Abstract
Despite the predominance of UVA relative to UVB in terrestrial sunlight, solar mutagenesis in humans and rodents is characterized by mutations specific for UVB. We have investigated the kinetics of repair of UVA- and UVB-induced DNA lesions in relation to mutagenicity in transgenic mouse fibroblasts irradiated with equilethal doses of UVA and UVB in comparison to SSL. We have also analyzed mutagenesis-derived carcinogenesis in sunlight-associated human skin cancers by compiling the published data on mutation types found in crucial genes in non-melanoma and melanoma skin cancers. Here, we demonstrate a resistance to repair of UVB-induced CPDs together with rapid removal of UVA-induced oxidized purines in the genome overall and in the cII transgene of SSL-irradiated cells. The spectra of mutation induced by both UVB- and SSL-irradiation in this experimental system are characterized by significant increases in relative frequency of C to T transitions at dipyrimidines, which are the established signature mutation of CPDs. This type of mutation is also the predominant mutation found in human non-melanoma and melanoma tumor samples in the TP53, CDKN2, PTCH, and protein kinase genes. The prevailing role of UVB over UVA in solar mutagenesis in our test system can be ascribed to different kinetics of repair for lesions induced by the respective UV-irradiation.
Keywords: DNA damage, mutations, skin cancer, sunlight, ultraviolet radiation
INTRODUCTION
Sunlight ultraviolet (UV) radiation is an established environmental physical carcinogen, which has been implicated in the etiology of human skin cancer (1, 2). Of the UV spectrum of sunlight, the UVC (<280 nm) is entirely absorbed by stratospheric oxygen (O2). The resulting decomposed O2 molecules undergo recombination to form ozone (O3), which in turn, blocks the majority of UVB (280–320 nm). Owing to its high penetrating efficiency, the UVA (>320–400 nm) can mostly pass through various atmospheric layers and reach the surface of the Earth (1, 3). Thus, the terrestrial sunlight UV is mainly comprised of UVA (~95%) and the remainder unabsorbed UVB (~5%) (1). Despite the disproportionally low presence of UVB in sunlight, its potent photocarcinogenicity accounts for the majority of solar UV-associated neoplasia (1, 3). Nonetheless, UVA is estimated to contribute to 10–20% of sunlight-induced carcinogenesis (4).
The UVB carcinogenicity is ascribed to the ability of this waveband to induce promutagenic DNA lesions, primarily cis-syn cyclobutane pyrimidine dimers (CPDs), and pyrimidine (6-4) pyrimidone photoproducts ((6-4)PPs) (5). The UVA is believed to trigger mutagenesis, among other modes of action, e.g. tumor promotion, through induction of photosensitization reactions that generate oxidative DNA lesions, particularly 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxo-dG) (5). Owing to their miscoding potentials, photodimeric CPDs and (6-4)PPs can induce single or tandem C to T transition mutations (6–9), whereas 8-oxo-dG can produce largely G to T transversion mutations (10, 11). The lesion specificity and signature mutations of the respective UV wavebands can be used for investigating “DNA-damage targeted mutagenicity” of sunlight. Correlating the types of induced DNA lesions and mutations consequent to solar UV irradiation can verify the contribution of UVA and UVB to mutagenesis, thereby, helping unravel the underlying mechanism of sunlight-induced carcinogenesis.
Transgenic rodent mutagenesis systems are invaluable models for correlation studies of DNA damage and mutagenesis at both genomic and single nucleotide level (12). Using transgenic Big Blue® mouse embryonic fibroblasts, we have shown a typical oxidative-DNA damage mediated mutagenicity of UVA in the lacI (13) and cII transgenes (14, 15). We have also demonstrated that addition of cellular photosensitizers, i.e., riboflavin (16) and delta-aminolevulinic acid (14), can intensify the observed UVA-induced DNA damage and mutagenesis, whereas inclusion of the antioxidant vitamin C can counteract the induced effects (16). The characteristic oxidative-DNA damage derived mutagenicity of UVA is, however, not prevalent in sunlight-induced mutagenesis, wherein single and tandem C to T transitions at dipyrimidine sites dominate the spectrum of induced mutations (17, 18). The latter points to an overruling involvement of CPDs and/or (6-4)PPs induced by UVB in solar mutagenesis. Mechanistically, however, the superseding role of UVB over UVA in sunlight-induced mutagenesis has not been explained yet.
Theoretically, a resistance to repair of UVB-induced lesions together with efficient removal of UVA-induced lesions may elucidate the predominance of characteristic UVB-induced mutations in solar mutagenesis (5). In the present study, we have tested this theory by comparing the formation and kinetics of repair of UVA- and UVB-induced lesions, as well as analyzing the mutagenicity of the respective UV wavebands in mouse cells. We have irradiated transgenic Big Blue® mouse embryonic fibroblasts with physiologically relevant doses of UVA and UVB, in comparison to that of simulated-sunlight UV (SSL). Subsequently, we have determined the formation and repair of oxidative DNA damage and dipyrimidine photolesions in the genome overall and specifically, in the cII transgene at nucleotide resolution level, as well as analyzed the cII mutant frequency and mutation spectrum. Furthermore, we have analyzed mutagenesis-derived carcinogenesis in sunlight-associated human skin cancers by compiling the published data on mutation types found in crucial genes in non-melanoma and melanoma skin cancers.
MATERIALS AND METHODS
Cell culture and UV irradiation
Early passage Big Blue® mouse embryonic fibroblasts were grown as monolayer at ~30–40% confluence in Dulbecco's Modified Eagle’s Medium supplemented with 10% fetal bovine serum. Prior to UV irradiation, the culture media were removed, and the cells were washed thoroughly with phosphate buffered saline (PBS). The culture dishes were filled with a 1 cm layer of PBS, placed on ice (with lids on), and then irradiated from above with UVA, UVB, and SSL (at ~5 cm distance). To ensure the sterility of cell cultures, it was inevitable to irradiate petri dishes through the lids, although this procedure could likely cause a reduction of fluence rate, most notably, in the case of UVB irradiation. Nonetheless, for all UV sources, we determined the fluence rate at the bottom surface of the irradiated culture dishes using a UVX radiometer (Ultraviolet Products, Upland, CA) placed under the respectively irradiated petri dishes (with lids on). This enabled us to adjust for the blocking effect of the lids during various types of UV-irradiation because we calculated the actual UV dose delivered to the cells after being filtered through the lids.
The UVA source was a Sellas Sunlight System (Medizinische Geräte GmbH; Gevelsberg, Germany) with an average fluence rate of 60 mW/cm2. The source exclusively emits long wave UVA (UVA1: 340–400 nm) and not UVA at the borderline with UVB. The UVB source consisted of three fluorescent tubes (Philips TL 20 W/12R) filtered through a cellulose acetate sheet, which cuts off wavelengths below 295 nm. The source has a peak spectral emission at 312 nm and an average fluence rate of 1.15 mW/cm2. The SSL source was an Oriel 1000 Watt Solar Simulator powered with an ozone-free Xenon Arc lamp (Model 81190, Newport, Stratford, CT). We installed an Atmospheric Attenuation Filter (Air Mass 1.0) in this solar simulator to correct the output emission to match the solar spectrum at ground level when the sun is directly overhead.
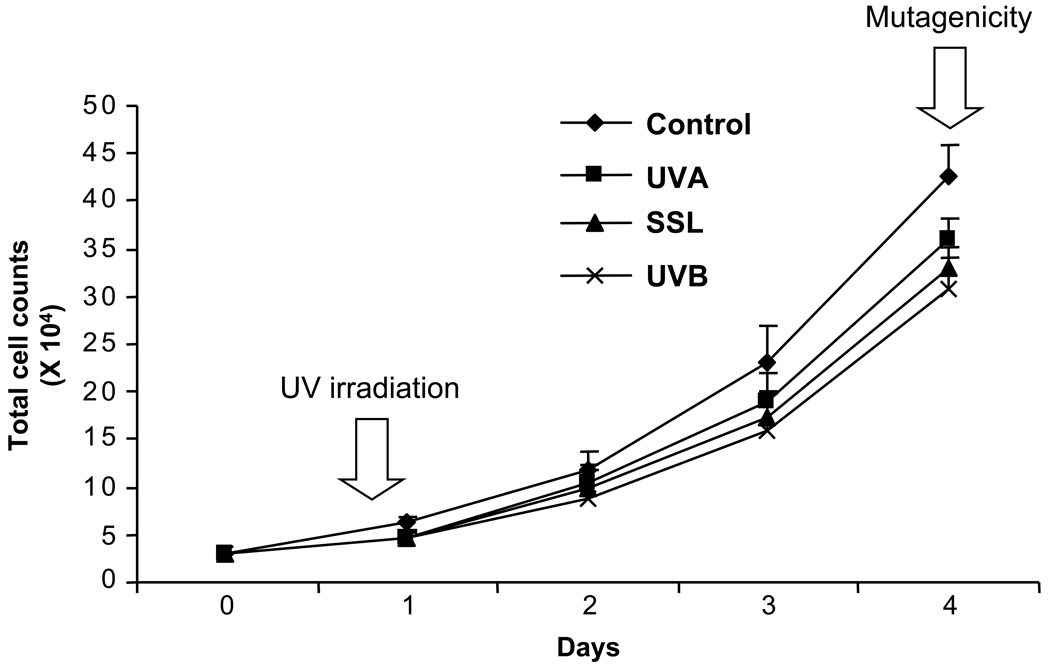
Obviously, only in viable and proliferative cells can the induced DNA damage be efficiently translated into mutations. Thus, in keeping with the objective of our study to examine DNA-damage targeted mutagenicity of UVA and UVB in comparison to SSL, we first determined biologically relevant doses of respective types of UV irradiation, which equally caused moderate cytotoxicity, i.e., 75% cell survival. Accordingly, we performed a series of preliminary tests in which increasing doses of UVA, UVB, or SSL-irradiation were administered to the cells, and the corresponding kill curves for the respective types of UV irradiation were established. Once the acceptable equilethal doses of UVA, UVB, and SSL-irradiation were determined, we then administered the respective doses of UV irradiation to a set of cell cultures at a similar cell density, and subsequently assessed the proliferative capacity of the cells at defined times post-irradiation. More specifically, we prepared a comparable set of cultures by seeding 3 × 104 cells per culture dish on the day prior to UV-irradiation (T = 0). Eight hours post-irradiation (T = 1) and every 24 hours thereafter until day four (T = 2, T = 3, and T= 4), we harvested a subset of cell cultures from all treatment groups and subsequently performed total cell counting. As shown in Figure 1, the total number of viable cells determined in triplicate cultures of mouse embryonic fibroblasts irradiated with UVA, UVB, or SSL-irradiation was comparable at all times post-irradiation.
Figure 1. Assessment of cell proliferation capacity in UV-irradiated mouse embryonic fibroblasts.
