Abstract
γ-Secretase activity is vital for the transmembrane cleavage of Notch receptors and the subsequent migration of their intracellular domains to the nucleus. Notch overexpression has been associated with breast, colon, cervical and prostate cancers. We tested the effect of three different γ-secretase inhibitors (GSIs) in breast cancer cells. One inhibitor (GSI1) was lethal to breast cancer cell lines at concentrations of 2 μM and above but had a minimal effect on the non-malignant breast lines. GSI1 was also cytotoxic for a wide variety of cancer cell lines in the NCI60 cell screen. GSI1 treatment resulted in a marked decrease in γ-secretase activity and downregulation of the Notch signalling pathway with no effects on expression of the γ-secretase components or ligands. Flow cytometric and western blot analyses indicated that GSI1 induces a G2/M arrest leading to apoptosis, through downregulation of Bcl-2, Bax and Bcl-XL. GSI1 also inhibited proteasome activity. Thus, the γ-secretase inhibitor GSI1 has a complex mode of action to inhibit breast cancer cell survival and may represent a novel therapy in breast cancer.
Keywords: γ-secretase, breast cancer, Notch, cell cycle arrest, proteasome, apoptosis
γ-Secretase is an aspartyl protease macromolecular complex comprising nicastrin (NCSTN), anterior pharynx-defective 1-a/b/c isoforms, the enzymic component presenilin-1/2 (PSEN) and presenilin enhancer protein-2 (PSENEN, Pen-2). By providing an internally hydrophilic environment within the plasma membrane (Lazarov et al, 2006), γ-secretase is responsible for cleaving more than 30 substrates, including amyloid precursor protein (APP), Notch (Mumm et al, 2000), ErbB4 (Ni et al, 2001), E-cadherin (Marambaud et al, 2002), CD44 (Lammich et al, 2002) and p75 (Schluesener et al, 1992), which regulate vital cell functions such as proliferation, cell cycle, cell adhesion and apoptosis. Interestingly, increased expression of Notch ligands, receptors, and/or downstream targets are highly associated in the pathogenesis of breast (Jones et al, 2004; Stylianou et al, 2006; Yamaguchi et al, 2008), brain (Purow et al, 2005), colon (Akiyoshi et al, 2008), cervical (Liu et al, 2007) pancreatic (Doucas et al, 2008) and skin cancers (Dang et al, 2006). Thus, the γ-secretase complex may be a potential therapeutic target in a wide array of carcinomas.
Notch signalling is initiated through the interactions between the plasma-embedded Notch heterodimer receptors and cell surface ligands (Jagged-1, -2, Delta-like -1, -2 and -4) present on adjacent cells (Lindsell et al, 1995). This results in a conformational change in Notch to reveal the site 2 cleavage site for metalloproteases (ADAM10, ADAM17), which leaves a 12 amino-acid stub of the Notch extracellular domain, required for subsequent recognition and cleavage by the γ-secretase complex (Brou et al, 2000; Mumm et al, 2000). γ-Secretase cleavage of Notch liberates the intracellular domain (NICD), which translocates to the nucleus where it binds to the CSL family of DNA-binding proteins (CBF1/RBPJ-κ in mammalian cells) (Tamura et al, 1995). The NCID–CBF1 complex initiates recruitment of transcriptional co-activators Mastermind-like-1, 2, 3, and p300 (Oswald et al, 2001) to enable gene transcription of Notch downstream targets. These include Hes, Hey (Iso et al, 2003) c-myc (Klinakis et al, 2006), p21 (Rangarajan et al, 2004) and cyclin D1 (Ronchini and Capobianco, 2001), which regulate proliferation, differentiation and apoptosis.
Notch activation has been observed in a number of malignancies. It was first reported when insertion of mouse mammary tumour virus within the Notch 4 locus incited tumour formation in mice (Gallahan et al, 1996). In humans, elevated Notch1 is observed in Ras-positive breast cancers (Weijzen et al, 2002), while both increased Notch1 and Jagged1 expression decreases patient survival (Jones et al, 2004; Reedijk et al, 2005; Dickson et al, 2007). In addition, Numb, a negative regulator of Notch signalling (McGill and McGlade, 2003), is significantly reduced in 50% of breast cancer tissues, inversely correlating with tumour size (Pece et al, 2004) and correlating with a poor prognosis (Colaluca et al, 2008). Inhibiting Notch signalling by overexpression of Numb reverts the transformed phenotype of MCF-7 cells (Stylianou et al, 2006), whereas silencing of Notch 3 inhibits proliferation and promotes apoptosis in ErbB2-negative breast cancer cell lines (Yamaguchi et al, 2008). Moreover, overexpression of Notch in non-tumourigenic breast epithelial cells induces transformation in vitro (Imatani and Callahan, 2000; Stylianou et al, 2006). Thus, deregulation of Notch signalling plays a significant role in tumorigenesis.
Synthetic γ-secretase inhibitors have been successful in treating Alzheimer's disease, where defective γ-secretase cleavage of the substrate molecule APP results in a longer Aβ42 variant of Aβ40 peptides, consequently leading to plaque formation (Lichtenthaler et al, 1997). In breast cancer, the γ-secretase inhibitor DAPT (N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester) has been shown to effectively reduce DCIS-induced mammosphere formation (Farnie et al, 2007). We therefore investigated the effects of commercially available γ-secretase inhibitors on the proliferation/survival of normal and malignant breast cancer cell lines, their effects on γ-secretase component expression and the possible mechanisms involved. We show here that inhibition of γ-secretase activity in breast cancer cell lines induces G2/M arrest and downregulation of antiapoptotic proteins leading to cell death.
