Abstract
Systemic Lupus Erythematosus (SLE) disproportionately affects minorities, such as Hispanic-Americans. Prevalence of SLE is 3–5 times higher in Hispanic Americans (HA) than European derived populations, and have more active disease at the time of diagnosis, with more serious organ system involvement. HA is an admixed population, it is possible that there is an effect of admixture on the relative risk of disease. This admixture can create substantial increase of linkage disequilibrium (LD) in both magnitude and range, which can provide a unique opportunity for admixture mapping. Main objectives of this study are to (a) estimate hidden population structure in HA individuals; (b) estimate individual ancestry proportions and its impact on SLE risk; (c) assess impact of admixture on ITGAM association, a recently identified SLE susceptibility gene; and (d) estimate power of admixture mapping in HA. Our dataset contained 1,125 individuals, of whom 884 (657 SLE cases and 227 controls) were self classified as HA. Using 107 unlinked ancestry informative markers (AIMs) we estimated hidden population structure and individual ancestry in HA. Out of 5,671 possible pair-wise LD, 54% were statistically significant, indicating recent population admixture. The best fitted model for HA was a four population model with average ancestry of European (48%), American-Indian (40%), African (8%) and a fourth population (4%) with unknown ancestry. We also identified significant higher risk associated with American-Indian ancestry (OR=4.84, P=0.0001, 95%CI=2.14—10.95) on overall SLE. We showed that ITGAM is associated as a risk factor for SLE (OR= 2.06, P=8.74×10−5, 95%CI=1.44–2.97). This association is not affected by population substructure or admixture. We have demonstrated that HA have great potential and are an 3 appropriate population for admixture mapping. As expected, the case-only design is more powerful than case-control design, for any given admixture proportion or ancestry risk ratio.
Keywords: SLE, Association, Hispanics, Admixture mapping, Hispanic-American, Population structure
INTRODUCTION
Systemic Lupus Erythematosus (SLE) is a multifaceted autoimmune disease with varying morbidity and mortality. Incidence of SLE reportedly varies between different ethnic groups, especially in minority populations. In fact, Hispanic-Americans (HA) show 3 to 5 times higher prevalence of disease than European derived populations (1, 2). Additionally, SLE tends to affect non-white populations more severely than whites, in terms of disease activity and clinical manifestations, with more frequent and more severe organ system involvement, which leads towards a lower probability of survival (3, 4, 5, 6). These disparities may arise from the interaction between genetic and non-genetic (environmental, socioeconomic-demographic, cultural and behavioral) factors.
HA are an admixed population formed by recent admixture, which may influence the relative risk of many complex diseases. Some complex diseases, for example type-2 diabetes and obesity are thought to show increased prevalence in HA population. It is possible that a recent genetic admixture has a role in increased disease prevalence for SLE. Therefore, understanding the ancestral components of study subjects is very important in any genetic study.
The HA can be extremely useful in admixture mapping, a recently developed method for mapping genes for complex traits. The idea of admixture mapping is simple (7, 8), near disease causing genes, most of the affected individuals will be enriched with the ancestry that has higher risk of having disease. Therefore, by comparing the proportion of ancestral chromosomes in a specific location to the average ancestry across the genome in affected individuals, it is possible to detect a region that may contain disease causal variants. Admixture mapping is based on the inherent difference in allele frequencies due to sections of the genome segregating together due to LD (9) and has the advantage of requiring a smaller number of markers and samples. The advantage of working with recently admixed populations is that association requires lower number of samples than “pure” populations. This procedure strongly depends on allele differences between ancestral populations, and thus works better for samples where disease risk can be associated to a particular ancestral population.
The main objectives of this study are to (a) estimate the hidden population structure of HA individuals; (b) estimate the individual ancestry proportions and its impact on SLE risk; (c) assess the impact of admixture on ITGAM, recently identified SLE susceptibility gene; (d) estimate the power of admixture mapping in HA.
