Summary
A wide range of genetically engineered murine models of type 2 diabetes have been created to try to understand the site of the primary defect in insulin action, and the relationship between insulin resistance and impaired β-cell function in diabetes. Genetic disruption of various aspects known to be important in diabetes has examined specific facets, including glucose sensing, transcription factors for the insulin gene, the insulin gene itself, insulin and insulin-like growth factor receptors, downstream signaling components and some mutations that increase insulin sensitivity. This article focuses on models that have given insight into insulin resistance and impaired insulin production, especially models that examine molecules involved in the signaling pathway downstream of insulin binding its receptor. These models recapitulate many features of human type 2 diabetes and, although they have emphasized the complexity of this disease, they offer numerous opportunities to characterize particular aspects and eventually fit them together to help delineate the human disease.
Keywords: diabetes, genetics, metabolism, mouse models, targeted mutagenesis
Introduction
According to most epidemiological studies, diabetes is becoming an epidemic throughout the world. Type 2 diabetes involves defects in peripheral insulin signaling and β-cell function;1 these abnormalities are genetically determined.2 The definition of insulin resistance has undergone significant changes in recent years.1 Early studies emphasized the role of nonoxidative glucose metabolism in muscle and lipolysis in adipose cells in the pathogenesis of diabetes. Recent data from genetically engineered mice indicate, however, that the liver, brain and pancreatic β-cell participate in the development of the disease at an earlier stage and in more ways than previously thought.1 The site of the primary defect in insulin action remains unclear, nonetheless, as does the relationship between insulin resistance and impaired β-cell function.
Understanding the pathophysiology of these combined defects is the ‘holy grail’ of endocrinology and should eventually lead to a better understanding of the disorder and the implementation of appropriate therapies. In this article, we discuss selected models of genetically engineered mice that have helped in the study of the pathogenetic mechanisms involved in type 2 diabetes. We cannot systematically review all relevant literature, but we will describe models that have advanced our understanding of the shared causes of insulin resistance and impaired insulin production, with a focus on genes in the insulin signaling pathway. In most cases we can only briefly touch upon the phenotype and we refer the reader to the original articles and prior reviews (for example Nandi et al.3). Although we cannot review all the mutations associated with impaired β-cell function and development, it bears emphasizing that effects on insulin biosynthesis, processing and transcription are also associated with β-cell failure.
Deletion of the Insulin Receptor
Although mutations of the insulin receptor gene (INSR) are rare as a cause of diabetes in humans, the compensatory hyperinsulinemia that is inevitably associated with common forms of insulin resistance leads to downregulation of the insulin receptor at the surface of the target cell. The simplest way to reproduce this abnormality and study its cellular consequence was to inactivate the insulin receptor in mice. Insr-null mice are born with slight growth retardation, become severely hyperglycemic and ketoacidotic and die within a few days, confirming the importance of the insulin receptor in postnatal fuel homeostasis.4 The growth retardation is associated with a marked increase in expression of insulin-like growth factor (Igf)-binding protein 1 (Igfbp-1). The main conclusion derived from this study was that the insulin receptor is the only mediator of insulin action, although the Igf-I receptor (encoded by Igf1r) normally contributes to insulin signaling by engaging in heterodimer formation with the insulin receptor.
More informative were the outcomes from models with tissue-specific deletion of Insr. This is a type of conditional mutation, by which genes can be knocked out in particular organs or cell types, or at different stage of development. The Cre–LoxP system can be used, in which the recombinase Cre removes sequences that are tagged by floxing (flanking the target sequence by inserting LoxP nucleotide sequences). Tissue-specific deletion of Insr has been achieved in the classic insulin-sensitive metabolically important tissues including liver, brown and/or white adipose tissue, muscle, brain, and the β-cell (see below); Insr deletion has also been engineered in the heart and vasculature.
Liver-specific Insr knockout (LIRKO) mice develop glucose intolerance with enhanced gluconeogenesis (Table 1), thus confirming the important role of insulin in controlling hepatic glucose production.5 They also exhibit hyperinsulinemia due to increased insulin production and reduced insulin clearance by the liver.
Table 1.
