Abstract
Carbon dioxide (CO2) elicits different olfactory behaviors across species. In Drosophila, neurons that detect CO2 are located in the antenna, form connections in a ventral glomerulus in the antennal lobe, and mediate avoidance. By contrast, in the mosquito these neurons are in the maxillary palps (MPs), connect to medial sites, and promote attraction. We found in Drosophila that loss of a microRNA, miR-279, leads to formation of CO2 neurons in the MPs. miR-279 acts through down-regulation of the transcription factor Nerfin-1. The ectopic neurons are hybrid cells. They express CO2 receptors and form connections characteristic of CO2 neurons, while exhibiting wiring and receptor characteristics of MP olfactory receptor neurons (ORNs). We propose that this hybrid ORN reveals a cellular intermediate in the evolution of species-specific behaviors elicited by CO2.
In insects, both the position of CO2 neurons and the behavior elicited by CO2 differ among species. For example, olfactory detection of CO2 through neurons positioned in or around the mouthparts of an insect, such as maxillary palps (MPs) and labial palps, correlates with feeding-related behaviors. Indeed, in some blood-feeding insects such as mosquitoes and tsetse flies, these neurons are harbored in the MPs and are important in locating hosts via plumes of CO2 that they emit (1–3). The hawkmoth, Manduca sexta, monitors nectar profitability of newly opened Datura wrightii flowers through CO2 receptor neurons located in their labial palps (4, 5). In these examples, CO2 acts as an attractant. Conversely, in Drosophila CO2 is a component of a stress-induced odor that triggers avoidance behavior (6). This repellent response is driven by antennal neurons expressing the CO2 receptor complex Gr21a-Gr63a (7, 8). How did these diverse behavioral responses to CO2 arise during insect evolution? We propose that this diversity emerged through multiple steps, including changes in cellular position (arising from elimination of CO2 neurons in one appendage and generation of these neurons in another) and changes in circuitry.
In the course of a genetic screen for mutants disrupting the organization of the olfactory system, we isolated a mutant (S0962−07) that resulted in the formation of ectopic Gr21a-expressing neurons in the MPs (Fig. 1A). Some 22 ± 1.5 (mean ± SEM) green fluorescent protein (GFP)–positive cells were observed in the mutant MP, whereas the number of antennal Gr21a olfactory receptor neurons (ORNs) was unaffected (Fig. 1B). In the wild type, Gr21a cell bodies were restricted to the antenna (Fig. 1A). The ectopic MP cells expressed both CO2 receptors (Gr21a and Gr63a) (Fig. 1C). Consistent with this finding, mutant cells conferred CO2 sensitivity to the MP (Fig. 1D). Staining the MP with an antibody to the pan-neuronal marker Elav revealed an increase of 21 ± 3.4 neurons in the mutant, which suggests that all ectopic neurons expressed Gr21a (fig. S1).
Fig. 1.
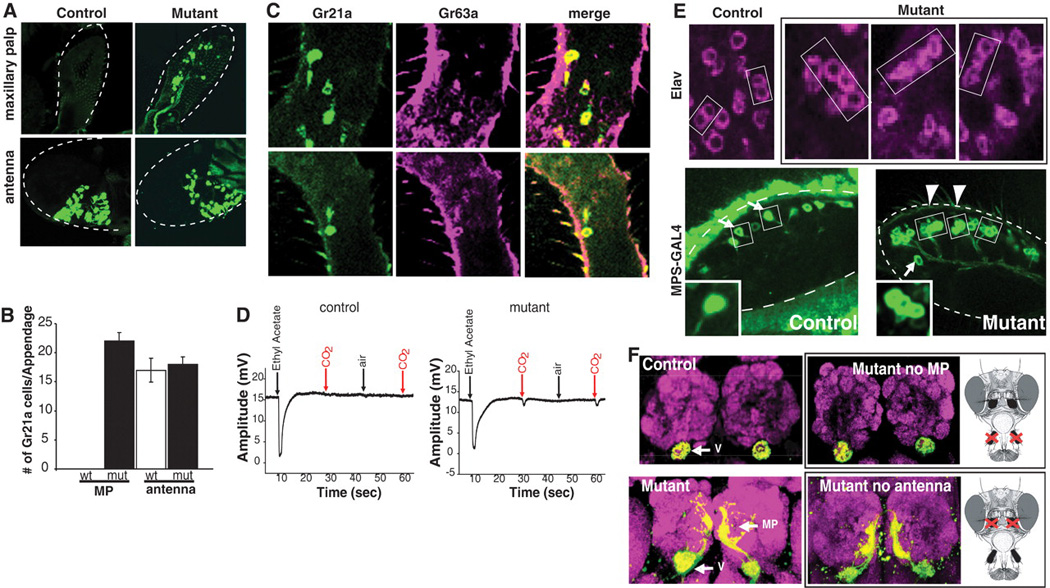
