Abstract
Genetic factors influence susceptibility to systemic lupus erythematosus (SLE). A recent family-based analysis in Caucasian and Chinese populations provided evidence for association of single-nucleotide polymorphisms (SNPs) in the complement receptor 2 (CR2/CD21) gene with SLE. Here we confirmed this result in a case-control analysis of an independent European-derived population including 2084 patients with SLE and 2853 healthy controls. A haplotype formed by the minor alleles of three CR2 SNPs (rs1048971, rs17615, rs4308977) showed significant association with decreased risk of SLE (30.4% in cases vs. 32.6% in controls, P = 0.016, OR = 0.90 [0.82-0.98]). Two of these SNPs are in exon 10, directly 5′ of an alternatively spliced exon preferentially expressed in follicular dendritic cells (FDC), and the third is in the alternatively spliced exon. Effects of these SNPs as well as a fourth SNP in exon 11 (rs17616) on alternative splicing were evaluated. We found that the minor alleles of these SNPs decreased splicing efficiency of exon 11 both in vitro and ex vivo. These findings further implicate CR2 in the pathogenesis of SLE and suggest that CR2 variants alter the maintenance of tolerance and autoantibody production in the secondary lymphoid tissues where B cells and FDCs interact.
Keywords: Alternative splicing, systemic lupus erythematosus, complement receptors, single-nucleotide polymorphisms, B cells, follicular dendritic cells
Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune disease affecting multiple organ systems that is characterized by circulating autoantibodies to nuclear antigens. Several well conducted studies have shown that there is a genetic component to lupus susceptibility and have specifically identified a number of susceptibility loci which are currently under investigation [reviewed in 1]. We recently identified a novel locus associated with lupus susceptibility at 1q32.2, which contains the complement receptor 2 (CR2) gene, and demonstrated the preferential transmission of a common three-single-nucleotide polymorphism (SNP) CR2 haplotype (rs3813946, rs1048971, rs17615) to affected offspring in Caucasian and Chinese lupus simplex families (P = 0.00001).2 Cr2 is also a major positional candidate gene in the murine Sle1c lupus susceptibility interval based on structural and functional alterations in its protein products.3 However, a recent case-control study in 509 cases and 964 controls of Japanese descent which included 7 CR2 SNPs including rs3813946, rs1048971, and rs17615 did not reveal a significant association of the analyzed CR2 SNPs with SLE.4
Human complement receptor 2 is encoded by a single gene containing 20 exons which is located at chromosome 1q32.2. The mature protein, expressed primarily on mature B cells and follicular dendritic cells (FDCs), exists as two known isoforms consisting of 15 or 16 repeating subunits termed short consensus repeats (SCRs) which form the extracellular domain. The two isoforms result from alternative splicing of exon 11 in the primary transcript.5-7 The factors that regulate the alternative splicing of this exon and the functional relevance of the different splice isoforms are not known, although the differential expression of the long isoform on follicular dendritic cells suggests a functional effect.8 CR2 binds C3d degradation products covalently bound to antigen in the process of complement activation, Epstein-Barr virus (EBV),9 the immunomodulatory protein CD23,10 and IFN-α11 Its cell- and stage-specific expression is controlled by proximal promoter sequences acting in conjunction with an intronic silencer.12-17 The results of several studies suggest that CR2 plays a major role in immunity [reviewed in 18].
SNP1 (rs3813946) of the lupus-associated CR2 haplotype, which is located in the 5′ untranslated region, modulates the transcriptional activity of CR2.2 It is not known whether SNP2 (rs1048971) and SNP3 (rs17615) of this haplotype, located in exon 10, alter gene function. However, since they are located directly upstream of an alternatively spliced cassette exon, they may affect alternative splicing of this exon, which could result in altered generation of the long 16 SCR isoform of CR2 relative to the short 15 SCR isoform. In addition, two tightly linked SNPs located within exon 11 (rs4308977 and rs17616) may also modulate splicing efficiency of this exon. We confirm here our previous association of rs3813946, rs1048971, and rs17615 with lupus susceptibility, extend our findings to show association of two additional CR2 SNPs with SLE, identify a protective CR2 haplotype containing these SNPs, and demonstrate the effects of the SNPs in exons 10 and 11 on the inclusion of exon 11 in the mature RNA transcript.
Results
Confirmation and fine mapping of genetic association of CR2 SNPs with SLE susceptibility
We previously reported association of a common haplotype containing the major allele of three CR2 SNPs [rs3813946 (+21, 5′UTR), rs1048971 (L592L, exon 10), and rs17615 (S639N, exon 10)] with risk of SLE in 258 Caucasian and 142 Chinese simplex families.2 To confirm and fine map this genetic association, an independent sample of 2084 SLE patients (including 519 with renal involvement and 1136 without) and 2853 healthy controls of European descent were genotyped using twelve SNPs spanning a 39 kb region of the CR2 gene from 0.6 kb upstream to 2.8 kb downstream of the gene (Figure 1). In addition to the five SNPs we previously tested [rs3813946, rs1567190 (haplotype-tagging SNP in intron 1), rs1048971, rs17615 and rs6540433 (A1061E in exon 18)], we selected five additional potentially functional SNPs [rs12135588 (at -616 in the promoter region), rs2063143 (intronic enhancer located in intron 2), rs4308977 (S663P in exon 11), rs9429940 (3′UTR, exon 20) and rs17045761 (3′ downstream, putative transcription factor binding site)], as well as two additional haplotype-tagging SNPs (rs12021671 in intron 18 and rs4618971 in 3′ downstream region). Single locus analysis showed allelic association of increased risk for SLE with the major alleles of three SNPs (rs1048971, P=0.025, OR=1.10 [1.01-1.20]; rs17615, P=0.010, OR=1.12 [1.03-1.22]; rs4308977, P=0.020, OR=1.11 [1.02-1.21]) (Table 1).
Figure 1.
CR2 SNP locations and haplotype blocks. The CR2 gene is composed of 20 exons, 19 of which are constitutively spliced into the mature RNA transcript,7 with exon 11 being an alternative cassette exon expressed primarily on FDCs.8 Twelve SNPs in the promoter region, exon 1 (5′UTR), intron 1, intron 2, exon 10, exon 11, exon 18, intron 18, exon 20 (3′UTR), and the 3′ downstream region were genotyped across the 39 kb region spanning the CR2 gene. Also shown is rs17616 (*) in exon 11, which was not genotyped but is in strong LD with rs4308977 (pairwise r2 = 0.89). R2 values of each SNP pair are depicted. Two haplotype blocks were constructed based on the strength of LD. The five SNPs used in the haplotypic association test (rs3813946, rs1048971, rs17615, rs4308977, rs6540433), enclosed in single line text boxes, are located in block 1. The SNPs in exon 10 (rs1048971, rs17615) and exon 11 (rs4308977 and rs17616) that were assessed for their effects on alternative splicing are in red font.
Table 1.
