Summary
Photocleavage of the polypeptide backbone is potentially a powerful and general method to activate or deactivate functional peptides and proteins with high spatial and temporal resolution. Here we show that 2-nitrophenylalanine is able to photochemically cleave the polypeptide backbone by an unusual cinnoline forming reaction. This unnatural amino acid was genetically encoded in E. coli, and protein containing 2-nitrophenylalanine was expressed and site specifically photocleaved.
Introduction
Natural proteolytic activities are involved in a wide array of biological processes including zymogen activation, protein degradation and the removal of signal peptides. The photochemical regulation of such processes is increasingly being used to probe cellular events [1, 2]. The ability to photochemically initiate protein cleavage would allow spatial and temporal control over the activity, lifetime, and localization of proteins both in vitro and in living cells. Indeed, it has been shown that the protein backbone can be photocleaved selectively if the unnatural amino acid 2-nitrophenylglycine (2-NPG) is incorporated into the protein [3, 4]. However, the production of proteins containing 2-NPG by current methods is problematic – biosynthesis using chemically aminoacylated suppressor tRNAs produces small, stoichiometric amounts of protein, and semisynthetic methods are generally restricted to substitutions at C- or N-terminal sites [5-7]. In order to further expand our ability to control proteolysis in vitro or directly in living cells we have genetically encoded the phenylalanine derivative, 2-nitrophenylalanine (2-NPA), in Escherichia coli (E. coli) in response to the amber codon, TAG. Upon irradiation at 365 nm, the 2-NPA residue photocleaves the protein backbone specifically at the site of incorporation.
Results
Photocleavage of a Model Peptide
We anticipated that incorporation of 2-NPA into proteins might allow photocleavage of the polypeptide backbone based on the photochemistry of (2-nitrophenyl)ethane derivatives[8-10] which like the more common 2-nitrobenzyl group [11, 12] are used as photochemical caging groups. Specifically, alkoxy substituents β to the nitrophenyl group undergo photochemical elimination reactions, suggesting that an amide group might behave similarly. To test this notion, model peptide 1 containing the 2-NPA group was synthesized (Figure 1). When peptide 1 (10 μM) was photolysed at 365 nm in phosphate buffered saline at pH 7.4 (PBS) photocleavage at the expected site was observed in greater than 95% yield. The quantum yield Φ of the photocleavage reaction was determined by chemical actinometry using 2-nitrobenzaldeyhyde as a known standard (Supplemental Data). The quantum yield at 365 nm was determined to be 0.07 ± 0.01. Analysis of the products of the photocleavage reaction by high resolution ESI-TOF mass spectrometry, and by 1H and 13C 1D and 2D NMR revealed two peptide fragments: a C-terminal carboxylate group, and an N-terminal cinnoline group (rather than the expected C-terminal amide and N-terminal substituted olefin). Based on the structure of the products and previous studies of 2-nitrophenylethane photolysis, we propose the putative mechanism outlined in Scheme 1. Upon photolysis, the nitrobenzyl group rearranges to the α-hydroxy-substituted nitrosophenyl group as expected. The nitroso group then undergoes an addition reaction with the N-terminal amide group [13] to generate the cyclic azo product 6. Subsequent hydrolysis of the activated carbonyl group affords the terminal cinnoline and carboxylate products. Support for this mechanism was obtained by photolysis of 1 in a solution of 30% methanol in PBS. Under these conditions, the methyl ester was formed in addition to the carboxylic acid hydrolysis product 2 (Figure S1 in Supplemental Data). Consistent with this mechanism, photolysis of a similar peptide where the amino terminus was unprotected gave several side products, which are likely the result of intramolecular cyclization reactions (Figures S2a-e).
Figure 1.
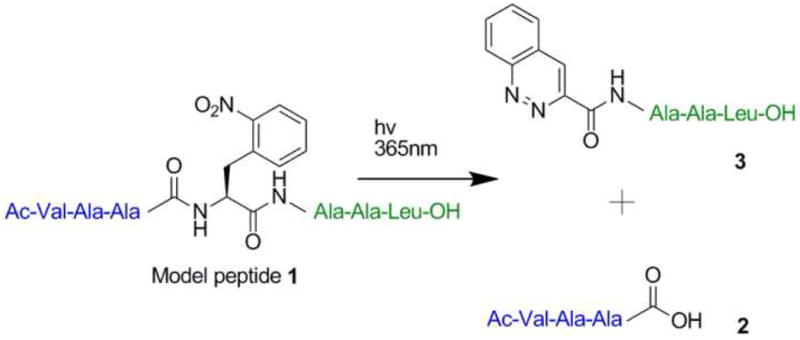
Photolysis of 1 leads to cleavage of the peptide backbone and generates a C-terminal carboxylate group and N-terminal cinnoline group
Scheme 1.