Effects of tissue-specific deletion of the insulin receptor in mice.
| Tissue with receptor deletion (strain) | Resulting (β-cell function | Insulin action | Effect on glycemic control |
|---|---|---|---|
| Liver (LIRKO) | Normal | Increased gluconeogenesis | Impaired glucose tolerance |
| Fat tissue (FIRKO) | Normal | Reduced inhibition of lipolysis and reduced glucose uptake | Normal |
| β-Cell (BIRKO) | Decreased β-cell mass; loss of first-phase insulin release | Normal | Age-dependent glucose intolerance |
| Muscle (MIRKO) | Normal | Severe muscle insulin resistance | Impaired glucose tolerance |
| Nestin-positive-cells (NIRKO) | Normal | Mild insulin resistance | Normal |
Abbreviations: BIRKO, β-cell insulin-receptor knockout; FIRKO, fat-tissue-specific insulin-receptor knockout; LIRKO, liver-specific insulin-receptor knockout; MIRKO, muscle-specific insulin-receptor knockout; NIRKO, nestin-positive-cell insulin-receptor knockout.
Insulin receptor deletion in fat-tissue-specific Insr knockout (FIRKO) mice has minimal effect on whole-body glucose homeostasis, even though insulin-mediated glucose uptake and inhibition of lipolysis are significantly reduced.6 Interestingly, the reduction in fat-cell development found in these mice is associated with increased lifespan.7 Brown-adipocyte-specific Insr knockout (BATIRKO) mice, on the other hand, have a reduction in β-cell mass and function; the etiology of this effect is unknown.8
The muscle-specific Insr knockout (MIRKO) mouse strain develops severe insulin resistance in skeletal muscle, but the effect is not associated with a diabetic phenotype; rather, the mutation results in increased insulin-induced adipocyte glucose uptake.9 The absence of a severe phenotype may be due to compensation by the Igf-I receptor or contraction-activated signaling pathways in skeletal muscle.
Nestin is an intermediate filament protein that serves as a marker for the central nervous system. Loss of the insulin receptor in nestin-positive-cell Insr knockout (NIRKO) mice leads to mild insulin resistance, obesity, hypertriglyceridemia and increased food intake (this increase is, however, limited to female mice). In hypothalamic nuclei, therefore, the insulin receptor apparently has an important role in insulin control of hepatic glucose production.10
Specific deletion of the insulin receptor in β-cell Insr knockout (BIRKO) mice causes a selective impairment in the first phase of glucose-stimulated insulin secretion, a phenotype reminiscent of that seen in patients with type 2 diabetes.11 This defect leads to age-dependent glucose intolerance and, in some mice, to overt diabetes. These data suggest that the insulin-resistant state in type 2 diabetics may, in part, also be responsible for the defect in insulin secretion that is seen in this disease. Similarly to Insr deletion, Igf1r ablation in β-cells affects insulin secretion and results in fasting hyperinsulinemia with impaired glucose tolerance (also see below).12,13 Interestingly, Insr ablation impairs compensatory β-cell hyperplasia in insulin resistance, indicating that insulin, acting through its receptor, is a proliferation factor for β-cells.14
Mutation of the Insulin-Like Growth Factor I Receptor
Complete deletion of the Igf-I receptor has a distinctive growth-retardation phenotype. Tissue-specific deletions of Igf1r have not resulted in severe metabolic derangements. Muscle-specific Igf1r knockout (MIGFRKO) mice did not show altered glucose homeostasis; nor, surprisingly, did mice with muscle-specific double-knockout of Igf1r and Insr.15 On the other hand, β-cell-specific deletion of the Igf1r (see above) affected glucose-stimulated insulin secretion by a reduction in expression of Slc2a2 (solute carrier family 2, member 2; previously termed glucose transporter 2 [Glut2]) and hexokinase; both of these molecules are important in β-cell glucose sensing.13 It is remarkable that the Igf1r mutation failed to affect β-cell proliferation, given that the Igf-I receptor is primarily a growth factor receptor. In fact, in β-cells the insulin receptor seems to be the main mediator of cellular proliferation signals, as indicated by conditional mutations of either the insulin receptor or the Igf-I receptor.14 These data are consistent with a model in which insulin is the primary β-cell growth factor.16
In contrast to these results, expression of a dominant-negative Igf1r (dnIgf1r)17 in skeletal muscle resulted in a model reminiscent of human type 2 diabetes—the MKR mouse strain. The dnIgf1r strain was engineered so that arginine substituted for lysine in the tyrosine-kinase-binding site and was expressed in skeletal muscle under the muscle creatine-kinase promoter. Hybrids formed between the dnIGF1r product and the endogenous insulin receptor or Igf-I receptor and resulted in trans-dominant inhibition of insulin and Igf signaling, with severe insulin resistance that led to hyperinsulinemia, hyperglycemia and hyper lipidemia. This model parallels the mouse model with muscle-specific deletion of the gene for Slc2a4 (solute carrier family 2, member 4; previously termed Glut4); this model also develops severe insulin resistance and as the mice age they develop diabetes.18
Mutations Affecting Insulin-Receptor Signaling Molecules
Deletion of insulin-receptor substrates
Insulin receptor substrate 1 (Irs1) was the first major substrate of the insulin receptor and the Igf-I receptor to be cloned. The major impact of Irs1 inactivation in mice is, however, intrauterine and post-natal growth retardation, associated with only mild insulin resistance (Table 2).19 Hyperplasia of β-cells allowed the mice to overcome the insulin resistance. Impairment of β-cell insulin secretion was, however, also seen, suggesting that insulin and/or Igf-I signaling through Irs1 is required for proper β-cell function. These data should not, however, be construed to indicate that Irs1 is unimportant for insulin signaling in peripheral tissues: mice with heterozygosity for deletions of both Insr and Irs1 demonstrated severe impairment of insulin action, and progeny homozygous for both deletions eventually developed diabetes;20 mice with combined heterozygosity for mutations in the genes encoding glucokinase and Irs1 also developed diabetes.20
Table 2.