Allellic association between CR2 SNPs and SLE in European-derived samples
| # | SNP | Position a | Relative Distance(kb) |
Functional Region |
Allele b |
Control (n=2853) |
SLE (n=2084) |
Non-LN (n=1136) |
LN (n=519) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | MAFC | MAF | P | OR [95% CI] | Pc | MAF | P | OR [95% CI] | Pc | MAF | P | OR [95% CI] | Pc | |||||
|
|
|
|
|
|
|
||||||||||||||
| 1 | rs12135588 | 2.06E+08 | 0.00 | Promotor | A | G | 0.034 | 0.031 | 0.541 | 1.07 [0.86-1.34] | 0.526 | 0.030 | 0.456 | 1.11 [0.84-1.47] | 0.643 | 0.037 | 0.623 | 1.09 [0.77-1.56] | 0.857 |
| 2 | rs3813946 * | 2.06E+08 | 0.64 | 5′UTR | A | G | 0.187 | 0.177 | 0.217 | 1.07 [0.96-1.19] | 0.144 | 0.187 | 0.985 | 1.00 [0.88-1.14] | 0.857 | 0.160 | 0.044 | 1.21 [1.00-1.46] | 0.052 |
| 3 | rs1567190 * | 2.06E+08 | 8.10 | Intron1 | A | G | 0.490 | 0.504 | 0.155 | 1.06 [0.98-1.15] | 0.122 | 0.507 | 0.176 | 1.07 [0.97-1.18] | 0.179 | 0.496 | 0.709 | 1.03 [0.90-1.17] | 0.857 |
| 4 | rs2063143 * | 2.06E+08 | 6.05 | Intron2 | G | A | 0.213 | 0.200 | 0.114 | 1.08 [0.98-1.20] | 0.089 | 0.204 | 0.399 | 1.05 [0.93-1.19] | 0.382 | 0.191 | 0.109 | 1.15 [0.97-1.36] | 0.098 |
| 5 | rs1048971 * | 2.06E +08 | 4.48 | Exon10 (L592L) | G | A | 0.369 | 0.347 | 0.025 | 1.10 [1.01-1.20] | 0.024 | 0.339 | 0.013 | 1.14 [1.03-1.26] | 0.018 | 0.353 | 0.319 | 1.07 [0.93-1.23] | 0.306 |
| 6 | rs17615 * | 2.06E +08 | 0.14 | Exon10 (S639N) | G | A | 0.328 | 0.304 | 0.010 | 1.12 [1.03-1.22] | 0.015 | 0.300 | 0.015 | 1.14 [1.03-1.27] | 0.020 | 0.305 | 0.155 | 1.11 [0.96-1.28] | 0.232 |
| 7 | rs4308977 § | 2.06E+08 | 0.44 | Exon10a (S663P) | A | G | 0.326 | 0.304 | 0.020 | 1.11 [1.02-1.21] | 0.029 | 0.301 | 0.027 | 1.13 [1.01-1.25] | 0.037 | 0.306 | 0.207 | 1.10 [0.95-1.27] | 0.273 |
| rs17616 d* | 2.06E+08 | 0.03 | Exon10a (R671H) | G | A | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| 8 | rs6540433 §* | 2.06E+08 | 6.47 | E xon18 (A 1062E ) | A | C | 0.158 | 0.168 | 0.170 | 1.08 [0.97-1.20] | 0.146 | 0.177 | 0.041 | 1.14 [1.01-1.30] | 0.046 | 0.163 | 0.704 | 1.04 [0.86-1.24] | 0.750 |
| 9 | rs12021671 § | 2.06E+08 | 4.75 | Intron18 | G | A | 0.362 | 0.362 | 0.972 | 1.00 [0.92-1.09] | 1.000 | 0.360 | 0.862 | 1.01 [0.91-1.12] | 1.000 | 0.367 | 0.767 | 1.02 [0.89-1.17] | 0.750 |
| 10 | rs9429940 § | 2.06E+08 | 4.77 | 3′UTR | G | A | 0.120 | 0.125 | 0.421 | 1.05 [0.93-1.19] | 0.588 | 0.131 | 0.173 | 1.11 [0.96-1.28] | 0.216 | 0.125 | 0.669 | 1.04 [0.85-1.28] | 0.750 |
| 11 | rs17045761 | 2.06E+08 | 0.44 | 3′ downstream | G | A | 0.047 | 0.051 | 0.352 | 1.09 [0.91-1.31] | 0.306 | 0.047 | 0.950 | 1.01 [0.80-1.27] | 0.857 | 0.051 | 0.554 | 1.10 [0.81-1.48] | 0.556 |
| 12 | rs4618971 § | 2.06E+08 | 2.71 | 3′ downstream | A | G | 0.222 | 0.228 | 0.465 | 1.04 [0.94-1.14] | 0.462 | 0.231 | 0.401 | 1.05 [0.94-1.18] | 0.556 | 0.230 | 0.555 | 1.05 [0.90-1.23] | 0.700 |
NCBI build 36.
1 represents the major allele and 2 is the minor allele.
Minor allele frequency.
No genotyping data.
Tag SNP.
Genotyped in previous study. 2
P values were calculated using Pearson’s chi square test and corrected using perm utation test ( Pc). Significant level is set at < 0.05.
The Hardy-Weinberg equilibrium Pvalue for each SNP is >0.01.
Our previous family-based study in which the three SNP haplotype containing the major allele of rs3813946, rs1048971 and rs17615 increased the risk for SLE in European-derived populations revealed a stronger association signal in the non-LN subset of SLE patients.2 Seeking confirmation in the current analysis, the SLE patients for whom clinical data was available were stratified by the presence or absence of lupus nephritis (LN)19 to assess the strength of association based on renal involvement. In the non-LN subset (n=1136), increased risk of disease was associated with the major alleles of rs1048971 (Pc=0.018, OR=1.14 [1.03-1.26]), rs17615 (Pc=0.020, OR=1.14 [1.03-1.27]) and rs4308977 (Pc=0.037, OR=1.13 [1.01-1.25]) and the minor allele of rs6540433 (Pc=0.046, OR=1.14 [1.01-1.30]) (Table 1). In the LN subset (n=519), only the major allele of rs3813946 appeared to be associated with increased risk of disease (P=0.044, OR=1.21 [1.00-1.46]); however, this association was no longer statistically significant after correction for the multiple tests performed (Pc=0.052) (Table 1).
Construction of SLE-associated CR2 haplotypes
SNP haplotypes were then constructed based on genotyping of the SNPs currently and previously2 associated with SLE (rs3813946, rs1048971, rs17615, rs4308977 and rs6540433), which resided in one 25kb block exhibiting linkage disequilibrium (Figure 1). As shown in Table 2, after stratifying SLE patients into LN and non-LN subsets, significant overall association of SNP haplotypes was observed only in the non-LN subset. Furthermore, in the non-LN subset, although rs3813946 did not show significant allelic association with SLE (Table 1), the three groups of SNP haplotypes containing this locus (G2, P=0.014; G3, P=0.015; G5, P=0.017) showed a stronger association than the two groups without this locus (G1, P=0.0496; G4, P=0.053).
Table 2.
Haplotypic association of CR2 SNPs conferring risk for SLE
| Group |
SNP |
Omnibus P |
||||||
|---|---|---|---|---|---|---|---|---|
| rs3813946 | rs1048971 | rs17615 | rs4308977 | rs6540433 | SLE | Non-LN | LN | |
| G1 | + | + | + | 0.054 | 0.0496 | 0.369 | ||
| G2 | + | + | + | 0.073 | 0.014 | 0.280 | ||
| G3 | + | + | + | + | 0.093 | 0.015 | 0.336 | |
| G4 | + | + | + | + | 0.095 | 0.053 | 0.555 | |
| G5 | + | + | + | + | + | 0.136 | 0.017 | 0.468 |
| Haplotype | Allele | FreqC | Freq | PS | OR | PM | Freq | PS | OR | PM | Freq | PS | OR | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||||
| G1-1 | A | A | G | 0.326 | 0.304 | 0.016 | 0.90[0.82-0.98] | 0.300 | 0.022 | 0.89[0.80-0.98] | 0.305 | 0.183 | 0.90[0.78-1.04] | |||
| G2-1 | A | A | A | 0.129 | 0.116 | 0.043 | 0.88[0.78-0.99 ] | 0.030 | 0.104 | 0.002 | 0.79[0.67-0.92] | 0.001 | 0.132 | 0.848 | 1.02[0.84-1.24] | |
| G2-2 | G | A | A | 0.198 | 0.188 | 0.197 | 0.93[0.84-1.03] | 0.196 | 0.757 | 0.98[0.86-1.10] | 0.173 | 0.066 | 0.85[0.71-1.01] | |||
| G1-2 | G | G | A | 0.633 | 0.655 | 0.023 | 1.10[1.02-1.20] | 0.662 | 0.014 | 1.14[1.03-1.26] | 0.649 | 0.307 | 1.08[0.94-1.24] | |||
| G2-3 | A | G | G | 0.631 | 0.653 | 0.023 | 1.10[1.02-1.20] | 0.005 | 0.660 | 0.013 | 1.14[1.03-1.26] | 0.002 | 0.647 | 0.307 | 1.08[0.94-1.24] | |
| G1-3 | A | G | A | 0.041 | 0.042 | 0.872 | 1.02[0.83-1.24] | 0.038 | 0.591 | 0.94[0.73-1.21] | 0.045 | 0.512 | 1.12[0.81-1.54] | |||
| G2-4 | A | A | G | 0.041 | 0.043 | 0.659 | 1.05[0.86-1.27] | 0.040 | 0.718 | 0.96[0.75-1.22] | 0.047 | 0.395 | 1.15[0.84-1.57] | |||
SNPs used to construct haplotypes are depicted as +.