Second passage mouse embryonic fibroblasts were seeded at a density of 3 × 104 cells per culture dish on the day prior to UV-irradiation (T = 0), and subsequently irradiated with equilethal (~75% cell survival) doses of UVA, UVB, and SSL in comparison with control as described in Materials and Methods. Eight hours post-irradiation (T = 1) and every 24 hours thereafter until day four (T = 2, T = 3, and T= 4), a subset of cell cultures from all treatment groups was harvested, and subsequently total cell counting was performed. The total number of viable cells was determined in triplicate cultures irradiated with respective types of UV-irradiation, and the results are averaged for all time points. Arrows indicate the defined times at which UV-irradiation and cell harvesting for mutagenicity experiments, respectively, were performed. Error bars = standard deviations.
For all comparative analyses, the equilethal doses of UVA, UVB, and SSL-irradiation administered to the cells were 18 J/cm2 for UVA (365 nm sensor), 14.4 J/cm2 for SSL (365 nm sensor) and 75.6 mJ/cm2 for UVB (310 nm sensor), which are all biologically relevant, i.e., equivalent to doses received by humans exposed to midday, clear sky, midsummer sun for a short period of time. Immediately after UV irradiation and at defined times (up to 24 hours post-treatment), the cells were harvested by trypsinization for evaluation of DNA damage and repair. Alternatively, the cells were cultured in complete growth medium for an additional 4 days, and afterward were analyzed for mutant frequency and mutation spectrum of the cII transgene. The 4-day growing period (expression time) is necessary for fixation of all mutations into the genome (12). At the time of harvesting, all cultured cells had undergone a few rounds of population doublings and reached full confluence. All experiments were conducted in triplicates.
Lesion-specific cleavage assays with UV damage endonuclease (UVDE), T4 endonuclease V (T4 Endo V), and formamidopyrimidine DNA glycosylase (Fpg)
The enzymatic digestion assays are based on the recognition and cleavage of specific types of DNA damage by specialized DNA repair enzymes, followed by gel electrophoresis. We used UVDE for cleavage of CPDs, and (6-4)PPs and their Dewar photoisomers combined (19–21), T4 Endo V for nicking specifically at CPDs (12), and Fpg for cutting of oxidized (ring-opened) purines (22, 23). Briefly, DNA was digested with an excess amount of UVDE (Trevigen, Gaithersburg, MD), T4 Endo V (Epicentre, Madison, WI), or Fpg (Trevigen) in buffers supplied by the manufacturers. After ethanol precipitation, the digests were loaded onto a 1.5% alkaline agarose gel, and run at 3.0 V/cm for 4 hours, with constant recirculation of the running buffer. The electrophoretic profiles were determined using standard ethidium bromide staining. Scanning was performed using a Quantity One image analyzer of the Bio-Rad Imaging Equipment (Bio-Rad Laboratories, Life Science Group, Hercules, CA).
Terminal transferase-dependent polymerase chain reaction (TD-PCR) and ligation-mediated polymerase chain reaction (LM–PCR)
The principles of TD-PCR are essentially similar to LM-PCR with the only differences being the primer extension and ligation steps (12). Methodologically, both techniques rely on the concept that DNA polymerase cannot synthesize past certain types of damage, i.e., bulky lesions, e.g., dipyrimidine photolesions, and single strand breaks. TD-PCR can be readily applied to all DNA templates that contain polymerase-blocking lesions, including CPDs and (6-4)PPs. Unlike TD-PCR, LM-PCR requires conversion of original DNA templates to single strand breaks with 5’- phosphate groups at the sites of lesion formation. To fulfill this requirement, we pre-treated the genomic DNA as follows, (I) for LM-PCR footprinting of oxidized (ring-opened) purines, we used Fpg digestion, which releases damaged bases to generate abasic sites, as well as cleaves the sugar-phosphate backbone to produce single strand DNA breaks with 5’- phosphate termini, and (II) for LM-PCR footprinting of CPDs, we used T4 Endo V digestion, which cleaves the glycosidic bond of the 5’-pyrimidine in a CPD and breaks the phosphodiester bond 3’- to the resulting abasic site, followed by Escherichia coli (E.coli) CPD photolyase reactivation, which detaches the dissociated 3’- pyrimidine, thereby, yielding single strand DNA with a normal base on the 5’- sugar-phosphate terminus (12) (see, ‘Supporting Information’).
cII Mutant frequency and mutation spectrometry determination
The λLIZ shuttle vectors, which contain the chromosomally integrated cII transgene, were recovered from the genomic DNA, and packaged into viable phage particles using the Transpack Packaging Kit (Stratagene, La Jolla, CA). After pre-adsorption of the phages to G1250 E. coli, the bacterial culture was grown on special TB1 agar plates. To select for cII mutants, the plates were incubated at 24°C for 48 hours. Alternatively, the plates were incubated under non-selective condition, i.e., 37°C overnight, to express both the wild type and mutant cII. Verification of all putative cII mutants was achieved by re-plating under the selective condition. The ratio of the number of verified mutant plaques to the total number of screened plaques denotes the cII mutant frequency. As recommended by the manufacturer (Stratagene), minimums of 3 × 105 rescued phages were screened for each experimental condition,. For mutation spectrometry, all verified mutant plaques were amplified by PCR using the “lambda select-cII sequencing primers” according to the manufacturer’s recommended protocol. The purified PCR products were then subjected to sequencing using a Big Dye terminator cycle sequencing kit and an ABI-3730 DNA Sequencer (ABI Prism, PE Applied BioSystems, Foster City, CA) (see, ‘Supporting Information’).
Statistical Analysis
Results are expressed as means ± SD. All variables in UV-irradiated versus control groups were compared by the Mann-Whitney Test. The entire mutation spectra and the specific types of mutation in the UV-irradiated versus control groups were compared by the hypergeometric test of Adams and Skopek (24) and chi-square test, respectively. All statistical tests were two-sided. Values of P < 0.05 were considered statistically significant.
RESULTS
Of relevance for the present study, culturing of mouse embryonic fibroblasts under physiologic O2 tension (3%) minimizes the burden of oxidative stress on the cells, and enhances their proliferation capacity (16). This leads to a significant reduction in the frequency of spontaneously derived mutations associated with in vitro aging, and an enhanced accumulation of induced mutations (16, 25, 26). Obviously, only in viable and replicating cells can the induced DNA lesions be efficiently translated into mutations. Therefore, we administered moderate but biologically relevant doses of UVA, UVB, and SSL at equilethal doses, leading to ~75% cell survival (see, Fig. 1).
DNA damage in the genome
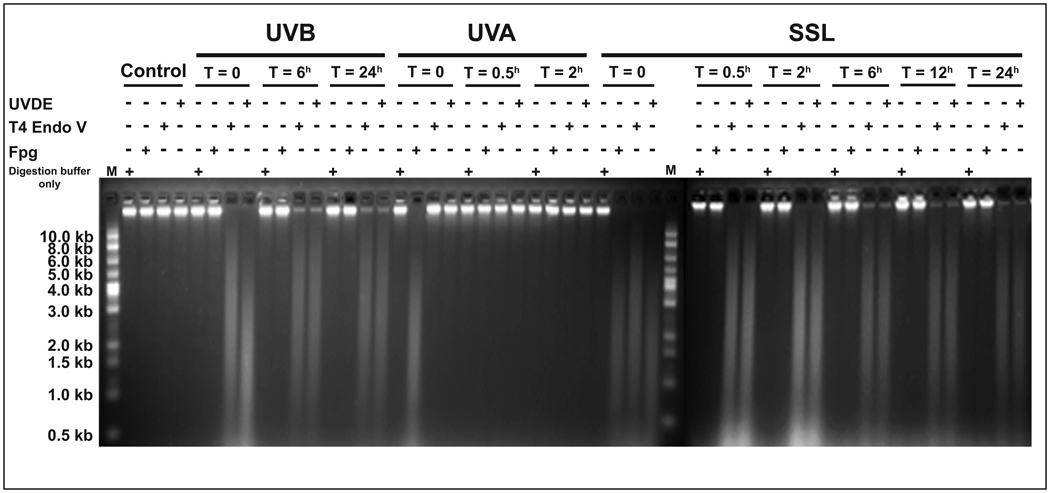
We used specific DNA repair enzymes in combination with alkaline agarose gel electrophoresis, to qualitatively assess different types of induced lesions in the genome overall of UV-irradiated cells. We used UVDE for determining CPDs and (6-4)PPs combined (19), T4 Endo V for quantifying CPDs only (12), and Fpg protein for detecting oxidized (ring-opened) purines, e.g., 8-oxo-dG (22, 23). As shown in Figure 2, both UVB- and SSL-irradiation produced UVDE-sensitive sites and to a lesser extent T4 Endo V-sensitive sites in the genome overall of irradiated cells. In both cases, the intensity of UVDE-sensitive sites equaled that of T4 Endo V-sensitive sites within six hours of irradiation. This indicates a complete removal of (6-4)PPs from the genome overall of UVB- and SSL-irradiated cells within this time period. Subsequently, equivalent levels of UVDE- and T4 Endo V-sensitive sites remained unchanged over 24 hours, which implies a persistence of CPDs in the genome overall throughout this time period. Conversely, UVA irradiation did not induce detectable UVDE-sensitive sites or T4 Endo V-sensitive sites in the genome overall of irradiated cells. However, the UVA- and SSL-irradiation both gave rise to Fpg-sensitive sites in the genomic DNA of irradiated cells. Remarkably, in both cases, the induced Fpg-sensitive sites were fully removed within thirty minutes of irradiation. Because Fpg recognizes sites of base loss (AP sites) in addition to base modifications (22, 23), the absence of AP sites in the genome overall of UVA-irradiated cells can be inferred from the absence of sites sensitive to T4 Endo V, since this latter enzyme would also recognize AP sites (27). UVB irradiation did not form appreciable levels of Fpg-sensitive sites in the genome overall of irradiated cells, however (see, Fig. 2).
Figure 2. Qualitative assessment of induced DNA damage in mouse genome.
Mouse embryonic fibroblasts were irradiated with equilethal (~75% cell survival) doses of UVA, UVB, and SSL in comparison with control as described in Materials and Methods. Genomic DNA was isolated and digested with specialized DNA repair enzymes, including (I) UVDE for detecting CPDs and (6-4)PPs combined, (II) T4 Endo V for determining CPDs only, and (III) Fpg for detecting oxidized (ring-opened) purines. The DNA digests were subjected to alkaline agarose gel electrophoresis, followed by standardized visualization procedure. For brevity, results at select time points are shown. (+) and (−) represent the presence and absence, respectively, of the indicated treatment conditions. In this assay, the frequency of lesions is roughly estimated from the central location of the most intense part of the smear but not from the length of the smear. Digestion buffer only = No enzyme was added to the reaction mix. M = molecular size marker.