Effects of deletions affecting insulin receptor signaling in mice.
| Protein deleted | Resulting β-cell function | Insulin action | Effect on glycemic control |
|---|---|---|---|
| Irs1 | Increased β-cell mass | Mild defects | Normal |
| Irs2 | Decreased β-cell mass | Impaired insulin action | Frank diabetes |
| Akt-2 | Normal | Impaired signaling | Hyperglycemia |
| Slc2a4 | Normal | Moderate insulin resistance | Hyperglycemia (in males) |
| Muscle-specific Slc2a4 | Normal | Very insulin-resistant | Impaired glucose tolerance; diabetes in a subset |
| Fat-specific Slc2a4 | Normal | Reduced suppression of gluconeogenesis | Impaired glucose tolerance; diabetes in a subset |
Abbreviations: Akt-2, RAC-β serine/threonine-protein kinase; Irs, insulin receptor substrate; Slc2a4, solute carrier family 2, member 4 (previously termed Glut4).
Interestingly, mice lacking Irs2 develop diabetes because of a combination of insulin deficiency and impaired insulin action. The lack of Irs2 severely impairs the β-cell's ability to overcome the insulin resistance, because these mice have impaired β-cell proliferation and increased β-cell apoptosis.21 These data raise the critical question of whether insulin resistance and β-cell failure are but two different aspects of the same pathophysiologic process. Interestingly, mutations of both Irs1 and Irs2 in liver come very close to recapitulating the complete Insr-knockout phenotype, suggesting three conclusions: firstly, Irs1 and Irs2 are the main substrates of insulin receptor signaling (Figure 1); secondly, their roles are overlapping, not distinct; and, thirdly, the liver is the primary site of metabolic regulation by insulin in rodents.22
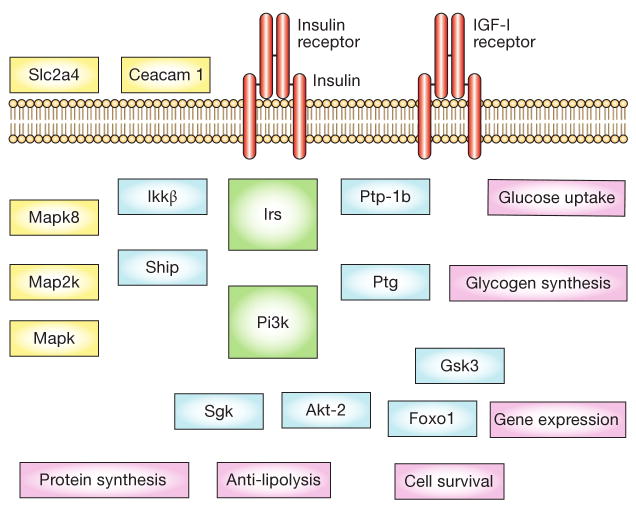
Figure 1.