Omnibus P represents the overall significance using all possible haplotypes.
FreqC represents the haplotype frequency of controls.
PS represents the significance of each haplotype.
PM is the meta-analysis P value resulting from combining the present case-control test and the previous TDT [In 258 SLE trios, G2-1: Transmitted:Untransmitted (T:U)=33.78:40.76, G2-3: T:U= 110.5:79.53; in 151 non-LN trios, G2-1: T:U=21.47:28.03, G2-3: T:U=67.05:41.06].
Haplotype group G1, constructed by three exonic SNPs (rs1048971, rs17615 and rs4308977), was comprised of two common haplotypes [G1-1, haplotype frequency in controls (FreqC) = 0.326; G1-2, FreqC = 0.633] and one rare haplotype (G1-3, FreqC < 0.05). Specific haplotypic association tests revealed that the haplotype G1-1, formed by the minor allele of each SNP, conferred a protective effect in the SLE group (P=0.016, OR=0.90 [0.82-0.98]) and in the stratified non-LN subset (P=0.022, OR=0.89 [0.80-0.98]). In contrast, the haplotype G1-2, formed by the major allele of each SNP, conferred risk in both groups (P=0.023, OR=1.10 [1.02-1.20] in SLE; P=0.014, OR=1.14 [1.03-1.26] in the non-LN subset).
Haplotype group G2, constructed by the regulatory SNP (rs3813946) located at the +21 position of the 5′ UTR and the two exonic SNPs (rs1048971, rs17615) used in group G1, was comprised of three common haplotypes (G2-1, FreqC = 0.129; G2-2, FreqC = 0.198; G2-3, FreqC = 0.631) and one rare haplotype (G2-4, FreqC < 0.05). Because SNP rs17615 and rs4308977 are in strong LD (r2=0.98) and are able to surrogate for each other, haplotype group G3 is equivalent to G2 and individual G3 haplotypes are not listed in Table 2. Specific haplotypic association tests showed that the protective effects conferred by the G1-1 haplotype containing exonic SNPs rs1048971, rs17615 and rs4308977 remained only in the presence of the major A allele of rs3813946 (G2-1: P=0.043, OR=0.88[0.78-0.99] in SLE and P=0.002, OR=0.79[0.67-0.92] in non-LN; G2-2: P=0.197, OR=0.93[0.84-1.03] in SLE and P=0.757, OR=0.98[0.86-1.10] in non-LN). The risk effects conferred by the G1-2 haplotype also remained in the presence of the major A allele of rs3813946 (G2-3: P=0.023, OR=1.10 [1.02-1.20] in SLE and P=0.013, OR=1.14[1.03-1.26] in non-LN), which was the allele that segregated most frequently with this haplotype.
Finally, haplotypes containing the SLE-associated SNP located in the transmembrane domain of CR2 (rs6540433) were analyzed. Although the omnibus p value of the G4 haplotype, in which this SNP was combined with the exonic SNPs rs1048971, rs17615, and rs4308977, was not significant (Table 2), the incorporation of the regulatory SNP rs3813946 to generate the G5 haplotype resulted in significant omnibus association in the non-LN subset (p = 0.017; Table 2). We performed conditional testing on this group of haplotypes to dissect whether the observed omnibus association was produced by multiple independent effects or could be explained by a single variant. As shown in Table 3, G5 is comprised of four common haplotypes (G5-1-G5-4) and one rare haplotype (G5-5). Specific haplotypic association testing showed haplotypes G5-1 (P=0.003, OR=0.79 [0.68-0.92]) and G5-3 (P=0.046, OR=1.14 [1.00-1.29]) contributed to the omnibus association result. However, omnibus significance could only be eliminated when conditioning on G5-1 (P=0.502), suggesting that G5-1 is the sole haplotype responsible for the omnibus association. Subsequently, we investigated the independent effects of each SNP in isolation, and found that only rs3813946 had an independent effect (P=0.026). However, if rs1048971, rs17615 and rs4308977 were combined in an independent effect test, significant association was observed (P=0.013), suggesting that these three exonic SNPs may jointly regulate biological functions of CR2. The significant signal remained when conditioning on either rs3813946 (P=0.006) or the combination of rs1048971, rs17615 and rs4308977 (P=0.045), supporting the results of the independent effect test that they represent two independent variants. There was no independent effect of rs6540433 (P=0.261), suggesting that the allelic association observed on this locus (Table 1) was caused by LD with upstream associated SNPs.
Table 3.
Conditional test on CR2 SNPs and haplotypes
| Haplotype | rs3813946 | rs1048971 | rs17615 | rs4308977 | rs6540433 | Freq | OR | P* | PSH |
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| G5-1 | A | A | A | G | A | 0.12 | 0.79 [0.68-0.92] | 0.003 | 0.502 |
| G5-2 | G | A | A | G | A | 0.20 | 0.99 [0.87-1.12] | 0.812 | 0.006 |
| G5-3 | A | G | G | A | C | 0.16 | 1.14 [1.01-1.29] | 0.046 | 0.036 |
| G5-4 | A | G | G | A | A | 0.48 | 1.05 [0.95-1.16] | 0.366 | 0.009 |
| G5-5 | A | A | G | A | A | 0.04 | 0.94 [0.73-1.21] | 0.597 | 0.007 |
|
|
|
||||||||
| PIE | 0.026 | 0.486 | NA | NA | 0.261 | P* is P value of specific haplotype. | |||
| 0.298 |
PSH is omnibus P value after conditioning on specific haplotype. |
||||||||
| 0.013 | |||||||||
|
| |||||||||
|
| |||||||||
| PSV | 0.006 | 0.101 | 0.071 | 0.071 | 0.033 | PIE is P value of each SNP in independent effect test. | |||
| 0.071 | |||||||||
| 0.045 |
PSV is omnibus P value after conditioning on specific SNP. |
||||||||
| 0.261 | |||||||||
Meta-analysis of SLE-associated CR2 haplotypes in European-derived populations
In our previous report, we showed that the three SNP haplotype containing the major alleles of rs3813946, rs1048971 and rs17615 increases the risk for SLE in European-derived populations, with a stronger association signal in the non-LN subset of SLE patients.2 We did not identify a protective CR2 haplotype in that analysis. However, integration of the haplotypic association results from the present case-control test and the previous transmission disequilibrium test (TDT) revealed a significant signal for both the risk and the protective CR2 haplotypes in the SLE group (G2-1, PM=0.005; G2-3, PM=0.03), with an even more robust result in the non-LN subset (G2-1, PM=0.001; G2-3, PM=0.002) (Table 2).
CR2 SNPs in exons 10 and 11 alter the splicing efficiency of exon 11
Taken together, these results suggest that two independent variants in CR2 contribute to risk of SLE. The major A allele of rs3813946 is associated with increased transcriptional activity of CR22 but the function of the three exonic SNPs (rs1048971, rs17615 and rs4308977) is as yet unknown. Two of these SNPs are located in exon 10 and one is located in exon 11 of the CR2 gene, which is alternatively spliced and found in a CR2 isoform preferentially expressed on follicular dendritic cells (Figure 2B).8 In addition, these SNPs are in tight linkage disequilibrium with a second SNP in exon 11 (rs17616; Figure 2B).