DNA damage in the cII transgene
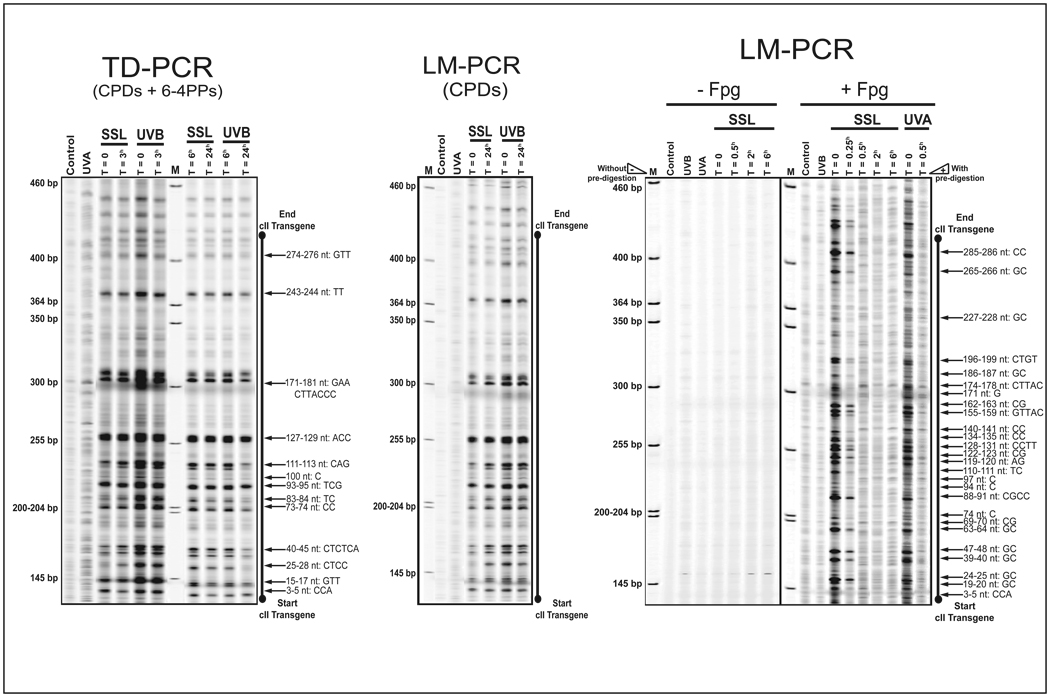
We used TD-PCR to map the formation of dipyrimidine photolesions (CPDs and (6-4)PPs combined) in the cII transgene of UV irradiated cells (12). As shown in Fig. 3a, both UVB- and SSL-irradiation produced CPDs and/or (6-4)PPs at similar locations along the cII transgene, with UVB-irradiation inducing relatively higher DNA damage. The UVA irradiation, however, did not form considerable level of dipyrimidine photolesions in the cII transgene. For the most part, the detected levels of damage consequent to UVB- and SSL-irradiation were decreased six hours post-irradiation. To precisely identify the type of dipyrimidine photolesions induced by UVB- and SSL-irradiation, we used successive T4 Endo V digestion and CPD photolyase reactivation coupled with LM-PCR, a genomic footprinting methodology for specific detection of CPDs (12). The resembling pattern of lesions detected by LM-PCR and TD-PCR, respectively, verified that a great number of dipyrimidine photolesions identified by TD-PCR, was indeed CPDs, which persisted over 24 hours post-UVB- and SSL-irradiation (Fig. 3b). By rule of elimination, the fraction of DNA damage that was repaired within six hours of UVB- and SSL-irradiation, as determined by TD-PCR, was presumably (6-4)PPs (see, Fig. 2).
Figure 3. Mapping of induced DNA damage in the cII transgene.
TD-PCR and LM-PCR footprinting of the full-length cII transgene was done using the genomic DNA of mouse embryonic fibroblasts irradiated with equilethal (~75% cell survival) doses of UVA, UVB, and SSL in comparison with control.
(a) Footprinting of dipyrimidine photolesions (CPDs and (6-4)PPs combined) by TD-PCR.
(b) Footprinting of CPDs by LM-PCR.
The LM-PCR bands migrate approximately 3 bases faster than the corresponding TD-PCR bands due to the addition of three riboguanosine triphosphate to all primer extension products in the latter method.
(c) Footprinting of oxidized (ring-opened) purines by LM-PCR. (+Fpg), with Fpg enzyme pre-digestion to quantify oxidized (ring-opened) purines; (−Fpg), without Fpg enzyme pre-digestion to quantify non-specific background bands, e.g., UV-induced single strand breaks or abasic sites and strand breaks, resulting form spontaneous depurination of DNA. For brevity, results at select time points are shown. bp = base pair; M = molecular size marker; nt = nucleotide position.
Furthermore, we used Fpg digestion in combination with LM-PCR to map the formation of oxidized (ring-opened) purines in the cII transgene of UV irradiated cells (12). As shown in Figure 3c, UVA- and SSL-irradiation induced comparable levels of oxidized (ring-opened) purines at similar positions along the cII transgene. In both cases, the induced oxidized (ring-opened) purines completely disappeared within thirty minutes of irradiation. Methodologically, in LM-PCR and TD-PCR, single strand DNA breaks may produce non-specific signals above background (12). To show the specificity of our footprinting analysis, we subjected the genomic DNA of all UV-irradiated cells to LM-PCR without enzymatic pre-digestion, a sensitive method to quantify single strand DNA breaks 16). In all UV-irradiated cells, we ruled out the possibility of formation of single strand DNA breaks in the cII transgene (Fig. 3c “-Fpg” panel). The latter is consistent with the lack of detection of single strand DNA breaks in the genome overall of UV-irradiated cells (Fig. 2, “Digestion buffer only” lanes; single strand DNA breaks manifest as smears in alkaline agarose gel electrophoresis).
cII Mutation analysis
UVA irradiation was weakly but significantly mutagenic to mouse embryonic fibroblasts as it elevated the cII mutant frequency 3.9-fold over background (P < 0.03) (Table 1). Conversely, SSL- and UVB-irradiation were extremely mutagenic as they increased the relative frequencies of cII mutants 43.3- and 99.3-fold, respectively, in the irradiated cells. To establish the spectra of induced and spontaneous mutations, we sequenced the DNA of cII mutants obtained from UV-irradiated and control cells. We randomly selected 110, 161, and 100 cII mutant plaques induced by UVB-, UVA-, and SSL-irradiation, respectively, in comparison to 154 spontaneously arisen control plaques. Of the respective number of plaques, 108, 152, 87, and 147 contained a minimum of one mutation in the cII transgene. As shown in Table 2, single mutations, mostly single base substitutions, predominated in all induced- and control mutation spectra. Also, tandem base substitutions, including the hallmark CC to TT transitions (6–9), occurred exclusively in UV-induced mutation spectra. Detailed mutation spectra induced by UVB-, UVA-, and SSL-irradiation as compared with spontaneous mutation spectrum are presented in Figure 4. The overall spectra of mutations induced by UVB-, UVA-, or SSL-irradiation were all significantly different from that of control (P < 0.001; Adams and Skopek test).
TABLE 1.
Mutant frequency of the cII transgene in Big Blue mouse embryonic fibroblasts irradiated with equilethal doses of UVA, UVB, and SSL or control
| Treatment | Total number of plaques (pfu*) |
Mutant plaques | Average mutant frequency (× 10−5)†,¶ |
|---|---|---|---|
| Control | 9,637,500 | 262 | 2.7 ± 0.7 |
| UVA | 2,903,500 | 305 | 10.82 ± 2.82 |
| UVB | 765,000 | 2,078 | 268.0 ± 10.7 |
| SSL | 555,000 | 656 | 117.0 ± 6.3 |
Plaque forming unit
Results are expressed as Mean ± SD
Each treatment condition was assayed at least 3 times and the average results are presented. To obtain sufficient number of cII mutant plaques for DNA sequencing, more assays were performed on samples with low mutant frequency, i.e., UVA-irradiated and control samples, as indicated by the respective high pfu numbers.
TABLE 2.
Types of UV-induced and spontaneously derived mutations in the cII transgene in Big Blue mouse embryonic fibroblasts irradiated with equilethal doses of UVB, UVA and SSL or control
| Mutation type | SSL | UVB | UVA | Control | ||
|---|---|---|---|---|---|---|
| Single mutation | 87 (100%) | 105 (97.2%) | 140 (97.9%) | 146 (99.3%) | ||
| Multiple mutations | 0 (0%) | 3 (2.8%) | 3 (2.1%) | 1 (0.7%) | ||
| Base substitution | Single | 85 (97.7%) | 101 (91.0%) | 128 (88.9%) | 125 (84.5%) | |
| Tandem | GG→AA or CC→TT | 2 (2.3%) | 5 (4.5%) | 2 (1.4%) | 0 (0%) | |
| GT→AG or CA→TC | 0 (0%) | 0 (0%) | 1 (0.7%) | 0 (0%) | ||
| AC→TT or TG→AA | 0 (0%) | 2 (1.8%) | 0 (0%) | 0 (0%) | ||
| AC→GA or TG→CT | 0 (0%) | 0 (0%) | 1 (0.7%) | 0 (0%) | ||
| AG→TA or TC→AT | 0 (0%) | 1 (0.9%) | 0 (0%) | 0 (0%) | ||
| AG→CA or TC→GT | 0 (0%) | 1 (0.9%) | 0 (0%) | 0 (0%) | ||
| Deletion | Single | 0 (0%) | 0 (0%) | 0 (0%) | 13 (8.8%) | |
| Multiple | 0 (0%) | 0 (0%) | 12 (8.3%) | 3 (2.0%) | ||
| Insertion | Single | 0 (0%) | 1 (0.9%) | 0 (0%) | 7 (4.7%) | |
| Multiple | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
Randomly selected cII mutant plaques, including 110, 161, and 100 plaques induced by UVB-, UVA-, and SSL-irradiation, respectively, in comparison to 154 control plaques were subjected to DNA sequencing. Of the respective number of plaques, 108, 152, 87, and 147 contained a minimum of one mutation in the cII transgene.