Ectopic CO2 neurons are formed in the MPs of S0962-07 mutants. (A) Gr21a expression in wild-type and mutant olfactory appendages. (B) Quantification of Gr21a-positive cells in the MP and antenna. (C) Gr63a (magenta, RNA antisense probe) and Gr21a (green, Gr21a-GAL4) are coexpressed in the MP. (D) Electropalpograms comparing the response to ethyl acetate, air, and CO2 in control and mutant flies. Contrary to lack of response from the control palps, 5 of 12 mutant MPs responded to CO2 (9). MPs recorded: n = 12 (control), n = 12 (mutant), P = 0.016. (E) Single confocal sections of MPs labeled with antibody to Elav (magenta) at 60 to 80 hours APF or with MPS-GAL4 and UAS-mCD8GFP (green) at 80 hours APF. Two neurons (Elav) or single ORNs (MPS-GAL4) are labeled in wild-type MP sensilla (arrows). Two additional neurons are observed in a subset of mutant sensilla (arrowheads and inset). (F) Mutant neurons in the MP target the V and medial glomeruli. Mutant flies without MP (upper right) and without antenna (lower right). Magenta, anti-NC82. In (A) and (E), dashed lines outline MPs.
In wild-type MPs, each sensillum contains two ORNs. By contrast, in the mutant MP sensilla, additional neurons expressing Elav and the general receptor Or83b were observed (Fig. 1E and fig. S1). This was also apparent when a MP ORN marker (MPS-GAL4) expressed in a subset of MP ORNs was used (Fig. 1E) (9). This marker labels single cells within a subset of wild-type MP sensilla (Fig. 1E, arrows); however, in mutant MPs, two additional neurons were observed (Fig. 1E, arrowheads), bringing the total number of neurons within these sensilla to four. Thus, the generation of ectopic Gr21a-Gr63a neurons is due to an increase in the number of neurons within sensilla rather than transformation of MP ORNs (fig. S1).
In the wild type, each class of adult ORNs sends projections from both antennae or MPs to the antennal lobe (AL). ORNs expressing same odorant receptors (ORs) typically form synapses in the same glomerulus within the AL (fig. S2) (10). CO2 neurons in the antenna target the V-glomerulus (Fig. 1F). To specifically assess the targeting of ectopic MP CO2 neurons, we examined flies where the antennae were surgically removed (Fig. 1F). We found that ectopic CO2 neurons targeted the V-glomerulus and other medial sites in the AL (Fig. 1F; see also below). The wiring specificity of antennal CO2 neurons in the mutants was identical to that in the wild type (Fig. 1F). Thus, the ectopic CO2 neurons in the MP target, at least in part, the same glomerulus innervated by the wild-type CO2 neurons in the antennae.
We mapped S0962−07 to a P-element insertion some 1 kb upstream of a microRNA, miR-279 (fig. S3). MicroRNAs (miRNAs) are small noncoding RNAs of about 22 nucleotides that bind to specific sequences of the 3′-untranslated region (3′UTR) of target genes and thereby repress gene expression posttranscriptionally. In recent years, miRNAs were implied in a variety of functions in the nervous system of different organisms (11). To assess whether miR-279 is responsible for the observed phenotype, we generated three small deletions that uncovered the miR-279 genomic region (fig. S3). These deletion mutants exhibited phenotypes indistinguishable from S0962-07 (fig. S3). The ectopic CO2 phenotype was rescued by a 3-kb fragment of genomic DNA encoding only miR-279 (fig. S3) (9). Thus, miR-279 is the gene disrupted in S0962-07 and must repress targets in the MP to inhibit ectopic CO2 neuron development.
