Abstract
BACKGROUND
The endothelial nitric-oxide synthase (NOS3) gene encodes the enzyme (eNOS) that synthesizes the molecule nitric oxide, which facilitates endothelium-dependent vasodilation in response to physical activity. Thus, energy expenditure may modify the association between the genetic variation at NOS3 and blood pressure.
METHODS
To test this hypothesis, we genotyped 11 NOS3 polymorphisms, capturing all common variations, in 726 men and women from the Medical Research Council (MRC) Ely Study (age (mean ± s.d.): 55 ± 10 years, body mass index: 26.4 ± 4.1 kg/m2). Habitual/non-resting energy expenditure (NREE) was assessed via individually calibrated heart rate monitoring over 4 days.
RESULTS
The intronic variant, IVS25+15 [G→A], was significantly associated with blood pressure; GG homozygotes had significantly lower levels of diastolic blood pressure (DBP) (−2.8 mm Hg; P = 0.016) and systolic blood pressure (SBP) (−1.9 mm Hg; P = 0.018) than A-allele carriers. The interaction between NREE and IVS25+15 was also significant for both DBP (P = 0.006) and SBP (P = 0.026), in such a way that the effect of the GG-genotype on blood pressure was stronger in individuals with higher NREE (DBP: −4.9 mm Hg, P = 0.02. SBP: −3.8 mm Hg, P = 0.03 for the third tertile). Similar results were observed when the outcome was dichotomously defined as hypertension.
CONCLUSIONS
In summary, the NOS3 IVS25+15 is directly associated with blood pressure and hypertension in white Europeans. However, the associations are most evident in the individuals with the highest NREE. These results need further replication and have to be ideally tested in a trial before being informative for targeted disease prevention. Eventually, the selection of individuals for lifestyle intervention programs could be guided by knowledge of genotype.
Hypertension, which affects 30–50% of adults in Europe and North America, is an established risk factor for cardiovascular disease.1 Improvements in blood pressure can be achieved in most individuals by increasing habitual physical activity levels.2 However, the extent to which blood pressure can be managed via exercise intervention varies markedly from one individual to the next, which may be due in part to genetic variation.3 Therefore, it is likely that the interaction between genetic and lifestyle factors determine blood pressure and the risk of developing hypertension. Genes that encode proteins involved in blood pressure homeostasis and that are sensitive to lifestyle changes such as diet and exercise may represent the most appropriate candidates to test hypotheses of gene–lifestyle interactions on hypertension risk.
The endothelial nitric-oxide synthase (NOS3) gene encodes the enzyme that synthesizes the molecule nitric oxide.4 In humans, endothelial nitric-oxide synthase (eNOS) is a major metabolic determinant of blood pressure, accounting for diastolic and systolic basal inhibition of ~30 mm Hg, as demonstrated in studies of eNOS blockade.5 Nitric oxide is released from endothelial cells and myocytes during the metabolism of energy and is, therefore, to varying degrees, continuously synthesized.4 Although NOS3 is basally expressed, elevations in energy expenditure markedly increase NOS3 activity,6 which accounts for some of the endothelium-dependent vasodilation associated with exercise.5 The mechanisms through which this is achieved include shear stress7 and AMP kinase–mediated NOS3 phosphorylation.8
Several recent studies in humans have described the interaction between NOS3 gene variants and exercise-related factors on cardiovascular traits such as blood pressure and coronary artery disease.9,10 In this study, we examined the interaction between habitual/non-resting energy expenditure (NREE) and11 NOS3 gene variants on blood pressure and hypertension risk in cross-sectional data from the Medical Research Council (MRC) Ely study.
METHODS
Study population
The MRC Ely Study11,12 is a prospective population-based cohort study of the etiology and pathogenesis of type 2 diabetes and related metabolic disorders. All participants in this study were white, living in the same geographical region in the east of England. This study includes 726 volunteers (298 men and 428 women), aged 31–73, from phase 2 of the MRC Ely study in whom the necessary genetic and phenotypic data were available. The study design, methods, and measurements have been described in detail elsewhere.13 Table 1 shows the clinical characteristics of the study participants (n = 726).
Table 1.