Proposed Mechanism of Photocleavage Reaction
Genetic Incorporation of 2-NPA into Proteins in E. coli
Given that photolysis of 2-NPA efficiently cleaves the polypeptide backbone, we next genetically encoded this unnatural amino acid in E. coli according to published procedures [14]. Briefly, an amber suppressor Methanococcus jannaschii and tyrosyl-tRNA synthetase pair which is orthogonal in E. coli was evolved to accept 2-NPA as a substrate instead of its natural substrate, tyrosine. A library of 6×109 active site mutants (in which residues Tyr32, Leu65, Ala67, His70, Phe108, Gln109, Tyr114, Asp158, Ile159, and Leu162 were randomized with NNK) was subjected to sequential rounds of positive and negative selection. Positive selection is based on the ability of aminoacyl-tRNA synthetases specific for 2-NPA to suppress an amber codon in the chloramphenicol acetyl transferase gene in the presence of 2-NPA and chloramphenicol. The negative selection is based on expression of a toxic barnase gene containing two amber stop codons in the absence of 2-NPA. Aminoacyl-tRNA synthetases that aminoacylate the amber suppressor tRNA with endogenous amino acids generate active barnase leading to cell death. After 3 rounds of selection, an aminoacyl-tRNA synthetase was isolated that incorporates 2-NPA with >95% fidelity based on SDS denaturing polyacrylamide gel analysis of the expression of an Asp61TAG mutant of bacteriophage T4 lysozyme (T4L) containing a C-terminal His6 tag in the presence and absence of 2-NPA (Figure S3a). ESI-TOF mass spectrometry confirmed incorporation of 2-NPA (Figure S3b). The overall yield was 2.1 mg/L after Ni-NTA and FPLC purification when cells were grown in minimal GMML media in the presence of 1 mM 2-NPA (the yield of wild type protein was 9.6 mg/L). The enzyme has 10 mutations in its active site; Y32G, L65H A67G, H70G, F108L, Q109S, Y114S, D158T, I159Y, and L162D. Six other aminoacyl-tRNA synthetases with a high degree of homology were also identified, but all displayed lower fidelity for 2-NPA incorporation (Table S1).
Expression and Photocleavage of a Model Protein, T4 Lysozyme
To determine whether incorporation of 2-NPA results in photocleavage of a folded protein, Ala82 in T4 lysozyme was substituted with 2-NPA. After Ni-NTA and subsequent FPLC purification, the protein was photolysed with >300 nm light in PBS buffer for varying periods of time and the products were then analyzed by SDS-PAGE (Figure 2). Two fragments corresponding to the photocleaved amino- and carboxy-terminal fragments of the parent protein migrated on SDS-PAGE at the expected molecular masses. A yield of 22 ± 5% cleavage was obtained after irradiation for 60 minutes as determined by optical densitometry of a coomassie-stained gel and confirmed by mass spectral analysis. The two fragments were unambiguously identified as photocleavage products of the parent protein by trypsin digestion and nano-flow LC tandem mass spectrometry (Figures S4a-b). Fragments containing the histidine tag were also identified by western blotting using anti-histidine tag antibodies. The photocleavage reaction of the folded protein did not afford near-quantitative cleavage as was seen with the model peptide. The most likely source of side products is intramolecular reactions with side chain amines or amides, as was observed upon photolysis of the unprotected model peptide.
Figure 2.

Photolysis of Ala82 (2-NPA) T4L in PBS solution using a 300 nm cut-off filter. Aliquots were taken at several time points, separated by SDS-PAGE and visualized by silver staining.
Photocleavage by Incorporation of 2-NPA at Various Residues of T4 Lysozyme
To determine if the position of the amino acid in the protein significantly affects the efficiency of photocleavage, we expressed several variants of T4L in which 2-NPA was incorporated at residues Lys60, Asp61, Lys65, Lys68, Asp72, Ala73 and Arg80. Photolysis of the purified proteins showed that all the proteins were cleaved, and that the placement of 2-NPA does somewhat affect the yield of photocleavage and quantity of side products produced (Figure 3), although there was no obvious correlation with local secondary structure or sequence. The Asp61TAG mutant afforded the most cleavage product with the least side product formation. The maximal yield of cleavage (30% ± 5%) was observed after 25 minutes of photolysis as determined by optical densitometry.
Figure 3.