Insulin receptor signaling cascades. Insulin binds the insulin receptor and activates a number of post-receptor cascades, many shared by the IGF-I receptor. The initial substrates that are involved include Irs molecules, followed by downstream protein kinases such as Sgk and Akt-2 that lead to cellular events that are mediated by other enzymes, such as glucose uptake (mediated by Akt-2), glycogen synthesis (Gsk3), gene expression (Foxo1, Mapk), cell survival, anti-lipolysis and protein expression. There are numerous modulators of these events including protein and lipid phosphatases (Ptp-1b and Ship), scaffolding proteins (Ptg), and even proteins that affect insulin signaling by affecting the internalization of the receptor complex (ceacam 1). Abbreviations: Akt-2, RAC-β serine/threonine-protein kinase; ceacam 1, carcinoembryonic antigen related cell adhesion molecule 1; Foxo1, forkhead box protein O1A; Gsk3, glycogen synthase kinase; IGF-I, insulin-like growth factor I; Ikkβ, inhibitor of nuclear factor κB; Irs, insulin receptor substrates; Map2k, Mapk kinase; Mapk, mitogen-activated protein kinase; Mapk8, mitogen-activated protein kinase 8 (previously termed Jnk1); Pi3k, phosphatidylinositol 3-kinase; Ptg, protein targeting to glycogen; Ptp-1b, protein tyrosine phosphatase1b; Sgk, serum/glucocorticoid regulated kinase; Ship, SH2-domain-containing inositol 5-phosphatase; Slc2a4, solute carrier family 2, member4 (previously termed Glut4).
In addition to their metabolic phenotype, the lifespan of mice with homozygous knockout of Irs2 was extended by about 20%.23 Mice lacking Irs2 in the brain have been generated by crossing a strain that expresses Cre in nestin-positive cells with a strain in which the Irs2 is floxed; heterozygous knockout of this gene in the brain caused reduced brain insulin signaling and, despite the insulin resistance and glucose intolerance that developed, these mice also showed increased lifespans.23 The increased longevity may be secondary to reduced brain insulin signaling; in contrast, lifespan reduction is associated with hyperinsulinemia and glucose intolerance.24,25 These studies therefore add support to the important role of insulin and Igf in longevity.26
Mouse models in which Irs3 or Irs4 were deleted did not demonstrate severely affected phenotypes,27,28 but when Irs1-knockout mice and Irs3-knockout mice are inter-crossed, deficiency of these two proteins causes lipoatrophy with insulin resistance, but without intrahepatic and intramuscular deposits of triglycerides.
Mutation of the RACβ serine/threonine-protein kinase (Akt-2) gene
Two mutant alleles of Akt2, which encodes RACβ serine/threonine-protein kinase (also known as protein kinase Akt-2 or protein kinase Bβ) have been generated in mice. In one report, mice lacking Akt2 showed hyperglycemia with impaired insulin signaling in muscle and adipocytes when allowed to feed normally.29 By contrast, in a second model the phenotype showed growth retardation, lipodystrophy and severe diabetes.30 Transgenic mice overexpressing constitutively active protein kinase Akt-2 in β-cells show increased islet cell mass and resistance to streptozotocin-induced diabetes. These data indicate that Akt2 is important in insulin action and β-cell function.
Deletion of the solute carrier family 2, member 4 (Glut4) gene
Mice with a complete Slc2a4 knockout develop moderate insulin resistance, which in male animals is associated with hyperglycemia in the fed state. Female mice do not develop hyperglycemia.31
Muscle-specific Slc2a4-knockout mice are insulin resistant and glucose intolerant, with decreased whole-body and insulin-stimulated muscle glucose uptake. Only a subset of the muscle-specific Slc2a4-knockout mice develop diabetes, suggesting that there are compensatory mechanisms that prevent the onset of hyperglycemia even when muscle glucose metabolism is reduced. Those that develop diabetes demonstrate insulin resistance in the liver and adipocytes, secondary to glucotoxicity and reversible by phloridzin.18
Some fat-specific Slc2a4-knockout mice develop diabetes associated with an impairment in insulin-induced suppression of hepatic gluconeogenesis, suggesting that the adipocyte defect affects other tissues secondarily, perhaps via chronic hyperinsulinemia or an as yet un defined circulating adipocytokines such as serum retinol-binding protein 4.32
Mutation of Protein Phosphatase 1 Regulatory Subunit 3c
Protein phosphatase 1 regulatory subunit 3C (also known as protein targeting to glycogen [Ptg]; encoded by Ppp1r3c) is a scaffolding protein that targets protein phosphatase 1 to enzymes involved in glycogen synthesis. Mice lacking one Ppp1r3c allele develop glucose intolerance and insulin resistance as they age. This effect is associated with depletion of tissue glycogen content due to a decrease in non oxidative glucose disposal and an apparent compensatory increase in muscle triglyceride content, resembling the findings in type 2 diabetes.33
Mutation of Ceacam 1
The hepatic membrane protein ceacam 1 (carcinoembryonic antigen related cell ad hesion mole cule 1) participates in the process of receptor-mediated insulin internalization and degradation. Transgenic mice expressing a dominant-negative ceacam 1 develop hyperinsulinemia and a metabolic syndrome of increased visceral adiposity and increased levels of tri glycerides and free fatty acids.34 These findings suggest that alterations in insulin clearance by the liver affect peripheral insulin sensitivity.