Materials and methods
Cell culture
Oestrogen receptor-α (ER)-positive (MCF-7, T47D, and ZR-75-1) and ER-negative (MDA-MB-231 and CAL-51) breast cancer cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% foetal calf serum (FCS) and 2 mM L-glutamine, 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin. Non-tumorigenic 226-L-U19 and 226-L-TS4 breast cell lines were kindly provided by Parmjit S Jat, Ludwig Institute for Cancer Research, London, and were cultured in DME/F12 medium supplemented with 10% FCS containing, 5 μg ml−1 insulin, 1 μg ml−1 hydrocortisone, 10 ng ml−1 epidermal growth factor, 10 ng ml−1 cholera toxin and 2 mM L-glutamine, 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin.
Effect of γ-secretase inhibitors on proliferation assays
The γ-secretase inhibitors, DAPT, Compound E ((2S)-2-{[(3,5-Difluorophenyl)acetyl]amino}-N-[(3S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-1,4-benzodiazepin-3-yl]propanamide) and γ-secretase inhibitor 1 (GSI1: z-Leu-Leu-Nle-CHO), were purchased from Merck Biosciences (Darmstadt, Germany). Cells (2.5 × 103) were seeded in triplicate and allowed to adhere overnight. The media was replaced with media containing the inhibitors diluted in dimethyl sulphoxide (DMSO). After 48 h, the media was removed and the cells were harvested and counted using a Coulter Counter (Beckman Coulter, Buckinghamshire, UK).
Sulphorhodamine B assay
The sulphorhodamine B (SRB) assay was used to screen for GSI1 cytotoxicity (Vichai and Kirtikara, 2006). The assay relies on the ability of SRB to bind to protein components of cells that have been fixed to tissue-culture plates by trichloroacetic acid (TCA). SRB is a bright-pink aminoxanthene dye with two sulphonic groups that bind to basic amino-acid residues under mild acidic conditions, and dissociate under basic conditions. As the binding of SRB is stoichiometric, the amount of dye extracted from stained cells is directly proportional to the cell mass. To determine the effect of GSI1 on cell number over time, SRB assays were performed as described. Cells were seeded with six replicates in flat-bottomed 96-well plates (3000 cells per well). The cells were allowed to adhere overnight, and then media containing GSI1 at different concentrations were added. One plate was assayed at this time point (time zero) and further plates were assayed at 2-day intervals until day 10. The cells were fixed by adding 100 μl per well of ice-cold 40% TCA to each well for 60 min. The plates were washed five times in running tap water and stained with 100 μl per well SRB reagent (0.4% w/v SRB (Sigma-Aldrich, Poole, UK)) in 1% acetic acid for 30 min. The plates were washed five times in 1% acetic acid and allowed to dry overnight. SRB was solubilised with 100 μl per well 10 mM Tris-base, shaken for 30 min and the optical density measured at 492 nm.
γ-Secretase activity assay
Cells (1.5 × 106) were treated with increasing concentrations of GSI1 (0.75, 2, and 5 μM) and γ-secretase activity was measured after 24 and 48 h using the R&D Systems (R&D, Minneapolis, MN, USA) γ-Secretase Activity Kit following the manufacturer's instructions. Briefly, cells were washed twice with ice-cold phosphate-buffered saline (PBS), harvested in the cell extraction buffer, and incubated on ice for at least 10 min. Lysates were centrifuged at 10 000 g for 1 min and supernatants were collected with a total protein yield of 0.5–1.0 mg ml−1. Pierce BCA Protein Assay (Pierce, Rockford, IL, USA) was used to determine the protein concentration in each sample. Protein (200 μg) was incubated with the γ-secretase fluorogenic substrate for 2 h at 37°C and fluorescence was measured at 355/460 nm.
CBF-1 reporter assay
The ability of the γ-secretase-cleaved NICD to bind to CBF1 and activate gene transcription was measured by the transfection of a reporter in which four copies of the wild-type CBF1-binding elements were cloned in front of a simian virus 40 promoter-driven luciferase gene (4xwtCBF1Luc) (Hsieh et al, 1996). Twenty-four hours after transfection, cells were treated with GSI1 and luciferase activity was measured 24 h later using a Promega Luciferase kit (Promega, Madison, WI, USA) following the manufacturer's recommendations.
National Cancer Institute Screen of γ-secretase inhibitor
The human tumour cell lines of the cancer-screening panel (NCI60) were cultured in RPMI 1640 medium containing 5% foetal bovine serum and 2 mM L-glutamine. Cells were inoculated into 96-well microtiter plates in 100 μl at plating densities ranging from 5000 to 40 000 cells per well, depending on the doubling time of individual cell lines. After cell inoculation, the microtiter plates were incubated at 37°C, 5% CO2 for 24 h before addition of experimental drugs. After 24 h, two plates of each cell line were fixed in situ with TCA to represent a measurement of the cell population for each cell line at the time of drug addition (Tz). GSI1 in complete medium containing 50 μg ml−1 gentamicin was added to the cell lines and the plates were incubated for an additional 48 h at 37°C, 5% CO2, and an SRB assay was performed. Using the seven absorbance measurements (time zero (Tz), control growth (C), and test growth in the presence of drug at the five concentration levels (Ti)), the percentage growth was calculated at each of the drug concentrations levels. Percentage growth inhibition was calculated as:
Three dose–response parameters were calculated for each experimental agent. Growth inhibition of 50% (GI50) was calculated as was the drug concentration resulting in total growth inhibition (TGI) is calculated from Ti=Tz. The LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared with that at the beginning) indicating a net loss of cells following treatment is calculated from [(Ti−Tz)/Tz] × 100=−50. Values were calculated for each of these three parameters if the level of activity is reached; however, if the effect is not reached or is exceeded, the value for that parameter is expressed as greater or less than the maximum or minimum concentration tested.