RESULTS
Demographics and characteristics of AIMs
Our HA sample included 657 independent SLE cases and 227 controls, where 102 were males and 782 were females. Samples were mainly of Mexican American descent (85% of cases and 56% controls), the remaining samples self identified as having ascent from Puerto Rico, and various other countries of Latin America (Table 1). There were no significant differences in allele frequency between countries of origin for cases and controls in our samples. We selected CEPH families with ancestry from northern and western Europe (CEU), and Yoruba in Ibadan, Nigeria (YRI), from the Hapmap v3 dataset (10), and Pima and Maya populations (referred as AI for American Indian) from the Human Genome Diversity Panel (HGDP) high-resolution genome-wide SNP dataset (11) as potential ancestral populations. We checked Fst, Gst values (12, 13), and allele frequency differences for these 3 populations and selected 107 autosomal markers as ancestral informative markers (AIMs). The mean ±se of Fst and Gst are 0.28±0.01 and 144.9±7.58, respectively. The inter-marker distance between 2 adjacent AIMs was at least 1 megabase (Mb). Markers with MAF<10% were not considered for AIMs detection
Table 1.
Demographic summary. Most of our samples were of Mexican descent, with a smaller proportion coming from Puerto Rico and other Latin-American countries.
| Status | Sample size | Gender | Country of origin | |||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Mexican American | Puerto Rico | Mexico | Other country | Unknown | ||
| Case | 657 | 66 | 591 | 498 | 59 | 59 | 17 | 24 |
| Control | 227 | 36 | 191 | 124 | 2 | 4 | -- | 97 |
| Total | 884 | 102 | 782 | 622 | 61 | 63 | 17 | 121 |
Estimation of the hidden population structure
Since HA are an admixed population, we expected to see population substructure in our samples. We performed Hardy-Weinberg equilibrium test (HWE) at all the AIMs, and 23 out of 107 (21.5%) AIMs among cases and 17 out of 107 (15.9%) among the controls showed significant deviation (P<0.05) from HWE, especially, towards excess homozygosity. Since population substructure leads to excess homozygosity, we should expect to see a trend toward the increase of homozygosity among the “null” markers (AIMs).
We employed pair-wise correlation (LD) between AIMs, and assessed for excess of association as a measure of recent admixture, as well as for the evidence of population substructure. Out of the 5671 possible pair-wise LD (r2) from 107 AIMs, 3066 (54%) were significant (P<0.05). When we include only those pair-wise LD where inter-marker distance >5Mb (N5Mb=541), 176 (32%) LD pairs were significant, thus demonstrating evidence of recent population admixture. Even more surprising was that 57% (Nsig=2890) of unlinked pairwise LD comparisons from different chromosomes (NUnlinked=5025) were significantly correlated; only 5% (NExpected=251) pair-wise LD between unlinked markers in populations with non-recent population admixture was expected.
Estimation of the individual ancestry proportions and its impact on SLE risk
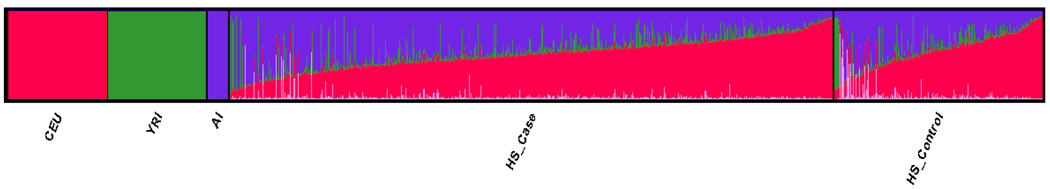
We found that HA admixture was best described by 4 population model (Figure 1). The case/control ancestry proportions of HA using population priors for the first 3 groups aligned with the CEU (49%/51%), AI (41%/35%), and YRI (8%/8%) (Table 2). An additional unknown “cluster 4” was detected with 3% and 5% ancestry proportions for cases and controls respectively (Table 2). It is noteworthy that ancestry estimates were consistent even without CEU, YRI, and AI as population priors (CEU=49%, AI=41%, YRI=6%, and cluster 4=4%), compared with overall proportions using priors (CEU=49%, AI=39%, YRI=8%, and cluster 4=4%) and were significantly correlated (Pearson’s r>99%). For individual ancestry proportions, there was a notable difference (41% vs. 35%, P=0.0001) between cases and controls in the AI Cluster (Figure 2).
