Abstract
Fanconi anemia (FA) is a genic disease resulting in bone marrow failure, high cancer risks, and infertility, and developmental anomalies including microphthalmia, microcephaly, hypoplastic radius and thumb. Here we present cDNA sequences, genetic mapping, and genomic analyses for the four previously undescribed zebrafish FA genes (fanci, fancj, fancm, and fancn, and show that they reverted to single copy after the teleost genome duplication. We tested the hypothesis that FA genes are expressed during embryonic development in tissues that are disrupted in human patients by investigating fanc gene expression patterns. We found fanc gene maternal message, which can provide Fanc proteins to repair DNA damage encountered in rapid cleavage divisions. Zygotic expression was broad but especially strong in eyes, central nervous system and hematopoietic tissues. In the pectoral fin bud at hatching, fanc genes were expressed specifically in the apical ectodermal ridge, a signaling center for fin/limb development that may be relevant to the radius/thumb anomaly of FA patients. Hatching embryos expressed fanc genes strongly in the oral epithelium, a site of squamous cell carcinomas in FA patients. Larval and adult zebrafish expressed fanc genes in proliferative regions of the brain, which may be related to microcephaly in FA. Mature ovaries and testes expressed fanc genes in specific stages of oocyte and spermatocyte development, which may be related to DNA repair during homologous recombination in meiosis and to infertility in human patients. The intestine strongly expressed some fanc genes specifically in proliferative zones. Our results show that zebrafish has a complete complement of fanc genes in single copy and that these genes are expressed in zebrafish embryos and adults in proliferative tissues that are often affected in FA patients. These results support the notion that zebrafish offers an attractive experimental system to help unravel mechanisms relevant not only to FA, but also to breast cancer, given the involvement of fancj (brip1), fancn (palb2) and fancd1 (brca2) in both conditions.
Keywords: genome duplication, disease model
1. Introduction
Fanconi anemia (FA; MIM# 227650) is a rare autosomal recessive disorder appearing at a frequency of about 3 per million and affecting approximately two thousand families in the United States. This devastating disease is characterized by developmental abnormalities in a number of organ systems and catastrophic bone marrow failure, often by five years of age. Bone marrow transplantation has become an effective therapy [1–6], but survivors experience increased susceptibility to squamous cell carcinomas of the head and neck [7,8]. Thirteen FA complementation groups have been identified and their genes cloned (FANCA, FANCB, FANCC, FANCD1 (BRCA2), FANCD2, FANCE, FANCF, FANCG (XRCC9), FANCI, FANCJ (BRIP1), FANCL, FANCM, and FANCN (PALB2)) [9–28]. FA proteins interact in a complex network that facilitates a DNA damage response leading to DNA repair. Cells exposed to DNA damaging agents or passing through the DNA synthesis phase of a normal cell cycle activate a Nuclear Core Complex consisting of eight FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM). The Nuclear Core Complex functions as an E3 ligase to trigger the monoubiquitination of the downstream proteins FANCI and FANCD2 (the ID complex) [21,29]. Monoubiquitination of the ID complex allows it to translocate to nuclear DNA repair foci containing BRCA1, histone H2AX, FANCD1 (BRCA2), FANCJ, FANCN and RAD51 [21,25,29,30]. In ways that are not fully understood, these protein complexes target to sites of DNA damage, which initiates DNA repair mechanisms. Biallelic mutation of any of the FA genes upstream of the ID complex disrupts ID monoubiquitination leading to the loss of nuclear foci and resulting genomic instability as reflected by the hypersensitivity of DNA to interstrand crosslinks (ICLs) caused by genotoxic agents such as Cisplatin, mitomycin C (MMC) and diepoxybutane (DEB) [31,32].
The sensitivity of FA cells to ICLs reflects defects in DNA repair mechanisms that lead to aberrant apoptosis, genomic instability, and cancer. Because the FA pathway intersects pathways for breast and other cancers [29,30], and is involved in the evolution of resistance to cancer chemotherapies [33,34], a better understanding of the mechanisms that lead to Fanconi anemia will be broadly applicable to the biology of malignancy. Despite major advances in our knowledge of the biochemistry of FA proteins, we still know little about the mechanisms by which loss-of-function mutations in the FA network impact the DNA damage response pathway and contribute to the heterogeneous clinical features found in FA patients (developmental defects, progressive onset of aplastic anemia, and increased predisposition to hematological malignancies and the formation of solid tumors). Furthermore, FA cells have other defects, including poor resistance to oxidative damage, premature telomere shortening, abnormal cell cycle kinetics, interaction with inflammation pathways, and hyperactivation of the MAPK pathway leading to overproduction of TNF-alpha [35–38].