Clinical characteristics of the study participants in the MRC Ely study
| Men (n = 298) | Women (n = 428) | |
|---|---|---|
| Characteristics | Mean ± s.d. | Mean ± s.d. |
| Age (years) | 55 ± 11 | 54 ± 10 |
| Body mass index (kg/m2) | 26.8 ± 3.2 | 26.5 ± 4.7 |
| Body fat (%) | 24.3 ± 4.7 | 37.6 ± 6.7 |
| Diastolic blood pressure (mm Hg) |
78.8 ± 10.9 | 74.4 ± 10.0 |
| Systolic blood pressure (mm Hg) |
129 ± 16 | 124 ± 16 |
| Non-resting energy expenditure (kJ/day) |
15047.2 ± 5817.8 | 11091.6 ± 4737.1 |
| Total serum cholesterol (mmol/l) |
5.9 ± 1.1 | 6.0 ± 1.1 |
| Serum triglycerides (mmol/l) | 1.4 ± 0.6 | 1.1 ± 0.6 |
| High-density lipoprotein cholesterol (mmol/l) |
1.3 ± 0.3 | 1.6 ± 0.4 |
| Low-density lipoprotein cholesterol (mmol/l) |
3.9 ± 1.0 | 3.8 ± 1.1 |
| Fasting blood glucose (mmol/l) | 5.0 ± 0.6 | 4.8 ± 0.5 |
| Fasting serum insulin | 41.6 ± 30.5 | 38.4 ± 26.6 |
| % of participants with obesity | 16.1 | 17.8 |
| % of participants with hypertension |
30.9 | 25.8 |
Ethical permission was granted by the Cambridgeshire Local Research Ethics Committee. All participants provided written informed consent.
Blood pressure and hypertension
Blood pressure was measured by trained personnel with the participant seated using an Accutorr automated sphygmomanometer (Datascope, Cambridge, UK). Blood pressure values were measured three times and the average of the measurements were considered for the study. Hypertension was defined as diastolic blood pressure (DBP) ≥90 mm Hg and/or systolic blood pressure (SBP) ≥140 mm Hg and/or the use of anti-hypertensive drugs.14 Participants self-reported the use of antihypertensive medications. There were 190 hypertensive participants, of whom 90 took antihypertensive drugs.
Anthropometry and physical activity measurement
Participants attended the laboratory after a 10-h overnight fast. Standard anthropometric data were obtained by trained observers with participants in lightweight clothing without shoes. Height and weight were measured using a rigid stadiometer and calibrated scales, respectively. Resistance was assessed using a standard bioimpedance technique (Bodystat, Isle of Man, UK). This device has previously been shown to be a valid and reliable15 measure of percentage body fat. Fat mass (FM) was calculated from percentage body fat, and body weight and fat free mass (FFM) was calculated as the difference between weight and FM.
NREE was assessed objectively in a subset of subjects (n = 652) using the Flex Heart Rate technique.16 This method has been shown to be reliable and valid for assessing energy expenditure when compared to the gold-standard methods of doubly labeled water and indirect calorimetry.16 Briefly, Flex Heart Rate technique involves the individual calibration of heart rate against energy expenditure assessed using indirect calorimetry at rest and during a graded exercise stress test. All methods for this study have been described in detail previously.17 In this analyses, NREE is defined as total free-living energy expenditure adjusted for FM and FFM. Since FM and FFM account for the vast majority of variation in basal metabolic rate, the residual variance represents all non-basal energy metabolism independent of adiposity, a potential confounding factor.