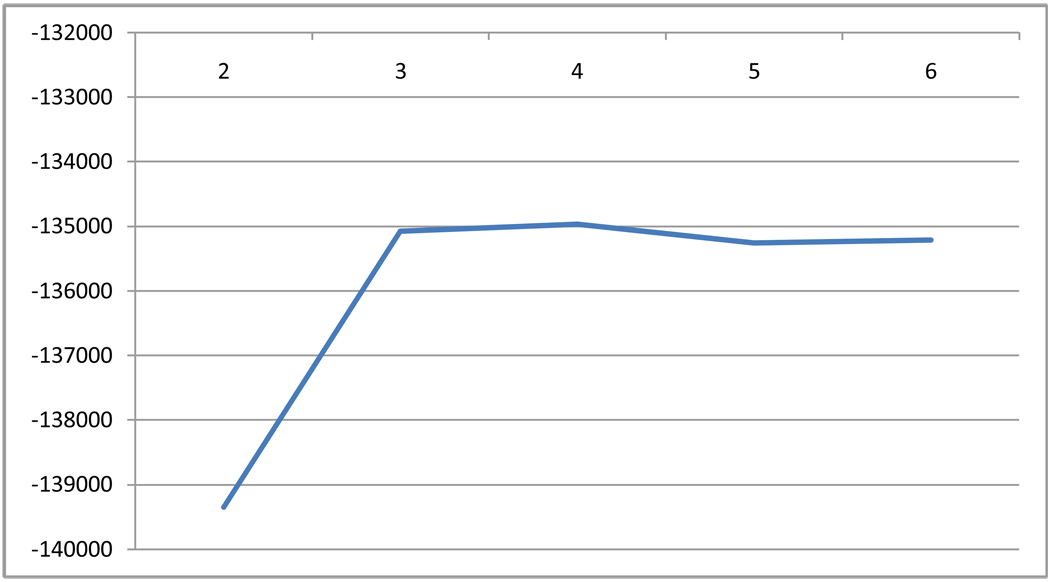
Figure 1.
Estimated likelihood for each number of population clusters inferred. Number of hidden populations (K) inferred is on the X-axis, and the estimated likelihood for each model is presented in the y-axis.
Table 2.
Estimated mean (±SE) proportions (%) of ancestry among HA under the best fitted (4 population) model .Cases and controls were treated independently to check for the differences of cluster membership between these 2 groups. Membership is compared with and without population priors; Pearson correlations were performed to align prior and no-prior clusters.
| Group (n) | CEU | AI | YRI | Cluster 4 | |
|---|---|---|---|---|---|
| With Prior | Case (n=657) | 51±0.71 | 35±0.64 | 8±0.47 | 5±0.37 |
| Control (n=227) | 49±0.61 | 41±0.64 | 8±0.4 | 3±0.2 | |
| All (n=884) | 49±0.64 | 39±0.64 | 8±0.44 | 4±0.27 | |
| No Prior | All (n=884) | 49±0.67 | 41±0.67 | 6±0.4 | 4±0.27 |
Figure 2.
Individual population structure for 884 Hispanic Americans is best described by 4 populations, CEU (red), YRI (green), AI (purple), and cluster 4(pink).
We also assessed whether individual ancestry is associated with overall disease phenotype by using logistic regression. We found that the AI ancestry was significantly associated with SLE risk (ORAI=4.84; 95%CI=2.14—10.95; P value=0.00018); cluster 4 although a small effect, was also significantly related to SLE (ORCluster4=0.03; 95% CI=0.01—0.2; P value=0.00028). Seldin et al. (14), also previously reported an increased OR of 7.94 for American Indian ancestry on SLE. However, CEU ancestry (ORCEU=0.44; 95% CI=0.2—1) and YRI ancestry (ORYRI=0.75; 95%CI 0.24—2.35) were not significantly associated with overall SLE risk.