Cell cycle analysis
Cells (3 × 105) were plated out and after 24 h the media was changed to one containing DMSO, or GSI1 at a concentration of 0.75, 1, or 5 μM. Both adherent and floating cells were harvested after 24 and 48 h, fixed with ice-cold 70% methanol, washed twice with 5 ml PBS, treated with 10 μg RNAse, and stained with 50 μg propidium iodide, and their DNA content was estimated by flow cytometry using a Beckton Dickinson FacsCanto flow cytometer (Beckton Dickinson, Franklin Lakes, NJ, USA) to determine the proportion of cells at each stage of the cell cycle.
RNA expression
Cells were lysed with 4 M guanidinium isothiocyanate and total RNA was extracted using RNAeasy (Qiagen Ltd, West Sussex, UK) according to the manufacturer's instructions. cDNA was synthesised from 2 μg of RNA with Moloney Murine Leukemia Virus reverse transcriptase and amplified by PCR on a LightCycler (Roche Diagnostics, Mannheim, Germany) using the LightCycler DNA FastStart SYBR Green 1 kit (Roche Diagnostics). Oligonucleotide sequences and PCR conditions are described in detail in Supplementary Table 1.
Protein expression
Cell lines were harvested and lysed in Lysis buffer (150 mM NaCl, 0.1% SDS, 5 mM EDTA, 10 mM Tris (pH 7.2), 1% Triton-X, and 1% deoxycholate), containing 1 mM phenylmethanesulphonylfluoride and protease inhibitor cocktail (Sigma-Aldrich P8340: AEBSF 104 mM, Aprotinin 0.08 mM, Leupeptin 2 mM, Bestatin 4 mM, Pepstatin A 1.5 mM, E-64 1.4 mM). Whole cell extracts were denatured at 100°C for 10 min and were electrophoresed on a 10% SDS–polyacrylamide gel. Primary antibodies Bcl-XL, Bcl-2, Bax and XIAP (Beckton Dickinson), Notch 1 intracellular domain (Abcam Plc, Cambridge, UK), and γ-tubulin (Sigma-Adrich) were incubated in 0.1% TBS-Tween overnight. Blots were visualised using chemiluminescence (ECL) (Amersham Bioscience, Buckinghamshire, UK), according to the manufacturer's instructions.
Proteasome activity
The inhibitory effect of GSI1 and MG132 (z-Leu-Leu-Leu-CHO) on the 20S proteasomal component (Millipore, Billerica, MA, USA) was determined in vitro after pre-incubation for 15 min at room temperature. Then, the enzyme-inhibitor mix was added to 1 × assay buffer (25 mM HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid, pH 7.5, 0.5 mM EDTA, 0.05% (v/v) NP-40 and 0.001% (w/v) SDS), and incubated with N-Succinyl-Leu-Leu-Val-Tyr-7-Amino-4-methylcoumarin (suc-LLVY-AMC, Millipore) for 75 min at 40°C. Three replicates were included per treatment. Fluoresence was measured at 380/460 nm.
The effect of GSI1 and MG132 on proteasomal activity in vivo was determined after treatment of MDA-MB-231 and MCF-7 cells (1 × 106) with the inhibitors for 4 h. Adherent cells were washed and scraped in cold PBS, collected and centrifuged for 5 min at 170 × g and 4°C. Cells were resuspended in 50 mM HEPES, pH 7.5, 5 mM EDTA, 150 mM NaCl and 1% Triton X-100, and incubated on ice for 30 min, with vortexing at 10 min intervals. Samples were centrifuged at 14 000 g for 15 min at 4°C, and the supernatant was collected. Lysates were incubated with 1 × assay buffer and 50 μM suc-LLVY-AMC at 40°C for 75 min. Three replicates were included per treatment. Fluorescence was measured at 380/460 nm.
Statistical analysis
Where indicated a t-test was performed (two-sided). Statistical significance was assumed when P<0.05.
Results
Downregulation of the Notch signalling pathway with GSI1 selectively affects the viability of breast cancer cells
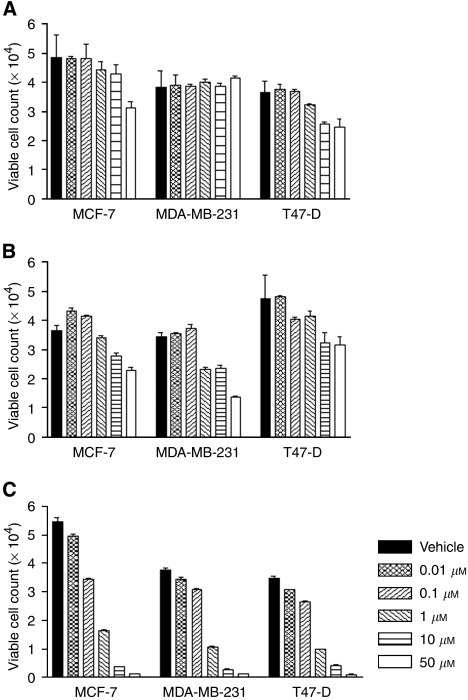
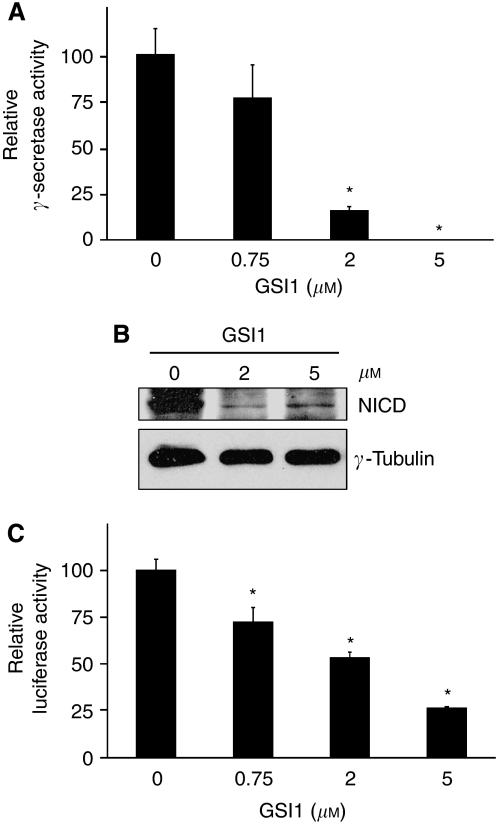
The breast cancer cell lines MDA-MB-231, T47D, and MCF-7 were treated with three γ-secretase inhibitors at concentrations in the range 0.01–50 μM. Compound E had no effect on the MDA-MB-231 cells and only reduced the proliferation of the other cell lines by less than 50% at a concentration of 50 μM (Figure 1A). The effect of DAPT on T47D and MCF-7 cell lines was comparable with the Compound E, but DAPT inhibited proliferation by approximately 50% in the MDA-MB-231 lines at 50 μM (Figure 1B). GSI1 had the most significant effect on all three cell lines at 1 μM reducing proliferation by approximately 80% (Figure 1C). GSI1 inhibited γ-secretase activity in MCF-7 cells in a dose-dependent manner and at 5 μM inhibition reached 100% (Figure 2A).