SNP selection and genotyping
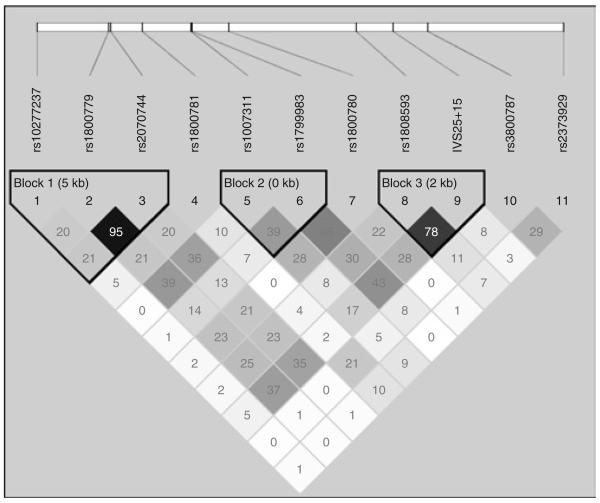
The study population was genotyped for 11 NOS3 gene variants. Five of these single-nucleotide polymorphisms (SNPs) (IVS2+42− rs1800781, IVS6−26− rs1007311, IVS11−30− rs1800780, E298D− rs1799983, and IVS25+15− rs891512) were identified by de novo polymorphism screening in a human diversity panel with 47 samples from four ethnically diverse populations and were genotyped, as described previously.18 Three SNPs (rs1800779, rs3800787, and rs2070744) were genotyped at the Wellcome Trust Sanger Institute, as described elsewhere.19 An additional six tagSNPs (rs10277237, rs1808593, rs2373929, rs3918227, rs3918186, and rs3918188) were selected from the dbSNP database to ensure full coverage of the common genetic variations in the NOS3 gene (5 kb upstream and downstream of the gene; r2 = 0.8; minor allele frequency >0.05; Supplementary Figure S1 online), according to the genetic variation reported for the CEU population of the International HapMap project Phase 2 release 21.20 The SNPs, rs3918227, rs3918186, and rs3918188, failed assay design and were not genotyped. The three tagSNPs selected from the HapMap capture 50% of the common genetic variation reported for the CEU population. Seven SNPs that were chosen in this study were not reported in the HapMap and were not in linkage disequilibrium with HapMap tagSNPs and therefore potentially capture part of the remaining 50% of the genetic variation in the CEU population. Figure 1 shows the pairwise linkage disequilibrium between SNPs from MRC Ely study.
Figure 1.
Pairwise linkage disequilibrium (LD) comparisons (r2 values shown in the squares) for the 11 SNPs in the nitric oxide synthase 3 (NOS3) gene identified in this study; LD blocks as defined by Gabriel method of haplotype analysis.
The additional three tagSNPs were genotyped by Custom TaqMan SNP Genotyping Assays (Applied Biosystems, Warrington, UK). The genotyping assays were carried out on 10 ng of genomic DNA in a 5 μl 384-well TaqMan assay using a PTC-225 Thermal Cycler (MJ Research, Watertown, MA). The ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Warrington, UK) was used for end point detection and allele calling. The genotyping success rate was >98%.
Statistical analysis
A likelihood ratio test was performed to assess whether the observed genotype frequencies were in Hardy–Weinberg equilibrium. The genotype frequencies and Hardy–Weinberg equilibrium P values of the 11 SNPs are represented in Supplementary Table S1 (online). Genotype frequencies of all SNPs were in Hardy–Weinberg equilibrium. Linkage disequilibrium between SNPs (r2 value) was estimated using Haploview V3.2 (http://www.broad.mit.edu/mpg/haploview).
Association analyses were conducted using SAS 9.1 for windows (SAS Institute, Cary, NC). The association between the NOS3 SNPs and blood pressure were tested using generalized linear models assuming an additive effect for each additional minor allele. Logistic regression analyses were performed to test the association of the NOS3 SNPs with the risk of hypertension. Interactions between SNPs and NREE (continuous trait) were tested by including interaction terms (NREE*SNPs) in the (generalized linear model or logistic) model. For clarity of presentation, the sample was stratified into sex-specific tertiles of NREE per kg body weight. All models were adjusted for age, sex, FM, and FFM. Interaction models included the main effects and were adjusted for age, sex, FM, and FFM. The association between IVS25+15 and blood pressure was adjusted for antihypertensive drug effect. DBPs and SBPs were log-transformed to obtain normal distributions; thus, geometric means are reported in the tables.
Haplotype blocks were defined using Gabriel's method, as implemented in Haploview (as shown in Figure 1). TagSNPs were selected using the pairwise tagging option (r2 = 0.8; minor allele frequency >0.05) and force-included the previously genotyped SNPs. We then used the HAPLOTYPE procedure in SAS/Genetics 9.13 to estimate the haplotype frequencies using the expectation–maximization algorithm. Haplotypes prevalent at ≥5% were retained for analyses. The coding of the haplotypes refers to the allele at each SNP (i.e., 1 for major allele and 2 for minor allele). Each number within the haplotype is ordered according to the SNPs' genomic location. Thus, the first number refers to allele ‘1’ or ‘2’ at the rs10277237 SNP, the second number refers allele ‘1’ or ‘2’ at the rs1800783 SNP, and so on for each haplotype block shown in the Figure 1. Supplementary Table S2 online shows the frequencies of the haplotypes that were frequent at ≥5% in this population. The association of these haplotypes with blood pressure was estimated by stepwise regression analysis. P < 0.05 was considered statistically significant. Power calculations were performed using Quanto v1.1.1 (http://hydra.usc.edu/gxe). Depending on the genotype studied, our analyses were powered at ≥80% to detect a difference of between 1.6–2.3 mm Hg for DBP and 2.5–3.5 mm Hg for SBP at a significance of 5% for each main effect model.