Impact of admixture on ITGAM association
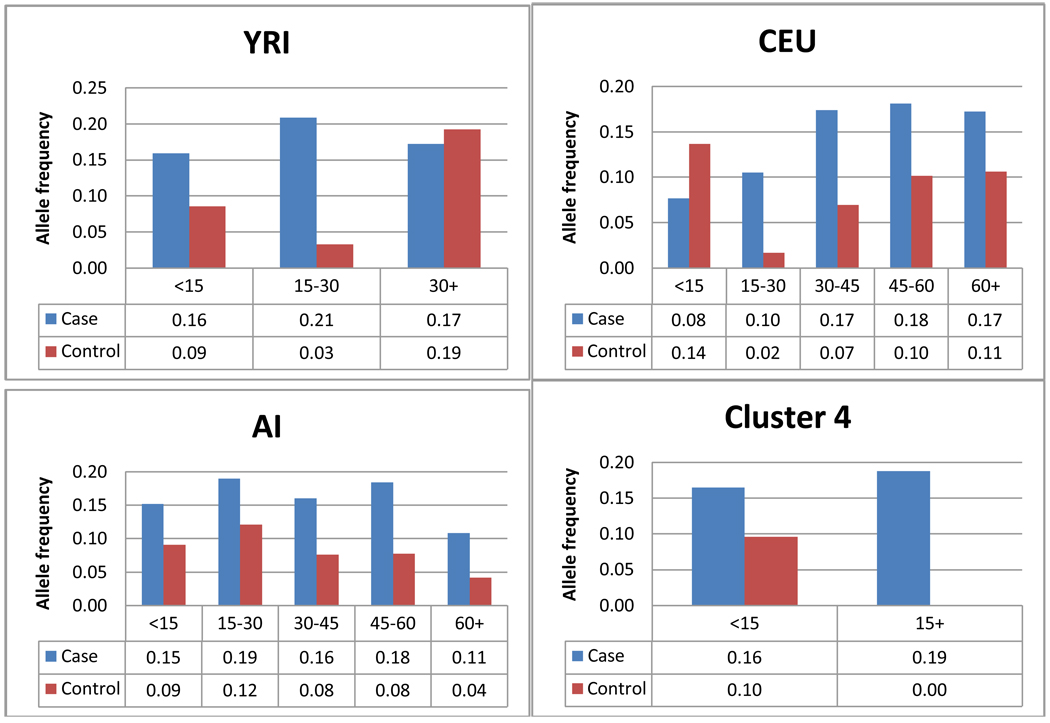
We selected ITGAM because of its known association to SLE (15). The most likely causal SNP was identified as exon-3 coding SNP rs1143679, that changes the amino acid arginine to histidine at position 77 (R77H). We found significant association of HA to SLE with uncorrected (P-value=8.74×10−5, OR= 2.06, 95%CI=1.44–2.97). We performed 3 correction procedures on the association of rs1143679: structured association test (SAT), covariate-adjusted, and ancestry matched corrections. The P-value for SAT was estimated = 1.56×10−4. In order to assess the effect of population structure on association of the selected marker we adjusted the association using the admixture proportion for each cluster (Table 3). P-values stemming from covariate-adjusted association were significant for all ancestries. Next we assessed whether there was a difference in allele frequencies across the distribution of ancestry proportion in cases and controls (Figure 3). We divided ancestry proportions into 5 intervals for CEU and AI (0–15%; 15%–30%; 30–45%; 45%–60%; and 60%+), 3 intervals for YRI (0–15%; 15%–30%; and 30%+), and 2 intervals for cluster 4 (<15%; 15%+). Intervals of different widths (10% and 25%) were also considered but results were similar (data not shown). Allele frequency for rs1143679, as expected, was different between cases and controls at each interval of ancestry proportion. Allele frequency for cases in CEU increased with ancestry proportion. On the contrary, allele frequency in AI cases decreased with ancestry proportion. Case allele frequency for YRI and cluster 4 were largely consistent throughout.
Table 3.
SNP rs11436769 association p-values corrected for populations structure using three correction methods: Structured association test, covariate adjusted and Ancestry matched. Four populations were identified, CEU, YRI, AI and cluster 4.
| Uncorrected | Corrected | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Structured Association Test | Admixture adjusted | Ancestry matched | ||||||||
| CEU | AI | YRI | Cluster 4 | CEU | AI | YRI | Cluster 4 | |||
| P-value | 8.74E-05 | 1.56E-04 | 4.94E-05 | 2.69E-05 | 6.88E-05 | 2.15E-04 | 5.91E-05 | 2.56E-05 | 5.44E-05 | 1.85E-04 |
| OR | 2.06 | -- | 2.13 | 2.20 | 2.10 | 2.00 | 2.02 | 2.11 | 2.02 | 1.91 |
| 95%CI | 1.44 –2.97 | -- | 1.48–3.08 | 1.52–3.19 | 1.46–3.04 | 1.38–2.88 | 1.41–2.90 | 1.47–3.02 | 1.41–2.89 | 1.34–2.72 |
Figure 3.
