Abstract
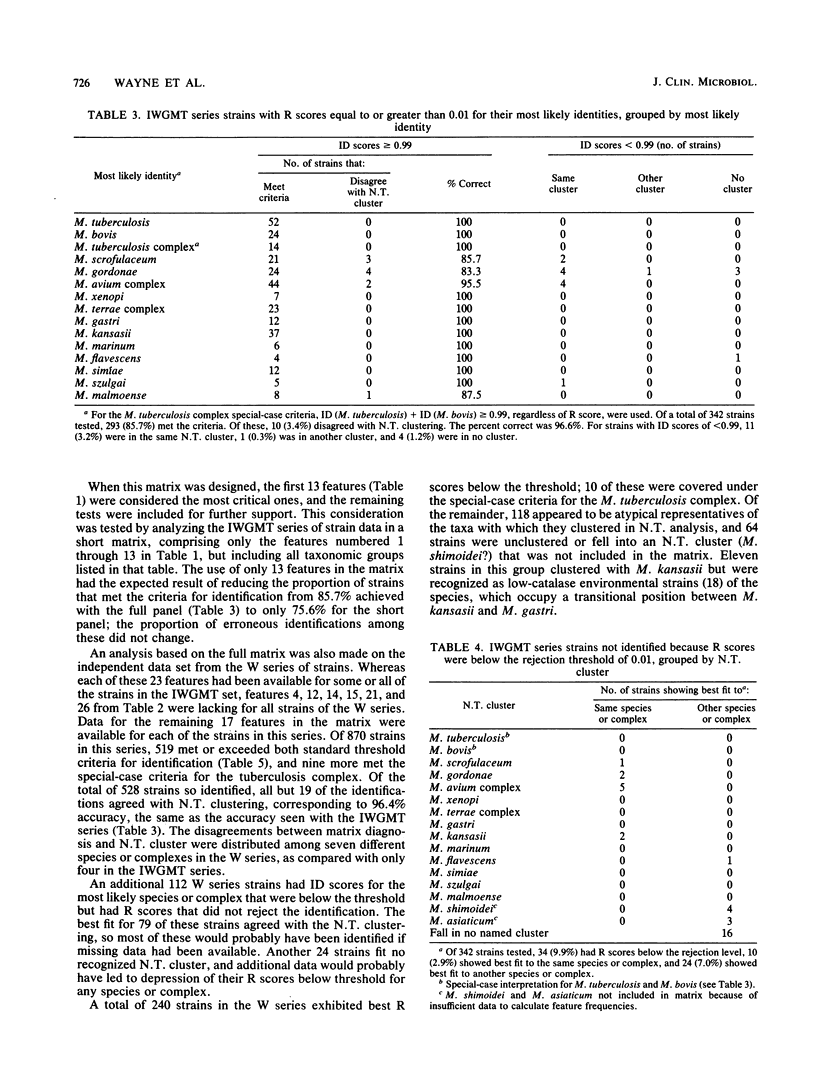
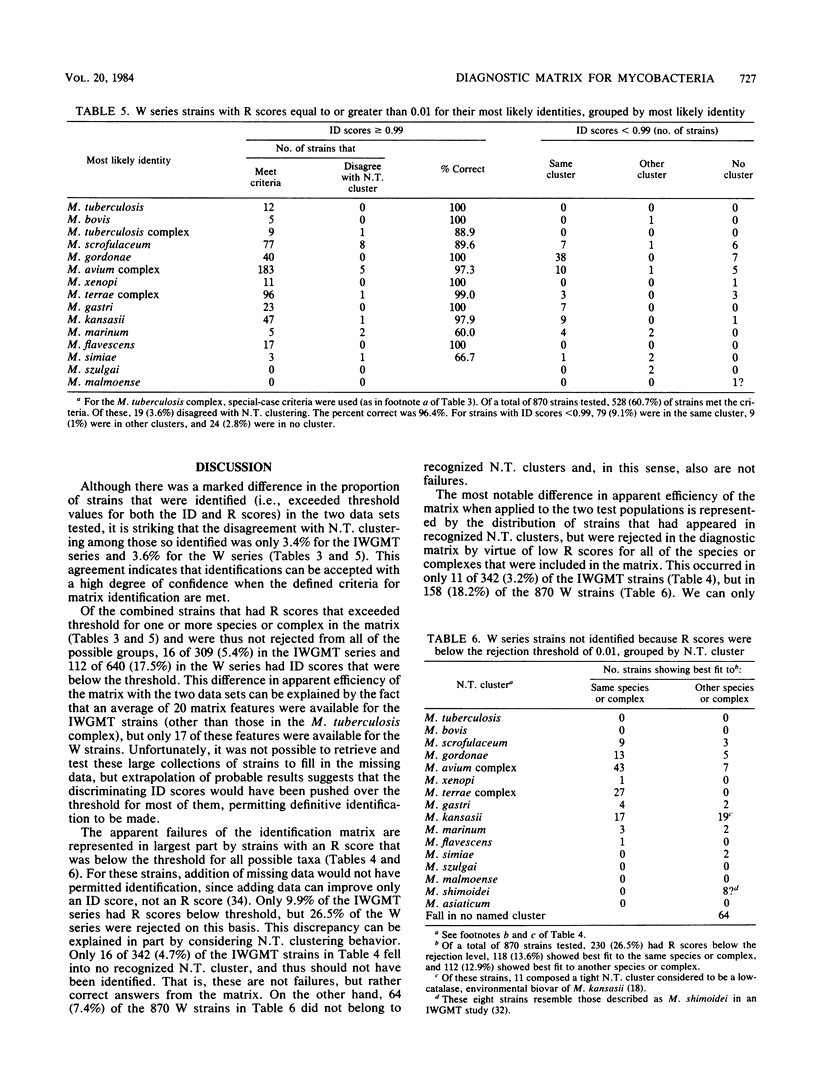
A probability matrix is presented for identification of slowly growing mycobacteria that are likely to be encountered in clinical laboratories. The matrix includes 23 features that are useful for identifying members of 14 species or species complexes. The computer program identifies strains as a function of the ID (identification) score, which measures the discrimination among possible alternative identifications, and the R (ratio) score, which measures the degree of fit to the most likely taxa. It is not necessary to employ all 23 tests when initiating an identification; the program will suggest additional tests to perform when a partial data set fails to yield a definitive identification. Two independent sets of cultures comprising a total of 1,212 strains were used to test the matrix. Correct diagnoses were based on clustering behavior in numerical taxonomic analysis with larger numbers of features. The probable efficiencies with the two sets were 94.2 and 83.4%, respectively, and the accuracy of the definitive identifications for both sets exceeded 95%. A discussion is presented of situations when it may be appropriate to override an R score that has caused the rejection of an identification and to thereby enhance the efficiency.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CHESTER S. T., BELL H. G. The management of the duodenal stump in gastric resection for technically difficult duodenal ulcer. West J Surg Obstet Gynecol. 1955 Nov;63(11):701–705. [PubMed] [Google Scholar]
- Dybowski W., Franklin D. A. Conditional probability and the identification of bacteria: a pilot study. J Gen Microbiol. 1968 Dec;54(2):215–229. doi: 10.1099/00221287-54-2-215. [DOI] [PubMed] [Google Scholar]
- Friedman R. B., Bruce D., MacLowry J., Brenner V. Computer-assisted identification of bacteria. Am J Clin Pathol. 1973 Sep;60(3):395–403. doi: 10.1093/ajcp/60.3.395. [DOI] [PubMed] [Google Scholar]