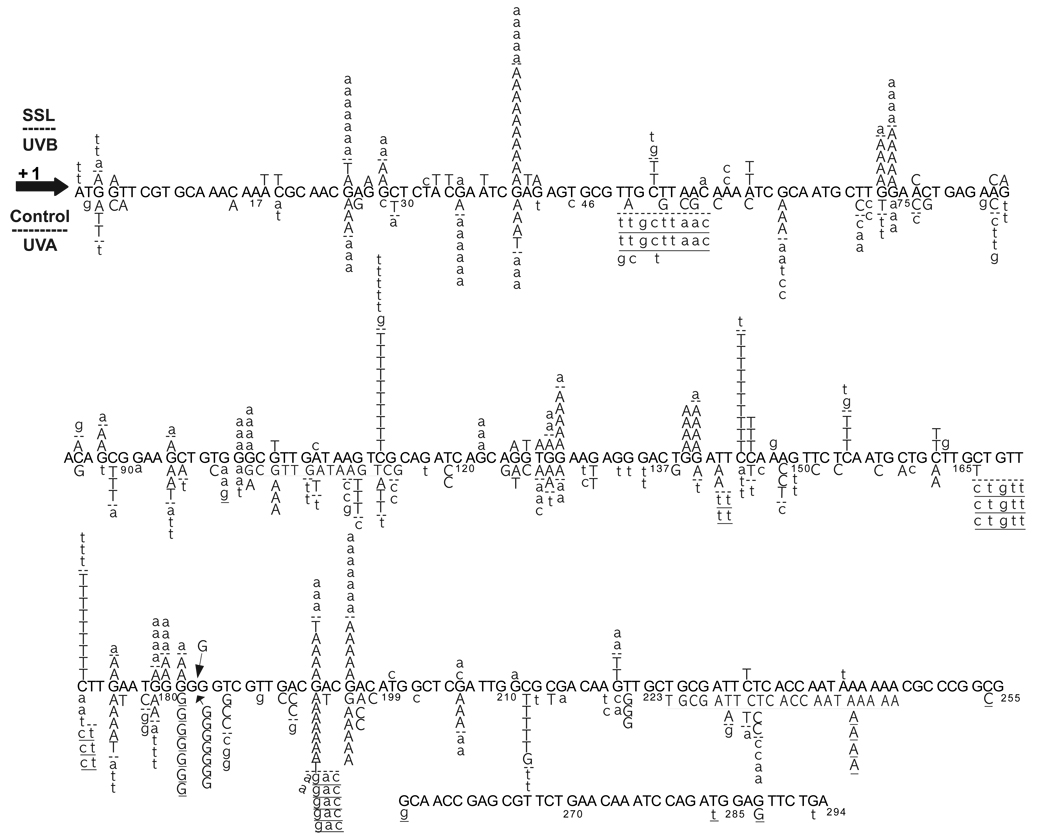
Figure 4. UV-induced and spontaneous cII mutation spectrometry.
Detailed mutation spectra of the cII transgene in mouse embryonic fibroblasts irradiated with equilethal (~75% cell survival) doses of UVB, UVA, and SSL in comparison to control. Mutations were quantified using the lambda transgenic shuttle vector recovery kit for Big Blue® rodents (Stratagene). Randomly selected cII mutant plaques, including 110, 161, and 100 plaques induced by UVB-, UVA, and SSL-irradiation, respectively, in comparison to 154 control plaques were subjected to DNA sequencing. Of the respective number of plaques, 108, 152, 87, and 147 contained a minimum of one mutation in the cII transgene. The UVB- and SSL-induced mutations are typed above the reference cII sequence, whereas the UVA-induced mutations and spontaneous control mutations are shown below the reference cII sequence. The UVB-induced mutations and spontaneous control mutations are typed in capital letters, whereas the SSL- and UVA-induced mutations are typed in small letters, the two are separated from one another, respectively, by (--). Data on UVA mutation spectrometry are adapted from Ref. (15). Substituted bases are in bold. Deleted bases are underlined; multiple deletions are continuously underlined. Inserted bases are shown with an arrow. Numbers below the bases are the nucleotide positions.
To specify the difference(s) between induced and control mutation spectra, we compared the frequency of each type of mutation, e.g., transitions, transversions, etc. in the respective mutation spectra. In the Big Blue system, the cII transgene is a non-transcribed gene (28). Thus, the strand bias of mutagenesis, a phenomenon caused by transcription-coupled DNA repair (TCR) (29), is unlikely to affect the spectrum of mutations produced in this transgene. Methodologically, therefore, it is appropriate to combine the strand mirror counterparts of all transitions (e.g., G to A + C to T) and transversions (e.g., G to T + C to A) when comparing the specific types of mutation between different treatment groups. As shown in Table 3, G:C to A:T transitions prevailed in the spectra of mutations produced by both UVB- and SSL-irradiation, i.e., 86.7% and 79.8% of all induced mutations, respectively, versus 39.2% in control; P < 0.001. The G:C to A:T transition mutations induced by UVB- and SSL-irradiation occurred (almost) entirely at dipyrimidine sites, i.e., 98% and 100%, respectively. The latter type of mutations accounted for 85% and 80%, respectively, of the observed increases in cII mutant frequency consequent to UVB- and SSL-irradiation.
TABLE 3.
Comparative mutation spectra of the cII transgene in Big Blue mouse embryonic fibroblasts irradiated with equilethal doses of UVB, UVA, and SSL or control
| Mutation type | Number of Mutations | % Mutations | Absolute Mutant Frequency (× 10−5) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSL | UVB | UVA | Control | SSL | UVB | UVA | Control | SSL | UVB | UVA | Control | |
| G:C→C:G | 5 | 0 | 8 | 6 | 5.6 | 0 | 5.7 | 4.1 | 6.6 | 0 | 0.6 | 0.1 |
| G:C→T:A | 3 | 6 | 36 | 19 | 3.4 | 5.0 | 25.7 | 12.8 | 3.9 | 13.4 | 2.8 | 0.4 |
| G:C→A:T | 71 | 104 | 43 | 58 | 79.8 | 86.7 | 30.7 | 39.2 | 93.3 | 232.3 | 3.3 | 1.1 |
| (at dipyrimidines)* | (71) | (102) | (38) | (43) | (79.8) | (85.0) | (27.1) | (29.1) | (93.3) | (227.8) | (2.9) | (0.8) |
| A:T→T:A | 3 | 7 | 11 | 10 | 3.4 | 5.8 | 7.9 | 6.8 | 3.9 | 15.6 | 0.9 | 0.2 |
| A:T→G:C | 3 | 0 | 13 | 15 | 3.4 | 0 | 9.3 | 10.1 | 3.9 | 0 | 1.0 | 0.3 |
| A:T→C:G | 4 | 2 | 14 | 17 | 4.5 | 1.7 | 10.0 | 11.5 | 5.3 | 4.5 | 1.0 | 0.3 |
| Del/Ins | 0 | 1 | 15 | 23 | 0 | 0.8 | 10.7 | 15.5 | 0 | 2.2 | 1.2 | 0.4 |
The number of G:C→A:T transition mutations occurring specifically at dipyrimidine sites is indicated into brackets. Data on UVA mutation spectrometry are adapted from Ref. (15). Del = deletion; Ins = insertion.
Recently, we have characterized the spectrum of mutations induced by UVA irradiation in both the cII (14, 15) and lacI(13) transgene of the same model system under standardized conditions. We have established that UVA irradiation predominantly induced single mutations, mainly single base substitutions, in both the cII and lacI transgene. Whereas the relative frequency of G:C to T:A transversions in the cII and lacI transgenes were significantly induced consequent to UVA irradiation, i.e., ~26% and 28% of all induced mutations in the respective genes, G:C to A:T transitions at dipyrimidine sites occurred almost equally in UVA-induced and control mutation spectra in both the cII and lacI transgene (13, 15).
Mutation spectrometry in human skin cancers
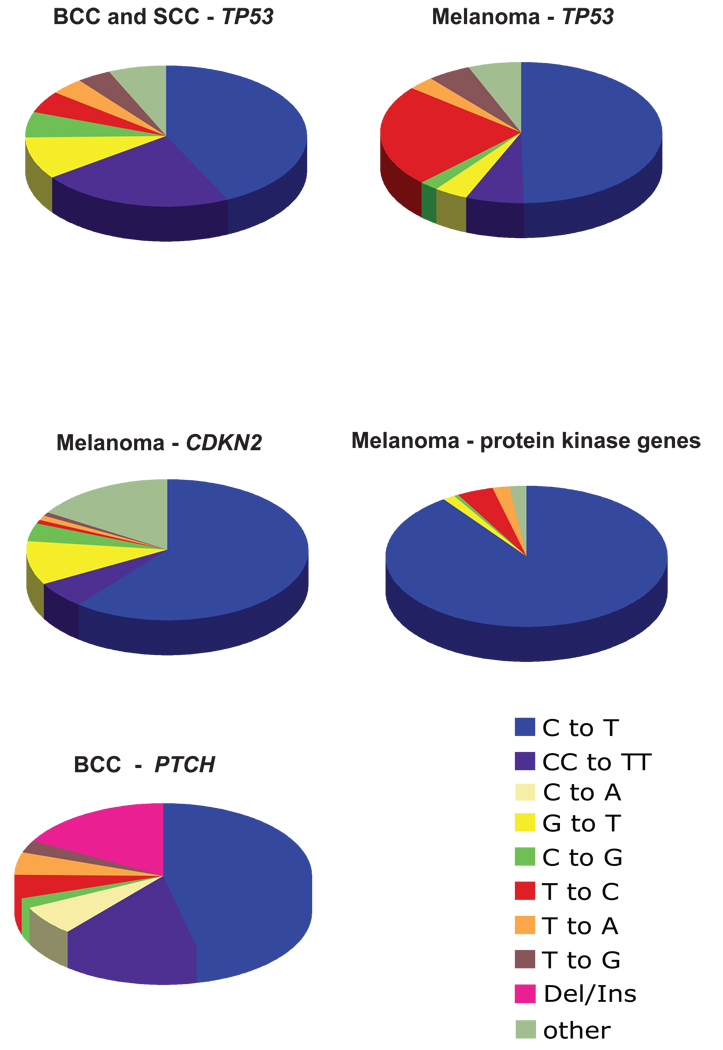
We have updated our knowledge of mutagenesis-derived carcinogenesis in sunlight-associated human skin cancers by performing a thorough literature search and compiling the published data on mutation types found in crucial genes in non-melanoma and melanoma skin cancers. We included relevant tumor suppressor genes in our mutation spectrometry data analysis and excluded oncogenes, such as BRAF or RAS from our analysis because mutations in the latter genes occur only at very few selected codon positions. i.e., BRAF codon V600(30) or RAS codons 12, 13, and 61 (31, 32), respectively. Data for the TP53 gene (n = 94 for melanomas and n = 482 for non-melanoma skin cancers) were obtained from the International Agency for Research on Cancer (IARC) mutation database (R11 version, October 2007) (33). Data for the CDKN2 gene (n = 99) (34–46) and PTCH gene (n = 183) (47–58) were derived from the published literature. Data for protein kinase genes (n = 146) were extracted from high throughput sequencing of cancer genomes (59).
As shown in Figure 5, C to T and CC to TT transition mutations predominate the mutation spectra found in the TP53 gene of human non-melanoma skin tumors (basal cell and squamous cell carcinomas), and in the TP53 (33), CDKN2 (34–46), and protein kinase genes (59) of melanomas. Furthermore, over 90% of such mutations occur exclusively at dipyrimidine sequences (33–46, 59). On the other hand, G to T transversions only constitute less than 10% of all mutations in all cases (33–46, 59). The single C to T transitions, tandem CC to TT transitions, and deletions/insertions account for ~47%, ~15%, and ~17%, respectively, of all mutations occurring in the PTCH gene in basal cell carcinomas.