Mutations Causing Lipodystrophy-Related Diabetes
Several models that ablated adipose tissue resulted in lipodystrophy-related diabetes. Among them are the A-ZIP/F strain and the aP2-SREBP1c strain, both of which have no adipocyte differentiation.35,36 The absence of adipocytes in both models resulted in severe insulin resistance associated with hypertriglyceridemia, fatty liver and increased levels of triglycerides in muscle. The absence of fat therefore results in triglyceride accumulation in other tissues that affects their responsiveness to insulin.
Mutation of the Insulin Gene
There are two insulin genes in rodents. In adult mice, insulin is synthesized from transcripts of the gene for insulin II (Ins2); however, ablation of either gene in mice is without consequences. In contrast, when both genes are knocked out, mice develop diabetic ketoacidosis and die within days of birth.37 These findings suggest that insulin signals exclusively through the insulin receptor, since the phenotypes of the two gene ablations (insulin or the insulin receptor) are indistinguishable. Interestingly, mutations affecting insulin processing are found in the Akita strain—a model in which diabetes develops spontaneously. This nonobese, insulin-deficient strain has a mutation in Ins2 that results in β-cell apoptosis and a phenotype resembling type 1 diabetes. The hyperglycemia results in glucose toxicity and secondary insulin resistance, with a picture resembling insulin-deficient, nonobese type 2 diabetes.38
Similar mutations of the insulin gene have now been found in diabetic patients,39 suggesting that abnormalities of insulin processing lead to impaired insulin production. Although these mutations account for only a small fraction of total diabetes cases, there are important implications of these findings for common forms of β-cell failure.
In recent years, the role of endoplasmic reticulum stress in the cellular pathogenesis of metabolic abnormalities has been highlighted by several studies.40,41 In this regard, it should be noted that alterations of the activity of eukaryotic translation initiation factor 2a (Eif2a)42 and Eif2α kinase 3 (also known as pancreatic Eif2α kinase [Perk]) lead to β-cell failure that is dependent on stress to the endoplasmic reticulum.43
Mutation of Glucokinase
Glucokinase is a glucose-phosphorylating enzyme that has a low dissociation constant and is expressed in pancreatic β-cells and liver. The β-cell glucokinase acts as a ‘glucose sensor’ by coupling glucose metabolism to insulin secretion. Mutations of glucokinase are associated with impaired insulin secretion in humans in a juvenile-onset form of diabetes known as maturity-onset diabetes of the young 2 (MODY2).44 Mouse models of gluco kinase gene defects include ubiquitous null mutations45 and conditional inactivation in β-cells or liver.46 These experiments suggest that, although β-cell glucokinase has a critical role in metabolic control and survival, the hepatic enzyme is important in regulating glycogen synthesis and insulin secretion.