People with mutations in some complementation groups that alter the Nuclear Core Complex experience greater cancer risks than others, which would not be expected if the only function of these proteins were to activate the ID complex. For some FA genes, even heterozygotes suffer an elevated risk of breast cancer (FANCD1 (BRCA2), FANCN (PALB2) and FANCJ (BRIP1). Understanding the network of interactions that unites different complementation groups into a single disease will benefit from studies that exploit model systems. The zebrafish model shares cellular, developmental, and genetic features with humans and provides advantages that facilitate experimentation. Clinically relevant studies designed to understand the complex web of interactions that unite the different complementation groups into a single disease could exploit a convenient, small vertebrate model such as zebrafish.
In this paper, we first report the identification and isolation of the four previously unidentified zebrafish orthologs of human FA genes, and then present gene expression data for all 13 complementation groups to address the mechanisms that cause FA to affect some organs but not others and to understand why different complementation groups have different phenotypes. Finally we evaluate zebrafish as a model for future FA research.
2. Material and methods
2.1. Genomic analysis
The Zv7 version of the zebrafish genome assembly (http://www.ensembl.org/Danio_rerio/index.html) was searched using human Fanconi proteins sequences. We further investigated sequences satisfying the reciprocal best BLAST “hit” (RBH) method for orthology [39]. Sequences from the genome assembly were used to design primers for 5′ and 3′ rapid amplification of cDNA ends (RACE, Advantage cDNA PCR Kit, Clontech, Inc., Palo Alto) using as template, 5′ or 3′ first strand zebrafish cDNA synthesized from mRNA of pooled embryos at 12, 24, and 48 hours post-fertilization (hpf). Some genes were too large for full-coverage by RACE, and so we performed standard amplification with forward and reverse gene-specific primers, using as template second strand cDNA from 60 hpf embryos. Amplified gene fragments were cloned using the TOPO Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA). FANC proteins were aligned using ClustalW [40]. Genomic structure was inferred using Genomescan (http://genes.mit.edu/genomescan/). Zebrafish fanc genes were mapped by single strand conformation analysis (SSCP) on the heat shock doubled haploid mapping panel [41]. For comparative mapping, putative orthologs were defined by RBH [39](Hirsh and Fraser, 2001). The human map locations of putative orthologs were obtained from Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi). Nomenclature rules specify human, mouse, and zebrafish genes as FANCA, Fanca, and fanca, respectively and human, mouse, and zebrafish proteins as FANCA, FANCA, and Fanca, respectively (http://zfin.org/zf_info/nomen.html). When indicating a vertebrate protein without respect to species, we use the nomenclature Fanca.
2.2. Expression analysis
For reverse transcriptase (RT) PCR, total RNA was extracted following instructions in the TRI REAGENT kit (Molecular Research Center Inc., TR-118) using pools of 40 to 50 embryos for each developmental stage. First strand of cDNA was generated from 2 µg of total RNA, using Superscript III Rnase H-reverse transcriptase (Invitrogen, #18080-044) and oligo(dT) primer. After deactivation of the reverse transcriptase by incubating the sample at 70°C for 15 minutes, RNA was degraded with RNase H (Biolabs, M0297S). A dilution 1:10 or 1:20 of the first strand cDNA (1:200 for actin) was then used to assay for gene expression by PCR. Gene specific oligonucleotide primers were: fanca, GCAGACCCGGAACAGCCCACAC/CAGCGCTGAATAATCCCGCAGACA; fancb, CGGCCCGTCTGCGGTGAAGA/GCCGCTGGAGAACTGAAGCCACAC; fancc, TGGAGGGGGCAGCAGTGAGC/TTGGTGGGGTGGTGGAAGAACG; fancd1, GGGCCAGAAAACACAGCAACTCAAA/GCACAGGCCCAGATAGCACTCG; fancd2, GCAGCGGGCGATCCACAAAGTC/GGCCATCCTCACTCGCTCCTTCAA, fance, CGGCCTCTCGCTGTTTGGTGAC/GCGGCCTGCAGTGATTTCTTGAG; fancf, CTGCTGCAGCGGAGCGTCTG/ATTTCACTGACACAATTTATTACTAAG; fancg, GCGCACTTTTGGCTGCTCTGTGTA/ACCACCGAGCAGCATAGCAGGAGA; fancj, CCAGAGCATCCAAACCACCTACAG/GTTGTTCCCCCGTGTCTTGTTCTC; fancl, GACGGCTTCATCACAGTGCTGGAAAA/GCCTTCAGCTGGAGTGTGCGAAACT; fancm, GAGCCCCGAGGACCAGGAG/GTGGGCGCCATGAAGACGA; actin, GAGAAGATCTGGCATCACACCTTC/GGTCTGTGGATACCGCAAGATTC.