RESULTS
Association between NOS3 gene polymorphism and blood pressure
We first carried out stepwise regression analyses for SBP and DBP separately, including all the 11 SNPs in the model along with age, sex, FM, and FFM. These analyses showed that, among the 11 SNPs, only IVS25+15 [G→A] SNP (rs891512) contributes significantly to variation in SBP (P = 0.014) and DBP (P = 0.009). Subsequent single-SNP analyses were performed for all SNPs, which confirmed the significant association of the IVS25+15 [G→A] SNP, showing that GG homozygotes had significantly lower levels of DBP ((additive: GG, 74.9 ± 0.5; GA, 76.8 ± 0.7; AA, 76.7 ± 1.9 mm Hg, P = 0.03) (dominant: GG, 74.9 ± 0.5; XA, 76.8 ± 0.6 mm Hg, P = 0.01)) and SBP ((additive: GG, 124.4 ± 0.7; GA, 127.2 ± 0.9; AA, 127.4 ± 2.8 mm Hg, P = 0.03) (dominant: GG, 124.4 ± 0.7; XA, 127.2 ± 0.9 mm Hg, P = 0.02)) when compared to A allele carriers (Table 2).
Table 2.
Blood pressure values (geometric mean ± s.e.) in each NOS3 IVS25+15 SNP genotype group
| Genotypes of IVS25+15 SNP | ||||
|---|---|---|---|---|
| Parameters | GG (n = 423) | GA (n = 244) | AA (n = 29) | P value |
| All participants | ||||
| Systolic blood pressure (mm Hg) |
124.4 ± 0.7 | 127.2 ± 0.9 | 127.4 ± 2.8 | 0.03 |
| Diastolic blood pressure (mm Hg) |
74.9 ± 0.5 | 76.8 ± 0.7 | 76.7 ± 1.9 | 0.03 |
| GG | XA (GA + AA) | |||
| Non-resting energy expenditure tertile 1 (physically inactive) | ||||
| (n = 117) | (n = 96) | |||
| Systolic blood pressure (mm Hg) |
126.5 ± 1.3 | 127.7 ± 1.3 | 0.67 | |
| Diastolic blood pressure (mm Hg) |
75.9 ± 0.9 | 75.9 ± 0.9 | 0.97 | |
| Non-resting energy expenditure tertile 2 (moderately active) | ||||
| (n = 122) | (n = 80) | |||
| Systolic blood pressure (mm Hg) |
123.9 ± 0.9 | 125.2 ± 1.3 | 0.36 | |
| Diastolic blood pressure (mm Hg) |
75.2 ± 0.8 | 76.7 ± 0.8 | 0.26 | |
| Non-resting energy expenditure tertile 3 (physically active) | ||||
| (n = 136) | (n = 71) | |||
| Systolic blood pressure (mm Hg) |
120.3 ± 1.1 | 125.2 ± 1.3 | 0.027 | |
| Diastolic blood pressure (mm Hg) |
72.9 ± 0.7 | 76.7 ± 1.5 | 0.020 | |
Interaction of NREE with the relationship between NOS3 gene polymorphism and blood pressure
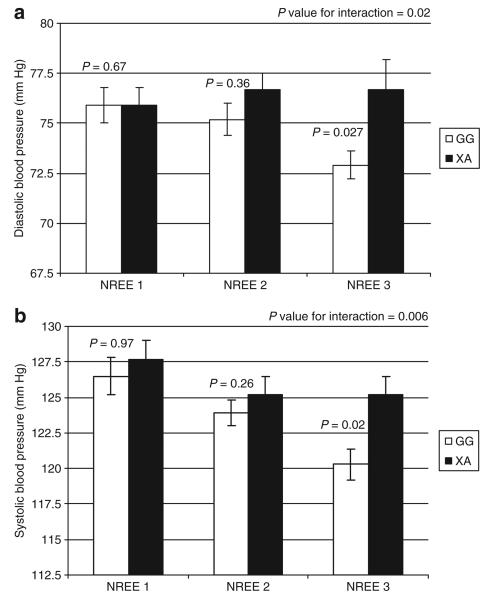
The association of the IVS25+15 [G→A] SNP with blood pressure was significantly modified by NREE (P for genotype × NREE interaction: DBP P = 0.006; SBP P = 0.02; Figure 2). Due to the low frequency of rare homozygotes (4.16%), and similarity in blood pressure of the A-allele carriers (XA), we used a dominant model and compared XA to G-allele homozygotes. The effect of the GG genotype on blood pressure was only observed in individuals in the highest tertile for NREE/kg; for DBP (mean ± s.e.m.): GG 72.9 ± 0.7 mm Hg vs. XA, 76.7 ± 1.5 mm Hg, P = 0.02), for SBP (GG, 120.3 ± 1.1 mm Hg vs. XA, 125.2 ± 1.3 mm Hg, P = 0.027) (Table 2), whereas no significant effect of the IVS25+15 [G→A] SNP on blood pressure was observed at the lower NREE levels.