Ancestry matched association to rs1143679. Bins were created for intervals of 15% for all ancestries, YRI was limited at 30%+ and cluster 4 was limited at 15%+. Allele frequencies are presented below each category.
We tested whether there was a difference in allele frequencies distribution across strata among all ancestry proportions and found that no heterogeneity present (CEU X2df=3 P-value=0.3; AI X2df=3 P-value= 0.89; YRI X2df=3 P-value= 0.17; cluster 4 X2df=3 P-value= 0.43). All three methods of association correction confirmed true association of rs1143679 in HA.
Power of admixture mapping in HA
We compared the number of samples required to detect association with a P-value of 10−5, with 80% statistical power in the case-only and case-control designs assuming complete extraction of individual ancestry. We found that for populations with higher degree of admixture required fewer individuals than less admixed populations. This number also decreases with increasing risk ratios. As expected, similar with previous studies (16,17), the number of samples required to achieve the same statistical power is lower in case-only designs (Table 4).
Table 4.
Sample size required for p = 10−5, power = 80%, affected only and case-control design (1:1).
| Locus specific ancestry risk | Affected Only | Case-Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Admixture Proportion | Admixture Proportion | |||||||||
| 10% | 20% | 30% | 40% | 50% | 10% | 20% | 30% | 40% | 50% | |
| 1.25 | 11638 | 9806 | 7471 | 6537 | 6276 | 46551 | 39223 | 29884 | 26149 | 25103 |
| 1.5 | 3525 | 1983 | 1511 | 1322 | 1269 | 14099 | 7931 | 6042 | 5287 | 5076 |
| 1.75 | 1850 | 1041 | 793 | 694 | 666 | 7401 | 4163 | 3172 | 2776 | 2665 |
| 2 | 1206 | 678 | 517 | 452 | 434 | 4824 | 2714 | 2068 | 1809 | 1737 |
DISCUSSION
Our analysis demonstrated that Hispanic Americans are a highly admixed population. Substantial population structure was evident when estimating both HWE within AIMs as well as LD between unlinked AIMs. Excess homozygosity in markers (21% of markers in cases and 16% of markers in controls) reinforces the conclusion that HA are an admixed population. There was a high proportion of LD in 29% of AIMs with inter-marker distances >5mB within chromosomes, there was also 65% significant LD in markers across chromosomes, and 57% significant LD between AIMs in different chromosomes.
HA admixture was best characterized by a 4 population model, the largest being closely related to the CEU group and the AI group. The 2 other clusters identified had less than 10% ancestry that correspond to the YRI cluster and an unidentified cluster (Figure 1). We believe this fourth cluster may be related to the different origins of African slaves in Latin-America. Indeed it is suggested that individuals of Spanish-speaking origin might have an Arabic or North-African component (18, 19). On close comparison of the ancestry proportion for K=3 and K=4, the proportion of CEU and AI ancestry are still the largest components of HA, and are very similar in estimates to the K=4 estimates. It is possible that the fourth cluster is related to substructure on the African ancestry component given the dual origin of the African origin slaves, namely Yoruban and Bantu (18, 19). It is also possible that there is an Asian population component that we have not taken into account. One unaddressed possibility is that the AI population was itself admixed, which would affect the estimation of the admixture proportion of the HA.
HA are among the most admixed population existing today, however, individual estimates of different ancestry vary substantially (Table 2). This feature makes it even more important to account for population structure as a confounding factor for disease association. The sign of the covariate estimates for the logistic regression model for risk of SLE indicated that there was a significant risk of SLE associated to the AI cluster, whereas there is an opposite effect coming from the fourth cluster. CEU and YRI clusters have SLE risk ratio close to 1 in our HA population. However, until a large-scale study to characterize admixture of AI populations is done it remains difficult to determine whether the Pima and Maya are the best representative ancestral populations to estimate HA admixture.