Figure 1.
Cytotoxic effect of γ-secretase inhibitors on breast cancer cell lines. Cells were treated with Compound E (A), DAPT (B), and GSI1 (C) for 48 h and their viability was determined using a Coulter counter. Data represent the average of three independent experiments ±s.d.
Figure 2.
GSI1 downregulates γ-secretase activity and the Notch pathway. (A) Relative γ-secretase activity in MCF-7 cells were treated with GSI1 for 24 h. (B) Western blot analysis of NICD in MCF-7 cells were treated with GSI1 for 24 h. (C) MCF-7 cells were transfected with a CBF1-luciferase reporter and the next day treated with GSI1 for 24 h. Values represent relative luciferase activity with respect to the mock-transfected and vehicle-treated cells. Bars indicate the average γ-secretase (A) or luciferase activity (C) ±s.d. of three independent experiments (* indicates P<0.05).
We confirmed that inhibition of γ-secretase by GSI1 downregulated the Notch pathway by directly detecting the NICD (which is cleaved by γ-secretase) with a specific antibody. Treatment of MCF-7 cells with 2 and 5 μM GSI1 decreased notably the levels of NICD (Figure 2B). It is well established that the Notch intracellular domain interacts with the transcriptional repressor CBF1 and abolishes CBF1-mediated repression. We used a surrogate for the activation of Notch by γ-secretase consisting of the transfection of a luciferase reporter containing CBF1-binding sites (Hsieh et al, 1996). In this system, activation of Notch is reflected by an increase in luciferase expression. Treatment of MCF-7 cells with GSI1 reduced the luciferase activity of the transfected reporter in a dose-dependent manner (Figure 2C). Interestingly, the effect of GSI1 on Notch-driven luciferase transcription was always lower than the corresponding inhibition of γ-secretase activity, probably due to a less than 100% transfection efficiency and activation of CBF1-Luciferase by Notch-independent mechanisms.
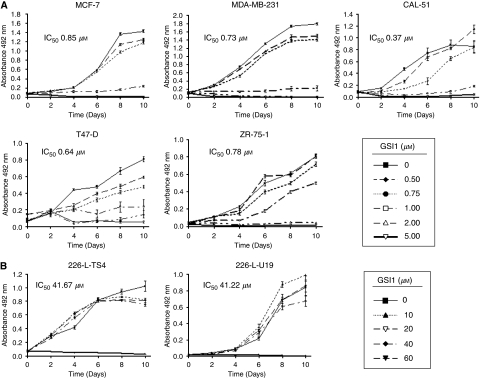
Sulphorhodamine B assays were then carried out on five tumorigenic and two non-tumorigenic breast cell lines to investigate the effects of GSI1 over a longer time frame. In MCF-7, MDA-MB-231, ZR-75-1, T47D and CAL-51 breast cancer cell lines 1 μM GSI1 and above resulted in cell death (range of IC50 values: 0.6–0.9 μM) (Figure 3A). No effect on the non-tumorigenic 226-L-U19 and 226-L-TS4 cell lines was seen in the range 0.5–40 μM, which showed IC50 values around 50 μM (Figure 3B).
Figure 3.
Cytotoxic effect of GSI1 on breast cancer cells. The effect of increasing concentrations of GSI1 in breast cancer cell lines (A) and non-tumorigenic breast cell lines (B) was determined by staining with SRB. Data represents the average ±s.d. of two independent experiments with six replicates at each time point. The IC50 for each cell line was determined from the corresponding dose–response curves (data not shown) and is indicated.
There was no effect on the expression of the γ-secretase components or ligands such as Notch, Jagged, and Numb in response to the GSI1 treatment (Supplementary Figure 1). Thus, the effect of GSI1 on breast cancer cells is not due to a differential expression of γ-secretase components or its downstream effectors.
Effect of GSI1 on the NCI 60 panel of cancer cell lines
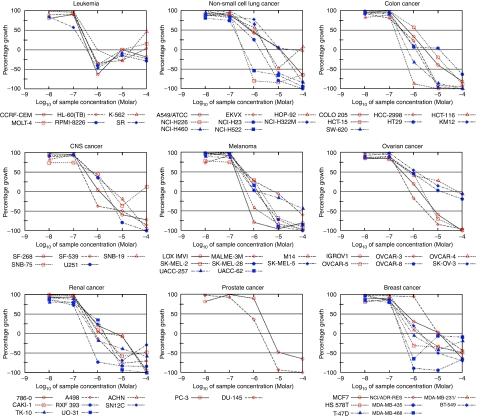
The effect of GSI1 (10 nm, 100 nM, 1, 10, and 100 μM) was tested, in duplicate, on the NCI panel of 60 cancer cell lines. A decrease in cell number was seen in all 60 cell lines with the mean log10 GI50 of −6.13±0.007 M (Figure 4). The mean log10 LC50 calculated for all of the cell lines was −4.41±0.011 M (Supplementary Table 2). When the screen was repeated the values obtained were −6.41±0.005 M for the log10 GI50, and −4.49±0.12 M for the log10 LC50.