Figure 5. Mutation spectra in human skin tumors.
The pie charts show the percentage of particular types of mutations found in the TP53 gene of human non-melanoma skin tumors (basal cell and squamous cell carcinomas), in the TP53, CDKN2, and protein kinase genes of melanomas, and in the PTCH gene of basal cell carcinomas. Data for the TP53 gene (n = 94 for melanomas and n = 482 for non-melanoma skin cancers) were obtained from the International Agency for Research on Cancer (IARC) mutation database (R11 version, October 2007) (33). Data for the CDKN2 gene (n = 99) (34–46) and PTCH gene (n = 183) (47–58) were derived from the published literature. The mutations found in protein kinase genes (n = 146) are derived from high throughput sequencing of cancer genomes (59).
DISCUSSION
Despite the predominance of mutagenic UVA in terrestrial sunlight UV, solar mutagenesis in humans and rodents is characterized by UVB-specific DNA damage and mutations (17, 18). The overshadowing of UVA by UVB, the minor UV component of sunlight, has not been mechanistically investigated, however. To study the overriding role of UVB over UVA in solar mutagenesis, we have examined the kinetics of repair of UVA- and UVB-induced lesions in relation to mutagenicity in mouse embryonic fibroblasts irradiated with equitoxic doses of UVA and UVB in comparison to SSL.
Qualitative assessment of DNA damage in the genome overall of UV-irradiated cells revealed that both UVB- and SSL-irradiation, but not UVA-irradiation, induced a combination of CPDs and (6-4)PPs. The induced (6-4)PPs were, however, repaired within six hours of both UVB- and SSL-irradiation. The CPDs formed by UVB- or SSL-irradiation remained persistent for at least 24 hours post-irradiation. A faster repair of (6-4)PPs relative to CPDs has been reported by others (60–66), and ascribed to a greater distortion or unwinding of the DNA helix by (6-4)PPs (67–69). The latter may facilitate the recognition and processing of (6-4)PPs by nucleotide excision repair (NER) machinery (70). Additionally, unlike CPDs that are formed almost equally in the core and linker regions of nucleosomes (71, 72), (6-4)PPs are predominantly formed in the latter regions, which are more accessible to complex DNA repair proteins (70, 73, 74). The observed persistence of CPDs in the genome of UV-irradiated cells is consistent with the known deficiency of rodent cells for carrying out NER for CPDs (75). We (76) and others (77–85) have previously shown a time-dependent resistance to repair of CPDs in normal human cells, as well.
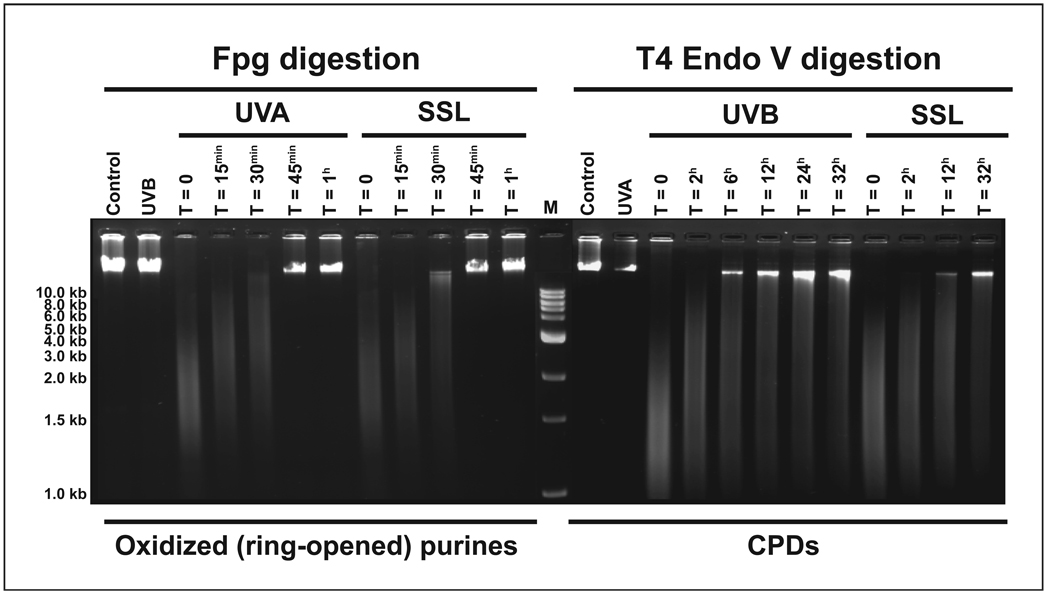
For verification purposes, we also performed a complementary experiment to establish the kinetics of repair of UV-induced DNA damage in human cells. Accordingly, we irradiated normal human skin fibroblasts under the same experimental conditions as those used for their counterpart mouse cells. As shown in Figure 6, we confirmed that, in agreement with previous findings reported by us (76) as well as by others (77–85), although repair of UVB- and SSL-induced CPDs is relatively more efficient in human cells as compared to mouse cells, a fraction of these lesions remains persistent in the genome of normal human cells over a full cell cycle, i.e., 32 hours.
Figure 6. Qualitative assessment of induced DNA damage in the genome of human cells.
Normal human skin fibroblasts were irradiated with equilethal (~75% cell survival) doses of UVA, UVB, and SSL in comparison with control as described in Materials and Methods. Genomic DNA was isolated and digested with Fpg for detecting oxidized (ring-opened) purines and T4 Endo V for determining CPDs. The DNA digests were subjected to alkaline agarose gel electrophoresis, followed by standardized visualization procedure. For brevity, results at select time points are shown. M = molecular size marker.
Conversely, UVA-irradiation failed to produce detectable dipyrimidine photolesions in the genome overall of irradiated cells. Instead, UVA- and SSL-irradiation alike formed significant levels of oxidized (ring-opened) purines in the genome of irradiated cells. In both cases, the induced oxidized (ring-opened) purines were completely removed from the genome within thirty minutes of irradiation. No significant formation of oxidized (ring-opened) purines was observed in the genome overall of UVB-irradiated cells, however. In confirmation, our TD-PCR footprinting verified an induction of dipyrimidine photolesions in the cII transgene consequent to UVB- and SSL-irradiation only. Also, comparative TD-PCR and LM-PCR analyses established a persistence of CPDs and an efficient repair of (6-4)PPs in the cII transgene consequent to UVB- and SSL-irradiation. Furthermore, LM-PCR footprinting of oxidized (ring-opened) purines showed a comparable formation of these lesions in the cII transgene after UVA- and SSL-irradiation only. The induced oxidized (ring-opened) purines were fully removed from the cII transgene within thirty minutes of UVA- and SSL-irradiation. In mammalian cells, base excision repair is known to rapidly restore the genome from a plethora of oxidative DNA damage (86–89). The fast repair of Fpg-sensitive sites found in our study is consistent with the previously reported repair rates of 8-hydroxyguanine in human keratinocytes and fibroblasts (90) and of oxidative purine modifications in Chinese hamster ovary cells (91), which showed half-lives of thirty minutes and 2–3 hours, respectively. Likewise, our supplementary experiment verified that, similar to the situation found in mouse fibroblasts, the Fpg-sensitive sites produced in the genomic DNA of UVA- and SSL-irradiated human cells are rapidly and efficiently repaired within 45 minutes post-irradiation (see, Fig. 6).
Determination of cII mutant frequency in UV-irradiated cells revealed that, in agreement with our previous findings (14–16), UVA-irradiation was weakly, yet, significantly mutagenic in the cII transgene. The relatively low mutagenicity of UVA-irradiation in the cII transgene can be attributed to the efficient removal of UVA-induced oxidized (ring-opened) purines from this gene. Presumably, a small fraction of UVA-induced lesions, which may have encountered replicative DNA polymerases before being subjected to base excision repair, has contributed to the observed mutagenicity of UVA-irradiation. In contrast, SSL-irradiation and UVB-irradiation showed potent mutagenicity in the cII transgene.
Mutation spectrometry analysis established a significantly different pattern of mutations induced by UVB- and SSL-irradiation from that of UVA-irradiation or control. The induced mutation spectra by UVB- and SSL-irradiation, respectively, showed remarkable resemblance to one another, however. The UVB- and SSL-induced mutation spectra were both characterized by significant increases in relative frequency of G:C to A:T transitions occurring (almost) exclusively at dipyrimidine sites, including a portion of hallmark CC to TT tandem transitions (6–9). These types of mutation are the signature mutations of dipyrimidine photolesions (6–9). We reaffirmed the major role of dipyrimidine photolesions in UVB- and SSL-irradiation induced mutagenesis by demonstrating that G:C to A:T transitions at pyrimidine dinucleotides accounted for 85% and 80% of the increases in cII mutant frequency consequent to the respective types of UV-irradiation. The characteristic G:C to T:A transversions induced by UVA-irradiation, however, were not distinguishable in SSL-irradiation induced mutation spectrum, presumably, due to the minor contribution of this type of mutation to the overall mutation load, i.e., 3.3%.
Thus far, diverging results have been reported on the types of DNA damage and mutations induced by UVA-irradiation (84, 85, 92–107). The discrepancies are likely to be due to the experimental variables used in different studies, including UVA source and dose, species and cell-types tested, and sensitivity of the applied measurement techniques. For instance, irradiation sources that emit contaminating UV wavebands, e.g., in the UVB range, are not suitable for characterizing UVA genotoxicity as they produce distorted induced mutation spectra by giving rise to UVB-specific photoproducts. Even a small contamination by UVB of a UVA source can be significant because per joule basis, UVB radiation is up to 50,000 times more genotoxic than UVA radiation (108). Woollons et al. (109) have demonstrated that 75% of all CPDs induced in human keratinocytes irradiated with a UVA sunlamp were caused by the 0.8% UVB component of this source. Likewise, the use of an irradiation source emitting a small but significant portion of UVB radiation resulted in a mutation spectrum predominated by single and tandem C→T transitions at dipyrimidine sites in the lacZ gene in mouse skin epidermis (102).
It is known that cells of varying species and types are differently resistant toward genotoxic and cytotoxic effects of UVA radiation (15, 16, 110). The differences may arise from various cellular contexts, e.g., photosensitizers and antioxidants contents, DNA repair capacity, and fidelity of DNA polymerase bypass, specific for each species and cell type (5, 111). To meet the sensitivity requirement of most available techniques for quantification of photo-induced DNA damages - especially at the level of nucleotide resolution - intense UVA irradiation is necessary to produce sufficient number of DNA lesions (84, 85, 99–101, 104). Such high doses of UVA irradiation may not necessarily be relevant for mutagenicity experiments because excessive doses of UVA may impede cell proliferation, thereby, precluding the DNA damage to be translated into mutation (5). The choice of dosing is also important for establishing DNA damage-targeted mutagenicity of UVA because irradiation dose per se may determine the type of induced DNA lesions (110). As we have recently shown, formation of DNA damage, in and of itself, is not sufficient to produce mutagenesis (112). Thus, it is imperative to investigate UVA-induced DNA damage and mutagenesis simultaneously and at biologically relevant doses (5).