Figure 2.
(a) Interaction of non-resting energy expenditure (NREE) (sex-specific tertiles of energy expenditure/kg) and IVS25+15 SNP on systolic blood pressure; (b) Interaction of NREE and IVS25+15 SNP on diastolic blood pressure.
NREE was negatively associated with DBP (P = 0.034) and SBP (P = 0.043), independently of the association of the IVS25+15 [G→A] genotype with blood pressure, i.e., blood pressure decreased by 0.019 ± 0.009 mm Hg (DBP) and 0.016 ± 0.008 mm Hg (SBP) for every 100 kJ increase in NREE.
Association between NOS3 gene polymorphism and hypertension
We also assessed the association between genotypes and hypertension. Consistent with the findings for blood pressure, we found that the odds ratio for hypertension for the individuals carrying the A allele at IVS25+15 was 1.695 (95% CI: 1.18–2.44, P = 0.004) compared to those without the A allele. We then stratified the cohort by sex-specific tertiles of NREE per kg body weight and reassessed the association between the IVS25+15 [G→A] genotype and hypertension risk within strata. In those with the highest levels of NREE, carriers of the XA genotype at IVS25+15 were significantly more likely to be hypertensive than non-carriers (P = 0.042), with 16 of 32 hypertensive individuals carrying the XA genotype (50%) compared to only 55 of 175 normotensive individuals carrying the XA genotype (31.4%). By contrast, there was no significant association between IVS25+15 [G→A] genotype frequencies and hypertension risk at lower NREE levels.
To exclude uncontrolled confounding, we examined the association between the IVS25+15 [G→A] SNP and several traditional risk factors (Supplementary Table S3 online). None of these risk factors showed association with the IVS25+15 SNP, suggesting that the observed association of this SNP with blood pressure and hypertension was not confounded by these risk factors.
DISCUSSION
In this study, we tested the association between NOS3 gene variants and blood pressure and hypertension in a population-based cohort of middle-aged white Europeans from the United Kingdom. Because NOS3 gene expression is sensitive to changes in energy expenditure,6 we also tested for interaction between genotype and NREE levels on blood pressure levels. We found that the common homozygous genotype at one of the loci (IVS25+15) was associated with lower blood pressure, but this protective effect was evident only in individuals with high energy expenditure. These results support a role for common variation at the NOS3 gene in the etiology of hypertension and suggest that high NREE may modify the protective effects of this variation on blood pressure homeostasis.
