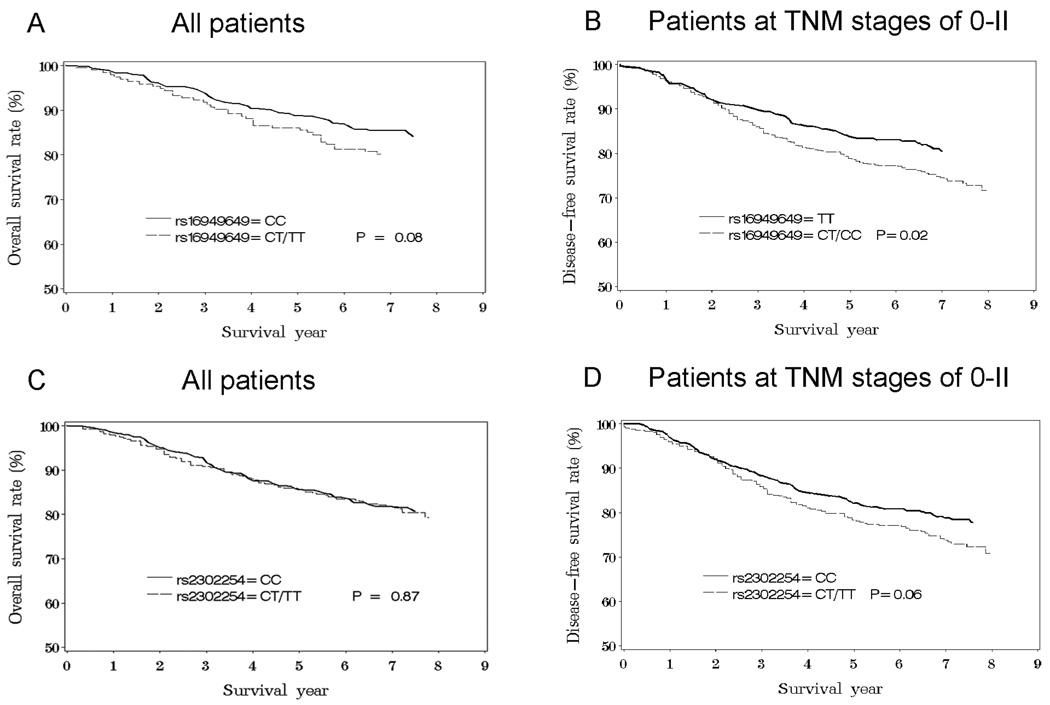
Results from association analyses for rs16949649 and rs2302254 and breast cancer prognosis outcomes are summarized in Table 2. Patients carrying the C allele in rs16949649 had a higher breast cancer-specific mortality (HR =1.4, 95% CI, 1.1–1.9) compared to patients with the TT genotype. This association was more pronounced in patients with an early-stage cancer (HR=1.7, 95% CI, 1.2–2.5) than those with a late-stage cancer (HR=1.1, 95% CI, 0.6–2.0). A similar pattern was observed for disease-free survival, although the association was slightly weaker than that for breast cancer-specific mortality. SNP rs2302254 was also associated with breast cancer prognosis, and the association was statistically significant for disease-free survival (HR=1.3, 95% CI, 1.0–1.6), particularly for patients with an early-stage cancer (HR=1.4, 95% CI, 1.1–1.8). The Kaplan – Meier survival curves are presented in Figure 1, showing that the survival function is consistent with the proportional hazard assumption.
Table 2.
