Abstract
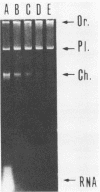
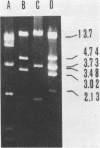
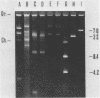
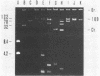
A rapid and simple plasmid isolation procedure was developed for the epidemiological analysis of plasmid-mediated antimicrobial resistance. By this method, plasmid DNAs ranging in molecular weight between 2.0 and 122 X 10(6) could be detected. Various bacteria, such as strains of the family Enterobacteriaceae, Pseudomonas aeruginosa, Haemophilus influenzae, and Staphylococcus aureus, could be analyzed. The plasmid DNA obtained could be directly used for restriction endonuclease analysis without further purification. In addition, this method made it possible to analyze several cultures at the same time.
Full text
PDF

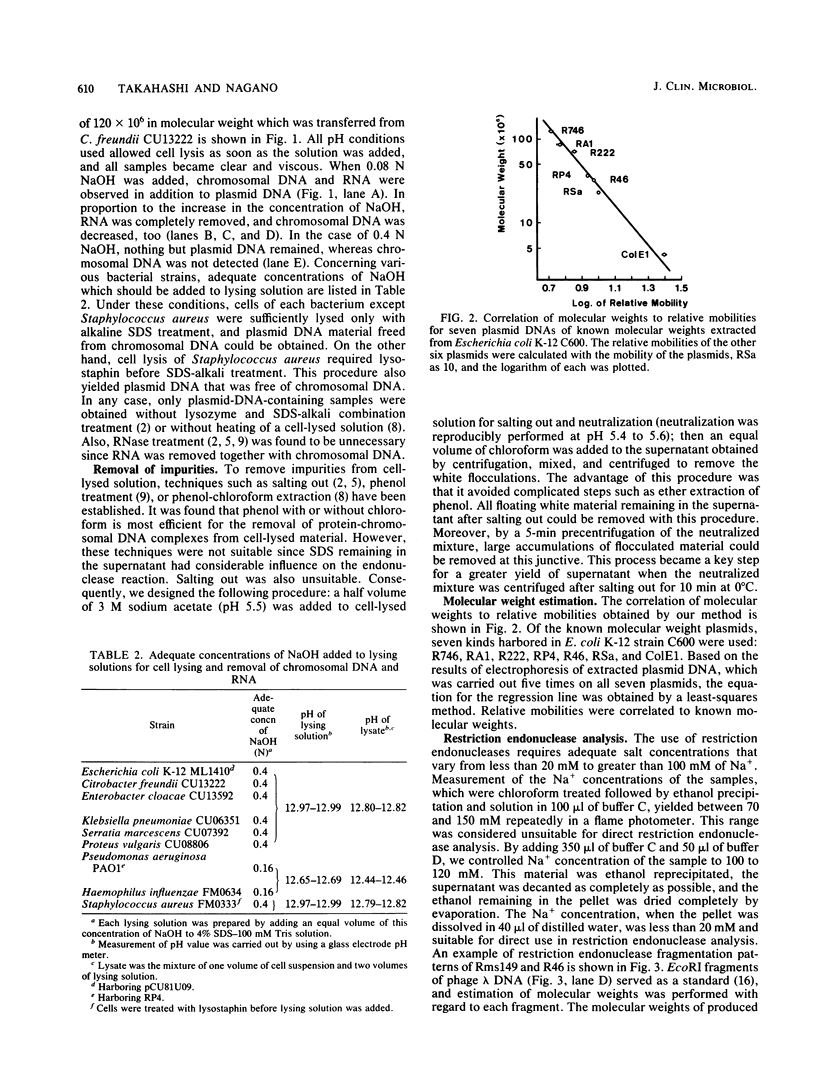

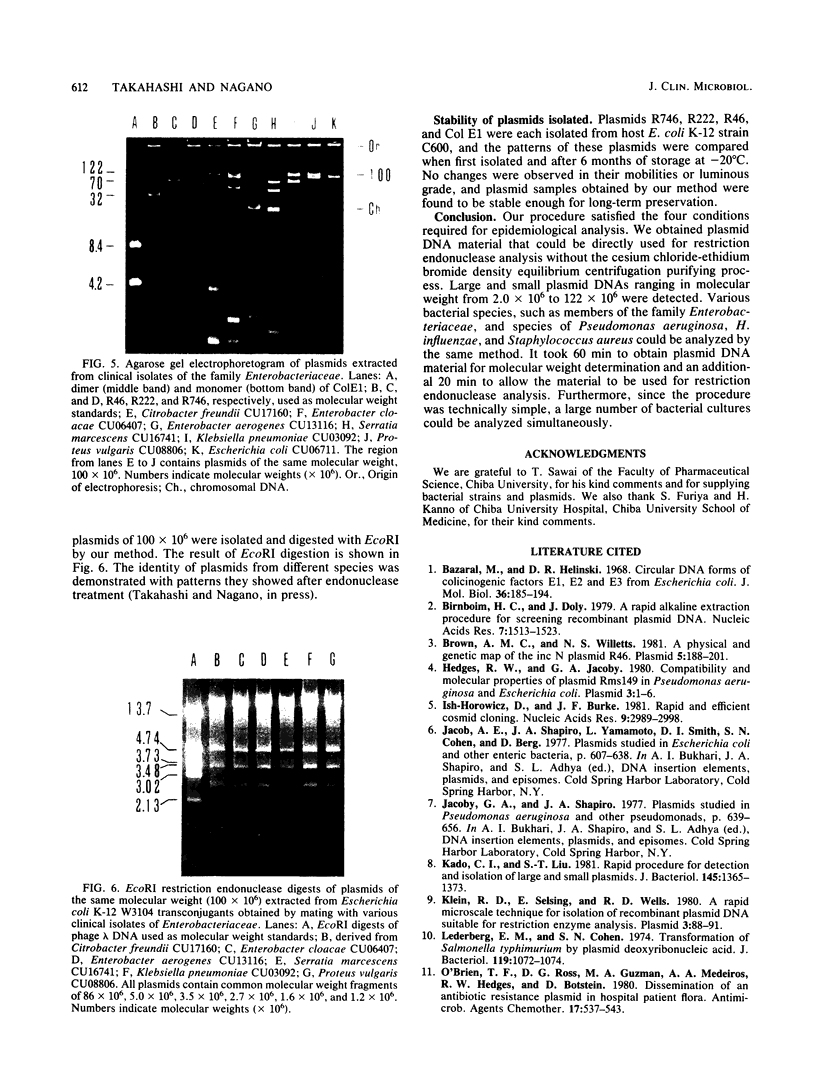

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. M., Willetts N. S. A physical and genetic map of the IncN plasmid R46. Plasmid. 1981 Mar;5(2):188–201. doi: 10.1016/0147-619x(81)90020-2. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacoby G. A. Compatibility and molecular properties of plasmid Rms 149 in Pseudomonas aeruginosa and Escherichia coli. Plasmid. 1980 Jan;3(1):1–6. doi: 10.1016/s0147-619x(80)90029-3. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T. F., Ross D. G., Guzman M. A., Medeiros A. A., Hedges R. W., Botstein D. Dissemination of an antibiotic resistance plasmid in hospital patient flora. Antimicrob Agents Chemother. 1980 Apr;17(4):537–543. doi: 10.1128/aac.17.