To assess whether miR-279 is expressed in the developing MPs, we generated transgenic flies carrying a transcriptional reporter construct (miR-279-GAL4). Expression was monitored in flies carrying this GAL4 construct and the reporter UAS-mCD8GFP (Fig. 2 and fig. S4). Around 40 to 50 hours after puparium formation (APF), large cells reminiscent of sensory organ precursors in other epithelia expressed miR-279 (Fig. 2 and fig. S4). At later stages, miR-279–expressing cells were found in clusters with smaller cells, some of which expressed neuronal markers (fig. S4). As ORNs matured, miR-279 expression was lost (fig. S4).
Fig. 2.
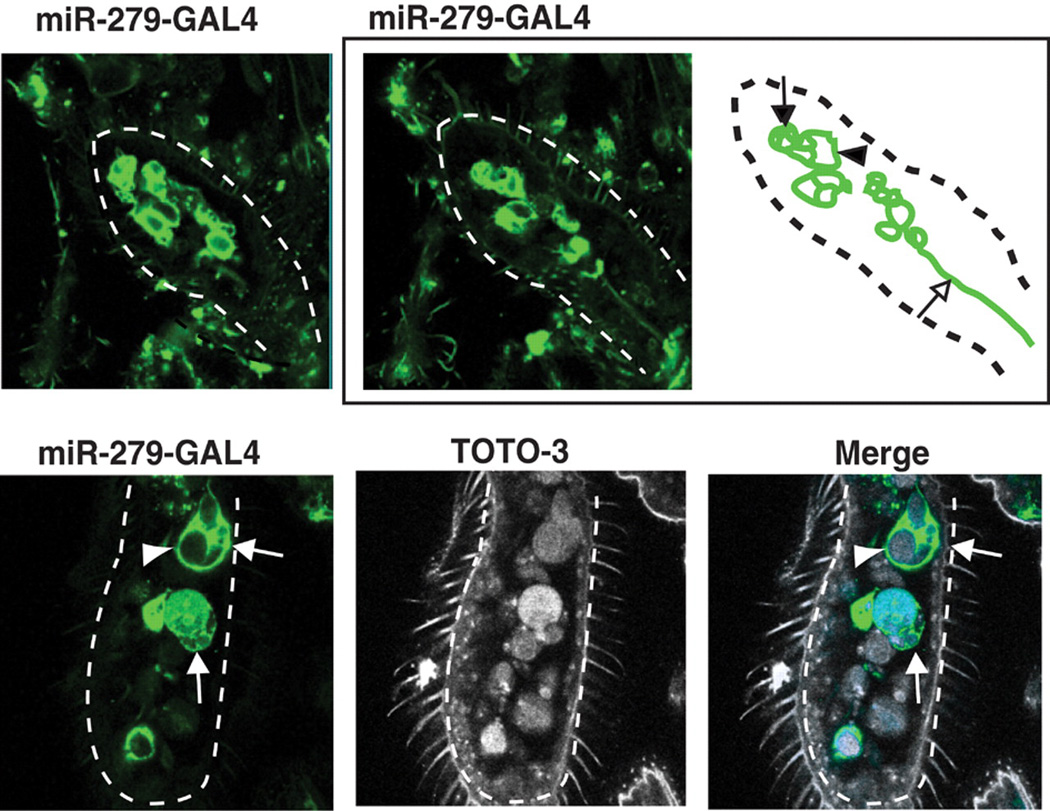
miR-279 is expressed in precursor cells in the developing MP. Expression of miR-279 was visualized with miR-279-GAL4 and UAS-CD8GFP (green). The arrowhead and arrow in both the schematic and the image panels point to a big cell and a cluster of small cells, respectively. The open arrow in the schematic points to a nerve fiber from one of the cell clusters. Nuclear counterstain TOTO-3 is used in the bottom panels. Dashed lines outline the developing MPs.