In situ hybridization to mRNA was used to detect fanc gene expression in whole-mounted embryos. For RNA probes, we in vitro transcribed clones linearized with TOPO/Not-1 and labeled them with digoxigenin-UTP using T3 RNA polymerase. Wild-type embryos (AB strain) were fixed in 4% paraformaldehyde at 41C for at least 2 days before dechorionating by hand using a dissecting microscope. In situ hybridizations were performed as described [42]with several individuals for each developmental stage [43]. The University of Oregon IACUC approved experiments for this study. Probes were: fanca: nucleotides 1–1529 of NM_001040635, including 94 nt of 5’UTR and exons 1–10; fancb: nucleotides 399–1127 on NM_001040636, exons 1–3; fancc: nucleotides 240–822 on NM_001040637, exons 1–7, excluding exon 4 (probe constructed from spice variant without exon 4); fancd1: nucleotides 681–1012 of NM_001110394, exons 7–10; fancd2: nucleotides 1928–2445 of NM_201341, exons 20–25; fance: nucleotides 643–1012 on NM_001040634, exons 2–10 plus 64 nt of 3’UTR; fancf: nucleotides 45–1128 of NM_001045234, including 20 nt of the 5’UTR, the entire single exon, and 41 nt of the 3’UTR; fancg: nucleotides 678–1064 of NM_205639, exons 5–7; fanci : nucleotides 3364–3910 of XM_001921104, exons 37–42; fancj: nucleotides 2048–2931 of NM_001110296, exons 12–18; fancl: nucleotides 90–809 of NM_212982, 54 nt of 5’UTR and exons 1–8; fancm: nucleotides 423- 2346 of NM_001113660, including exons 1–10 and additional exons not yet annotated because of incomplete genomic sequence; fancn: nucleotides 225–975 of XM_001919731, exons 2–8.
3. Results
3.1. Does zebrafish contain an intact FA network?
To exploit zebrafish for FA research, we must identify similarities and differences between the FA networks of zebrafish and human. The first evidence that zebrafish possesses an FA network functionally similar to human was the isolation and sequencing of fancd2 cDNA [44], followed by the identification of fancg with its multiple TPR motifs conserved with human [45]. These results suggested that zebrafish may possess a functional FA network.
Evidence that zebrafish has a full FA genetic system came with the isolation and characterization of cDNAs and/or genomic BAC clones for zebrafish orthologs for the remaining human FA genes known at the time (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, and FANCL) [46]. The sequence of coding regions showed that zebrafish FA proteins often share low levels of overall identity with the human genes -- for example, Fancd1, Fancb, Fancf, and Fance proteins have only 21–28% amino acid identities between the zebrafish and human proteins, but some regions within these proteins were more highly conserved, suggesting regions of functional significance. Proof that the zebrafish genes are in fact orthologs of the human genes comes from three independent data sets. First, the intron/exon organization of the zebrafish/human ortholog pairs is nearly identical. Second, amino acid hydrophobicity plots are extensively correlated, showing overall conservation of protein shape. Third, comparative synteny analysis showed that the neighbors of zebrafish fanc genes are generally orthologs of human FANC gene neighbors. This shows that chromosome regions containing fanc/FANC genes have been evolutionarily conserved for the 450 million years that separate zebrafish and human from their last common ancestor [47].
Four human FA genes have been discovered since the original genomic work on zebrafish (FANCI [20,22], FANCJ [23,24], FANCM [13]) and FANCN [26,48,49]. We report here the isolation and characterization of the zebrafish orthologs of these remaining FA genes.