Mutation of Pancreatic and Duodenal Homeobox 1
The homeodomain transcription factor PDX1 (pancreatic and duodenal homeobox 1) is expressed at an early stage of pancreatic morphogenesis in uncommitted pancreatic precursor cells and is then restricted to β-cells. Homozygous null mutations of PDX1 in humans have been associated with pancreatic agenesis,47 whereas haploinsufficiency causes MODY448 and is associated with some forms of type 2 diabetes.49,50 Consistent with the human data, complete ablation of Pdx1 in mice results in arrested pancreatic development. Specific disruption of Pdx1 in β-cells results in diabetes as a consequence of impaired expression of several genes required for proper β-cell function, including those encoding insulin, Glut2 and amyloid polypeptide.51 PDX1 therefore has a critical role in pancreatic differentiation and in the maintenance of β-cell function. For example, recent evidence implicates Pdx1 in islet remodeling through apoptosis,37,52 a physiologic process that represents a potential site of failure in patients with type 2 diabetes.53
Mutation of Insulin Gene Transcription Factors
Neurogenic differentiation 1
Neurogenic differentiation 1 (Neurod1; also known as Beta2) is a basic helix-loop-helix protein expressed in pancreatic endocrine cells, intestine, and brain. In β-cells, it has been shown to activate transcription of the insulin gene. Ablation of Neurod1 expression in mice results in lethal perinatal diabetes, associated with a drastic reduction in β-cell number. In addition to a reduction in β-cell numbers, islet number is also reduced and the surviving islets show aberrant development, indicating a role for Neurod1 in islet morphogenesis.54
Hepatocyte nuclear factor 4α
In humans, mutations of the transcription factor hepatocyte nuclear factor 4α (HNF-4α) cause MODY1.55 Mice lacking Hnf-4α show a complex phenotype, however, including defective hepatocyte differentiation during embryogenesis.56,57 Conditional ablation of Hnf-4α in β-cells, on the other hand, results in hyperinsulinemia and impaired glucose tolerance. In vitro studies show an impaired response to glucose and sulfonylureas in Hnf-4α-deficient β-cells, associated with a partial reduction in the levels of the potassium-channel subunit Kir6.2 (ATP-sensitive inward rectifier potassium channel subunit 11).58
Hepatocyte nuclear factor 1
Mutations of the gene encoding HNF-1α cause MODY3,59 whereas mutations in HNF-1β lead to MODY5.44 In either instance, patients display impaired glucose-stimulated insulin secretion. Mice lacking Hnf-1α develop hyperglycemia and impaired insulin secretory responses to glucose and arginine, possibly because of defective β-cell glycolytic signaling;60 these findings support the view that Hnf-1α is required for maintenance of normal β-cell function.
The underlying defect in MODY5 is less well understood. Ablation of Hnf-1β in the whole mouse results in embryonic lethality. In contrast, mice with conditional knockout of Hnf-1β in β-cells have impaired glucose tolerance and reduced insulin secretion in response to glucose, but not to arginine. Paradoxically, this molecular phenotype is associated with increased levels of Hnf-1α and Pdx1 mRNA, but decreased Hnf-4 mRNA.61
Gene Deletions Associated with Increased Insulin Sensitivity
Forkhead box protein O1A
Forkhead box protein O1A (Foxo1) has an important role in insulin control of hepatic glucose production by regulating transcription of the genes encoding glucose-6-phosphatase and phospho-enolpyruvate carboxykinase. Insulin activation of Akt2 results in nuclear exclusion of Foxo1 and inhibits the actions of Foxo1. Haploinsufficiency of the Foxo1 gene restores insulin sensitivity and rescues the diabetic phenotype in insulin-resistant mice by reducing hepatic expression of gluconeogenetic genes and increasing adipocyte expression of insulin-sensitizing genes.62 Conditional Foxo1 knockout in liver results in impaired hormonal regulation of glucose production, indicating that Foxo1 is important for signaling involving cyclic AMP or insulin.63
Mice lacking Irs2 develop β-cell failure. Haplo-insufficiency of Foxo1 reverses β-cell failure in Irs2-knockout mice through partial restoration of β-cell proliferation and increased expression of the pancreatic transcription factor Pdx1.64
Tyrosine-protein phosphatase, non-receptor type 1
Tyrosine-protein phosphatase, non-receptor type 1 (previously termed protein tyrosine phosphatase 1b [Ptp-1b]; encoded by Ptpn1) is a tyrosine phosphatase that inhibits insulin action. Deletion of Ptpn1 in mice is associated with increased insulin sensitivity as seen with the increased insulin receptor phosphorylation in muscle and liver. This leads to improved glucose tolerance, and the mice are refractory to obesity and insulin resistance otherwise induced by a high-fat diet.65
Phosphatidylinositol 3-kinase regulatory subunits
Phosphatidylinositol 3-kinase (Pi3k) is considered critical in insulin signaling to the glucose uptake pathways. Surprisingly, therefore, mice lacking the α isoform of the p85 subunit show increased insulin sensitivity and hypoglycemia due to increased glucose transport in skeletal muscle and adipocytes.66 Initially it was concluded that there was compensatory binding of other subunits of Pi3k receptor (especially p50α); however, mice lacking all p85 splice isoforms are also hypoglycemic and insulin-sensitive, and the exact mechanism is yet to be defined.