NME1 gene variants and breast cancer survival
| Marker | Breast cancer mortality |
Disease -free survival a |
||||
|---|---|---|---|---|---|---|
| cases | events | HR (95% CI) b | cases | events | HR (95% CI) b | |
| All patients | ||||||
| rs16949649 | ||||||
| TT | 382 | 63 | 1.0 (ref) | 365 | 91 | 1.0 (ref) |
| CC/CT | 751 | 160 | 1.4 (1.1–1.9) | 728 | 213 | 1.3 (1.0–1.6) |
| CC | 213 | 42 | 1.4 (0.9–2.0) | 207 | 53 | 1.0 (0.7–1.4) |
| CT | 538 | 118 | 1.4 (1.1–2.0) | 521 | 160 | 1.3 (0.8–1.6) |
| rs2302254 | ||||||
| CC | 631 | 125 | 1.0 (ref) | 608 | 165 | 1.0 (ref) |
| CT/TT | 500 | 99 | 1.2 (0.9–1.5) | 483 | 140 | 1.3 (1.0–1.6) |
| CT | 411 | 82 | 1.2 (0.9–1.6) | 398 | 121 | 1.3 (1.1–1.7) |
| TT | 89 | 17 | 1.1 (0.7–1.8) | 85 | 19 | 1.0 (0.6–1.6) |
| Patients with TNM stage 0–II | ||||||
| rs16949649 | ||||||
| TT | 314 | 36 | 1.0 (ref) | 304 | 57 | 1.0 (ref) |
| CC/CT | 622 | 107 | 1.7 (1.2–2.5) | 610 | 156 | 1.5 (1.1–2.1) |
| CC | 179 | 30 | 1.6 (1.0–2.6) | 177 | 42 | 1.3 (0.9–2.0) CT |
| CT | 443 | 77 | 1.8 (1.2–2.6) | 433 | 114 | 1.6 (1.2–2.2) |
| rs2302254 | ||||||
| CC | 504 | 72 | 1.0 (ref) | 491 | 103 | 1.0 (ref) |
| CT/TT | 429 | 71 | 1.3 (0.9–1.8) | 420 | 110 | 1.4 (1.1–1.8) |
| CT | 351 | 56 | 1.3 (0.9–1.8) | 343 | 93 | 1.5 (1.1–2.0) |
| TT | 78 | 15 | 1.3 (0.7–2.2) | 77 | 17 | 1.0 (0.6–1.7) |
| Patients with TNM stage III–IV | ||||||
| rs16949649 | ||||||
| TT | 44 | 18 | 1.0 (ref) | 38 | 23 | 1.0 (ref) |
| CC/CT | 80 | 38 | 1.1 (0.6–2.0) | 69 | 41 | 1.1 (0.6–2.0) |
| CC | 19 | 9 | 1.3 (0.5–2.9) | 15 | 8 | 1.1 (0.5–2.6) |
| CT | 61 | 29 | 1.0 (0.5–2.0) | 54 | 33 | 1.1 (0.6–2.0) |
| rs2302254 | ||||||
| CC | 82 | 37 | 1.0 (ref) | 73 | 43 | 1.0 (ref) |
| CT/TT | 43 | 20 | 0.9 (0.5–1.6) | 35 | 22 | 1.1 (0.6–2.0) |
| CT | 37 | 18 | 0.9 (0.5–1.7) | 32 | 20 | 1.0 (0.6–1.8) |
| TT | 6 | 2 | 1.0 (0.2–4.5) | 3 | 2 | 3.9 (0.9–18.1) |
Defined as risk of breast cancer relapse, metastasis, and death.
Adjusted by age, education, TNM stage, radio therapy, chemotherapy, tamoxifen use, ER and PR status. Patients who died of causes other than breast cancer were excluded in the breast cancer mortality analyses.
Fig. 1.
Kaplan – Meier survival analyses with breast cancer-specific death (A, C) and disease-free survival (B, D) among breast cancer patients.
Using genotype data from these two SNPs, we derived haplotypes for study patients. Three of the four possible haplotypes (in the order of rs16949649/rs2302254), TC (57.18%), CC (25.71%) and CT (16.85%) accounted for more than 99.7% among all patients. The adjusted HRs for haplotype CC was 1.3 (95% CI, 1.0–1.6)_and CT was 1.1 (95% CI, 0.9–1.4), for breast cancer death when compared to the common haplotype TC (data not shown in tables). Such associations were more evident among patients with an early-stage cancer, TNM stage 0-II, with adjusted HRs for overall survival of 1.4 (95% CI, 1.0–2.0) for patients carrying the CC haplotype and 1.3 (95% CI, 1.0–1.7) for patients carrying the CT haplotype compared to those with the TC haplotype. The CC and CT haplotypes were also associated with disease-free survival. However, the associations were not statistically significant (data not shown).