While it is generally agreed upon that the UVB portion of sunlight is responsible for the induction of non-melanoma skin cancers, i.e. basal cell and squamous cell carcinomas, there have been suggestions that UVA is involved in the formation of melanoma. For example, UVA is capable of inducing melanoma-like lesions in opossums (113) and in certain fish species (114). In a recent meta-analysis of data linking UVA tanning bed exposure to melanoma risk, exposure to sun-beds before 35 years of age significantly increased the risk of melanoma, based on 7 informative studies (summary relative risk, 1.75; 95% CI, 1.35–2.26) (115). Our compilation of published data on the types of mutations found in human skin tumors revealed that C to T and CC to TT transitions clearly predominate the mutation spectra for both non-melanoma and melanoma skin cancers in the TP53 gene, and for melanomas in the CDKN2 and in protein kinase genes. In the same set of samples, G to T transversions, which would be a hallmark of UVA-induced mutagenesis, were generally rare and accounted for only 10% or less of the tumor associated mutations (Fig. 5). Also, single and tandem C to T transitions combined constitute approximately 62% of all mutations occurring in the PTCH gene in basal cell carcinomas. These results obtained with human tumor specimens are completely in line with the data obtained in our experimental system in which we showed that sunlight-induced mutagenesis is characterized by UVB but not UVA induced mutations.
We acknowledge that to simultaneously detect DNA damage/repair and mutagenesis, we have used a transcriptionally inactive reporter gene of a mouse model system (116). In humans, however, DNA-damage targeted mutagenesis in endogenous cancer-relevant genes might be more complex, e.g., because of influential factors such as TCR, which has been shown to preferentially remove DNA lesions from the transcribed strand of actively expressed genes (29, 62, 63, 117). Admittedly, therefore, caution should be taken in interpreting data obtained from in vitro or in vivo model systems because these systems lack ‘complete’ comparability to humans (12). Nonetheless, if used properly, both systems can provide invaluable information, which may help unravel many aspects of human carcinogenesis (28, 118).
We (119, 120) and others (121) have verified the utility of in vitro model systems for establishing DNA damage-targeted mutagenicity of carcinogens by demonstrating that treatment of normal human bronchial epithelial cells with a tobacco-derived carcinogen, benzo[a]pyrene diol epoxide (B[a]PDE), results in DNA damage formation at lung cancer mutational hotspots in the RAS oncogenes and TP53 tumor suppressor gene. Similarly, solar UV irradiation in vitro has been shown to induce DNA damage at distinctive nucleotide positions along cancer-related genes, which co-localize with skin cancer mutational hotspots established in the respective target genes in vivo (12). More specifically, we have shown that B[a]PDE can form DNA lesions at specific nucleotide positions along the lacI and cII transgenes in Big Blue mouse embryonic fibroblasts, which correspond to the sites of B[a]PDE-induced mutations in the respective transgenes (122). The patterns of B[a]PDE-induced DNA adduction and mutagenesis in both transgenes perfectly mirrored those found in the TP53 gene in smoking-related lung cancer (122). Likewise, we have demonstrated DNA adduct-targeted mutagenicity of other smoke-derived carcinogens, e.g., N-hydroxy-4-acetylaminobiphenyl and dibenzo[a,l]pyrene, in the same model system (123, 124).
In conclusion, the present study is an attempt to address the question of the etiology of sunlight-induced mutagenesis in mammalian cells, specifically in mouse cells. Here, we have unequivocally shown a resistance to repair of UVB-induced CPDs together with rapid removal of UVA-induced lesions in the cII transgene of mouse embryonic fibroblasts, which can mechanistically explain the predominance of characteristic UVB-induced mutagenesis in SSL-irradiated mouse cells.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institute of Environmental Health Sciences (ES06070) to G.P.P.
ABBREVIATIONS
- (6-4)PP
pyrimidine (6-4) pyrimidone photoproducts
- 8-oxo-dGs
8-oxo-7,8-dihydro-2’-deoxyguanosines
- B[a]PDE
benzo[a]pyrene diol epoxide
- CPDs
cis-syn cyclobutane pyrimidine-dimers
- E.coli
Escherichia coli
- Fpg
formamidopyrimidine DNA glycosylase
- LM-PCR
ligation-mediated polymerase chain reaction
- NER
nucleotide excision repair
- SSL
simulated sunlight
- T4 Endo V
T4 endonuclease V
- TCR
transcription-coupled DNA repair
- TD-PCR
terminal transferase-dependent polymerase chain reaction
- UV
ultraviolet
REFERENCES
- 1.de Gruijl FR. Photocarcinogenesis: UVA vs. UVB radiation. Skin Pharmacol Appl Skin Physiol. 2002;15:316–320. doi: 10.1159/000064535. [DOI] [PubMed] [Google Scholar]
- 2.Woodhead AD, Setlow RB, Tanaka M. Environmental factors in nonmelanoma and melanoma skin cancer. J Epidemiol. 1999;9:S102–S114. doi: 10.2188/jea.9.6sup_102. [DOI] [PubMed] [Google Scholar]
- 3.Setlow RB. The wavelengths in sunlight effective in producing skin cancer: a theoretical analysis. Proc Natl Acad Sci U S A. 1974;71:3363–3366. doi: 10.1073/pnas.71.9.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelfkens G, de Gruijl FR, van der Leun JC. Ozone depletion and increase in annual carcinogenic ultraviolet dose. Photochem Photobiol. 1990;52:819–823. doi: 10.1111/j.1751-1097.1990.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 6.Brash DE, Seetharam S, Kraemer KH, Seidman MM, Bredberg A. Photoproduct frequency is not the major determinant of UV base substitution hot spots or cold spots in human cells. Proc Natl Acad Sci U S A. 1987;84:3782–3786. doi: 10.1073/pnas.84.11.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YC, Maher VM, Mitchell DL, McCormick JJ. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascucci B, Versteegh A, van Hoffen A, van Zeeland AA, Mullenders LH, Dogliotti E. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J Mol Biol. 1997;273:417–427. doi: 10.1006/jmbi.1997.1268. [DOI] [PubMed] [Google Scholar]
- 9.Otoshi E, Yagi T, Mori T, Matsunaga T, Nikaido O, Kim ST, Hitomi K, Ikenaga M, Todo T. Respective roles of cyclobutane pyrimidine dimers, (6-4)photoproducts, and minor photoproducts in ultraviolet mutagenesis of repair-deficient xeroderma pigmentosum A cells. Cancer Res. 2000;60:1729–1735. [PubMed] [Google Scholar]
- 10.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 11.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C-->T.A transversions in simian kidney cells. Proc Natl Acad Sci U S A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besaratinia A, Pfeifer GP. Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis. 2006;27:1526–1537. doi: 10.1093/carcin/bgi311. [DOI] [PubMed] [Google Scholar]
- 13.Kim SI, Pfeifer GP, Besaratinia A. Mutagenicity of ultraviolet A radiation in the lacI transgene in Big Blue mouse embryonic fibroblasts. Mutat Res. 2007;617:71–78. doi: 10.1016/j.mrfmmm.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besaratinia A, Bates SE, Synold TW, Pfeifer GP. Similar mutagenicity of photoactivated porphyrins and ultraviolet A radiation in mouse embryonic fibroblasts: involvement of oxidative DNA lesions in mutagenesis. Biochemistry. 2004;43:15557–15566. doi: 10.1021/bi048717c. [DOI] [PubMed] [Google Scholar]
- 15.Besaratinia A, Synold TW, Xi B, Pfeifer GP. G-to-T transversions and small tandem base deletions are the hallmark of mutations induced by ultraviolet a radiation in mammalian cells. Biochemistry. 2004;43:8169–8177. doi: 10.1021/bi049761v. [DOI] [PubMed] [Google Scholar]
- 16.Besaratinia A, Kim SI, Bates SE, Pfeifer GP. Riboflavin activated by ultraviolet A1 irradiation induces oxidative DNA damage-mediated mutations inhibited by vitamin C. Proc Natl Acad Sci U S A. 2007;104:5953–5958. doi: 10.1073/pnas.0610534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumaz N, Drougard C, Sarasin A, Daya-Grosjean L. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1993;90:10529–10533. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanjilal S, Pierceall WE, Cummings KK, Kripke ML, Ananthaswamy HN. High frequency of p53 mutations in ultraviolet radiation-induced murine skin tumors: evidence for strand bias and tumor heterogeneity. Cancer Res. 1993;53:2961–2964. [PubMed] [Google Scholar]
- 19.Yajima H, Takao M, Yasuhira S, Zhao JH, Ishii C, Inoue H, Yasui A. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. Embo J. 1995;14:2393–2399. doi: 10.1002/j.1460-2075.1995.tb07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno S, Iwai S, Takao M, Yasui A. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res. 1999;27:3096–3103. doi: 10.1093/nar/27.15.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avery AM, Kaur B, Taylor JS, Mello JA, Essigmann JM, Doetsch PW. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:2256–2264. doi: 10.1093/nar/27.11.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boiteux S, O'Connor TR, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. Embo J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadet J, Ravanat JL, Martinez G, Medeiros M, Di Mascio P. Singlet Oxygen Oxidation of Isolated and Cellular DNA: Product Formation and Mechanistic Insights. Photochem Photobiol. 2006;82:1219–1225. doi: 10.1562/2006-06-09-IR-914. [DOI] [PubMed] [Google Scholar]
- 24.Adams WT, Skopek TR. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987;194:391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- 25.Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2003;2:287–294. doi: 10.1046/j.1474-9728.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 26.Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. Mutant frequencies and spectra depend on growth state and passage number in cells cultured from transgenic lacZ-plasmid reporter mice. DNA Repair (Amst) 2006;5:52–60. doi: 10.1016/j.dnarep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- 28.Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 30.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 31.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 32.van Elsas A, Zerp SF, van der Flier S, Kruse KM, Aarnoudse C, Hayward NK, Ruiter DJ, Schrier PI. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am J Pathol. 1996;149:883–893. [PMC free article] [PubMed] [Google Scholar]