The beneficial effects of exercise on blood pressure homeostasis have been widely reported.21,22 Although the NOS3 gene is a strong biological candidate for the regulation of blood pressure, the evidence supporting a role for common variation at the NOS3 gene in the development of hypertension is less well defined.23-26 NOS3 gene polymorphisms have been associated with hypertension and coronary artery disease in people from Japan,25 Singapore,26 and Korea,27 while no association was observed in people from China28 and Australia.29 It is possible that these ethnic-specific findings could result from genuine genetic differences between populations caused by different founder effects, statistical fluctuations where some of the findings are false, or gene–environment interactions, such as differences in NREE. The polymorphism for which we observed associations with blood pressure in this study (IVS25+15) was not studied in relation to blood pressure in any of the previously published population; hence, there are no studies against which the results of this study can be compared. The etiological relevance of the IVS25+15 SNP in blood pressure regulation is poorly understood. It is plausible that IVS25+15 could act as a marker for other functional polymorphisms at NOS3 or have some intrinsic functional significance; although this variant is in the intronic region, it could affect mRNA stability and enzyme levels by affecting splicing.30 The IVS25+15 variant may also tag SNPs in genes other than NOS3; for example, several other potentially atherogenic genes are located within the same chromosomal region as NOS3 (Ch.7q36). These include the insulin-induced gene 1 (ref. 31) and fatty acid-binding protein5-like 3.32
Thus far, all existing published studies of gene-physical activity interaction in hypertension have suffered from low statistical power, either because the sample size is too small and/or because the characterization of physical activity is imprecise. A strength of this study is that NREE was assessed objectively using individually calibrated heart-rate monitoring against directly measured energy expenditure, whereas the majority of existing studies of this nature have used subjective measures such as questionnaires and interviews, which are less precise and are susceptible to response bias. Nonetheless, this study is underpowered to detect small associations (<1.6 for DBP and <2.5 for SBP). Thus, we cannot be certain that all our negative findings are truly negative. On the other hand, all participants in this study were white, living in the same geographical region in the east of England. Although this is generally perceived to be an ethnically homogenous population, we cannot completely exclude the possibility that underlying genetic heterogeneity exists. However, provided that individuals with non-European ancestry are excluded, the extent of population stratification in the British population is generally modest.33 Moreover, the findings we report here have moderate P values, which will need to be confirmed in replication studies and, most importantly, will need to be supported by mechanistic studies.
In summary, given the defined biological role that NOS3 plays in the regulation of blood pressure, the observations that the IVS25+15 SNP is associated with blood pressure and hypertension, both directly and through effect modification by physical activity, are highly plausible. Although physical activity does not seem to be effective in lowering blood pressure of A-allele carriers, it may still be beneficial to other metabolic traits. Our findings need to be further replicated and ideally tested in a trial before being informative for targeted disease prevention. Eventually, the selection of individuals for lifestyle intervention programs could be guided by knowledge of genotype.
Supplementary Material
Acknowledgments
We are grateful to the volunteers in the MRC Ely Study, who gave their time to take part in this study. The MRC Ely Study was funded by the MRC and Wellcome Trust (to N.J.W.). P.W.F was supported in part via grants from Novo Nordisk (370579201) and the Swedish Diabetes Association (DIA2006-013). I.B. is funded by the Wellcome Trust. We thank Naheed Rana for her assistance in haplotype analysis.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/ajh
Disclosure: The authors declared no conflict of interest.
References