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C., Swenson C. D., Owens L. M., Smith A. L. Characterization of chloramphenicol-resistant Haemophilus influenzae. Antimicrob Agents Chemother. 1980 Oct;18(4):610–615. doi: 10.1128/aac.18.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski P. L., Peterson B. C., Gerding D. N., Cleary P. P. Physical characterization of ten R plasmids obtained from an outbreak of nosocomial Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 1979 Apr;15(4):616–624. doi: 10.1128/aac.15.4.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Currie C. A. Endemic gentamicin resistance R factors on a spinal cord injury unit. J Clin Microbiol. 1983 Aug;18(2):236–241. doi: 10.1128/jcm.18.2.236-241.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantulavanich S., Olexy V. M., Prasad T. R., Bird T. J., Talanda-Fath C., Grieble H. G., Farrand S. K. An R plasmid of broad host-range, coding for resistance to nine antimicrobial agents endemic in Gram-negative nosocomial isolates. J Med Microbiol. 1981 Nov;14(4):371–380. doi: 10.1099/00222615-14-4-371. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Tompkins L. S., Plorde J. J., Falkow S. Molecular analysis of R-factors from multiresistant nosocomial isolates. J Infect Dis. 1980 May;141(5):625–636. doi: 10.1093/infdis/141.5.625. [DOI] [PubMed] [Google Scholar]
- Totten P. A., Vidal L., Baldwin J. N. Penicillin and tetracycline resistance plasmids in Staphylococcus epidermidis. Antimicrob Agents Chemother. 1981 Sep;20(3):359–365. doi: 10.1128/aac.20.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]