We next sought to identify the target gene(s) responsible for the miR-279 mutant phenotype. About 205 potential target mRNAs of miR-279 were previously predicted (12, 13). One of the strongest candidates for miR-279 regulation is Nerfin-1. The Nerfin-1 3′UTR contains multiple miR-279 binding sites (Fig. 3F) and encodes a transcription factor expressed in neuronal precursors and transiently in nascent neurons in the embryonic central nervous system (14). Nerfin-1 protein appeared in miR-279–positive cells between 50 and 60 hours APF (Fig. 3A). Nerfin-1 and miR-279 gradually redistributed, generating complementary expression patterns. Cells with high levels of Nerfin-1 expressed low levels of miR-279 and vice versa (Fig. 3, B and C, and fig. S5).
Fig. 3.
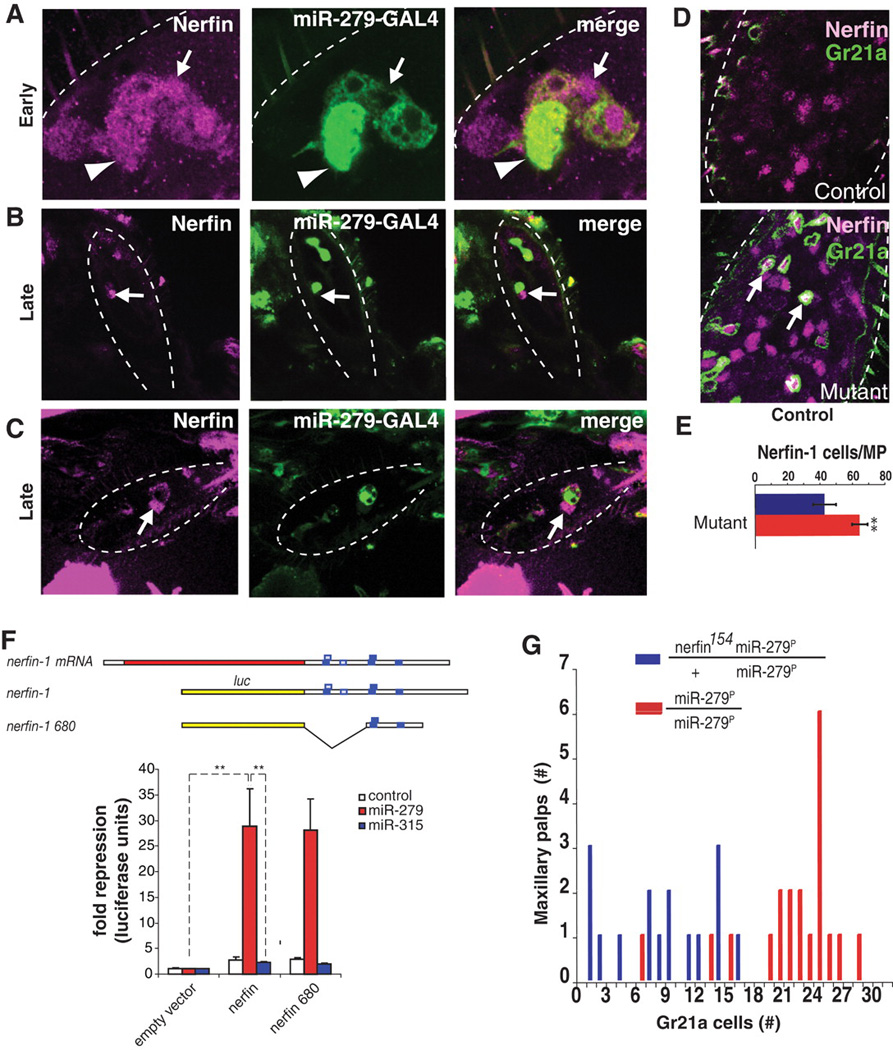
Nerfin-1 is a target of miR-279. (A to C) Expression pattern of Nerfin-1 (magenta) and miR-279 (green, see Fig. 2) in developing MPs at early (A) and later [(B) and (C)] stages. (D) Nerfin-1 (magenta) is expressed in ectopic CO2 neurons (green) in the mutant MPs (arrows). (E) Quantification of Nerfin-1–positive nuclei in wild-type and mutant MPs at 60 to 80 hours APF. MPs scored: wild type, n = 7; mutant n = 9; **P < 0.001. (F) miR-279 inhibits nerfin-1 expression in cultured Drosophila S2 cell lines (**P < 0.001). (G) nerfin-1 is a dominant suppressor of miR-279 (P < 0.001).