SH2-domain-containing inositol 5-phosphatase
SH2-domain-containing inositol 5-phosphatase dephosphorylates phosphoinositol and, not surprisingly, ablation of expression of this protein therefore enhances insulin action through the Pi3k pathway, leading to neonatal demise from hypoglycemia.67
Mitogen-activated protein kinase 8
Serine phosphorylation of Irs proteins and insulin receptors is a physiological mechanism providing negative feedback control of insulin signaling; for example, phosphorylation of Ser307 is associated with decreased interaction of signaling molecules with the insulin receptor. The activity of mitogen-activated protein kinase 8 (also known as stress-activated protein kinase Jnk1), on the other hand, results in this serine phosphorylation secondary to activation by cytokines and free fatty acids, leading to insulin resistance. Mice lacking mitogen-activated protein kinase 8 seem to be protected against insulin resistance caused genetically (by mutation of the obese gene) or environmentally (by diet).68
Inhibitor of nuclear factor κB kinase subunit β
Overexpression of the inhibitor of nuclear factor κB kinase subunit β (IKKβ; encoded by IKBKB) in cultured cells leads to altered insulin action, whereas inhibition of this gene improves insulin resistance. Ikbkb-knockout mice do not develop insulin resistance during a lipid infusion or when subjected to high-fat feeding.69
Conclusions
Type 2 diabetes is an extremely complex disorder. Although tissue insulin resistance and β-cell dysfunction are essential components of the disorder, our understanding of the mechanisms involved in their etiology is still in complete. Genetically engineered mice have given us insights into these mechanisms, but have also opened up new questions regarding the inter action between signaling pathways within cells and between the various tissues involved in metabolic homeostasis. This short and necessarily incomplete review has emphasized the importance of some of these pathways in the pathogenesis of the disease, but it should leave the reader with the distinct feeling that there are multiple new avenues of research available.
Key Points.
The pathophysiology of type 2 diabetes mellitus includes β-cell dysfunction and insulin resistance
Mouse models that have specific gene-deletions are useful for studying disease processes such as diabetes
Footnotes
Competing interests: The authors declared no competing interests.
Contributor Information
Derek LeRoith, D LeRoith is Chief of the Division of Endocrinology, Diabetes and Bone Disease, Department of Medicine, Mount Sinai School of Medicine, New York, NY, USA.
Domenico Accili, D Accili is Professor of Medicine and Director, Columbia Diabetes and Endocrinology Research Center, Naomi Berrie Diabetes Center, University College of Physicians and Surgeons, New York.
References
- 1.Accili D. Lilly lecture 2003: the struggle for mastery in insulin action: from triumvirate to republic. Diabetes. 2004;53:1633–1642. doi: 10.2337/diabetes.53.7.1633. [DOI] [PubMed] [Google Scholar]
- 2.Permutt MA, Hattersley AT. Searching for type 2 diabetes genes in the post-genome era. Trends Endocrinol Metab. 2000;11:383–393. doi: 10.1016/s1043-2760(00)00329-5. [DOI] [PubMed] [Google Scholar]
- 3.Nandi A, et al. Mouse models of insulin resistance. Physiol Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 4.Accili D, et al. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 5.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 6.Bluher M, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 7.Bluher M, et al. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 8.Guerra C, et al. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. J Clin Invest. 2001;108:1205–1213. doi: 10.1172/JCI13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 10.Bruning JC, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 11.Kulkarni RN, et al. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni RN, et al. β-Cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter β-cell mass. Nat Genet. 2002;31:111–115. doi: 10.1038/ng872. [DOI] [PubMed] [Google Scholar]
- 13.Xuan S, et al. Defective insulin secretion in pancreatic β cells lacking type 1 IGF receptor. J Clin Invest. 2002;110:1011–1019. doi: 10.1172/JCI15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada T, et al. Insulin receptors in β-cells are critical for islet compensatory growth response to insulin resistance. Proc Natl Acad Sci U S A. 2007;104:8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laustsen PG, et al. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol Cell Biol. 2007;27:1649–1664. doi: 10.1128/MCB.01110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Accili D. A kinase in the life of the β cell. J Clin Invest. 2001;108:1575–1576. doi: 10.1172/JCI14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez AM, et al. Functional inactivation of the IGF-I and insulin receptors in skeletal muscle causes type 2 diabetes. Genes Dev. 2001;15:1926–1934. doi: 10.1101/gad.908001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zisman A, et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med. 2000;6:924–928. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 19.Tamemoto H, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 20.Bruning JC, et al. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88:561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 21.Kubota N, et al. Disruption of insulin receptor substrate 2 causes type 2 diabetes because of liver insulin resistance and lack of compensatory β-cell hyperplasia. Diabetes. 2000;49:1880–1889. doi: 10.2337/diabetes.49.11.1880. [DOI] [PubMed] [Google Scholar]