Figure 2.
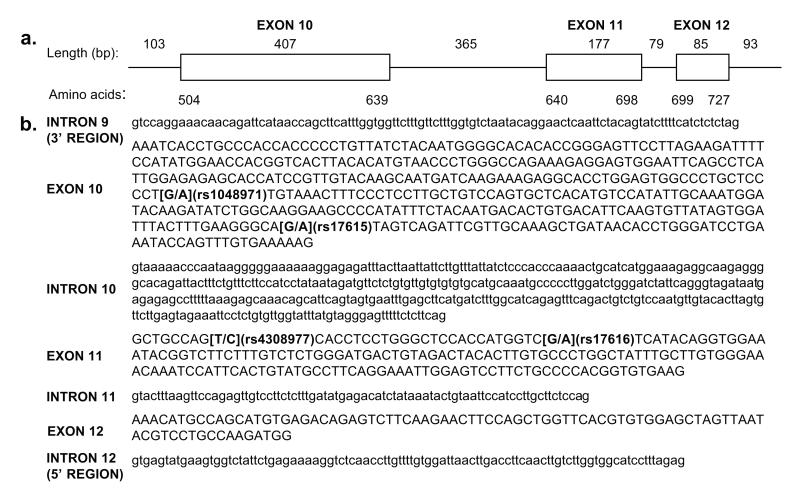
Structure of CR2 minigene containing exon 11. (A) The CR2 minigene introduced in the KpnI site of the pL53In exon trapping vector contains 103 nucleotides of intron 9, the entire exon 10, intron 10, exon 11, intron 11, and exon 12 sequences, and 93 nucleotides of intron 12. Corresponding amino acid residues are indicated below the minigene. Exon 10 transcribes SCR 9 and 10 of the mature CR2 protein, exon 11 transcribes SCR 11, and exon 12 transcribes the first half of SCR 12. (B) The sequence of the CR2 minigene is shown divided into introns and exons. The SNPs in exons 10 and 11 that are studied here are shown in bold with rs designations and with the major allele listed first.
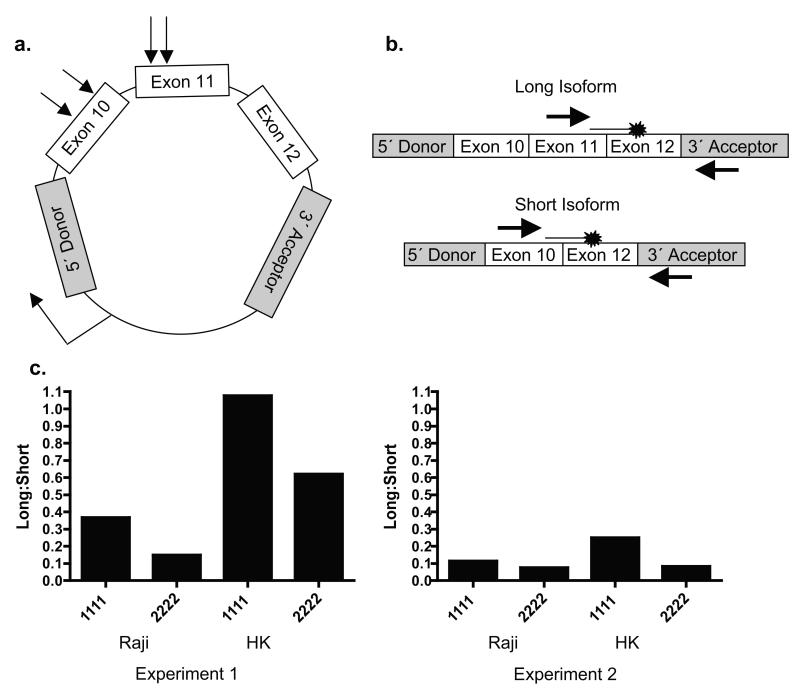
To evaluate an effect of these four SNPs on the splicing efficiency of exon 11, an exon trapping system was used. As the genomic DNA segment extending from exon 10 to exon 12 encompasses only 1114 nucleotides (Figure 2A), we amplified this region including flanking DNA from intron 9 and intron 12 from a BAC clone containing human CR2 (RP11-35C1) and cloned it into the exon trapping vector pL53In.20 The unique KpnI site in pL53In used for cloning is located within the second intron of the rat preproinsulin gene and is flanked by the second exon of the gene containing a 5′ splice donor site and the third exon containing a 3′ splice acceptor site and polyadenylation site, allowing splice variants to be generated that contain the rat 5′ and 3′ preproinsulin exons surrounding the trapped CR2 exons (Figure 3A). Vectors containing either the major or the minor alleles at all four SNPs in exons 10 and 11, representing the two haplotypes found in >95% of controls (G1-1 and G1-2, Table 2), were transiently transfected into the Raji lymphoblastoid B cell line and the HK FDC cell line 21 to allow splicing to occur in the two cellular environments in which CR2 is expressed, since tissue-specific factors are known to influence alternative splicing.22, 23
Figure 3.
Analysis of CR2 exon trapping in Raji and HK cells. (A) The CR2 minigene containing the minor or major alleles at the SNPs in exons 10 and 11 (shown by arrows) was cloned into the pL53In vector so that the CR2 exons are flanked by the second and third exons of the rat preproinsulin gene (in grey). Transcription of the minigene after transfection is driven by the RSV LTR (90° arrow). (B) Quantitative RT-PCR strategy to measure relative levels of vector-derived trapped CR2 exons. The primers to detect trapping of the long isoform are located in exon 11 of CR2 and exon 3 of the rat preproinsulin gene, and the probe spans exons 11 and 12 of CR2. The primers to detect trapping of the short isoform are located in exon 10 of CR2 and exon 3 of the rat preproinsulin gene, and the probe spans exons 10 and 12 of CR2. (C) Relative amounts of vector-derived isoform mRNA in Raji and HK cells. After 24 hours, RNA was prepared from transfected cells and subjected to RT-PCR. In both cell lines, the amount of vector-derived long isoform mRNA relative to short isoform mRNA was decreased in cells transfected with the minor allele construct. Data shown are the results of two independent experiments.
After 24 hours, the relative amount of vector-derived mRNA transcripts including or excluding exon 11 was determined by quantitative reverse transcriptase (RT)-PCR (Figure 3B). The presence of the major alleles at all four SNPs correlated with a higher ratio of long isoform transcripts relative to short isoform transcripts in both Raji and HK transfectants (Experiment 1: 0.37 vs 0.15 for Raji, 1.08 vs 0.62 for HK; Experiment 2: 0.11 vs 0.08 for Raji, 0.25 vs 0.08 for HK; Figure 3C), suggesting that either the major alleles increase the splicing efficiency of exon 11 or the minor alleles decrease it.