We chose ITGAM to test the effect of admixture on disease association. We used 3 correction methods to address the effect of population stratification. The ITGAM SNP, rs1143679, is already known to be associated with SLE (15). We showed that this association remained significant with SLE by all correction methods. For ancestry matched correction, we noticed that the allele frequency of cases increased with CEU ancestry proportion, indicating that the risk locus for rs1143679 is associated to CEU ancestry. The same allele frequency decreased for AI ancestry. Thus, admixture corrections to association P-values could shed light on locus-specific disease origin of risk alleles to a given ancestry.
One very important characteristic to consider when dealing with admixed populations is the reduced number of samples required to find significant effects on affected only and case-control studies (Table 4). For any statistical power/risk ratio combination, the number of samples required for a case-only or a case-control study is significantly different under different admixture proportions. This is because the case-only test compares the local ancestry proportion (which is moderately variable) with the genome-average ancestry which is known more accurately. On the other hand, case-control test compares 2 local ancestry proportions (1 in case and 1 from controls), both of which are variable. For example, with a 2-fold increased risk ratio, using a case-only design in a population with 50% admixture proportion, we would need 434 cases, whereas if admixture proportion were 10% then 1,206 cases are needed (Table 4). On the other hand, with a case-control (1:1 ratio) design, the number of required samples, for the same combination of admixture proportion and locus risk, increases four-fold (1,737 for 50% admixture and 4,824 for 10% admixture). These results are similar to other previous studies (16,17). However, these sample sizes are substantially different from the results that Tian et al. (2007) (20) found by simulation. This difference could be due to the way the simulations were performed. In their simulations Tian et al (20) assumed the actual information content extracted from his used AIMs, whereas the other studies (16,17), including the present study, assumed that the AIMs were completely informative about extracting the ancestry information.
The present study suffers from some shortcoming. The precise estimation of an individual ancestry is highly dependent on (i) choice and size of ancestral populations, (ii) number of AIMs, and (iii) precise estimation of parental allele frequencies. If the size of the parental population is small, then the precise estimation of allele frequencies from parental populations is not accurate. Although CEU and YRI samples were not small, sample size of AI was small (N=24) for getting accurate estimates. Also, we only used 107 autosomal AIMs, so we might have not estimated the precise individual ancestry for each individual.
However, we began this study to investigate the degree of admixture among self-reported HA individuals. Our objective was to gain a better understanding of the influence of ancestry on genetics of SLE by estimating the population structure of HA populations with SLE as a group and as individuals. We have found that genetic ancestry proportions can vary significantly within HA group. Using a panel of Ancestry informative markers (AIMs) we characterized the admixture pattern, dynamics, extent of LD, and relationships between individual ancestry and SLE within our Hispanic sample. We assessed SLE association with ITGAM SNP, rs1143679, and evaluated the effect of population substructure on observed association. We also studied the feasibility of admixture mapping in HA and demonstrated great power in case-only design.
Recent discoveries in gene identification in complex traits, for example, non-diabetic end-stage renal disease (21), and prostate cancer (22) in African Americans (AA), are provocative and can be a proof of principle for admixture mapping. This was possible due to availability of panels of highly informative AIMs for AA. Panels of AIMs are now also available for conducting whole-genome admixture mapping in HA (23, 24, 25). Therefore, our HA are a valuable resource that has a great potential for SLE gene identification using whole-genome admixture mapping approach.
MATERIAL AND METHODS
Subjects
We used 4 prior populations, CEU (NCEU=109) and YRI (NYRI=108) unrelated samples from Hapmap (10), and Pima and Mayas from the Human Genome Diversity Panel (HGDP) high-resolution genome-wide SNP dataset (NAI=24) (11). Our study included 884 unrelated Hispanic individuals. There were 657 individuals SLE cases, defined as fulfilling at least 4 of the 11 revised ACR disease criteria (26, 27), and 227 unaffected controls. Medical records were obtained for all SLE participants. All indentifying information was stripped and approval was obtained from the respective Internal Review Boards and the collaborating institutions. Informed consent was obtained for all individuals. DNA extraction and quality control has been described elsewhere (15).