3.1.1. FANCI
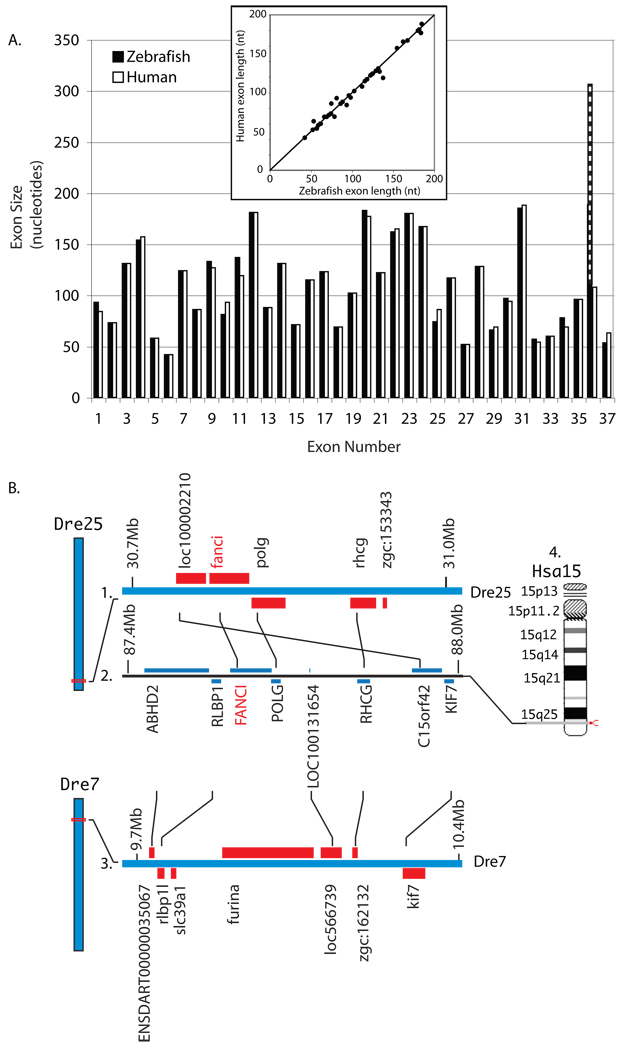
A tBLASTn search of the zebrafish genome with the human FANCI protein sequence showed evidence for fanci on Danio rerio linkage group 25 (Dre25, NW_001878626), with significant hits for 28 human exons. A Fanci protein model (XP_001921139) derived by automated identification contains 1,367 amino acids, slightly longer than human FANCI isoform-1 (NP_001106849) with 1,328 amino acids. Alignment of the zebrafish protein model to human FANCI isoform-1 revealed 56% overall amino acid identity, making this the most highly conserved FA protein sequence in zebrafish/human comparisons [46]. Exons 1–35 of zebrafish Fanci and human FANCI are similar in size (Fig. 1A). A plot of the length of each zebrafish exon vs. the length of its human orthologous exon (insert Fig. 1A) has a correlation coefficient r of 0.97. Our search of zebrafish ESTs, however, suggested at least two splice variants for the carboxy-terminal end. The common variant (e.g. EST EB929584) contains two additional exons, for a total of 37, the same as in human. Other ESTs (e.g. AL922844), however, contain a long carboxy-terminal sequence that translates to 101 amino acids. (Figure 1A). Using primers designed from Zv7, we cloned and sequenced an 802bp fragment of fanci from embryonic cDNA (submitted to GenBank as accession FJ032296). The zebrafish Fanci protein predicted from our cDNA is identical to human FANCI for the LVLRK motif (human residues 519–523), which contains lysine K523, the site of monoubiquitination [22]. At the site of four human FANCI missense mutations (http://chromium.liacs.nl/LOVD2/FANC/home.php?select_db=FANCI), the wild-type residues are identical in zebrafish at two sites (G422G and R1285R), similar at one site (L289F) and non-conserved with respect to the wild-type human protein only at one site (C750N).
Figure 1.