To test whether Nerfin-1 is up-regulated in miR-279 mutants, we stained mutant MPs with antibodies to Nerfin-1. We found 22 ± 4.8 additional Nerfin-1–expressing cells in miR-279 mutant MPs relative to controls (Fig. 3E). This is similar to the number of ectopic CO2 neurons in the MP (Fig. 1B). The vast majority of CO2 ORNs in the MP expressed Nerfin-1 (Fig. 3D and fig. S5). Thus, the expression pattern of Nerfin-1 protein in the wild type and in mutant MPs is consistent with nerfin-1 mRNA being a target for miR-279 in vivo.
To determine whether miR-279 directly binds to nerfin-1 3′UTR and inhibits its expression, we used a luciferase reporter assay in cultured cells. The luciferase-coding region was fused to the full-length nerfin-1 3′UTR, which contains four conserved 8-nucleotide oligomer target sites for miR-279 (15), as well as to a subregion containing three of these sites (Fig. 3F). Luciferase activity of both nerfin-1 sensor constructs was strongly repressed when cells were cotransfected with miR-279 (Fig. 3F). By contrast, the activity of either nerfin-1 sensor was unaffected by noncognate miR-315. Antisense oligomers directed against the miR-279 core sequence specifically relieved nerfin-1 reporter repression (fig. S6). Thus, we conclude that nerfin-1 is a direct target of miR-279.
We next assessed whether Nerfin-1 down-regulation by miR-279 inhibits the development of CO2 neurons in the MPs. To do this, we reduced the level of nerfin-1 by half genetically in a miR-279 mutant background. This decreased the number of CO2 neurons in the MP relative to miR-279 mutants (Fig. 3G), providing strong in vivo evidence that miR-279 is necessary to down-regulate Nerfin-1 in MPs during normal development. Nerfin-1 up-regulation alone was not sufficient to generate a miR-279–like phenotype (fig. S7). Taken together, these findings suggest that miR-279 down-regulates Nerfin-1 and other targets to prevent CO2 neuron development in the MPs.