Genotyping
Cases and controls were genotyped in Illumina custom designed high-density array. Our study was a part of a large-scale genotyping project shared by multiples independent investigators as described elsewhere (15, 28). Alleles with MAF < 10% were eliminated as were markers with less than 90% call rate. All markers were set on the plus strand to be consistent about the alleles across among the ancestral populations and the study population. Markers allele frequency and missingness was calculated with PLINK (29).
Selection of AIMs
For each AIM, we estimated the marker information by both Fst (12) and Gst (13) values. Fst is defined as the proportion of the total genetic variance in a subpopulation relative to the total genetic variance (12). Gst (13) is defined as gene differentiation relative to the total population. These values were calculated in R and FSTAT (30). We selected 107 AIMs, with mean Fst =0.3 ±0.1 (ranges from 0.04 to 0.59) and mean Gst=145±78.4 (ranges from 19.3 to 387.9). The minimum inter-marker distance between AIMs was at least 1Mb. In Caucasian populations, the extent of LD is generally limited to regions smaller than 100 kb (31, 32), which would be still shorter in Africans (and expected to be in American-Indians). Therefore, the correlations that arise between linked markers in Hispanic-Americans are likely due to the result of recent admixture (33).
Population structure analysis
We calculated allele frequencies and counts using PLINK (29), with these values we calculated HWE and estimated excess of homozigosity (34). We detected population structure using AIMs (35, 36), described as the markers with the highest ability to discriminate between populations. Data sets were merged using PLINK (29). We calculated and plotted LD of these 107 markers with R library GenABEL (37)
Population structure was analyzed with STRUCTURE (36) which implements a model-based clustering method for inferring population sub-structure using AIMs. Assumed populations were K=2 to 6. The best fitting K was assessed using the criteria from Evanno (38).
Detection and Correction of Association
Case-control allelic association was carried out in PLINK. We used each detected cluster as a covariate for structural corrections. We recorded odds-ratios and p-values for all markers with and without covariate.
We tested each cluster as a logistic covariate for association to SLE to determine the relative risk of a particular ethnicity to SLE. Interactions between SNPs and each cluster were assessed with PLINK (29). We tested the differences between minor allele frequency of rs1143679 for 5 intervals of ancestral proportion (0%–15%; 15%–30%; 30%–45%; 45%–60% and 60%+) in CEU and AI; we used only the 3 first intervals for YRI and 2 for cluster 4. Heterogeneity was tested using Chi-squared test using REVMAN (38). Corrections to association P-values were performed by structure association in STRUCTURE, with covariates as case-control logistic regression in PLINK (29) and SAS (40), and meta-analysis was performed by using REVMAN(39).
Power analysis
To estimate the relative efficiency of the admixture mapping in this Hispanic population, we conducted an analytical power analysis, for case-only and case-control design at a single locus under different admixture proportions varying from 10%–50%. For the sample size estimation, we have used the expected information calculations presented in Table 1 in Hoggart et al. (16). We used the expected information from two gametes with admixture proportion, θ, where the affected-only expected information is: and the case-control (with case-control ratio p/(1−p)) expected information is:
Therefore, by definition, for a case-control ratio of 1:1, the expected information for case-control design is 0.25 times smaller than the case-only design. The required sample size under a specific type I error and a locus specific ancestry risk λ (we used λ=1.25 to 2) the Hoggart’s equation 4 (in ref. 16) is:
where, n is the number of samples required to obtain Z1−b (0.84) of 80% power, with a significant level Z1−a (4.26) of 10−5, and V0 is the expected information from two gametes. It is important to note that in our power calculations we did not use simulations to incorporate the effect of coverage of the AIMs as done by Tian et al (20). Also, we are estimating power at a single locus assuming a “two population” admixture model.
AKNOWLEDGEMENTS
We wish to thank the patients and their families for their cooperation and blood samples. We also like to acknowledge the support from LFRR staffs, especially, Gail Bruner, Jennifer Kelly, Dr. Jashua Ojwang, Dr. Ken Kaufman for conducting a large scale genotyping project from where the current samples were genotyped. Additionally we would like to thank Dr Judith James and Dr Gary Gilkerson for providing genotypic data and some Hispanic samples used in this study. This study was made possible by funding from RO1A1063622, PO1AR049084, P30AR053483 and P20RR020143.