- 22.Dong X, et al. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J Clin Invest. 2006;116:101–114. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taguchi A, et al. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 24.Barbieri M, et al. Glucose regulation and oxidative stress in healthy centenarians. Exp Gerontol. 2003;38:137–143. doi: 10.1016/s0531-5565(02)00153-5. [DOI] [PubMed] [Google Scholar]
- 25.Miller RA, et al. Big mice die young: early life body weight predicts longevity in genetically heterogeneous mice. Aging Cell. 2002;1:22–29. doi: 10.1046/j.1474-9728.2002.00006.x. [DOI] [PubMed] [Google Scholar]
- 26.Bonkowski MS, et al. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci U S A. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu SC, et al. Insulin receptor substrate 3 is not essential for growth or glucose homeostasis. J Biol Chem. 1999;274:18093–18099. doi: 10.1074/jbc.274.25.18093. [DOI] [PubMed] [Google Scholar]
- 28.Fantin VR, et al. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and glucose homeostasis. Am J Physiol Endocrinol Metab. 2000;278:E127–E133. doi: 10.1152/ajpendo.2000.278.1.E127. [DOI] [PubMed] [Google Scholar]
- 29.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB β) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 30.Garofalo RS, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB β. J Clin Invest. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz EB, et al. Cardiac and adipose tissue abnormalities but not diabetes in mice deficient in GLUT4. Nature. 1995;377:151–155. doi: 10.1038/377151a0. [DOI] [PubMed] [Google Scholar]
- 32.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 33.Crosson SM, et al. PTG gene deletion causes impaired glycogen synthesis and developmental insulin resistance. J Clin Invest. 2003;111:1423–1432. doi: 10.1172/JCI17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poy MN, et al. CEACAM1 regulates insulin clearance in liver. Nat Genet. 2002;30:270–276. doi: 10.1038/ng840. [DOI] [PubMed] [Google Scholar]
- 35.Moitra J, et al. Life without white fat: a transgenic mouse. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimomura I, et al. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duvillie B, et al. Phenotypic alterations in insulin-deficient mutant mice. Proc Natl Acad Sci U S A. 1997;94:5137–5140. doi: 10.1073/pnas.94.10.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong EG, et al. Nonobese, insulin-deficient Ins2Akita mice develop type 2 diabetes phenotypes including insulin resistance and cardiac remodeling. Am J Physiol Endocrinol Metab. 2007;293:E1687–E1696. doi: 10.1152/ajpendo.00256.2007. [DOI] [PubMed] [Google Scholar]
- 39.Stoy J, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci U S A. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51(Suppl 3):S455–S461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 42.Zhang P, et al. The PERK eukaryotic initiation factor 2 α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk–/– mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 44.Froguel P, Velho G. Molecular genetics of maturity-onset diabetes of the young. Trends Endocrinol Metab. 1999;10:142–146. doi: 10.1016/s1043-2760(98)00134-9. [DOI] [PubMed] [Google Scholar]
- 45.Grupe A, et al. Transgenic knockouts reveal a critical requirement for pancreatic β cell glucokinase in maintaining glucose homeostasis. Cell. 1995;83:69–78. doi: 10.1016/0092-8674(95)90235-x. [DOI] [PubMed] [Google Scholar]
- 46.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 47.Stoffers DA, et al. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 48.Stoffers DA, et al. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 49.Hani EH, et al. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104:R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macfarlane WM, et al. Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest. 1999;104:R33–R39. doi: 10.1172/JCI7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahlgren U, et al. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson JD, et al. Increased islet apoptosis in Pdx1+/– mice. J Clin Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler AE, et al. β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 54.Naya FJ, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 56.Chen WS, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 57.Li J, et al. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta RK, et al. The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 60.Pontoglio M, et al. Defective insulin secretion in hepatocyte nuclear factor 1α-deficient mice. J Clin Invest. 1998;101:2215–2222. doi: 10.1172/JCI2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, et al. Selective deletion of the Hnf1β (MODY5) gene in β-cells leads to altered gene expression and defective insulin release. Endocrinology. 2004;145:3941–3949. doi: 10.1210/en.2004-0281. [DOI] [PubMed] [Google Scholar]
- 62.Nakae J, et al. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 63.Matsumoto M, et al. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Kitamura T, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic β cell growth. J Clin Invest. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elchebly M, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 66.Terauchi Y, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 α subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 67.Clement S, et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 68.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 69.Yuan M, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkβ. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]