When analyzing the axonal projections of the CO2 neurons in the MPs, we observed that these neurons targeted one or more medial glomeruli in addition to the V-glomerulus, the target of antennal CO2 neurons (Fig. 1F and fig. S2). These medial glomeruli are normally innervated by MP Or42a and Or59c ORNs. Double-labeling experiments revealed that mutant neurons also coexpressed Or42a and Or59c, but not other MP ORs (Fig. 4A). Analysis of subsets of MP ORNs also revealed that Or42a and Or59c classes each showed an approximate increase of 10 cells in the MPs, whereas others were unaffected (Fig. 4, B and C). These results indicate that the ectopic CO2 neurons are formed as additional cells within Or42a and Or59c sensilla and are hybrid in identity. They express ORs and exhibit wiring characteristics of two classes of neurons.
Fig. 4.
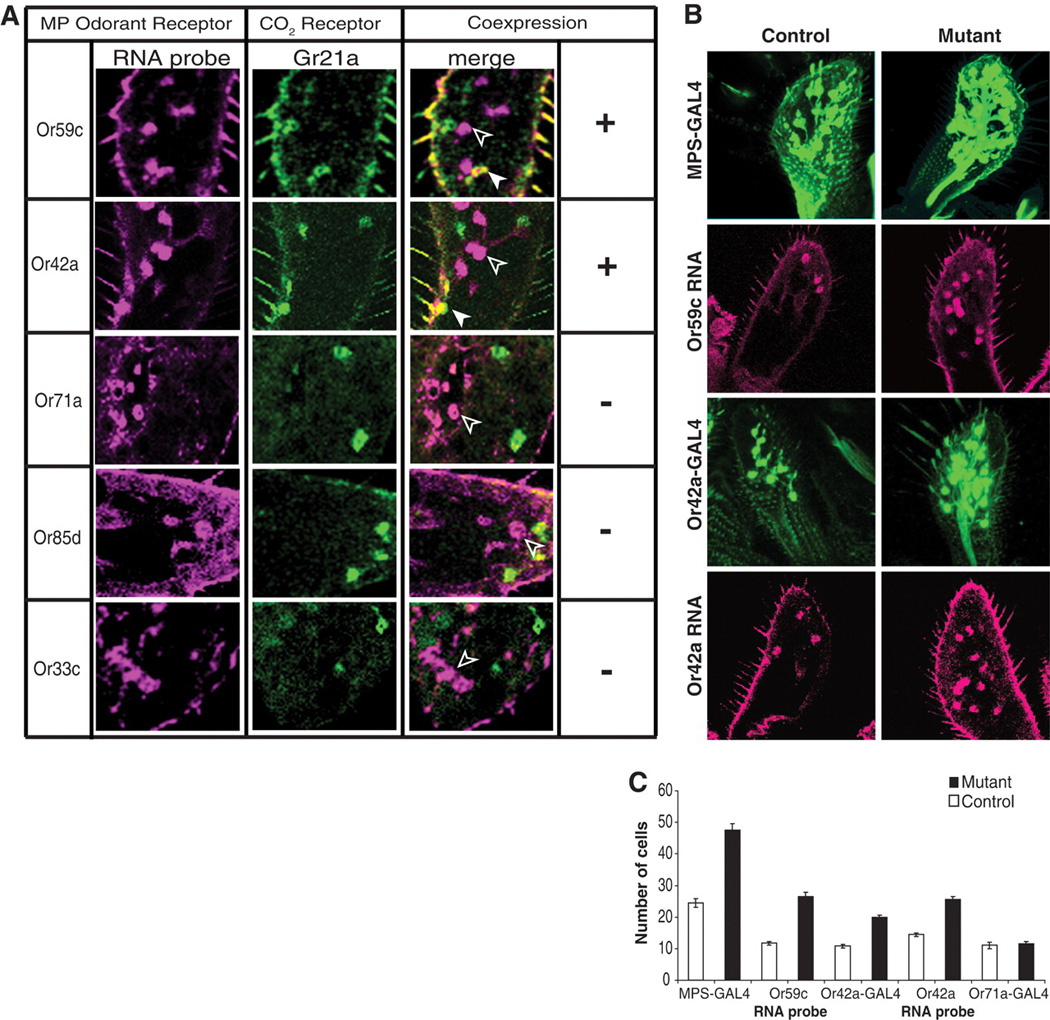
Ectopic neurons exhibit mixed sensory identity. (A) MPs of mutant flies labeled with RNA antisense probes (magenta) and with Gr21a-GAL4 and UAS-mCD8GFP (green). Or59c and Or42a transcript (magenta) overlaps partly with Gr21a-expressing cells (green, solid arrowhead). Cells only positive for Or59c or Or42a are labeled only in magenta (open arrowhead). (B and C) MPs of mutant flies contain more Or42a- and Or59c-expressing cells. (B) Labeling of MP ORNs with GAL4 reporter constructs (Or42a-GAL4 and MPS-GAL4) or Or59c or Or42a RNA probe. (C) Quantification of the data from (B). Total increase in the number of cells in mutants using MPS-GAL4 driver corresponds to the number of ectopic CO2 neurons (see Fig. 1B).
It is interesting that the loss of miR-279 generates a CO2 neuron within a sensillum harboring four neurons in the MP (Fig. 1E and fig. S1), given that the antennal CO2 sensilla in Drosophila are the only sensilla in the olfactory system to harbor four ORNs (16). Because miR-279 acts within the precursor cells in the MP to prevent Nerfin-dependent formation of olfactory neurons, this observation raises the intriguing possibility that positioning of CO2 neurons on different olfactory appendages might have evolved through changes at the level of precursor cell development. Thus, the evolutionary elimination of CO2 neurons from MP sensilla might have required decreasing the number of cells with neuronal identities through down-regulation of Nerfin-1 by miR-279.