- Friedman R., MacLowry J. Computer identification of bacteria on the basis of their antibiotic susceptibility patterns. Appl Microbiol. 1973 Sep;26(3):314–317. doi: 10.1128/am.26.3.314-317.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUBICA G. P., POOL G. L. Studies on the catalase activity of acid-fast bacilli. I. An attempt to subgroup these organisms on the basis of their catalase activities at different temperatures and pH. Am Rev Respir Dis. 1960 Mar;81:387–391. doi: 10.1164/arrd.1960.81.3.387. [DOI] [PubMed] [Google Scholar]
- Lapage S. P., Bascomb S., Willcox W. R., Curtis M. A. Identification of bacteria by computer: general aspects and perspectives. J Gen Microbiol. 1973 Aug;77(2):273–290. doi: 10.1099/00221287-77-2-273. [DOI] [PubMed] [Google Scholar]
- Meissner G., Schröder K. H., Amadio G. E., Anz W., Chaparas S., Engel H. W., Jenkins P. A., Käppler W., Kleeberg H. H., Kubala E. A co-operative numerical analysis of nonscoto- and nonphotochromogenic slowly growing mycobacteria. J Gen Microbiol. 1974 Aug;83(2):207–235. doi: 10.1099/00221287-83-2-207. [DOI] [PubMed] [Google Scholar]
- Murphy D. B., Hawkins J. E. Use of urease test disks in the identification of mycobacteria. J Clin Microbiol. 1975 May;1(5):465–468. doi: 10.1128/jcm.1.5.465-468.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamura M. Identification of mycobacteria. Tubercle. 1967 Dec;48(4):311–338. doi: 10.1016/s0041-3879(67)80040-0. [DOI] [PubMed] [Google Scholar]
- WAYNE L. G., DOUBEK J. R. CLASSIFICATION AND IDENTIFICATION OF MYCOBACTERIA. II. TESTS EMPLOYING NITRATE AND NITRITE AS SUBSTRATE. Am Rev Respir Dis. 1965 May;91:738–745. doi: 10.1164/arrd.1965.91.5.738. [DOI] [PubMed] [Google Scholar]
- WAYNE L. G., DOUBEK J. R., RUSSELL R. L. CLASSIFICATION AND IDENTIFICATION OF MYCOBACTERIA. I. TESTS EMPLOYING TWEEN 80 AS SUBSTRATE. Am Rev Respir Dis. 1964 Oct;90:588–597. doi: 10.1164/arrd.1964.90.4.588. [DOI] [PubMed] [Google Scholar]