Evaluation of functionality of NME1 variants
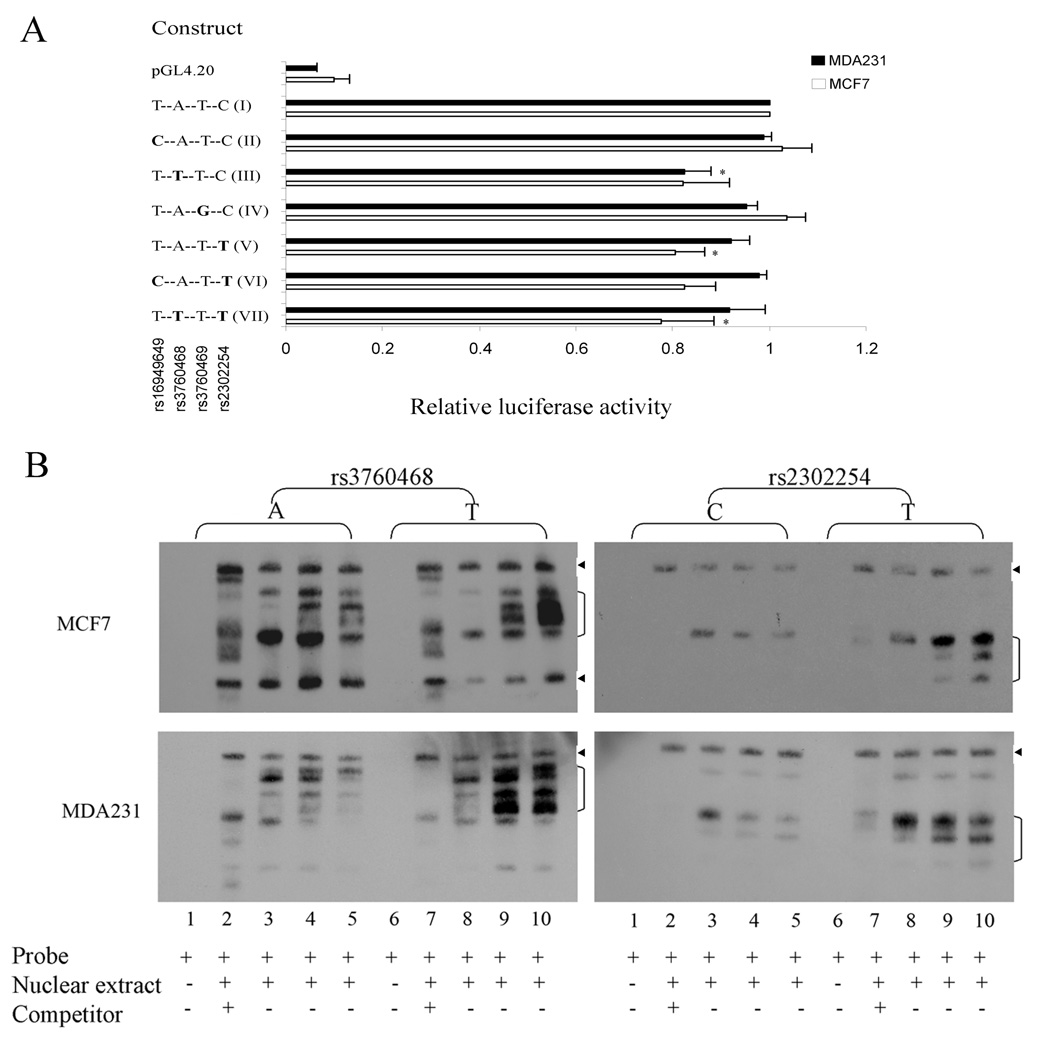
To evaluate whether these two SNPs affect NME1 gene expression, we conducted an in vitro NME1 promoter/luciferase assay in the metastatic MDA231 and non-metastatic MCF7 breast cancer cells. We generated several luciferase reporter constructs containing different variant alleles and haplotypes (in the order of rs16949649/rs3760468/rs3760469/rs2302254) in the promoter region of the NME1 gene, spanning from upstream of 1900 base pair to ATG translation start codon. The variant allele(s) were introduced by site-directed mutagenesis. MDA231 and MCF7 breast cancer cells were transiently transfected with these constructs. The luciferase assay results showed that rs2302254 minor allele (construct V, T-A-T-T) reduced NME1 promoter activity by about 20% (0.80 ± 0.06 relative to major allele T-A-T-C, construct I) in the MCF7 cells (p<0.05) and rs16949649 minor allele (construct II, C-A-T-C) does not have any apparent effects on NME1 promoter when compared to their major allele (construct I, T-A-T-C) (Fig. 2A). However, the minor allele (construct III, T-T-T-C) in the rs3760468, a SNP in close proximity to and in strong LD with rs16949649, reduces NME1 promoter activity by 18% compared to the major allele containing promoter in both MCF7 (0.82 ± 0.09, p=0.06) and MDA231 (0.82 ± 0.05, p<0.05) cells. We also examined possible interactive effects of those SNPs by examining NME1 promoter harboring different haplotype sequences (construct VI, C-A-T-T and construct VII, T-T-T-T) in breast cancer cells and found no apparent interaction of these two SNPs (Fig. 2A).
Fig. 2.
Characterization of NME1 promoter polymorphisms in breast cancer cells. A, Schematic of NME1 promoter/luciferase reporter constructs and relative luciferase activities of those constructs in MDA231 and MCF7 cells. Each value (relative to wild type) represents mean ± s.d. of five experiments. Statistical analysis was done using student’s t test. * P < 0.05 compared with wild-type construct (I).