Although we hypothesize that relocation of CO2 ORNs to different appendages was important in the evolution of differences in CO2 sensing, additional mechanisms must have evolved to modify the neural circuitry to alter species-specific behaviors in response to CO2. The ectopic CO2 neurons are hybrid cells, which express additional receptors (Or59c or Or42a) and also target medial glomeruli, typically innervated by wild-type ORNs expressing these ORs. This is particularly interesting given that CO2 neurons in mosquitoes connect to medial glomeruli, driving an attractive response (17–19). We speculate that this hybrid cell represents an evolutionary intermediate on a path leading to species-specific CO2 behavior (20). Perhaps suppressing the expression of Or59c or Or42a ORs could convert this hybrid cell to one dedicated only to CO2 reception. The nature of the behavioral output to CO2 (i.e., attraction versus repulsion) by this cell, however, may be dictated by altering the wiring specificity to one site or the other (medial versus ventral, respectively). More generally, we propose that natural selection can work on such an evolutionary intermediate to generate different combinations of OR, wiring, and cellular positional specificities, depending on the insects′ environmental needs. This may in turn lead to novel olfactory responses to different odorants, or to the same odorant in different species.
Supplementary Material
References and Notes
- 1.Kellogg FE. J. Insect Physiol. 1970;16:99. doi: 10.1016/0022-1910(70)90117-4. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 2.Grant AJ, O′Connell RJ. Ciba Found. Symp. 1996;200:233. doi: 10.1002/9780470514948.ch17. [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 3.Bogner F. Physiol. Entomol. 1992;17:1992. [UC-eLinks] [Google Scholar]
- 4.Thom C, Guerenstein PG, Mechaber WL, Hildebrand JG. J. Chem. Ecol. 2004;30:1285. doi: 10.1023/b:joec.0000030298.77377.7d. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 5.Bogner F, Boppre M, Ernst KD, Boeckh J. J. Comp. Physiol. A. 1986;158:741. doi: 10.1007/BF01324818. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 6.Suh GS, et al. Nature. 2004;431:854. doi: 10.1038/nature02980. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 7.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Nature. 2007;445:86. doi: 10.1038/nature05466. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 8.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3574. doi: 10.1073/pnas.0700079104. [Abstract/Free Full Text] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See supporting material on Science Online.
- 10.Vosshall LB. Curr. Opin. Neurobiol. 2000;10:498. doi: 10.1016/s0959-4388(00)00111-2. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 11.Kosik KS, Krichevsky AM. Neuron. 2005;47:779. doi: 10.1016/j.neuron.2005.08.019. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 12.Stark A, Brennecke J, Russell RB, Cohen SM. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [CrossRef] [Medline][UC-eLinks] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [Medline][UC-eLinks] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzin A, Brody T, Moore AW, Odenwald WF. Dev. Biol. 2005;277:347. doi: 10.1016/j.ydbio.2004.09.027. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15. doi: 10.1016/j.cell.2004.12.035. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 16.Couto A, Alenius M, Dickson BJ. Curr. Biol. 2005;15:1535. doi: 10.1016/j.cub.2005.07.034. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 17.Anton S, et al. Arthropod Struct. Dev. 2003;32:319. doi: 10.1016/j.asd.2003.09.002. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 18.Distler P, Boeckh J. J. Exp. Biol. 1997;200:1873. doi: 10.1242/jeb.200.13.1873. [Abstract] [DOI] [PubMed] [Google Scholar]
- 19.Ignell R, Dekker T, Ghaninia M, Hansson BS. J. Comp. Neurol. 2005;493:207. doi: 10.1002/cne.20800. [CrossRef] [ISI] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 20.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Nature. 2007;445:383. doi: 10.1038/nature05451. [CrossRef] [Medline][UC-eLinks] [DOI] [PubMed] [Google Scholar]
- 21.We thank L. Vosshall, B. Dickson, W. Odenwald, R. Klein, G. Tavosanis, J. Carlson, and D. Anderson for providing reagents and comments on experiments; W. Tom, A. Lorenze, and P. Alcala for technical assistance; A. Acker-Palmer, T. Suzuki, G. Tavosanis, R. Klein, and members of the laboratories for comments on the manuscript; A. Luke for helping with miR-279 deletions; and Y.-T. Chou for cloning the luciferase sensors. Supported by the Jane Coffin Childs Memorial Fund and a National Research Service Award (P.C.); EMBO and the Human Frontiers Science Program (I.G.K.); the Leukemia and Lymphoma Foundation, the Burroughs Wellcome Foundation, and the V-Foundation for Cancer Research (E.C.L.); and NIH grant DC006485 (S.L.Z.). G.S.B.S. is an HHMI Associate; S.L.Z. is an HHMI Investigator.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