- 1.Wolf-Maier K, Cooper RS, Banegas JR, Giampaoli S, Hense HW, Joffres M, Kastarinen M, Poulter N, Primatesta P, Rodriguez-Artalejo F, Stegmayr B, Thamm M, Tuomilehto J, Vanuzzo D, Vescio F. Hypertension prevalence and blood pressure levels in 6 European countries, Canada, and the United States. JAMA. 2003;289:2363–2369. doi: 10.1001/jama.289.18.2363. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AP. Aerobic and resistive exercise modify risk factors for coronary heart disease. Med Sci Sports Exerc. 1989;21:669–674. doi: 10.1249/00005768-198912000-00008. [DOI] [PubMed] [Google Scholar]
- 3.An P, Borecki IB, Rankinen T, Perusse L, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. Evidence of major genes for exercise heart rate and blood pressure at baseline and in response to 20 weeks of endurance training: the HERITAGE family study. Int J Sports Med. 2003;24:492–498. doi: 10.1055/s-2003-42011. [DOI] [PubMed] [Google Scholar]
- 4.Marsden PA, Schappert KT, Chen HS, Flowers M, Sundell CL, Wilcox JN, Lamas S, Michel T. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–293. doi: 10.1016/0014-5793(92)80697-f. [DOI] [PubMed] [Google Scholar]
- 5.Gamboa A, Shibao C, Diedrich A, Choi L, Pohar B, Jordan J, Paranjape S, Farley G, Biaggioni I. Contribution of endothelial nitric oxide to blood pressure in humans. Hypertension. 2007;49:170–177. doi: 10.1161/01.HYP.0000252425.06216.26. [DOI] [PubMed] [Google Scholar]
- 6.Hambrecht R, Adams V, Erbs S, Linke A, Krankel N, Shu Y, Baither Y, Gielen S, Thiele H, Gummert JF, Mohr FW, Schuler G. Regular physical activity improves endothelial function in patients with coronary artery disease by increasing phosphorylation of endothelial nitric oxide synthase. Circulation. 2003;107:3152–3158. doi: 10.1161/01.CIR.0000074229.93804.5C. [DOI] [PubMed] [Google Scholar]
- 7.Boo YC, Sorescu G, Boyd N, Shiojima I, Walsh K, Du J, Jo H. Shear stress stimulates phosphorylation of endothelial nitric-oxide synthase at Ser1179 by Akt-independent mechanisms: role of protein kinase A. J Biol Chem. 2002;277:3388–3396. doi: 10.1074/jbc.M108789200. [DOI] [PubMed] [Google Scholar]
- 8.Searles CD. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression. Am J Physiol Cell Physiol. 2006;291:C803–C816. doi: 10.1152/ajpcell.00457.2005. [DOI] [PubMed] [Google Scholar]
- 9.Kimura T, Yokoyama T, Matsumura Y, Yoshiike N, Date C, Muramatsu M, Tanaka H. NOS3 genotype-dependent correlation between blood pressure and physical activity. Hypertension. 2003;41:355–360. doi: 10.1161/01.hyp.0000051500.02578.6d. [DOI] [PubMed] [Google Scholar]
- 10.Rankinen T, Rice T, Perusse L, Chagnon YC, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. NOS3 Glu298Asp genotype and blood pressure response to endurance training: the HERITAGE family study. Hypertension. 2000;36:885–889. doi: 10.1161/01.hyp.36.5.885. [DOI] [PubMed] [Google Scholar]
- 11.Forouhi NG, Luan J, Hennings S, Wareham NJ. Incidence of Type 2 diabetes in England and its association with baseline impaired fasting glucose: The Ely study 1990-2000. Diabet Med. 2007;24:200–207. doi: 10.1111/j.1464-5491.2007.02068.x. [DOI] [PubMed] [Google Scholar]
- 12.Franks PW, Bhattacharyya S, Luan J, Montague C, Brennand J, Challis B, Brage S, Ekelund U, Middelberg RP, O'Rahilly S, Wareham NJ. Association between physical activity and blood pressure is modified by variants in the G-protein coupled receptor 10. Hypertension. 2004;43:224–228. doi: 10.1161/01.HYP.0000109319.63240.08. [DOI] [PubMed] [Google Scholar]
- 13.Wareham NJ, Byrne CD, Williams R, Day NE, Hales CN. Fasting proinsulin concentrations predict the development of type 2 diabetes. Diabetes Care. 1999;22:262–270. doi: 10.2337/diacare.22.2.262. [DOI] [PubMed] [Google Scholar]
- 14.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, Roccella EJ. Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. National Heart, Lung, and Blood Institute; National High Blood Pressure Education Program Coordinating Committee. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 15.Sun SS, Chumlea WC, Heymsfield SB, Lukaski HC, Schoeller D, Friedl K, Kuczmarski RJ, Flegal KM, Johnson CL, Hubbard VS. Development of bioelectrical impedance analysis prediction equations for body composition with the use of a multicomponent model for use in epidemiologic surveys. Am J Clin Nutr. 2003;77:331–340. doi: 10.1093/ajcn/77.2.331. [DOI] [PubMed] [Google Scholar]
- 16.Livingstone MB, Prentice AM, Coward WA, Ceesay SM, Strain JJ, McKenna PG, Nevin GB, Barker ME, Hickey RJ. Simultaneous measurement of free-living energy expenditure by the doubly labeled water method and heart-rate monitoring. Am J Clin Nutr. 1990;52:59–65. doi: 10.1093/ajcn/52.1.59. [DOI] [PubMed] [Google Scholar]