CR2 SNPs in exons 10 and 11 alter the splicing efficiency of exon 11 in primary B cells
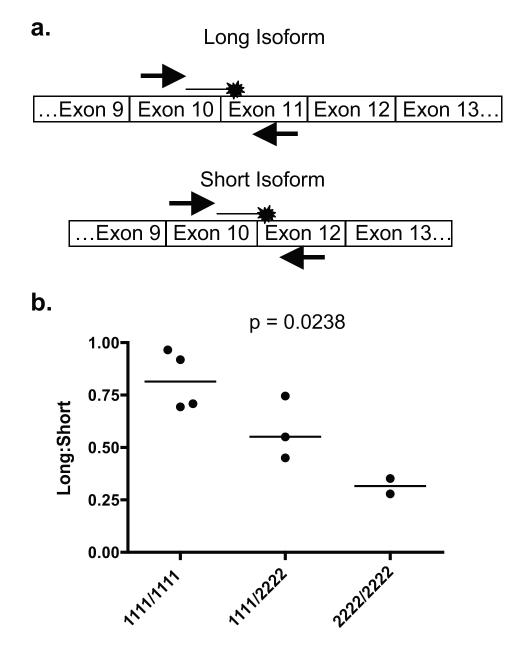
Next, RNA was obtained from purified B cells derived from healthy human subjects who were genotyped for the presence of the major or minor allele at each SNP in exon 10 and 11. The relative amount of long and short isoform mRNA was determined by quantitative RT-PCR (Figure 4A). A higher ratio of long isoform relative to short isoform mRNA was found in individuals homozygous for the major alleles at all four SNPs compared to individuals homozygous for the minor alleles (median ratio homozygous major, 0.81, median ratio heterozygous 0.55, median ratio homozygous minor 0.32; p = 0.0238; Figure 4B). These data are consistent with those obtained in the exon splicing system, and suggest that the major alleles for these SNPs support the inclusion of exon 11 in the mature RNA transcript whereas the minor alleles favor its exclusion.
Figure 4.
Analysis of CR2 isoform mRNA in primary B cells. (A) Quantitative RT-PCR strategy to measure relative levels of long and short isoform mRNA in primary B cells. The primers to detect the long isoform are located in exons 10 and 11 and the primers to detect the short isoform are located in exons 10 and 12, with both probes flanking their respective exon-exon junctions. (B) Average quantity of long isoform CR2 mRNA relative to average quantity of short isoform CR2 mRNA in primary B cells in human subjects genotyped for the SNPs in exons 10 and 11. Quantities of long or short isoform CR2 mRNA were determined in triplicate for each subject, averaged, and normalized to the average quantity of actin mRNA for that subject.
Discussion
Complement receptor 2 was first shown to be a strong candidate gene for lupus susceptibility in the NZM2410 mouse model of lupus, in which the Cr2 gene products were structurally and functionally altered.3 Next, a family-based analysis of Caucasian and Chinese populations demonstrated a significant association between the major alleles of a 3 SNP haplotype of CR2 (rs3813946, rs1048971, rs17615) and lupus susceptibility.2 We now confirm the association of these three CR2 SNPs, and identify two additional CR2 SNPs (rs4308977, rs6540433) significantly associated with SLE susceptibility. Three of these SNPs (rs1048971, rs17615, rs4308977) span two exons and modulate the splicing efficiency of the second exon, providing a potential mechanism for their effects in SLE. The function of these SNPs appears to be modified by a fourth SNP, rs3813946, which alters CR2 transcription.2
Although our original family-based study identified only a risk CR2 haplotype, our current analysis reveals both a protective and a risk CR2 haplotype. The protective haplotype, comprised of the major allele of rs3813946 and the minor alleles of rs1048971, rs17615, and rs4308977, was likely not identified in the previous family-based study because its frequency is much lower than that of the risk haplotype (0.124 vs 0.64) and the power of the previous TDT analysis was not sufficient to allow its detection. The major alleles of rs1048971, rs17615, and rs4308977 demonstrate both allelic and haplotypic association with increased risk of disease in our current analysis, as shown in Tables 1 and 2. However, the association of these alleles with risk in conjunction with the minor allele (G5-3) but not the major allele for the transmembrane SNP rs6540433 (G5-4) suggests that the minor allele of rs6540433 may be the true causal variant in this extended haplotype, although an independent effect of this locus was not supported by the haplotype-based conditional test.
The protective CR2 haplotype we identify here contains two independent interacting variants. The first variant contains the minor alleles of rs1048971, rs17615, and rs4308977, which decrease the splicing efficiency of the alternatively spliced exon 11, resulting in increased relative amounts of the short CR2 isoform. The second variant contains the major allele of the regulatory SNP rs3813946, which is associated with increased CR2 transcription. This haplotype is protective only in individuals that do not have renal disease, perhaps because of specific effects on non-renal disease manifestations; alternatively, other stronger risk alleles may obscure the protective effects of this haplotype in patients who develop renal disease.
The extent of the role of alternative splicing of gene products in generating human diversity has only recently begun to be realized. Approximately 95% of multiexon genes undergo alternative splicing, with a mean of 7 such events per gene.24 The tissue-specific restriction of certain splice variants has been described and is determined by cis-regulatory elements,23 suggesting that the function of each isoform depends on the environment in which it is expressed. Furthermore, biological functions have been described for many of the alternatively spliced mRNA isoforms identified to date.25 Interindividual variation in isoforms as a result of SNPs located in splicing regulatory motifs may occur in up to 21% of alternatively spliced genes,26 and the effects of these polymorphisms on splicing efficiency is believed to contribute significantly to disease severity and susceptibility [reviewed in 27].
The parameters that control splice site selection in pre-mRNA include splice site strength, the presence or absence of splicing regulators, RNA secondary structure, the exon/intron architecture, and the process of pre-mRNA synthesis itself.28 The SNPs located in exons 10 and 11 of the CR2 gene would not be anticipated to alter splice site strength based on their location. Furthermore, they would not affect intron size, which influences the likelihood that a downstream exon will be alternatively spliced. However, they may alter the cis-acting regulatory RNA sequence elements termed exonic splicing enhancers (ESE) and silencers (ESS), which are common and can influence splice site selection. In addition, the alteration in nucleotide sequence as a result of these variants could affect the secondary structure of the pre-RNA and interfere with or modulate splice site recognition.29 Finally, since intron removal is physically and temporally linked to RNA transcription, differences in the structure of the promoter or rate of transcription can influence splice site selection [reviewed in 30]. This is particularly interesting in light of the fact that the lupus-associated polymorphism in the 5′UTR of the CR2 gene (rs3813946) (Table 1) alters transcriptional activity,2 which could in turn affect the splicing efficiency of exon 11 and may explain why this SNP appears to amplify the protective effect associated with the SNPs in exons 10 and 11.
These data suggest that the SNPs in exons 10 and 11 of the CR2 gene alter lupus susceptibility because of their differential effects on inclusion of exon 11 in the mature CR2 mRNA. The SCR encoded by exon 11 contains two putative N-linked glycosylation sites,5 so can be predicted to alter the structure of the CR2 protein in which it is expressed. This may result in either increased or decreased avidity for CR2 ligands, including C3d, EBV, CD23, and IFN-α, which could have effects on B cell and FDC function. The differential expression of the longer CR2 isoform on FDCs suggests that it may play an important role in antigen trapping, and its decreased expression in individuals with the protective minor allele haplotype may result in reduced affinity maturation of autoantibodies and fewer long-lived autoreactive plasma cells in individuals prone to develop lupus. Furthermore, the tissue-specificity of the long isoform does not appear to occur at the level of mRNA, as we and others31 have demonstrated both long and short isoform mRNA in primary B cells from healthy human subjects. However, if the long isoform is not expressed at the protein level in B cells,8 its increased generation in individuals expressing the major allele haplotype for the SNPs in exons 10 and 11 might result in a relative deficiency of the CR2 protein on B cells, a phenotype that has been associated with increased autoimmunity in animal models of lupus32, 33 and that has been described in patients with SLE.34-36 Although the specific alleles at these SNPs may alter the secondary and tertiary structure of the CR2 protein with the concomitant functional effects associated with this, it has been suggested that the primary pathogenic effect for many exonic SNPs is at the level of alterative splicing.27
Our results may differ from those of a Japanese case-control study that did not confirm our previous association of 3 CR2 SNPs (rs3813946, rs1048971, rs17615) with SLE for several reasons. The Japanese study included only 509 cases with lupus and 964 controls; therefore the sample size may have lacked sufficient power to demonstrate an effect of the SNPs studied. In addition, they did not include the SNPs in exon 11 and the transmembrane domain (rs4308977, rs6540433) in their analysis. However, at this time we cannot rule out the possibility that SNPs in CR2 are associated with lupus in European-derived and Chinese populations but do not contribute to lupus susceptibility in Japanese.