Fanci. A.) Comparison of orthologous exons for zebrafish fanci (white) and human FANCI (black). Striped exon indicates length of zebrafish exon 36 splice variant. Insert: Plot of the length of each zebrafish exon vs. the length of its human orthologous exon. The diagonal indicates equality. B. Conserved syntenies for FANCI. B1.) A portion of zebrafish linkage group Dre25. B2.) A portion of human chromosome 15 (Hsa15). FANCI and POLG are nearest neighbors in human and their orthologs are nearest neighbors in zebrafish. B3.) A portion of Dre7, showing the duplicated region of Hsa15 lacking a fanci gene. Duplicated co-orthologs of RHCG (called rhcg and zgc:162132) lie on Dre25 and Dre7. For three genes, only one duplicate is shown and the other duplicates are separated from the illustrated genomic segments by inversions: FURIN (furina, Dre7_9,888,462bp and furinb, Dre25_5,717,183bp), ABHD2 (ENSDARG00000025797, Dre7_9,734,476bp and zgc:153750, Dre25_9,887,349bp), and RLBP1 (rlbp1, Dre25_9878310bp and rlbp1l, Dre7_9758124bp). B4.) Hsa15 showing the location of FANCI.
To test for conservation of the fanci/FANCI genomic region, we investigated conserved syntenies. Immediately adjacent to fanci in zebrafish linkage group 25 (Dre25) lies plog, and rhcg (Fig. 1B1) and the orthologs of these three genes on human chromosome 15 (Hsa15q26.1) are also neighbors in the same order with the addition of a single unverified predicted sequence (Fig. 1B2, B4). This conserved synteny data shows that the chromosome segment containing fanci/FANCI has remained intact for at least the last 450 million years.
After the divergence of the ray fin fish lineage (which includes zebrafish and other teleosts) and the lobe fin fish lineage (which includes coelacanths and humans) a genome duplication event occurred in the ray fin fish lineage before the explosive diversification of teleosts about 300 million years ago [50–53]. Within a few tens of millions of years after the duplication event, most duplicates reverted to singleton status, but today’s teleosts retain duplicates of about 30% of human genes. Due to this genome duplication, zebrafish has two copies of the FANCI-containing region of Hsa15, one on Dre25 (Fig. 1B1) and the other on Dre7 (Fig. 1B3). These two duplicate chromosome segments have copies of RHCG (shown in the figure) as well as FURIN, ABHD2, and RLBP1, which are separated from the segments shown in the figure by inversions. In human, RLBP1, FANCI, POLG, and LOC100131654 are adjacent; in comparison, on zebrafish chromosome Dre7, the positions of the flanking genes rlbp1l and loc566739 were maintained but the position of fanci and polg was taken by other genes (furina and slc39a1) due to chromosome rearrangements.
3.1.2. FANCJ
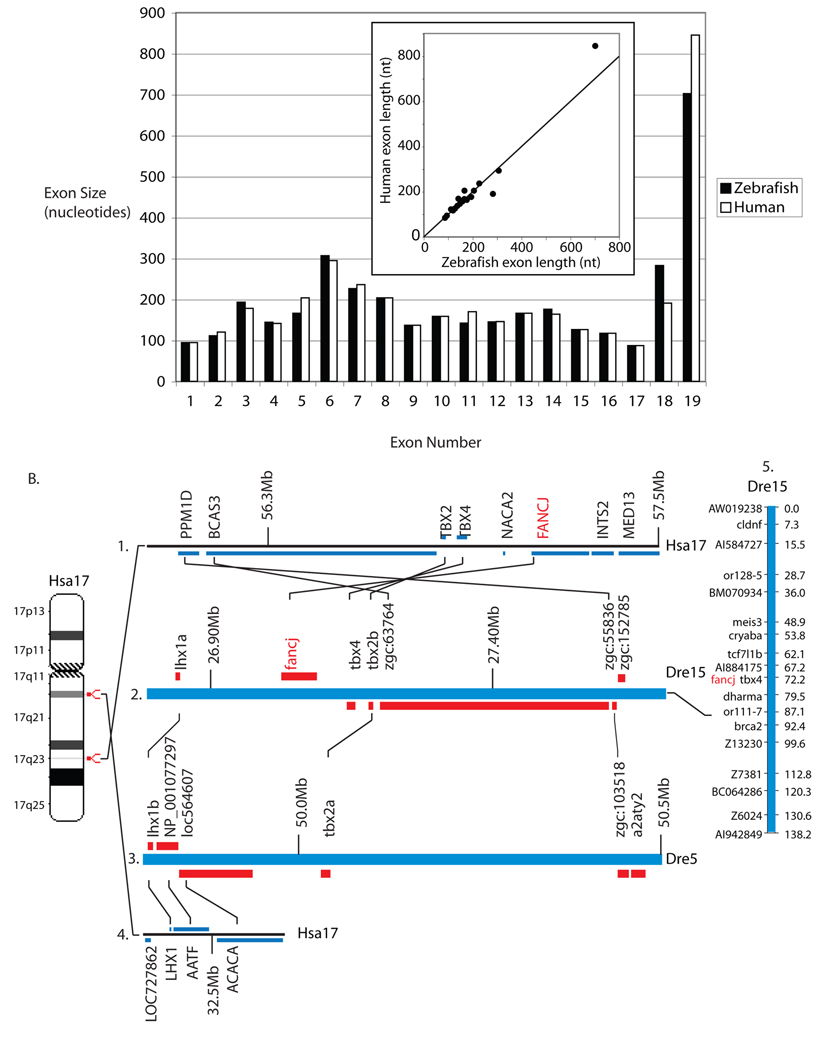