Photolysis of various T4L mutant proteins containing 2-NPA at the indicated sites. Polypeptides of the expected sizes are observed.
Discussion
The 2-NPA group is able to photochemically cleave the polypeptide backbone when chemically or biosynthetically incorporated into a protein. By analyzing the products of the photolysis of a model peptide, we identified an unusual photocleavage reaction which generates a C-terminal carboxylate group and N-terminal cinnoline ring. Thus the mechanism of peptide bond cleavage is distinct from that of 2-NPG. The ability to genetically encode 2-NPA expands the range of in vitro and in vivo experiments to which the photocleavable amino acid can be applied. The yield of photocleavage is less than 50%, likely due to intramolecular cyclization products of reactive intermediates. Thus the current methodology will likely be useful for generating active proteins from photocaged precursors (e.g., zymogen activation, release of a biologically active peptides, etc.), but will be less useful for the inactivation of proteins in cells (unless the side products are also inactive). The synthetase that we have developed to accept 2-NPA should also be useful as a template to genetically encode amino acids with enhanced photochemical properties such as the dimethoxy substituted analogue which has a higher quantum yield and longer wavelength excitation. Attempts to genetically encode 2-NPG in E. coli have not yet been successful despite several efforts with distinct orthogonal tRNA/aminoacyl-tRNA synthetase pairs. In addition the 2-NPA is a close analog of phenylalanine and would be expected to minimally perturb protein structure, dynamics, and function, in contrast to the β-branched 2-NPG residue which might be expected to affect the conformation of the polypeptide backbone.
Significance
We have demonstrated that 2-nitrophynlalanine, when incorporated chemically or biosynthetically into a polypeptide is able to site specifically cleave the polypeptide backbone upon irradiation with 365 nm light. In a model peptide the photocleavage reaction yields a C-terminal carboxylate group and N-terminal cinnoline group as the major cleavage products in excellent overall yields. A maximum cleavage efficiency of ∼30% was achieved when the 2-NPA group was biosynthetically incorporated into proteins, which is likely due to competing intramolecular cyclization reactions with nearby nucleophilic groups. Because 2-NPA is a close analogue of phenylalanine, its incorporation into peptides or proteins is expected to minimally perturb their structure and function. Thus the 2-NPA can be used to activate or inactivate biologically active peptides with high temporal and spatial resolution, both in vitro and in living cells [15]. In the case of proteins (where the photocleavage efficiency will typically be lower), this methodology will be most useful for generating biologically active species from inactive precursors, rather than directly inactivating proteins where a significant change in activity will be more difficult to quantify.
Experimental Procedures
General
All chemicals were obtained from commercial sources and used without further purification. NMR data was collected on a Bruker DRX-600 instrument with chemical shifts recorded relative to residual solvent peaks of (CD3)2SO (1H 2.50ppm, 13C 39.52ppm). Mass spectra were acquired at the Scripps Center for Mass Spectrometry (La Jolla, CA) and at the Genomics Institute of the Novartis Research Foundation (La Jolla, CA). Standard molecular biology techniques[16] were used throughout. DH10b E. coli was used for routine cloning, DNA propagation, library selections, and protein expression.
Peptide Photocleavage
2-Nitrophenylalanine as the FMOC protected derivative was obtained from CSPS Pharmaceuticals, Inc. (San Diego, California). The model peptide 1 was synthesized by Anaspec (San Jose, California) and purified by HPLC with a C18 column (SUPELCO, Discovery, 250 mm × 4.6 mm, 5 μm) using a water : 5-95% acetonitrile gradient over 60 minutes with both solvents containing 0.05% formic acid. A solution of peptide 1 (10 μM) in phosphate buffered saline pH 7.4 (PBS), was photolyzed using an Oriel 500W Mercury arc lamp fitted with a water filled/water cooled liquid filter. The beam was filtered through a 365 nm band-pass filter (FWHM 10 nm, Asahi Spectra USA Inc.) and irradiated in a quartz cuvette with a 5.0 mm pathlength at room temperature. In order to collect enough material for NMR characterization, 10 mg was photolysed for 40 minutes in a large Pyrex dish atop a DNA illumination table (Fotodyne, Fotoprep) fitted with a 302 nm light source. The solution was concentrated by rotary evaporation and the products separated by HPLC as described above for the purification of the intact peptide.