- WAYNE L. G. Two varieties of Mycobacterium kansasii with different clinical significance. Am Rev Respir Dis. 1962 Nov;86:651–656. doi: 10.1164/arrd.1962.86.5.651. [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Andrade L., Froman S., Käppler W., Kubala E., Meissner G., Tsukamura M. A co-operative numerical analysis of Mycobacterium gastri, Mycobacterium kansasii and Mycobacterium marinum. J Gen Microbiol. 1978 Dec;109(2):319–327. doi: 10.1099/00221287-109-2-319. [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Dietz T. M., Gernez-Rieux C., Jenkins P. A., Käppler W., Kubica G. P., Kwapinski J. B., Meissner G., Pattyn S. R., Runyon E. H. A co-operative numerical anaysis of scotochromogenic slowly growing mycobacteria. J Gen Microbiol. 1971 Jun;66(3):255–271. doi: 10.1099/00221287-66-3-255. [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Doubek J. R. Diagnostic key to mycobacteria encountered in clinical laboratories. Appl Microbiol. 1968 Jun;16(6):925–931. doi: 10.1128/am.16.6.925-931.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G. Microbiology of tubercle bacilli. Am Rev Respir Dis. 1982 Mar;125(3 Pt 2):31–41. doi: 10.1164/arrd.1982.125.3P2.31. [DOI] [PubMed] [Google Scholar]
- Wayne L. G. Phenol-Soluble Antigens from Mycobacterium kansasii, Mycobacterium gastri, and Mycobacterium marinum. Infect Immun. 1971 Jan;3(1):36–40. doi: 10.1128/iai.3.1.36-40.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974 Jan;109(1):147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- Willcox W. R., Lapage S. P., Bascomb S., Curtis M. A. Identification of bacteria by computer: theory and programming. J Gen Microbiol. 1973 Aug;77(2):317–330. doi: 10.1099/00221287-77-2-317. [DOI] [PubMed] [Google Scholar]
- Wolinsky E. Nontuberculous mycobacteria and associated diseases. Am Rev Respir Dis. 1979 Jan;119(1):107–159. doi: 10.1164/arrd.1979.119.1.107. [DOI] [PubMed] [Google Scholar]