REFERENCES
- 1.Alarcon GS, McGin G, Jr, Petri M, Reveille JD, Ramsey-Goldman R, Kimberly RP for the PROFILE Study Group. Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus. 2002;11:95–101. doi: 10.1191/0961203302lu155oa. [DOI] [PubMed] [Google Scholar]
- 2.McKeigue PM. Prospects for Admixture Mapping of Complex Traits. Am J Hum Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nature Reviews Genetics. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty R, Weiss KM. Frequencies of complex diseases in hybrid populations. Am J Phys Anthropol. 1986;70:489–503. doi: 10.1002/ajpa.1330700408. [DOI] [PubMed] [Google Scholar]
- 5.Escalante A, Rincon ID. Epidemiology and impact on rheumatic disorders in the US Hispanic population. Curr Opin Rheumatol. 2001;13:104–110. doi: 10.1097/00002281-200103000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Reveille JD, Moulds JM, Ahn C, Friedman AW, Baethge B, Roseman J, et al. Systemic Lupus Erythematosus in three ethnic groups. I. The effects of HLA Class II, C4, and CR1 alleles, socioeconomic factors, and ethnicity at disease onset. Arthritis Rheum. 1998;41:1161–1172. doi: 10.1002/1529-0131(199807)41:7<1161::AID-ART4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Molina JF, Molina J, Garcia G, Gharavi AE, Wilson WA, Espinoza LR. Ethnic differences in the clinical expression of Systemic Lupus Erythematosus: A comparative study between African-Americans and Latin Americans. Lupus. 1997;6:63–67. doi: 10.1177/096120339700600109. [DOI] [PubMed] [Google Scholar]
- 8.Petri M, Perez-Gutthann S, Longenecker JC, Hochberg M. Morbidity of Systemic Lupus Erythematosus: role of race and socioeconomic status. Am J Med. 1991;91:345–353. doi: 10.1016/0002-9343(91)90151-m. [DOI] [PubMed] [Google Scholar]
- 9.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 11.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 12.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 13.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 14.Seldin MF, Qi L, Scherbarth HR, Tian C, Ransom M, Silva G, et al. Amerindian ancestry in Argentina is associated with increased risk for Systemic Lupus Erythematosus. Genes and Immunity. 2008;9:389–393. doi: 10.1038/gene.2008.25. [DOI] [PubMed] [Google Scholar]
- 15.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 16.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montana G, Pritchard JK. Statistical Tests for Admixture Mapping with Case-Control and Cases-Only Data. Am J Hum Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sans M. Admixture Studies in Latin America: From the 20th to the 21th century. Human Biology. 2000;72:155–177. [PubMed] [Google Scholar]
- 19.Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 20.Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, et al. A Genomewide Single-Nucleotide Plymorphism Panel for Mexican American Admixture Mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Shaad Y, Man L, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Gen. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiman CA, Patterson V, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Gen. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, Silva G, Belmont JW, Hanson RL, Knowler WC, Gregersen PK, Ballinger DG, Seldin MF. A genomewide single–nucleotide–polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, et al. A Genomewide Admixture Map for Latino Populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, et al. A Genomewide Admixture Mapping Panel for Hispanic/Latino Populations. AMJHG. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of Systemic Lupus Erythematosus Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 27.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of Systemic Lupus Erythematosus. Arthritis Rheum. 1997;40:1725–1734. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 28.Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN). Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown Kf, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goudet J. FSTAT (Version 1.2): A computer program to calculate F- statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. Structure of Haplotype Blocks in the Human Genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature. 411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 33.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, et al. Population stratification confounds genetic association of studies among Latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falush D, Stephens M, Pritchard JK. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard J, Stephens M, Donelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 38.Evanno G, Regnaut S, J Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 39.Review Manager (RevMan) [Computer program] Version 5.0. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration. 2008 [Google Scholar]
- 40.SAS [Computer program] Version 9.1. SAS Institute. 2008:23. [Google Scholar]