Figure 4.
Cytotoxic effect of GSI1 on the NCI60 panel of cancer cell lines. The cytotoxic effect of GSI1 (10, 100 nM, 1, 10, 100 μM) on cancer cells was determined by a SRB assay after 24 h of drug treatment. Although NCI/ADR-RES data are shown within the breast cancer cell panel, recent evidence suggests that this is an ovarian cancer multidrug-resistant line derived from OVCAR-8 (Liscovitch and Ravid, 2007). Results shown are representative of two independent experiments.
When the breast cancer cell lines only were examined the mean log10 GI50 were −6.23±0.063 and −6.58±0.059 M (on the second run the MDA-MB-468 cell line was included). The mean values of log10 LC50 were −5.12±0.078 and −4.86±0.12 M in runs 1 and 2, respectively. GSI1 had a negligible effect on cell number at 100 nM, but at concentration of 1 μM and higher it had an inhibitory effect (Figure 4). Interestingly, the multidrug-resistant cell line NCI/ADR-RES, resistant to adriamycin and other P-glycoprotein substrates, was less sensitive to GSI1 than the other breast cancer cell lines tested and than most other cancer cell lines. At 1 μM, GSI1 did not show any decrease in cell number, which was only noticeable at 10 μM (Figure 4). Thus, GSI1 inhibits the proliferation of many different cancer cell lines corresponding to a variety of solid tumours and leukaemia.
GSI1 induces a G2/M arrest resulting in apoptosis
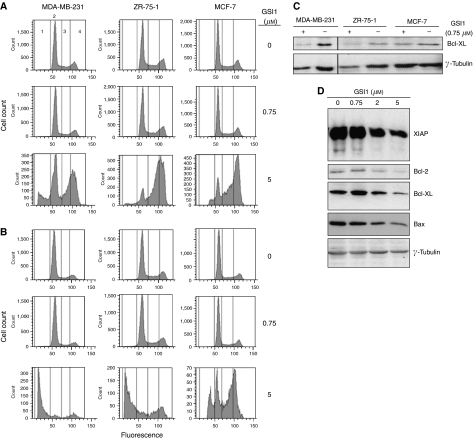
We observed a marked increase in the percentage of cells in G2/M arrest after 24 h incubation with 5 μM GSI1 (Figure 5A). After 48 h treatment, a large proportion of the ZR-75-1 and MDA-MB-231 cells were apoptotic (58 and 77%, respectively) and only a small percentage were in G2/M arrest (Figure 5B). Seventeen percent of MCF-7 cells were apoptotic after treatment with 5 μM GSI1 (Figure 5B), but there was a substantial increase in the percentage of cells in G2/M arrest. Media replacement experiments indicated that when fresh medium was added to MDA-MB-231 cells previously treated with GSI1 for 48 h, the cells recovered their proliferative capacity when concentration was up to 1 μM. However, at higher concentrations no further proliferation was observed (data not shown). When the levels of the antiapoptotic protein Bcl-XL were monitored in MDA-MB-231, ZR-75-1, and MCF-7 cells, we found a downregulation due to GSI1 treatment (0.75 μM for 48 h) in the two former cell lines (Figure 5C). However, GSI1 treatment in MDA-MB-231 and ZR-75-1 cell lines also downregulated the level of γ-tubulin (Figure 5C) as well as other proteins tested as loading controls (eIF4E, RPLP0, ribosomal protein S6, among others). We then monitored the levels of the antiapoptotic proteins XIAP, Bcl-2, Bax and Bad after treatment with increasing doses of GSI1 and found that their levels decreased progressively from 2 to 5 μM GSI1 (Figure 5D). Thus, GSI1 triggers apoptosis in breast cancer cells by downregulating the expression of antiapoptotic proteins.
Figure 5.
GSI1 induces G2/M cell cycle arrest and triggers apoptosis in breast cancer cells. (A, B) Cell cycle analyses of breast cancer cell lines treated with GSI1 for 24 h (A) and 48 h (B). Cells were stained with propidium iodide and gated according to their fluorescence to differentiate cell cycle phases: 1, apoptotic cells; 2, cells in G0/G1; 3, cells in S phase; 4, cells in G2-M. Data shown are a representative of two independent experiments. (C, D) Western blot analyses were used to verify the modulation of the apoptotic response due to GSI1 treatment. (C) Three different breast cancer cell lines treated with a single GSI1 concentration (0.75 μM) for 48 h. (D) MCF-7 cells treated for 48 h with increasing concentrations of GSI1. Tubulin was used as a loading control. Note the slight effect of GSI1 on tubulin levels in MDA-MB-231 and ZR-75-1 cells (C).
GSI1 inhibits proteasome activity both in vivo and in vitro but has less cytotoxic effect on breast cancer cells than MG132
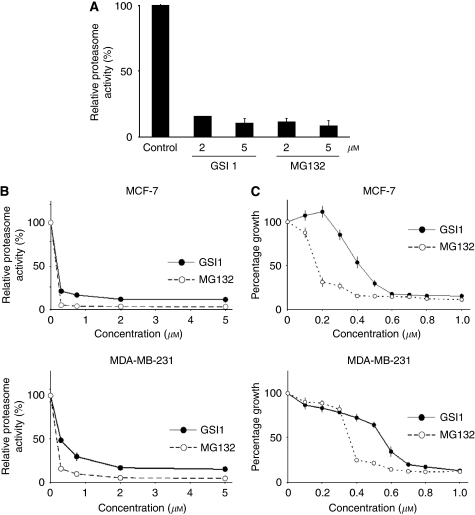
As GSI1 (z-Leu-Leu-Nle-CHO) is chemically and structurally similar to proteasomal inhibitor MG132 (z-Leu-Leu-Leu-CHO), the possibility of GSI1 affecting proteasomal activity was explored. Proteasomal activity was severely reduced (80-90%) upon incubation treatment of the isolated proteasome 20S proteolytic core particle subunit with either compound in vitro (Figure 6A). Similarly, a strong inhibitory effect was observed in vivo (Figure 6B), although MG132 was marginally more efficient than GSI1. However, the cytotoxic effect of both compounds on both MCF-7 and MDA-MB-231 cells was markedly different (Figure 6C), MG132 showing a stronger cytotoxic effect than GSI1 (between 1.5- and 2-fold). Thus, despite their similar chemical structure and protesome inhibition, GSI1 and MG132 affect the growth of breast cancer cells differently.