Quantum Yield of Peptide Photolysis
The quantum yield was determined by chemical actinometry using 2-nitrobenzylaldehyde as a standard. [17]. Actinometry was performed in low optical density mode due to the poor solubility of the peptide in an aqueous PBS. Both peptide and 2-nitrobenzaldehyde solutions were 10 μM in PBS. An Oriel 500W Mercury arc lamp fitted with a water filled/water cooled liquid filter was used as a light source, and the beam was filtered through a 365 nm band-pass filter (FWHM 10 nm, Asahi Spectra USA Inc.) and irradiated in a quartz cuvette with a 5.0 mm pathlength. The time-course for photoreaction of the peptide and 2-nitrobenzaldehyde was followed by HPLC analysis using a Waters 2690 separations module equipped with a C18 column (SUPELCO, Discovery, 250 mm × 4.6 mm, 5 μm) and a Waters 996 photodiode array detector. Pump A delivered H2O/0.1% formic acid, and pump B delivered acetonitrile/0.1% formic acid. The components were separated by a linear gradient over 25 minutes from 1 to 90% buffer B. Each sample was analyzed 5 times.
Genetic Selection of a Mutant Aminoacyl-tRNA Synthetase Specific for 2-NPA
A library of M. jannaschii TyrRS synthetase mutants was constructed as previously described [18] where residues Tyr32, Leu65, Ala76, His70, Phe108, Gln109, Tyr114, Asp158, Ile159, and Leu162 were randomized (NNK). After 3 positive and three negative rounds of selection [18], aminoacyl-tRNA synthetases with the sequences listed below (Table S1) were enriched. Clone 1 has the highest fidelity for incorporation of 2-NPA.
Bacteriophage T4 Lysozyme Mutagenesis
A C-terminal, hexahistidine tagged, bacteriophage T4 lysozyme gene (containing N2D, C54T, C97A mutations) was amplified from pLeiT4L-wt [19] and inserted into the vector pBAD/JYAMB [20] using the NcoI and KpnI restriction sites, thereby exchanging the myoglobin gene for that of T4 lysozyme and creating pBAD-T4L-wt. Standard site directed mutagenesis was used to create several mutants of the T4L gene (Stratagene, Quikchange II). The primers used for mutagenesis are given in Supplemental Data. All plasmid constructs were verified by sequencing.
Protein Expression and Purification
Bacteriophage T4 lysozyme containing 2-NPA at position 61 was expressed in E. coli DH10B cells harboring the two plasmids pBAD-T4L-61TAG and pBK-TyrRS-2NPA (clone 1). A 1 L GMML culture containing 50 μg/mL kanamycin, 50 μg/mL ampicillin and 2-NPA (1 mM) was grown at 37°C to an OD600 of 0.5. The culture was then induced by adding arabinose to a final concentration of 0.05% and incubated with shaking at 30°C for 18 hrs. Cells were collected by centrifugation, and the His-tagged T4 lysozyme was purified by Ni-NTA affinity chromatography under native conditions according to the manufacturer's protocol (QIAexpressionist, Qiagen). Imidazole was removed by several rounds of concentration (Amicon Ultra-15, Millipore) and dilution in MES buffer A (MES buffer A: 50 mM MES, 20 mM NaCl, 1 mM EDTA, pH 6.3; MES buffer B: same as buffer A, except containing 500 mM NaCl). The protein was then purified by FPLC using a MonoS column (Amersham Pharmacia Biotech) with a gradient of 0 to 95% MES buffer B in MES buffer A over 35 minutes. The protein was then exhaustively dialyzed against PBS (3000 MWCO Slide-A-Lyzer Dialysis Cassette, Pierce) to give a final yield of 2.1 mg/L.
Determination of Protein Photocleavage Products
12.5 μg of protein T4L containing 2-NPA at position 82 was photolysed (>300 nm) in 1 mL of PBS at room temperature. Photolysis products were separated by SDS-PAGE (4-12%, Bis-Tris, Invitrogen) and stained (GelCode Blue, Pierce). Protein bands were excised and analyzed by LC/MS/MS after in-gel trypsin digestion, and characteristic molecular ions unambiguously showed that the lower molecular weight bands corresponded to the N- and C-terminal fragments of the parent protein.
Protein Photocleavage Quantitation
Optical densitometry was used to determine the extent of protein photocleavage. Briefly, a standard curve of optical density versus the amount of protein per band [as determined by the Bradford method (BCA Protein Assay Kit, Pierce)] was established. The protein samples were subjected to SDS-PAGE (4-12% Bis-Tris, Invitrogen), stained for 4 hours (GelCode Blue, Pierce), and destained for 2 hours with several washings with milliQ water. The optical density of each band was measured (ChemiDOC XRS, QuantityOne software). In order to quantify photocleavage, several 12.5 μg samples of T4L containing 2-NPA at position 61 were photolyzed (>300 nm) in 1 mL of 10-fold diluted PBS for varying amounts of time (0, 5, 10, 15, 30, and 60 minutes). The samples were concentrated to 25 μL, analyzed on SDS-PAGE and the optical density of the protein bands was measured in the same fashion as the standard curve samples.