Most cases of systemic lupus erythematosus are believed to result from the effects of common gene variants individually associated with low penetrance of disease in combination with environmental factors that trigger disease development. Using both a family-based2 and case-control study, we have shown that five CR2 variants are associated with SLE. One variant alters CR2 transcription,2 and three others modulate the ratio of CR2 splice isoforms that are expressed differentially at the protein level in B cells and FDCs. The fifth polymorphism, located in the transmembrane domain and resulting in a nonconservative amino acid change of glutamic acid to alanine, may affect receptor signaling, perhaps via effects on CD19 co-association with CR2.37 Combinatorial effects of these SNPs may modulate the penetrance or severity of disease phenotypes, as has been described in atypical cystic fibrosis 38 and in animal models.39-41 These findings provide additional evidence that CR2 contributes to human lupus susceptibility and suggest several mechanisms for this effect, some of which may be novel targets for therapeutic intervention.
Materials and Methods
Human subjects
DNA samples used for the case-control association study were collected from individuals of European ancestry by institutes in the United States, the United Kingdom and Sweden, the majority of which were previously used in a collaborative study [The International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) and Lupus Large Association Study (LLAS)].42 All SLE patients met the American College of Rheumatology criteria for the classification of SLE.19 All subjects were enrolled after informed consent had been obtained under the auspices of the appropriate authority at each institution.
To ensure that only samples from unrelated and unique individuals were used in further analyses, genotypes of 500 SNPs with minor allele frequencies greater than 0.40 were compared for each sample. Samples from a total of 534 individuals that were duplicated or related were removed. To reduce the impact of population substructure, principle component analysis (PCA) was performed using 18,201 SNPs genotyped in the LLAS data set. A total of 213 samples that violated the assumption of sample homogeneity based on the PCA were trimmed. Data cleaning resulted in the final data set with a modest inflation (λGC<1.12), which consisted of 2084 SLE patients (226 male, 1858 female) and 2853 healthy controls (928 male, 1925 female). Renal status was known for 1655 SLE patients, 519 of whom were diagnosed with lupus nephritis and 1136 who were not.19
RNA samples used for quantitative analysis of CR2 isoform mRNA were collected from healthy non-smoking adults between 18 and 60 years of age who had no family history of autoimmune disease, were on no medications, and did not drink more than 1 alcoholic beverage daily on average. All subjects were enrolled after informed consent had been obtained. This study was approved by the Colorado Multiple Institutional Review Board.
Genotyping
For the case-control study, DNA samples were assembled at the Oklahoma Medical Research Foundation (OMRF). DNA quantity was measured using PicoGreen (Molecular Probes). 250 ng of DNA from each individual was genotyped on the BeadStation 500GX (Illumina, San Diego, CA, USA) using the Infinium II platform at the Lupus Genetics Studies Unit of the OMRF. In brief, the flow process of genotyping included whole-genome amplification, fragmentation, hybridization, single-base extension using fluorescence-labeled nucleotides and imaging. Within the 39 kb CR2 gene, 12 SNPs with minor allele frequencies > 0.03 and inter-SNP distance of > 25 bp, that either tagged major haplotypes in the HapMap database or were potentially functional, were selected for genotyping. To avoid the bias introduced by genotyping, stringent quality control was applied to each SNP included in the subsequent analyses: 1) Overall missing genotype less than 2% (with the exception of rs3813946, 6.2%); 2) No significant differences in the proportions of missing genotype data between cases and controls (p>0.05); 3) Allele frequencies of controls statistically consistent with expectations for ethnicity-matched HapMap samples (p>0.01).
Genotyping of subjects for analysis of CR2 isoform mRNA was performed using commercial genotyping kits for rs1048971, rs17615, rs4308977, and rs17616 (Taqman® SNP Genotyping Assays, Applied Biosystems, Foster City, CA, USA). Real time rtPCR was performed using the Applied Biosystems 7000 Real-Time PCR System (Applied Biosystems) at the Barbara Davis Center DERC Molecular Core.
Sample collection and processing
Peripheral blood was collected in heparinized syringes and mononuclear cells (PBMCs) were isolated over Ficoll-Paque (Sigma-Aldrich, St. Louis, MO, USA). DNA for genotyping was purified from PBMCs using the QIAamp DNA Mini Kit (QIAgen, Valencia, CA, USA). B cells were purified from PBMCs using the Easy Sep Human B Cell Enrichment Kit (95-99% CD19+; StemCell Technologies, Vancouver, BC, Canada) and RNA was processed immediately from B cells using the RNeasy Plus Mini Kit (QIAgen). Quality check and quantification of RNA was performed using the Agilent 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA) at the University of Colorado Cancer Center Microarray Core Facility. RNA and DNA samples were stored at -70°C prior to use.
Plasmid construction and site-directed mutagenesis
The region of the human CR2 gene spanning from intron 9 to intron 12 was amplified from a BAC clone containing human CR2 (RP11-35C1; Invitrogen, Carlsbad, CA, USA) using the following PCR primers which contain unique KpnI restriction sites at the 5′ and 3′ ends of the product: 5′- GCATGGTACCGTCCAGGAAACAACAG-3′ and 5′- GCATGGTACCCTCTAAAGGATGCCAC-3′. The purified PCR product was digested with KpnI and subcloned into the KpnI site of the exon trap vector, pL53In20 (kind gift of Dr. Raul Torres, National Jewish Health and University of Colorado Denver, Denver, CO, USA), which contains the LTR of Rous Sarcoma Virus (RSV) which functions as a strong promoter. The KpnI site of this vector is flanked by functional splice donor and acceptor sites of the rat preproinsulin gene and a polyadenylation site. This construct contained the minor allele at each of the four SNPs in exons 10 and 11. Site-directed mutagenesis was performed (QuikChange Multi Site Directed Mutagenesis Kit, Stratagene, La Jolla, CA, USA) to create a second vector containing the major allele for each SNP. Successful mutagenesis was confirmed by sequencing.
DNA Sequencing
DNA templates were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction kit version 1.1 (Applied Biosystems part number 4337451) by modification of the standard protocol. In the case of difficult templates, the Ready Mix was combined with an aliqout of the dGTP BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems part number 4307175). The standard sequencing thermo-cycling parameters were as follows: denaturation for 5 min at 94°C, followed by 30 cycles of denaturation at 96°C for 10 sec, annealing at 50°C for 5 sec, and extension/termination at 60°C for 4 min, followed by incubation at 10°C until the samples were processed. Residual dye-labeled dideoxynucleotides (dye-terminators) were removed from the cycle-sequencing reaction products using the paramagnetic bead technology (CleanSEQ, part number 000136) from Agencourt Bioscience Corporation (Beverly, MA, USA) and a modification of the manufacturer’s recommended protocol. The products were sequenced on a fluorescent capillary automated sequencer with a 50 cm long, 48-capillary array containing POP7 polymer (Hitachi 3730 Genetic Analyzer; Applied Biosystems). Analyses of DNA sequences were done with Sequencing Analysis version 3.3 (Applied Biosystems) and alignment of DNA sequences were done with Sequencher 3.1 (Gene Codes Corporation, Ann Arbor, MI, USA).
Cell Culture and Transfection
The follicular dendritic cell line, HK, was kindly provided by Dr. Yong Choi (Ochsner Clinic Foundation, New Orleans, LA, USA). The Raji B cell line was obtained from American Type Culture Collection (Manassas, VA, USA). Both lines were maintained in complete RPMI with 10% fetal calf serum. Plasmids were transfected into the cell lines using the Cell Line Nucleofector Kit V (Amaxa Biosystems, Gaithersburg, MD, USA). 2 X 106 Raji cells or 1 X 106 HK cells were transfected with each construct. Raji cells were transfected using the Amaxa Nucleofector program M-013 and HK cells were transfected using the Amaxa Nucleofector program T-030. After transfection, cells were plated in 1 ml of complete RPMI with 10% fetal calf serum in either a 12 well tissue culture plate (Raji cells) or a 6 well tissue culture plate (HK cells). After 24 hours, cells were harvested and total RNA was isolated using the QIAgen RNeasy Plus Mini Kit (QIAgen).