Figure 6.
GSI1 inhibits proteasome activity both in vivo and in vitro but has less cytotoxic effect on breast cancer cells than MG132. (A) Inhibition of proteasome activity in vitro. The 20S proteasomal component was pre-incubated with GSI1, MG132, or vehicle for 15 min at room temperature, and then the proteolysis of suc-LLVY-AMC was monitored by fluorimetry. One unit of activity was defined as the amount of protein producing one μmol of AMC per minute at 40°C. Data represent the average ±s.d. of two experiments. (B) Effect of both GSI1 and MG132 on proteasome activity in cell extracts from breast cancer cells. Both drugs where added to the culture medium of breast cancer cells, and after 4 h, proteasome activity was determined in cell extracts by proteolysis of suc-LLVY-AMC. Data represent the average ±s.d. of three experiments. (C) Cytotoxic effect of GSI1 and MG132 on breast cancer cells. The effect of increasing drug concentrations on breast cancer cells was determined by staining with SRB after 48 h. Data represent the average ±s.d. of two independent experiments with six replicates at each time point.
Discussion
In this study we endeavoured to determine whether the γ-secretase complex, which has an integral role in signalling of Notch, is a potential therapeutic target in breast cancer. For this we tested the effect of commercial γ-secretase inhibitors on breast cell lines. The three γ-secretase inhibitors tested had markedly different effects on different breast cancer cell lines. Both DAPT and Compound E are chemical inhibitors of the complex. DAPT reduces ductal carcinoma in situ mammosphere formation (Farnie et al, 2007) and pancreatic cancer cell growth (Kimura et al, 2007). Another γ-secretase inhibitor, MRK-003, reduces tumour cell proliferation, inhibits serum independence, and induces apoptosis of lung cancer cell lines in vitro and in vivo using mouse xenograft models (Konishi et al, 2007). Thus, the γ-secretase complex is now becoming an accepted target in cancer therapy, in particular, with regard to Notch signalling (Shih and Wang, 2007).
Differential responses between tumourigenic and non-tumourigenic cell lines may be explained by differential expression of Numb, a negative regulator of the Notch pathway, and NICD. It has been shown that non-tumourigenic cells express Numb but not NICD (Stylianou et al, 2006) indicating that, as expected, the Notch pathway is not activated in non-cancerous cells. Conversely, cancer cells have Numb downregulated, NICD upregulated and the Notch pathway activated (Stylianou et al, 2006), and are sensitive to the cytotoxic effect of GSI1 by its effect on the Notch pathway.
We show here that γ-secretase inhibition promotes a cell cycle arrest at G2/M, which further triggers the apoptotic response. Expression of cyclin B1, which controls the G2/M checkpoint, can be regulated by the Notch pathway (through putative CBF-1-binding elements in its promoter). Breast cancer cells in which the Notch pathway has been targeted, either by an inhibitor of γ-secretase or by Notch-1 RNAi, downregulate cyclin B1 and suffer G2/M arrest (Rizzo et al, 2008). In addition, in MCF-7 cells another γ-secretase inhibitor triggers the DNA damage response with the concomitant upregulation of the cell cycle regulators, p53 and p21, which may promote defective cell division, consequently abrogating antiapoptotic mechanisms (Alimirah et al, 2007). We observed a dose-dependent downregulation of Bcl-2, Bax, Bad and XIAP upon GSI1 treatment (Figure 5) and a corresponding dose-dependent activation of caspase 3/7 in MDA-MB-231 cells (data not shown). Increased apoptosis upon treatment with a γ-secretase inhibitor has also been observed in Kaposi sarcoma, multiple myeloma (Nefedova et al, 2008), melanoma (Leggas et al, 2004) and tongue carcinoma (Yao et al, 2007). This may be indicative of a possible mechanism through which inhibition of γ-secretase modulates decreased viability, as observed in the comprehensive NCI screen.
Notch and APP are probably the best-studied γ-secretase substrates, and we have shown that GSI1 treatment downregulates the Notch pathway in breast cancer cells. However, as γ-secretase acts upon a large variety of substrates, it is likely that the cytotoxic effect of GSI1 upon cells will be due to the downregulation of several downstream targets involved in vital cell functions, which ultimately affect the survival of cancer cells.
Interestingly, NCI/ADR-RES, multidrug-resistant cells overexpressing the ABC transporter ABCB1 (P-glycoprotein) (Raguz et al, 2008) and prostate PC-3 cells, are the least susceptible of all the cells in the NCI-60 cell panel. As PC-3 cells express also P-glycoprotein (Rao et al, 2005), and considering the molecular structure of GSI1, it is possible that GSI1 will be a substrate of P-glycoprotein (Crivori et al, 2006). This would make GSI1 less effective in cancers from tissues expressing P-glycoprotein such as those from colon or the adrenal gland, or in those which have acquired P-glycoprotein-mediated drug resistance (Burger et al, 2003).