B, Electrophoretic mobility shift assays. Nuclear protein extracts from MCF7 (top panels) and MDA-MB-231 (bottom panels) breast cancer cells were incubated with biotin-labeled probes corresponding to the major allele (lanes 1 to 5) or the minor allele (lanes 6 to 10) of rs3760468 (left panels) and rs2302254 (right panels) in the absence or presence of competitors. Lanes 1 and 6, no nuclear protein extract; 2 and 7, competitor in 200 fold molar excess; 3 and 8 (5 mM MgCl2), 4 and 9 (2.5 mM MgCl2), 5 and 10 (1.25 mM MgCl2), no competitor. Arrowhead, nonspecific DNA-protein complexes bands.
To investigate whether the sequence at the rs3760468 and rs2302254 SNP sites interact with nuclear proteins of breast cancer cells and if so, whether a single nucleotide change in these sites can alter the protein-DNA interactions, we performed electrophoretic mobility shift assays. In these assays, oligonucleotide probes corresponding to the major or minor allele were incubated with nuclear protein extracts from MDA-231 and MCF7 breast cancer cells. The results showed that T minor allele in SNP rs2302254 resulted in altered DNA-protein complexes intensity in both MCF7 and MDA231 cells. Similarly, T minor allele in SNP rs3760468 altered DNA-protein complexes intensity in MDA231 cells, while it altered both DNA-protein complexes intensity and patterns in MCF7 cells (Fig. 2B). In contrast, the difference between major and minor alleles do not alter nonspecific DNA-protein complexes bands which were unaffected by the presence of large amounts of unlabeled competitors.
Discussion
The NME1 gene was identified on the basis of its reduced expression in highly metastatic murine melanoma cell lines (2). The molecular mechanisms of NME1-mediated metastasis suppression are emerging (13;14), but are still largely unknown. It has been reported that NME1 protein and mRNA expression in tumor specimens are inversely correlated with metastatic potential in a variety of tumor types, including breast, melanoma, gastric, ovarian, cervical, and hepatocellular carcinomas (3). Little is known about the prognostic effect of the functional polymorphisms of the NME1 gene in breast cancer except one study showed an intronic EcoR1 polymorphism was not associated with breast cancer in a small sample size of Mexican patients (15).