Reverse Transcription and Amplification of cDNA
For analysis of RNA, first-strand cDNA was prepared by reverse transcription using 150 ng of total RNA from transfected cell lines or 36 ng of total RNA from primary B cells in a volume of 20 μl (2.5 μM of random hexamer primers, 20 units of RNase inhibitor, 50 units of Maloney murine leukemia virus reverse transcriptase, 1X PCR Buffer II, 5 mM MgCl2, 1 mM dATP, 1 mM dCTP, 1 mM dGTP, 1 mM dTTP, all from Applied Biosystems) at 42°C for 30 minutes. DNA/RNA hybrids were denatured at 99°C for 5 minutes and samples were stored at 4°C.
To measure exogenous long isoform CR2 transcripts in the transfected cells, the primer pair consisted of an oligonucleotide spanning nucleotides 2110-2131 of CR2 cDNA sequence (sense primer, 5′-TGTATGCCTTCAGGAAATTGGA-3′), located in exon 11, and an oligonucleotide located in the rat preproinsulin exon containing the 3′ splice acceptor and polyadenylation site (anti-sense primer, 5′-CAGTTGTGCCACCCATCTTG-3′), and the probe consisted of an oligonucleotide spanning the junction of exon 11 and exon 12 of CR2 [6FAM (6-carboxyfluorescein reporter dye) — ACGGTGTGAAGAAAC - MGBNFQ (molecular-groove binding non-fluorescent quencher)]. To measure exogenous short isoform CR2 transcripts in the transfected cells, the primer pair consisted of an oligonucleotide spanning nucleotides 1944-1965 of the CR2 cDNA sequence (sense primer, 5′-CACCTGGGATCCTGAAATACCA-3′), located in exon 10, and a second oligonucleotide located in the rat preproinsulin exon containing the 3′ splice acceptor and polyadenylation site (anti-sense primer, 5′-CTCCACCCAGCTCCAGTTGT-3′), and the probe consisted of an oligonucleotide spanning the junction of exon 10 and exon 12 of CR2 (6FAM-CATGTTTCTTTTTCACAAAC-MGBNFQ). Serial dilutions of cDNA synthesized from cells transfected with the minor allele construct were used to create a standard curve for each primer/probe set. Relative amounts of vector-derived transcripts including or excluding exon 11 were determined using the efficiency-corrected ΔCt method.
To measure endogenous long isoform CR2 transcripts, the primer pair consisted of oligonucleotides spanning nucleotides 1938-1962 of the CR2 cDNA sequence (sense primer, 5′-TGATAACACCTGGGATCCTGAAATA-3′), located in exon 10, and oligonucleotides spanning nucleotides 2038-2014 of the CR2 cDNA sequence (anti-sense primer, 5′-AGAAGACCGTATTTCCACCTGTATG-3′), located in exon 11, and the probe consisted of an oligonucleotide spanning the junction of exon 10 and exon 11 (6FAM-CTGGCAGCCTTTTT-MGBNFQ). To measure endogenous short isoform CR2 transcripts, the primer pair consisted of oligonucleotides spanning nucleotides 1944-1965 of the CR2 cDNA sequence (sense primer, 5′-CACCTGGGATCCTGAAATACCA-3′), located in exon 10, and oligonucleotides spanning nucleotides 2189-2165 of the CR2 cDNA sequence (anti-sense primer, 5′-TCTTGAAGACTCTGTCTCACATGCT-3′), located in exon 12, and the probe consisted of an oligonucleotide spanning the junction of exon 10 and exon 12 (6FAM-CATGTTTCTTTTTCACAAAC-MGBNFQ). To measure β-actin transcripts as an endogenous reference, a commercial kit was used (Applied Biosystems). Serial dilutions of cDNA amplified from Raji RNA were used to create a standard curve for each primer/probe set. The efficiency-corrected ΔCt method was used to determine the relative amounts of long isoform and short isoform CR2 transcripts normalized to β-actin transcripts.
Five μl of the cDNA synthesis reaction was used for PCR amplification in a 25-μl final reaction volume (12.5 μl 2X Applied Biosystems Universal Master Mix, 0.3 μM each oligonucleotide primer, and 0.05 μM TaqMan probe). Samples were analyzed in triplicate. Real time rtPCR was performed using the Applied Biosystems 7000 Real-Time PCR System (Applied Biosystems) at the Barbara Davis Center DERC Molecular Core using the following program: 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds then 60°C for 1 minute.
Statistical Analysis
The threshold of Hardy-Weinberg equilibrium p value is set at >0.01. Allelic association tests in the case-control and family-based studies were calculated using Pearson’s χ2 test and transmission disequilibrium test (TDT), respectively. The adaptive permutation method of Plink43 was used to correct the significance results obtained in the association tests. Linkage disequilibrium values among pairs of SNPs were calculated by Haploview 4.1 (www.broad.mit.edu/mpg/haploview/index.php). Confidence intervals were used to define haplotype blocks. Odds ratios and corresponding 95% confidence intervals for case-control data were estimated using the Taylor series method. To integrate the results from case-control studies and the TDT, the method described by Kazeem and Farrall44 was adopted, in which the odds ratios for TDT data were obtained using the proportion of transmitted and non-transmitted alleles from heterozygous parents of affected individuals. Standard E-M algorithms were used to impute haplotypes. Logistic regression was used to perform conditional haplotype-based tests. Both haplotypic association analyses and conditional tests were conducted by Plink. CR2 mRNA levels between CR2 genotypes were compared using the Kruskal-Wallis test. Statistical analyses were performed using InStat (GraphPad, San Diego, CA, USA). P values <0.05 were considered to indicate statistical significance.
Acknowledgements
We thank Dr. Raul Torres (National Jewish Health and University of Colorado Denver School of Medicine) for providing the pL53In vector, Dr. Yong Choi (Ochsner Clinic Foundation) for providing the HK FDC line, and Carissa Homme and Lauren Kuhlman (University of Colorado Denver School of Medicine, Aurora, CO, USA) for technical assistance. We also thank Dr. Peter Gregersen (Feinstein Institute for Medical Research, Manhassat, NY, USA) for contributing DNA samples from control subjects to LLAS, and the Wake Forest University Health Sciences Center for Public Health Genomics for assistance with the principal component analysis of the genotyping data from LLAS1. The pL53In constructs were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core (http://loki.uchsc.edu), which is supported by the NIH/NCI Cancer Core Support Grant (P30 CA046934). The quality of the RNA samples prepared from healthy human subjects was evaluated in the University of Colorado Cancer Center Microarray Core. Quantitative RT-PCR was performed in the DERC Molecular Biology Core Facility, which is supported by National Institutes of Health (NIH) grant P30 DK57516. Other support for this work included grants from the American College of Rheumatology Research and Education Foundation, the Alliance for Lupus Research, the US Department of Veterans Affairs, Kirkland Scholar/Hospital for Special Surgery and Rheuminations, and NIH (NIAID-DAIT-BAA-05-11, N01 AR12253, N01 AR62277, P01 AR049084, P20 RR015577, P20 RR020143, P30 AR053483, P50 AR48940, R01 DE015223, R01 AI31584, R01 AI070983R01 AR42460, and R37 AI24717.