- 33.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 34.Castellano M, Pollock PM, Walters MK, Sparrow LE, Down LM, Gabrielli BG, Parsons PG, Hayward NK. CDKN2A/p16 is inactivated in most melanoma cell lines. Cancer Res. 1997;57:4868–4875. [PubMed] [Google Scholar]
- 35.Flores JF, Walker GJ, Glendening JM, Haluska FG, Castresana JS, Rubio MP, Pastorfide GC, Boyer LA, Kao WH, Bulyk ML, Barnhill RL, Hayward NK, Housman DE, Fountain JW. Loss of the p16INK4a and p15INK4b genes, as well as neighboring 9p21 markers, in sporadic melanoma. Cancer Res. 1996;56:5023–5032. [PubMed] [Google Scholar]
- 36.Fujimoto A, Morita R, Hatta N, Takehara K, Takata M. p16INK4a inactivation is not frequent in uncultured sporadic primary cutaneous melanoma. Oncogene. 1999;18:2527–2532. doi: 10.1038/sj.onc.1202803. [DOI] [PubMed] [Google Scholar]
- 37.Herbst RA, Gutzmer R, Matiaske F, Mommert S, Kapp A, Weiss J, Arden KC, Cavenee WK. Further evidence for ultraviolet light induction of CDKN2 (p16INK4) mutations in sporadic melanoma in vivo. J. Invest. Dermatol. 1997;108:950. doi: 10.1111/1523-1747.ep12296624. [DOI] [PubMed] [Google Scholar]
- 38.Kumar R, Lundh Rozell B, Louhelainen J, Hemminki K. Mutations in the CDKN2A (p16INK4a) gene in microdissected sporadic primary melanomas. Int. J. Cancer. 1998;75:193–198. doi: 10.1002/(sici)1097-0215(19980119)75:2<193::aid-ijc5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 39.Lamperska K, Karezewska A, Kwiatkowska E, Mackiewicz A. Analysis of mutations in the p16/CDKN2A gene in sporadic and familial melanoma in the Polish population. Acta Biochim. Pol. 2002;49:369–376. [PubMed] [Google Scholar]
- 40.Piccinin S, Doglioni C, Maestro R, Vukosavljevic T, Gasparotto D, D'Orazi C, Boiocchi M. p16/CDKN2 and CDK4 gene mutations in sporadic melanoma development and progression. Int. J. Cancer. 1997;74:26–30. doi: 10.1002/(sici)1097-0215(19970220)74:1<26::aid-ijc5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Platz A, Ringborg U, Lagerlof B, Lundqvist E, Sevigny P, Inganas M. Mutational analysis of the CDKN2 gene in metastases from patients with cutaneous malignant melanoma. Br. J. Cancer. 1996;73:344–348. doi: 10.1038/bjc.1996.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollock PM, Pearson JV, Hayward NK. Compilation of somatic mutations of the CDKN2 gene in human cancers: non-random distribution of base substitutions. Genes Chromosomes Cancer. 1996;15:77–88. doi: 10.1002/(SICI)1098-2264(199602)15:2<77::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 43.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz A, Puig S, Lynch M, Castel T, Estivill X. Retention of the CDKN2A locus and low frequency of point mutations in primary and metastatic cutaneous malignant melanoma. Int. J. Cancer. 1998;76:312–316. doi: 10.1002/(sici)1097-0215(19980504)76:3<312::aid-ijc4>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 45.Smith-Sørensen B, Hovig E. CDKN2A (p16INK4A) somatic and germline mutations. Human Mutat. 1996;7:294–303. doi: 10.1002/(SICI)1098-1004(1996)7:4<294::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 46.Walker GJ, Flores JF, Glendening JM, Lin AH, Markl ID, Fountain JW. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer. 1998;22:157–163. doi: 10.1002/(sici)1098-2264(199806)22:2<157::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 47.D'Errico M, Calcagnile A, Canzona F, Didona B, Posteraro P, Cavalieri R, Corona R, Vorechovsky I, Nardo T, Stefanini M, Dogliotti E. UV mutation signature in tumor suppressor genes involved in skin carcinogenesis in xeroderma pigmentosum patients. Oncogene. 2000;19:463–467. doi: 10.1038/sj.onc.1203313. [DOI] [PubMed] [Google Scholar]
- 48.Evans T, Boonchai W, Shanley S, Smyth I, Gillies S, Georgas K, Wainwright B, Chenevix-Trench G, Wicking C. The spectrum of patched mutations in a collection of Australian basal cell carcinomas. Hum Mutat. 2000;16:43–48. doi: 10.1002/1098-1004(200007)16:1<43::AID-HUMU8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Gailani MR, Stahle-Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Unden AB, Dean M, Brash DE, Bale AE, Toftgard R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 50.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 51.Heitzer E, Lassacher A, Quehenberger F, Kerl H, Wolf P. UV fingerprints predominate in the PTCH mutation spectra of basal cell carcinomas independent of clinical phenotype. J Invest Dermatol. 2007;127:2872–2881. doi: 10.1038/sj.jid.5700923. [DOI] [PubMed] [Google Scholar]
- 52.Kim MY, Park HJ, Baek SC, Byun DG, Houh D. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J Dermatol Sci. 2002;29:1–9. doi: 10.1016/s0923-1811(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 53.Ratner D, Peacocke M, Zhang H, Ping XL, Tsou HC. UV-specific p53 and PTCH mutations in sporadic basal cell carcinoma of sun-exposed skin. J Am Acad Dermatol. 2001;44:293–297. doi: 10.1067/mjd.2001.112361. [DOI] [PubMed] [Google Scholar]
- 54.Reifenberger J, Wolter M, Knobbe CB, Kohler B, Schonicke A, Scharwachter C, Kumar K, Blaschke B, Ruzicka T, Reifenberger G. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 55.Unden AB, Holmberg E, Lundh-Rozell B, Stahle-Backdahl M, Zaphiropoulos PG, Toftgard R, Vorechovsky I. Mutations in the human homologue of Drosophila patched (PTCH) in basal cell carcinomas and the Gorlin syndrome: different in vivo mechanisms of PTCH inactivation. Cancer Res. 1996;56:4562–4565. [PubMed] [Google Scholar]
- 56.Wolter M, Reifenberger J, Sommer C, Ruzicka T, Reifenberger G. Mutations in the human homologue of the Drosophila segment polarity gene patched (PTCH) in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1997;57:2581–2585. [PubMed] [Google Scholar]
- 57.Xie J, Johnson RL, Zhang X, Bare JW, Waldman FM, Cogen PH, Menon AG, Warren RS, Chen LC, Scott MP, Epstein EH., Jr Mutations of the PATCHED gene in several types of sporadic extracutaneous tumors. Cancer Res. 1997;57:2369–2372. [PubMed] [Google Scholar]
- 58.Zhang H, Ping XL, Lee PK, Wu XL, Yao YJ, Zhang MJ, Silvers DN, Ratner D, Malhotra R, Peacocke M, Tsou HC. Role of PTCH and p53 genes in early-onset basal cell carcinoma. Am J Pathol. 2001;158:381–385. doi: 10.1016/S0002-9440(10)63980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas DC, Okumoto DS, Sancar A, Bohr VA. Preferential DNA repair of (6-4) photoproducts in the dihydrofolate reductase gene of Chinese hamster ovary cells. J Biol Chem. 1989;264:18005–18010. [PubMed] [Google Scholar]
- 61.Mitchell DL, Brash DE, Nairn RS. Rapid repair kinetics of pyrimidine(6-4)pyrimidone photoproducts in human cells are due to excision rather than conformational change. Nucleic Acids Res. 1990;18:963–971. doi: 10.1093/nar/18.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vreeswijk MP, van Hoffen A, Westland BE, Vrieling H, van Zeeland AA, Mullenders LH. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J Biol Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 63.van Hoffen A, Venema J, Meschini R, van Zeeland AA, Mullenders LH. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. Embo J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell DL, Byrom M, Chiarello S, Lowery MG. Effects of chronic exposure to ultraviolet B radiation on DNA repair in the dermis and epidermis of the hairless mouse. J Invest Dermatol. 2001;116:209–215. doi: 10.1046/j.1523-1747.2001.01192.x. [DOI] [PubMed] [Google Scholar]
- 65.van der Wees CG, Vreeswijk MP, Persoon M, van der Laarse A, van Zeeland AA, Mullenders LH. Deficient global genome repair of UV-induced cyclobutane pyrimidine dimers in terminally differentiated myocytes and proliferating fibroblasts from the rat heart. DNA Repair (Amst) 2003;2:1297–1308. doi: 10.1016/j.dnarep.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Courdavault S, Baudouin C, Sauvaigo S, Mouret S, Candeias S, Charveron M, Favier A, Cadet J, Douki T. Unrepaired cyclobutane pyrimidine dimers do not prevent proliferation of UV-B-irradiated cultured human fibroblasts. Photochem Photobiol. 2004;79:145–151. doi: 10.1562/0031-8655(2004)079<0145:ucpddn>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 67.Franklin WA, Doetsch PW, Haseltine WA. Structural determination of the ultraviolet light-induced thymine-cytosine pyrimidine-pyrimidone (6-4) photoproduct. Nucleic Acids Res. 1985;13:5317–5325. doi: 10.1093/nar/13.14.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao SN, Kollman PA. Conformations of deoxydodecanucleotides with pyrimidine (6-4)-pyrimidone photoadducts. Photochem Photobiol. 1985;42:465–475. doi: 10.1111/j.1751-1097.1985.tb01596.x. [DOI] [PubMed] [Google Scholar]
- 69.Pearlman DA, Holbrook SR, Pirkle DH, Kim SH. Molecular models for DNA damaged by photoreaction. Science. 1985;227:1304–1308. doi: 10.1126/science.3975615. [DOI] [PubMed] [Google Scholar]
- 70.Szymkowski DE, Lawrence CW, Wood RD. Repair by human cell extracts of single (6-4) and cyclobutane thymine-thymine photoproducts in DNA. Proc Natl Acad Sci U S A. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitchell DL, Nguyen TD, Cleaver JE. Nonrandom induction of pyrimidine-pyrimidone (6-4) photoproducts in ultraviolet-irradiated human chromatin. J Biol Chem. 1990;265:5353–5356. [PubMed] [Google Scholar]
- 72.Gale JM, Smerdon MJ. UV induced (6-4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochem Photobiol. 1990;51:411–417. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 73.Koehler DR, Courcelle J, Hanawalt PC. Kinetics of pyrimidine(6-4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol. 1996;178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buterin T, Hess MT, Gunz D, Geacintov NE, Mullenders LH, Naegeli H. Trapping of DNA nucleotide excision repair factors by nonrepairable carcinogen adducts. Cancer Res. 2002;62:4229–4235. [PubMed] [Google Scholar]
- 75.Ruven HJ, Seelen CM, Lohman PH, van Kranen H, van Zeeland AA, Mullenders LH. Strand-specific removal of cyclobutane pyrimidine dimers from the p53 gene in the epidermis of UVB-irradiated hairless mice. Oncogene. 1994;9:3427–3432. [PubMed] [Google Scholar]
- 76.Tu Y, Bates S, Pfeifer GP. Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J Biol Chem. 1997;272:20747–22055. doi: 10.1074/jbc.272.33.20747. [DOI] [PubMed] [Google Scholar]