We found, for the first time, that genetic polymorphisms in the NME1 gene promoter region were associated with breast cancer survival in a large cohort of 1134 Chinese patients. The minor C allele in SNP rs16949649 was associated with higher breast cancer-specific mortality (HR =1.4, 95% CI =1.1–1.9), and the association was more evident in patients with an early stage cancer (HR=1.7, 95% CI =1.2–2.5). The minor T allele in SNP rs2302254 was associated with disease-free survival (HR=1.3, 95% CI, 1.0–1.6). The reasons for a stronger association with early-stage cancer than late-stage cancer are unknown. However, it is possible that the prognosis of late-stage cancer is determined primarily by characteristics of a tumor, such as tumor grades and somatic changes. The sample size for late-stage cancer in our study is small and thus we can not exclude the possibility that our study is not well–powered enough to evaluate the association in patients with a late-stage cancer.
We also investigated the potential underlying mechanisms of the positive association observed in our association analysis of breast cancer patients through in vitro biochemical analyses. The results from in vitro functional analysis showed both minor alleles in SNPs rs3760468 and rs2302254 reduced NME1 promoter activity in MDA-MB-231 or MCF7 breast cancer cells. This suggests that the reduced expression of the NME1 gene in breast cancer tissues in some patients may be a result of these genetic polymorphisms. It was reported that the deletion of a 544bp AvrII fragment, which contains SNPs rs16949649, rs3760468, and rs3760469, resulted in a 20% increase of NME1 promoter activity in MCF7 cells and deletion of the 195bp Nhe-XbaI fragment, which contains SNP rs2302254, abolishes NME1 promoter activity (16). These data are consistent with our findings, implicating the presence of regulatory elements in the polymorphic region of NME1 promoter. The altered NME1 promoter activity could be the result of differential affinity of transcription machinery to the underlying SNPs. The electrophoretic mobility shift assays further confirmed our hypothesis that minor T alleles in both SNPs rs3760468 and rs2302254 alter DNA-nuclear protein(s) interactions (Fig. 2B). Thus, it is possible that inhibitory nuclear protein(s) selectively bind to the minor allele to repress NME1 transcription, or the minor allele containing DNA sequences has less affinity to the transcription machinery and results in a relatively weaker NME1 transcription. This perhaps is one of the mechanisms through which the NME1 genetic variants influence breast cancer survival. A database search (http://www.cbrc.jp/research/db/TFSEARCH.html) for transcription factor binding sites showed that the variant sequences at the rs3760468 and rs2302254 sites have a high degree of similarity with several consensus elements recognized by transcription factors differentially. Those transcription factors include HSF, Kr, ADR1, Oct-1, Ftz, C/EBP and Nkx-2. Although these putative transcription factors or their associated proteins have not been confirmed by biological experiments to be involved in the regulation of NME1 expression, it is possible that some of them might have differential binding capacity to the variant sequences of NME1 SNPs.
A number of factors have been reported to be related to breast cancer prognosis, such as histologic features of the tumor, including tumor size, histologic grade, nodal status and lymphovascular invasion, hormone receptor status, HER-2 status and Ki67 expression (17–19). However, these known clinical prognostic markers have proven insufficient in predicting the risk for metastasis. Gene expression profiles of tumor tissues have shown great promise for predicting the metastatic potential of breast cancers (20;21). However, it is not practical to apply this technology in routine clinical care (14). Studies that evaluate novel prognostic biomarkers, such as genetic polymorphisms, may provide valuable information toward the design of cost-efficient and practical approaches to identify high risk patients for personalized medical care. Recent studies conducted in mouse models of mammary tumors have demonstrated that the genetic background from which a tumor arises can have a significant effect on the ability of the tumor to metastasize (22). Gene profiling data has further demonstrated that genetic background significantly influences gene expression, including the metastasis signature genes (23). These findings suggest the possibility that person-to-person genetic variability could predispose individuals to cancer metastasis. Identification and characterization of these polymorphisms may have significant implications for predicting clinical prognosis and aid the development of personalized treatment for recurrent disease.