We also showed that GSI1 is a potent proteasomal inhibitor, and may inhibit breast cancer cell proliferation through dual targets of the γ-secretase complex and the proteasome. Whether GSI1 targets each specifically, or inhibition occurs through sequential consequence remains uncertain. It is tempting to speculate that aldehyde-based compounds, such as GSI1 and MG132, may be able to bind interchangeably between the two complexes. Although a low-resolution electron microscopy model of the γ-secretase complex has recently been described (Lazarov et al, 2006), only the complete high-resolution X-ray structure of the γ-secretase complex may suggest putative shared 3D regions to which aldehyde-based inhibitors bind. In addition, it is possible that cross-talk between the γ-secretase complex and the proteasome exists, as PSEN may be cleaved into the active C- and N-terminal fragments by the proteasome (Massey et al, 2005). Furthermore, it has been suggested that the γ-secretase complex can act in a similar way to the proteasome, as they can recognise, capture substrates and feed them through the proteolytic-containing cavity of their respective complexes to produce functional cleaved fragments (Kopan and Illagan, 2004).
The cytotoxic effect of GSI1 and MG132 on breast cancer cells is different (Figure 6C). As GSI1, MG132 induces G2/M arrest and apoptosis with loss of Bcl-2 expression (Yan et al, 2007), thus the therapeutic use of proteasome inhibitors are also being considered in cancer treatment (Voorhees et al, 2003; Sato et al, 2008). Whether the uptake of these compounds by breast cancer cells or their half-lives (either in the culture medium or intracellularly) is different, and thus modulate their potency, remains to be established.
Two facts are important from a potential future clinical application of GSI1: (1) the inhibitor is effective in triggering cell death of cancer cell lines at much lower concentrations than those required in non-tumorigenic cell lines, and (2) its effect on breast cancer cells is irrespective of their ER status. This last point is particularly important as triple-negative breast tumours (ER-, progesterone receptor-, and Her-2-negative), which are particularly aggressive and have a poor prognosis, are currently only treated with traditional chemotherapy (Haffty et al, 2006; Cleator et al, 2007). Interestingly, γ-secretase inhibitors have the potential of increasing sensitivity and efficacy of other chemotherapeutic drugs as well as hormonal and targeted therapeutic agents. For example, synergistic treatment of breast cancer cells with a γ-secretase inhibitor and trastuzamub (Osipo et al, 2008) or Tamoxifen (Rizzo et al, 2008) was more effective in reducing proliferation than the individual treatments. The combined treatments of γ-secretase inhibitors and chemotherapeutic agents have illustrated a greater extent of antiproliferative effects and/or apoptosis in multiple myeloma (Nefedova et al, 2008), T-cell acute lymphoblastic leukaemia (De Keersmaecker et al, 2008), and colon cancer cell lines (Akiyoshi et al, 2008).
Overall these data indicate that γ-secretase is a potential therapeutic target in breast cancer.
Acknowledgments
We would like to acknowledge funding from Breast Cancer Research Trust (RB, MJS), Cancer Research UK (AF), and the Experimental Cancer Medicine Centre (SR). We also thank Parmjit S Jat for providing cell lines, Diane Watling for flow cytometry advice and Laki Buluwela, Adam Paige, and Mauritius Kleijnen for helpful discussions.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, Koga K, Nakashima H, Sato N, Tanaka M, Katano M (2008) Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology 134: 131–144 [DOI] [PubMed] [Google Scholar]
- Alimirah F, Panchanathan R, Davis FJ, Chen J, Choubey D (2007) Restoration of p53 expression in human cancer cell lines upregulates the expression of Notch1: implications for cancer cell fate determination after genotoxic stress. Neoplasia 9: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell 5: 207–216 [DOI] [PubMed] [Google Scholar]
- Burger H, Foekens JA, Look MP, Meijer-van Gelder ME, Klijn JGM, Wiemer EAC, Stoter G, Nooter K (2003) RNA expression of breast cancer resistance protein, lung resistance-related protein, multidrug resistance-associated proteins 1 and 2, and multidrug resistance gene 1 in breast cancer: correlation with chemotherapeutic response. Clin Cancer Res 9: 827–836 [PubMed] [Google Scholar]
- Cleator S, Heller W, Coombes RC (2007) Triple-negative breast cancer: therapeutic options. Lancet Oncol 8: 235–244 [DOI] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP (2008) NUMB controls p53 tumour suppressor activity. Nature 451: 76–80 [DOI] [PubMed] [Google Scholar]
- Crivori P, Reinach B, Pezzetta D, Poggesi I (2006) Computational models for identifying potential P-glycoprotein substrates and inhibitors. Mol Pharm 3: 33–44 [DOI] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO (2006) Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878 [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, Bekaert S, Vandenberghe P, Odero MD, Marynen P, Cools J (2008) In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica 93: 533–542 [DOI] [PubMed] [Google Scholar]
- Dickson BC, Mulligan AM, Zhang H, Lockwood G, O’Malley FP, Egan SE, Reedijk M (2007) High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol 20: 685–693 [DOI] [PubMed] [Google Scholar]
- Doucas H, Mann CD, Sutton CD, Garcea G, Neal CP, Berry DP, Manson MM (2008) Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol 97: 63–68 [DOI] [PubMed] [Google Scholar]
- Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ (2007) Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst 99: 616–627 [DOI] [PubMed] [Google Scholar]
- Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH (1996) Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res 56: 1775–1785 [PubMed] [Google Scholar]
- Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D (2006) Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 24: 5652–5657 [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD (1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol Cell Biol 16: 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imatani A, Callahan R (2000) Identification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell lines. Oncogene 19: 223–231 [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194: 237–255 [DOI] [PubMed] [Google Scholar]
- Jones C, Mackay A, Grigoriadis A, Cossu A, Reis-Filho JS, Fulford L, Dexter T, Davies S, Bulmer K, Ford E, Parry S, Budroni M, Palmieri G, Neville AM, O’Hare, MJ, Lakhani SR (2004) Expression profiling of purified normal human luminal and myoepithelial breast cells: identification of novel prognostic markers for breast cancer. Cancer Res 64: 3037–3045 [DOI] [PubMed] [Google Scholar]
- Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T (2007) Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci 98: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A (2006) Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci USA 103: 9262–9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP (2007) Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res 67: 8051–8057 [DOI] [PubMed] [Google Scholar]
- Kopan R, Illagan MXG (2004) γ-secretase: proteasome of the membrane? Nat Rev Mol Cell Biol 5: 499–504 [DOI] [PubMed] [Google Scholar]
- Lammich S, Okochi M, Takeda M, Kaether C, Capell A, Zimmer AK, Edbauer D, Walter J, Steiner H, Haass C (2002) Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Abeta-like peptide. J Biol Chem 277: 44754–44759 [DOI] [PubMed] [Google Scholar]
- Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H (2006) Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci USA 103: 6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggas M, Adachi M, Scheffer GL, Sun D, Wielinga P, Du G, Mercer KE, Zhuang Y, Panetta JC, Johnston B, Scheper RJ, Stewart CF, Schuetz JD (2004) Mrp4 confers resistance to topotecan and protects the brain from chemotherapy. Mol Cell Biol 24: 7612–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Ida N, Multhaup G, Masters CL, Beyreuther K (1997) Mutations in the transmembrane domain of APP altering gamma-secretase specificity. Biochemistry 36: 15396–15403 [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G (1995) Jagged: a mammalian ligand that activates Notch1. Cell 80: 909–917 [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Ravid D (2007) A case study in misidentification of cancer cell lines: MCF-7/AdrR cells (re-designated NCI/ADR-RES) are derived from OVCAR-8 human ovarian carcinoma cells. Cancer Lett 245: 350–352 [DOI] [PubMed] [Google Scholar]
- Liu J, Ye F, Chen H, Lu W, Zhou C, Xie X (2007) Expression of differentiation associated protein Hes1 and Hes5 in cervical squamous carcinoma and its precursors. Int J Gynecol Cancer 17: 1293–1299 [DOI] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK (2002) A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J 21: 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey LK, Mah AL, Monteiro MJ (2005) Ubiquilin regulates presenilin endoproteolysis and modultes γ-secretase components, Pen-2 and nicastrin. Biochem J 391: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA and McGlade CJ (2003) Mammalian numb proteins promote Notch 1 receptor ubiquitination and degradation of the Notch 1 intracellular domain. J Biol Chem 278: 23196–23203 [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R (2000) A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell 5: 197–206 [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Sullivan DM, Bolick SC, Dalton WS, Gabrilovich DI (2008) Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood 111: 2220–2229 [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G (2001) gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294: 2179–2181 [DOI] [PubMed] [Google Scholar]
- Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, Miele L (2008) ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene 27: 5019–5032 [DOI] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM (2001) p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol 21: 7761–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP (2004) Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol 167: 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, Maric D, Eberhart CG, Fine HA (2005) Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res 65: 2353–2363 [DOI] [PubMed] [Google Scholar]
- Raguz S, Randle RA, Sharpe ER, Foekens JA, Sieuwerts AM, Meijer-van Gelder ME, Melo JV, Higgins CF, Yague E (2008) Production of P-glycoprotein from the MDR1 upstream promoter is insufficient to affect the response to first-line chemotherapy in advanced breast cancer. Int J Cancer 122: 1058–1067 [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA (2004) Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6: 171–183 [DOI] [PubMed] [Google Scholar]
- Rao PS, Govindarajan R, Mallya KB, West W, Rao US (2005) Characterization of a new antibody raised against the NH2 terminus of P-glycoprotein. Clin Cancer Res 11: 5833–5839 [DOI] [PubMed] [Google Scholar]
- Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE (2005) High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res 65: 8530–8537 [DOI] [PubMed] [Google Scholar]
- Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, Hao L, Yao K, Rajan P, Hicks C, Siziopikou K, Selvaggi S, Bashir A, Bhandari D, Marchese A, Lendahl U, Qin JZ, Tonetti DA, Albain K, Nickoloff BJ, Miele L. (2008) Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res 68: 5226–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ (2001) Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol Cell Biol 21: 5925–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Rajendra E, Ohta T (2008) The UPS: a promising target for breast cancer treatment. BMC Biochem 9(Suppl 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluesener HJ, Koeppel C, Jung S (1992) Multidrug transport in human autoimmune T line cells and peripheral blood lymphocytes. Immunopharmacology 23: 37–48 [DOI] [PubMed] [Google Scholar]
- Shih I-M, Wang TL (2007) Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res 67: 1879–1882 [DOI] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K (2006) Aberrant activation of notch signaling in human breast cancer. Cancer Res 66: 1517–1525 [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol 5: 1416–1423 [DOI] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K (2006) Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 1: 1112–1116 [DOI] [PubMed] [Google Scholar]
- Voorhees PM, Dees EC, O’Neil B, Orlowski RZ (2003) The proteasome as a target for cancer therapy. Clin Cancer Res 9: 6316–6325 [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L (2002) Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med 8: 979–986 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Ohwada S, Tatsuta K, Inoue J, Semba K, Watanabe S (2008) NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res 68: 1881–1888 [DOI] [PubMed] [Google Scholar]
- Yan XB, Yang DS, Gao X, Feng J, Shi ZL, Ye Z (2007) Caspase-8 dependent osteosarcoma cell apoptosis induced by proteasome inhibitor MG132. Cell Biol Int 31: 1136–1143 [DOI] [PubMed] [Google Scholar]
- Yao J, Duan L, Fan M, Wu X (2007) Gamma-secretase inhibitors exert antitumor activity via down-regulation of Notch and Nuclear factor kappa B in human tongue carcinoma cells. Oral Dis 13: 555–563 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.