Supplementary Material
Acknowledgments
We thank Eric Brustad, Dan Groff, Bill Webb, and M.G. Finn for assistance and helpful discussion. This work is supported by NIH grant R01 GM62159 (P.G.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young DD, Deiters A. Photochemical control of biological processes. Organic & Biomolecular Chemistry. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 2.Pelliccioli AP, Wirz J. Photoremovable protecting groups: reaction mechanisms and applications. Photochemical & Photobiological Sciences. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 3.England PM, Lester HA, Davidson N, Dougherty DA. Site-specific, photochemical proteolysis applied to ion channels in vivo. Proceedings of the National Academy of Sciences, USA. 1997;94:11025–11030. doi: 10.1073/pnas.94.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endo M, Nakayama K, Kaida Y, Majima T. Design and Synthesis of Photochemically Controllable Caspase-313. Angewandte Chemie International Edition. 2004;43:5643–5645. doi: 10.1002/anie.200460889. [DOI] [PubMed] [Google Scholar]
- 5.Pellois JP, Hahn ME, Muir TW. Simultaneous Triggering of Protein Activity and Fluorescence. J Am Chem Soc. 2004;126:7170–7171. doi: 10.1021/ja0499142. [DOI] [PubMed] [Google Scholar]
- 6.Pellois JP, Muir TW. A Ligation and Photorelease Strategy for the Temporal and Spatial Control of Protein Function in Living Cells. Angewandte Chemie International Edition. 2005;44:5713–5717. doi: 10.1002/anie.200501244. [DOI] [PubMed] [Google Scholar]
- 7.Bosques CJ, Imperiali B. Photolytic Control of Peptide Self-Assembly. J Am Chem Soc. 2003;125:7530–7531. doi: 10.1021/ja035360b. [DOI] [PubMed] [Google Scholar]
- 8.Walbert S, Pfleiderer W, Steiner UE. Photolabile Protecting Groups for Nucleosides: Mechanistic Studies of the 2-(2-Nitrophenyl)ethyl Group. Helvetica Chimica Acta. 2001;84:1601–1611. [Google Scholar]
- 9.Bühler S, Lagoja I, Giegrich H, Stengele KP, Pfleiderer W. New Types of Very Efficient Photolabile Protecting Groups Based upon the [2-(2-Nitrophenyl)propoxy]carbonyl (NPPOC) Moiety. Helvetica Chimica Acta. 2004;87:620–659. [Google Scholar]
- 10.Hasan A, Stengele KP, Giegrich H, Cornwell P, Isham KR, Sachleben RA, Pfleiderer W, Foote RS. Photolabile protecting groups for nucleosides: Synthesis and photodeprotection rates. Tetrahedron. 1997;53:4247–4264. [Google Scholar]
- 11.Bley F, Schaper K, Görner H. Photoprocesses of Molecules with 2-Nitrobenzyl Protecting Groups and Caged Organic Acids. Photochemistry and Photobiology. 2008;84:162–171. doi: 10.1111/j.1751-1097.2007.00215.x. [DOI] [PubMed] [Google Scholar]
- 12.Momotake A, Lindegger N, Niggli E, Barsotti RJ, Ellis-Davies GC. The nitrodibenzofuran chromophore: a new caging group for ultra-efficient photolysis in living cells. Nat Meth. 2006;3:35–40. doi: 10.1038/nmeth821. [DOI] [PubMed] [Google Scholar]
- 13.Ayyangar NR, Naik SN, Srinivasan KV. A novel synthesis of unsymmetrical azo aromatics inaccessible by diazo-coupling reaction. Tetrahedron Letters. 1989;30:7253–7256. [Google Scholar]
- 14.Wang L, Brock A, Herberich B, Schultz PG. Expanding the Genetic Code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 15.Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Habor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 17.Kuhn Hj, Braslavsky SE, Schmidt R. Chemical actinometry (IUPAC Technical Report) Pure Appl Chem. 2004;76:2129. [Google Scholar]
- 18.Xie J, Schultz PG. An expanding genetic code. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. A Genetically Encoded Boronate-Containing Amino Acid13. Angewandte Chemie International Edition. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. Generation of a Bacterium with a 21 Amino Acid Genetic Code. J Am Chem Soc. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