- 17.Ekelund U, Brage S, Franks PW, Hennings S, Emms S, Wareham NJ. Physical activity energy expenditure predicts progression toward the metabolic syndrome independently of aerobic fitness in middle-aged healthy Caucasians: the medical research council Ely study. Diabetes Care. 2005;28:1195–1200. doi: 10.2337/diacare.28.5.1195. [DOI] [PubMed] [Google Scholar]
- 18.Barroso I, Luan J, Middelberg RP, Harding AH, Franks PW, Jakes RW, Clayton D, Schafer AJ, O'Rahilly S, Wareham NJ. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol. 2003;1:E20. doi: 10.1371/journal.pbio.0000020. Erratum in: PLoS Biol 2003; 1:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franks PW, Luan J, Barroso I, Brage S, Gonzalez Sanchez JL, Ekelund U, Rios MS, Schafer AJ, O'Rahilly S, Wareham NJ. Variation in the eNOS gene modifies the association between total energy expenditure and glucose intolerance. Diabetes. 2005;54:2795–2801. doi: 10.2337/diabetes.54.9.2795. [DOI] [PubMed] [Google Scholar]
- 20.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbert JA, Silagy CA, Finucane P, Withers RT, Hamdorf PA, Andrews GR. The effectiveness of exercise training in lowering blood pressure: a meta-analysis of randomised controlled trials of 4 weeks or longer. J Hum Hypertens. 1997;11:641–649. doi: 10.1038/sj.jhh.1000509. [DOI] [PubMed] [Google Scholar]
- 22.Whelton SP, Chin A, Xin X, He J. Effect of aerobic exercise on blood pressure: a meta-analysis of randomized, controlled trials. Ann Intern Med. 2002;136:493–503. doi: 10.7326/0003-4819-136-7-200204020-00006. [DOI] [PubMed] [Google Scholar]
- 23.Bonnardeaux A, Nadaud S, Charru A, Jeunemaitre X, Corvol P, Soubrier F. Lack of evidence for linkage of the endothelial cell nitric oxide synthase gene to essential hypertension. Circulation. 1995;91:96–102. doi: 10.1161/01.cir.91.1.96. [DOI] [PubMed] [Google Scholar]
- 24.Hunt SC, Williams CS, Sharma AM, Inoue I, Williams RR, Lalouel JM. Lack of linkage between the endothelial nitric oxide synthase gene and hypertension. J Hum Hypertens. 1996;10:27–30. [PubMed] [Google Scholar]
- 25.Miyamoto Y, Saito Y, Kajiyama N, Yoshimura M, Shimasaki Y, Nakayama M, Kamitani S, Harada M, Ishikawa M, Kuwahara K, Ogawa E, Hamanaka I, Takahashi N, Kaneshige T, Teraoka H, Akamizu T, Azuma N, Yoshimasa Y, Yoshimasa T, Itoh H, Masuda I, Yasue H, Nakao K. Endothelial nitric oxide synthase gene is positively associated with essential hypertension. Hypertension. 1998;32:3–8. doi: 10.1161/01.hyp.32.1.3. [DOI] [PubMed] [Google Scholar]
- 26.Moe KT, Lim ST, Wong P, Chua T, Desilva DA, Koh TH, Wong MC, Chin-Dusting J. Association analysis of endothelial nitric oxide synthase gene polymorphism with primary hypertension in a Singapore population. J Hum Hypertens. 2006;20:956–963. doi: 10.1038/sj.jhh.1002096. [DOI] [PubMed] [Google Scholar]
- 27.Kim IJ, Bae J, Lim SW, Cha DH, Cho HJ, Kim S, Yang DH, Hwang SG, Oh D, Kim NK. Influence of endothelial nitric oxide synthase gene polymorphisms (−786T>C, 4a4b, 894G>T) in Korean patients with coronary artery disease. Thromb Res. 2007;119:579–585. doi: 10.1016/j.thromres.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Q, Su SY, Chen SF, Li B, Gu DF. Association study of the endothelial nitric oxide synthase gene polymorphisms with essential hypertension in northern Han Chinese. Chin Med J (Engl) 2006;119:1065–1071. [PubMed] [Google Scholar]
- 29.Granath B, Taylor RR, van Bockxmeer FM, Mamotte CD. Lack of evidence for association between endothelial nitric oxide synthase gene polymorphisms and coronary artery disease in the Australian Caucasian population. J Cardiovasc Risk. 2001;8:235–241. doi: 10.1177/174182670100800408. [DOI] [PubMed] [Google Scholar]
- 30.Pagenstecher C, Wehner M, Friedl W, Rahner N, Aretz S, Friedrichs N, Sengteller M, Henn W, Buettner R, Propping P, Mangold E. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet. 2006;119:9–22. doi: 10.1007/s00439-005-0107-8. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Takaishi K, Cook W, McCorkle SK, Unger RH. Insig-1 “brakes” lipogenesis in adipocytes and inhibits differentiation of preadipocytes. Proc Natl Acad Sci USA. 2003;100:9476–9481. doi: 10.1073/pnas.1133426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li WD, Dong C, Li D, Garrigan C, Price RA. A genome scan for serum triglyceride in obese nuclear families. J Lipid Res. 2005;46:432–438. doi: 10.1194/jlr.M400391-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.