- 77.Ford JM, Lommel L, Hanawalt PC. Preferential repair of ultraviolet light-induced DNA damage in the transcribed strand of the human p53 gene. Mol Carcinog. 1994;10:105–109. doi: 10.1002/mc.2940100208. [DOI] [PubMed] [Google Scholar]
- 78.Tung BS, McGregor WG, Wang YC, Maher VM, McCormick JJ. Comparison of the rate of excision of major UV photoproducts in the strands of the human HPRT gene of normal and xeroderma pigmentosum variant cells. Mutat Res. 1996;362:65–74. doi: 10.1016/0921-8777(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 79.Kobayashi T, Takeuchi S, Saijo M, Nakatsu Y, Morioka H, Otsuka E, Wakasugi M, Nikaido O, Tanaka K. Mutational analysis of a function of xeroderma pigmentosum group A (XPA) protein in strand-specific DNA repair. Nucleic Acids Res. 1998;26:4662–4668. doi: 10.1093/nar/26.20.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Therrien JP, Drouin R, Baril C, Drobetsky EA. Human cells compromised for p53 function exhibit defective global and transcription-coupled nucleotide excision repair, whereas cells compromised for pRb function are defective only in global repair. Proc Natl Acad Sci U S A. 1999;96:15038–15043. doi: 10.1073/pnas.96.26.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riou L, Eveno E, van Hoffen A, van Zeeland AA, Sarasin A, Mullenders LH. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 82.Emmert S, Kobayashi N, Khan SG, Kraemer KH. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc Natl Acad Sci U S A. 2000;97:2151–2216. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mathonnet G, Leger C, Desnoyers J, Drouin R, Therrien JP, Drobetsky EA. UV wavelength-dependent regulation of transcription-coupled nucleotide excision repair in p53-deficient human cells. Proc Natl Acad Sci U S A. 2003;100:7219–7224. doi: 10.1073/pnas.1232161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Courdavault S, Baudouin C, Charveron M, Canguilhem B, Favier A, Cadet J, Douki T. Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations. DNA Repair (Amst) 2005;4:836–844. doi: 10.1016/j.dnarep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci U S A. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pflaum M, Boiteux S, Epe B. Visible light generates oxidative DNA base modifications in high excess of strand breaks in mammalian cells. Carcinogenesis. 1994;15:297–300. doi: 10.1093/carcin/15.2.297. [DOI] [PubMed] [Google Scholar]
- 87.Spencer JP, Jenner A, Aruoma OI, Cross CE, Wu R, Halliwell B. Oxidative DNA damage in human respiratory tract epithelial cells. Time course in relation to DNA strand breakage. Biochem Biophys Res Commun. 1996;224:17–22. doi: 10.1006/bbrc.1996.0977. [DOI] [PubMed] [Google Scholar]
- 88.Grishko VI, Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage in nuclear DNA sequences with different transcriptional activities. Mutat Res. 1997;384:73–80. doi: 10.1016/s0921-8777(97)00017-7. [DOI] [PubMed] [Google Scholar]
- 89.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Errico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. New functions of XPC in the protection of human skin cells from oxidative damage. Embo J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hollenbach S, Dhenaut A, Eckert I, Radicella JP, Epe B. Overexpression of Ogg1 in mammalian cells: effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis. 1999;20:1863–1868. doi: 10.1093/carcin/20.9.1863. [DOI] [PubMed] [Google Scholar]
- 92.Drobetsky EA, Turcotte J, Chateauneuf A. A role for ultraviolet A in solar mutagenesis. Proc Natl Acad Sci U S A. 1995;92:2350–2354. doi: 10.1073/pnas.92.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sage E, Lamolet B, Brulay E, Moustacchi E, Chateauneuf A, Drobetsky EA. Mutagenic specificity of solar UV light in nucleotide excision repair-deficient rodent cells. Proc Natl Acad Sci U S A. 1996;93:176–180. doi: 10.1073/pnas.93.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeMarini DM, Shelton ML, Stankowski LF., Jr Mutation spectra in Salmonella of sunlight, white fluorescent light, and light from tanning salon beds: induction of tandem mutations and role of DNA repair. Mutat Res. 1995;327:131–149. doi: 10.1016/0027-5107(94)00179-9. [DOI] [PubMed] [Google Scholar]
- 95.Robert C, Muel B, Benoit A, Dubertret L, Sarasin A, Stary A. Cell survival and shuttle vector mutagenesis induced by ultraviolet A and ultraviolet B radiation in a human cell line. J Invest Dermatol. 1996;106:721–728. doi: 10.1111/1523-1747.ep12345616. [DOI] [PubMed] [Google Scholar]
- 96.Palmer CM, Serafini DM, Schellhorn HE. Near ultraviolet radiation (UVA and UVB) causes a formamidopyrimidine glycosylase-dependent increase in G to T transversions. Photochem Photobiol. 1997;65:543–549. doi: 10.1111/j.1751-1097.1997.tb08602.x. [DOI] [PubMed] [Google Scholar]
- 97.Ananthaswamy HN, Fourtanier A, Evans RL, Tison S, Medaisko C, Ullrich SE, Kripke ML. p53 Mutations in hairless SKH-hr1 mouse skin tumors induced by a solar simulator. Photochem Photobiol. 1998;67:227–232. doi: 10.1562/0031-8655(1998)067<0227:mihshm>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 98.Izumizawa Y, Yang SJ, Negishi T, Negishi K. DNA lesion and mutagenesis induced in phageM13mp2 by UVA, UVB and UVC irradiation. Nucleic Acids Symp Ser. 2000:73–74. doi: 10.1093/nass/44.1.73. [DOI] [PubMed] [Google Scholar]
- 99.Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E. Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J Biol Chem. 2000;275:26732–26742. doi: 10.1074/jbc.M001450200. [DOI] [PubMed] [Google Scholar]
- 100.Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 102.Ikehata H, Kudo H, Masuda T, Ono T. UVA induces C-->T transitions at methyl-CpG-associated dipyrimidine sites in mouse skin epidermis more frequently than UVB. Mutagenesis. 2003;18:511–519. doi: 10.1093/mutage/geg030. [DOI] [PubMed] [Google Scholar]
- 103.Agar NS, Halliday GM, Barnetson RS, Ananthaswamy HN, Wheeler M, Jones AM. The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations: a role for UVA in human skin carcinogenesis. Proc Natl Acad Sci U S A. 2004;101:4954–4959. doi: 10.1073/pnas.0401141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Courdavault S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat Res. 2004;556:135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 105.Kappes UP, Runger TM. No major role for 7,8-dihydro-8-oxoguanine in ultraviolet light-induced mutagenesis. Radiat Res. 2005;164:440–445. doi: 10.1667/rr3434.1. [DOI] [PubMed] [Google Scholar]
- 106.Kozmin S, Slezak G, Reynaud-Angelin A, Elie C, de Rycke Y, Boiteux S, Sage E. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006;126:667–675. doi: 10.1038/sj.jid.5700093. [DOI] [PubMed] [Google Scholar]
- 108.de Gruijl FR, Sterenborg HJ, Forbes PD, Davies RE, Cole C, Kelfkens G, van Weelden H, Slaper H, van der Leun JC. Wavelength dependence of skin cancer induction by ultraviolet irradiation of albino hairless mice. Cancer Res. 1993;53:53–60. [PubMed] [Google Scholar]
- 109.Woollons A, Kipp C, Young AR, Petit-Frere C, Arlett CF, Green MH, Clingen PH. The 0.8% ultraviolet B content of an ultraviolet A sunlamp induces 75% of cyclobutane pyrimidine dimers in human keratinocytes in vitro. Br J Dermatol. 1999;140:1023–1030. doi: 10.1046/j.1365-2133.1999.02899.x. [DOI] [PubMed] [Google Scholar]
- 110.Besaratinia A, Synold TW, Chen HH, Chang C, Xi B, Riggs AD, Pfeifer GP. DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 umor suppressor gene. Proc Natl Acad Sci U S A. 2005;102:10058–10063. doi: 10.1073/pnas.0502311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 112.Kim SI, Pfeifer GP, Besaratinia A. Lack of mutagenicity of acrolein-induced DNA adducts in mouse and human cells. Cancer Res. 2007;67:11640–11647. doi: 10.1158/0008-5472.CAN-07-2528. [DOI] [PubMed] [Google Scholar]
- 113.Ley RD. Ultraviolet radiation A-induced precursors of cutaneous melanoma in Monodelphis domestica. Cancer Res. 1997;57:3682–3684. [PubMed] [Google Scholar]
- 114.Setlow RB, Grist E, Thompson K, Woodhead AD. Wavelengths effective in induction of malignant melanoma. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6666–6670. doi: 10.1073/pnas.90.14.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.International Agency for Research on Cancer Working Group on artificial ultraviolet (UV) light and skin cancer. The association of use of sunbeds with cutaneous malignant melanoma and other skin cancers: A systematic review. Int J Cancer. 2007;120:1116–1122. doi: 10.1002/ijc.22453. [DOI] [PubMed] [Google Scholar]
- 116.Jakubczak JL, Merlino G, French JE, Muller WJ, Paul B, Adhya S, Garges S. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci U S A. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lommel L, Hanawalt PC. Increased UV resistance of a xeroderma pigmentosum revertant cell line is correlated with selective repair of the transcribed strand of an expressed gene. Mol Cell Biol. 1993;13:970–976. doi: 10.1128/mcb.13.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dutt A, Wong KK. Mouse models of lung cancer. Clin Cancer Res. 2006;12:4396s–4402s. doi: 10.1158/1078-0432.CCR-06-0414. [DOI] [PubMed] [Google Scholar]
- 119.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 120.Smith LE, Denissenko MF, Bennett WP, Li H, Amin S, Tang M, Pfeifer GP. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 121.Feng Z, Hu W, Chen JX, Pao A, Li H, Rom W, Hung MC, Tang MS. Preferential DNA damage and poor repair determine ras gene mutational hotspot in human cancer. J Natl Cancer Inst. 2002;94:1527–1536. doi: 10.1093/jnci/94.20.1527. [DOI] [PubMed] [Google Scholar]
- 122.Yoon JH, Smith LE, Feng Z, Tang M, Lee CS, Pfeifer GP. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 2001;61:7110–7117. [PubMed] [Google Scholar]
- 123.Besaratinia A, Bates SE, Pfeifer GP. Mutational signature of the proximate bladder carcinogen N-hydroxy-4-acetylaminobiphenyl: inconsistency with the p53 mutational spectrum in bladder cancer. Cancer Res. 2002;62:4331–4338. [PubMed] [Google Scholar]
- 124.Yoon JH, Besaratinia A, Feng Z, Tang MS, Amin S, Luch A, Pfeifer GP. DNA damage, repair, and mutation induction by (+)-Syn and (−)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxides in mouse cells. Cancer Res. 2004;64:7321–7328. doi: 10.1158/0008-5472.CAN-04-1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.