References
- 1.Tsao BP. Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol. 2004;16:513–21. doi: 10.1097/01.bor.0000132648.62680.81. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Boackle SA, Hanvivadhanakul P, Ulgiati D, Grossman JM, Lee Y, et al. Association of a common complement receptor 2 haplotype with increased risk of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2007;104:3961–6. doi: 10.1073/pnas.0609101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, et al. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity. 2001;15:775–85. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- 4.Miyagawa H, Yamai M, Sakaguchi D, Kiyohara C, Tsukamoto H, Kimoto Y, et al. Association of polymorphisms in complement component C3 gene with susceptibility to systemic lupus erythematosus. Rheumatology (Oxford) 2008;47:158–64. doi: 10.1093/rheumatology/kem321. [DOI] [PubMed] [Google Scholar]
- 5.Weis JJ, Toothaker LE, Smith JA, Weis JH, Fearon DT. Structure of the human B lymphocyte receptor for C3d and the Epstein-Barr virus and relatedness to other members of the family of C3/C4 proteins. JExpMed. 1988;167:1047–66. doi: 10.1084/jem.167.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MD, Cooper NR, Tack BF, Nemerow GR. Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes. ProcNatlAcadSciUSA. 1987;84:9194–8. doi: 10.1073/pnas.84.24.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisaku A, Harley JB, Frank MB, Gruner BA, Frazier B, Holers VM. Genomic organization and polymorphisms of the human C3d/Epstein-Barr virus receptor. J Biol Chem. 1989;264:2118–25. [PubMed] [Google Scholar]
- 8.Liu Y-J, Xu J, de Bouteiller O, Parham CL, Grouard G, Djossou O, et al. Follicular dendritic cells specifically express the long CR2/CD21 isoform. J Exp Med. 1997;185(1):165–70. doi: 10.1084/jem.185.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci USA. 1984;81:4510–4. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aubry J-P, Pochon S, Graber P, Jansen KU, Bonnefoy J-Y. CD21 is a ligand for CD23 and regulates IgE production. Nature. 1992;358:505–7. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 11.Delcayre AX, Salas F, Mathur S, Kovats K, Lotz M, Lernhardt W. Epstein Barr virus/complement C3d receptor is an interferon alpha receptor. EMBO J. 1991;10:919–26. doi: 10.1002/j.1460-2075.1991.tb08025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vereshchagina LA, Tolnay M, Tsokos GC. Multiple transcriptional factors regulate the inducible expression of the human complement receptor 2 promoter. J Immunol. 2001;166:6156–63. doi: 10.4049/jimmunol.166.10.6156. [DOI] [PubMed] [Google Scholar]
- 13.Tolnay M, Vereshchagina LA, Tsokos GC. NF-kappaB regulates the expression of the human complement receptor 2 gene. J Immunol. 2002;169:6236–43. doi: 10.4049/jimmunol.169.11.6236. [DOI] [PubMed] [Google Scholar]
- 14.Ulgiati D, Holers VM. CR2/CD21 proximal promoter activity is critically dependent on a cell type-specific repressor. J Immunol. 2001;167:6912–1919. doi: 10.4049/jimmunol.167.12.6912. [DOI] [PubMed] [Google Scholar]
- 15.Ulgiati D, Pham C, Holers VM. Functional analysis of the human complement receptor 2 (CR2/CD21) promoter: characterization of asal transcriptional mechanisms. J Immunol. 2002;168:6279–85. doi: 10.4049/jimmunol.168.12.6279. [DOI] [PubMed] [Google Scholar]
- 16.Makar KW, Pham CTN, Dehoff MH, O’Connor SM, Jacobi SM, Holers VM. An intronic silencer regulates B lymphocyte cell- and stage-specific expression of the human complement receptor type 2 (CR2, CD21) gene. J Immunol. 1998;160:1268–78. [PubMed] [Google Scholar]
- 17.Makar KW, Ulgiati D, Hagman J, Holers VM. A site in the complement receptor 2 (CR2/CD21) silencer is necessary for lineage specific transcriptional regulation. Int Immunol. 2001;13:657–64. doi: 10.1093/intimm/13.5.657. [DOI] [PubMed] [Google Scholar]
- 18.Holers VM. Complement receptors and the shaping of the natural antibody repertoire. Springer Semin Immun. 2005;26:405–23. doi: 10.1007/s00281-004-0186-y. [DOI] [PubMed] [Google Scholar]
- 19.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 20.Auch D, Reth M. Exon trap cloning: using PCR to rapidly detect and clone exons from genomic DNA fragments. Nucleic Acids Research. 1990;18:6743–4. doi: 10.1093/nar/18.22.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H-S, Zhang X, Choi YS. Activation and proliferation of follicular dendritic cell-like cells by activated T lymphocytes. J Immunol. 1994;153:2951–61. [PubMed] [Google Scholar]
- 22.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castle JC, Zhang C, Shah JK, Kulharni AV, Kalsotra A, Cooper TA, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–25. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 25.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Research. 2001;29:2850–9. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–31. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 27.Wang G-S, Cooper TA. Splicing in disease: disruption of he splicing code and the decoding machinery. Nature Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 28.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–5. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 29.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genetics. 2007;3:2147–55. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 31.Illges H, Braun M, Peter HH, Melchers I. Analysis of the human CD21 transcription unit reveals differential splicing of exon 11 in mature transcripts and excludes alternative splicing as the mechanism causing solubilization of CR2. Mol Immunol. 1997;34(10):683–93. doi: 10.1016/s0161-5890(97)00099-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Jiang N, Deppong C, Singh J, Dolecki G, Mao D, et al. A role for the Cr2 gene in modifying autoantibody production in systemic lupus erythematosus. J Immunol. 2002;169:1587–92. doi: 10.4049/jimmunol.169.3.1587. [DOI] [PubMed] [Google Scholar]
- 33.Prodeus AP, Georg S, Shen L-M, Pozdnyakova OO, Chu L, Alicot EM, et al. A critical role for complement in the maintenance of self-tolerance. Immunity. 1998;9:721–31. doi: 10.1016/s1074-7613(00)80669-x. [DOI] [PubMed] [Google Scholar]
- 34.Wilson JG, Ratnoff WD, Schur PH, Fearon DT. Decreased expression of the C3b/C4b receptor (CR1) and the C3d receptor (CR2) on B lymphocytes and of CR1 on neutrophils of patients with systemic lupus erythematosus. Arth Rheum. 1986;29:739–47. doi: 10.1002/art.1780290606. [DOI] [PubMed] [Google Scholar]
- 35.Levy E, Ambrus J, Kahl L, Molina H, Tung K, Holers VM. T lymphocyte expression of complement receptor 2 (CR2/CD21): a role n adhesive cell-cell interactions and dysregulation in a patient with systemic lupus erythematosus (SLE) Clin Exp Immunol. 1992;90:235–44. doi: 10.1111/j.1365-2249.1992.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquart HV, Svendsen A, Rasmussen JM, Nielsen CH, Junker P, Svehag S-E, et al. Complement receptor expression and activation of the complement cascade on B lymphocytes from patients with systemic lupus erythematosus (SLE) Clin Exp Immunol. 1995;101:60–5. doi: 10.1111/j.1365-2249.1995.tb02277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto AK, Martin DR, Carter RH, Klickstein LB, Ahearn JM, Fearon DT. Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med. 1993;178:1407–17. doi: 10.1084/jem.178.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner B, Truninger K, Sanz J, Schaller A, Gallati S. The role of common single-nucleotide polymorphisms on exon 9 and exon 12 skipping in nonmutated CFTR alleles. Hum Mutat. 2004;24:120–9. doi: 10.1002/humu.20064. [DOI] [PubMed] [Google Scholar]
- 39.Cornall RJ, Cyster JG, Hibbs ML, Dunn AR, Otipoby KL, Clark EA, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 40.Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA. 2000;97:6670–5. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giles BM, Tchepeleva SN, Kachinski JJ, Ruff K, Croker BP, Morel L, et al. Augmentation of NZB autoimmune phenotypes by the NZW-derived Sle1c lupus susceptibility interval. J Immunol. 2007;178:4667–75. doi: 10.4049/jimmunol.178.7.4667. [DOI] [PubMed] [Google Scholar]
- 42.The International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool et for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. 2005;69:329–35. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]