In summary, we found that the NME1 gene promoter polymorphisms were associated with cancer survival among breast cancer patients. These associations are supported by in vitro data generated in our study showing the functional significance of these genetic variants. These results are consistent with the role of the NME1 gene in cancer metastasis. Our results, if confirmed in future studies, may have significant implications in identifying high risk patients for personalized treatment and follow-up care.
Acknowledgments
We thank Regina Courtney and Qing Wang for technical assistance in genotyping, Brandy Venuti for assistance in the preparation of this article, and all of the study participants and research staff of the Shanghai Breast Cancer Study for their support. This research was supported by NIH grants RO1 CA64277 and RO1 CA90899 from the National Cancer Institute.
Financial Support: This research was supported by NIH grants RO1 CA64277 and RO1 CA90899 from the National Cancer Institute.
Reference
- 1.Liotta LA, Stetler-Stevenson WG. Principles of molecular cell biology of cancer: cancer metastasis. In: DeVita SHV, Rosenberg SA, editors. Cancer: Principles and Practice of Oncology. Philadelphia: Lippincott Co.; 1993. pp. 134–149. [Google Scholar]
- 2.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 3.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 4.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmieri D, Horak CE, Lee JH, Halverson DO, Steeg PS. Translational approaches using metastasis suppressor genes. J Bioenerg Biomembr. 2006;38:151–161. doi: 10.1007/s10863-006-9039-9. [DOI] [PubMed] [Google Scholar]
- 6.Leone A, Flatow U, King CR, et al. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- 7.Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene. 1993;8:2325–2333. [PubMed] [Google Scholar]
- 8.Ouatas T, Salerno M, Palmieri D, Steeg PS. Basic and translational advances in cancer metastasis: Nm23. J Bioenerg Biomembr. 2003;35:73–79. doi: 10.1023/a:1023497924277. [DOI] [PubMed] [Google Scholar]
- 9.Boissan M, Wendum D, rnaud-Dabernat S, et al. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–845. doi: 10.1093/jnci/dji143. [DOI] [PubMed] [Google Scholar]
- 10.Gao YT, Shu XO, Dai Q, et al. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Shu XO, Cui Y, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–5019. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 12.Shu XO, Gao YT, Cai Q, et al. Genetic polymorphisms in the TGF-beta 1 gene and breast cancer survival: a report from the Shanghai Breast Cancer Study. Cancer Res. 2004;64:836–839. doi: 10.1158/0008-5472.can-03-3492. [DOI] [PubMed] [Google Scholar]
- 13.Horak CE, Lee JH, Elkahloun AG, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–7246. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 14.Hunter KW, Crawford NP. Germ line polymorphism in metastatic progression. Cancer Res. 2006;66:1251–1254. doi: 10.1158/0008-5472.CAN-05-3705. [DOI] [PubMed] [Google Scholar]
- 15.Rubio SA, Martinez SE, Corona JS, et al. EcoRI polymorphism of the metastasis-suppressor gene NME1 in Mexican patients with breast cancer. Breast Cancer Res Treat. 2006;96:159–161. doi: 10.1007/s10549-005-9072-0. [DOI] [PubMed] [Google Scholar]
- 16.Ouatas T, Clare SE, Hartsough MT, De La RA, Steeg PS. MMTV-associated transcription factor binding sites increase nm23-H1 metastasis suppressor gene expression in human breast carcinoma cell lines. Clin Exp Metastasis. 2002;19:35–42. doi: 10.1023/a:1013897022827. [DOI] [PubMed] [Google Scholar]
- 17.Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. 2005;16:1723–1739. doi: 10.1093/annonc/mdi352. [DOI] [PubMed] [Google Scholar]
- 18.de AE, Cardoso F, de CG, Jr., Colozza M, et al. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–1513. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes DF. Prognostic and predictive factors revisited. Breast. 2005;14:493–499. doi: 10.1016/j.breast.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 21.van ' V, Dai H, van d V, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 22.Park YG, Zhao X, Lesueur F, et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet. 2005;37:1055–1062. doi: 10.1038/ng1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Crawford N, Lukes L, Finney R, Lancaster M, Hunter KW. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]