Using human FANCJ as a query in a tBLASTn search of the zebrafish genome, we identified matches for 14 of the 19 human exons on zebrafish chromosome 15 (Fig. 2A). We confirmed the location of fancj on chromosome 15 (Fig. 2B) by genetic mapping on the heat shock double haploid meiotic mapping panel [41]. We used data from the zebrafish genome project to design primer sets for standard PCR and RACE using zebrafish embryonic cDNA as template. Cloning and sequencing of overlapping fragments yielded a full length coding sequence for zebrafish fancj (GenBank submission EF088194). We compared our cDNA sequence by BLASTn to publicly available zebrafish EST sequences and found perfect matches between the 5’ and 3’ ends (CO360039 and BQ132722, respectively), thus corroborating our cDNA sequencing and gene model. The predicted fancj translation product contains 1,218 amino acid residues with 47% overall identity to human. The fancj coding sequence is distributed on 19 genomic exons, the same number, size, and approximate length as human FANCJ exons (Figure 2A). A plot of the length of each zebrafish exon vs. the length of its human orthologous exon (insert Fig. 2A) has a correlation coefficient r of 0.99. Zebrafish Fancj contains the helicase superfamily C-terminal domain, which is 79% identical to the corresponding domain in human FANCJ. Six missense mutations found in human FANCJ patients are listed in the Fanconi anemia database (http://chromium.liacs.nl/LOVD2/FANC/home.php?select_db=FANCJ). Zebrafish has the same amino acid as human at five of these sites (R251, Q255, H396, W647, and R707) and one (A349) is biochemically similar (S) in zebrafish.
Figure 2.
Fancj. A) Comparison of orthologous exons for zebrafish fancj (white) and human FANCJ (black). Insert: Plot of the length of each zebrafish exon vs. the length of its human orthologous exon. The diagonal indicates equality. B1.) A portion of Hsa17 including FANCJ. B2.) A portion of Dre15 including fancj. The order of five orthologs in a row including FANCJ/fancj is conserved between human and zebrafish genomes. B3.) A portion of Dre5 containing duplicates of genes near fancj (lhx1b/lhx1a; tbx2a/tbx2b; and zgc:103518/zgc:55836) but not fancj. B4.) Part of Hsa17 orthologous to a portion of Dre5. B5.) We mapped fancj to Dre15 using primers CCCCTGTAAAGCGTATCCCTCTCA and TTGCAATAACAGACAGAATAGATGGACTCA (NW_001877562 nucleotides 1506166–1506522).
Not only is fancj similar to FANCJ in sequence and exon structure, but also the chromosomal neighborhood is highly conserved. Four of the five genes to the right of FANCJ on Hsa17 are conserved adjacent to fancj in zebrafish chromosome Dre15 in the same order (Fig. 2B1 and 2). These results show continuity of chromosomal synteny in this region since the divergence of teleost and mammalian genomes. Two genes flanking fancj (lhx1a and tbx2b) are both duplicated on Dre5 (Fig. 2B3), a legacy of the teleost genome duplication event. The relative position of fancj on Dre5 is occupied by the orthologs of two genes adjacent to LHX1, the human ortholog of zebrafish lhx1a (Fig. 2B4). Thus, after the genome duplication, the fancj gene was retained on Dre15 but lost from Dre5, while some flanking genes were retained in duplicate on both chromosomes.
3.1.3. FANCM
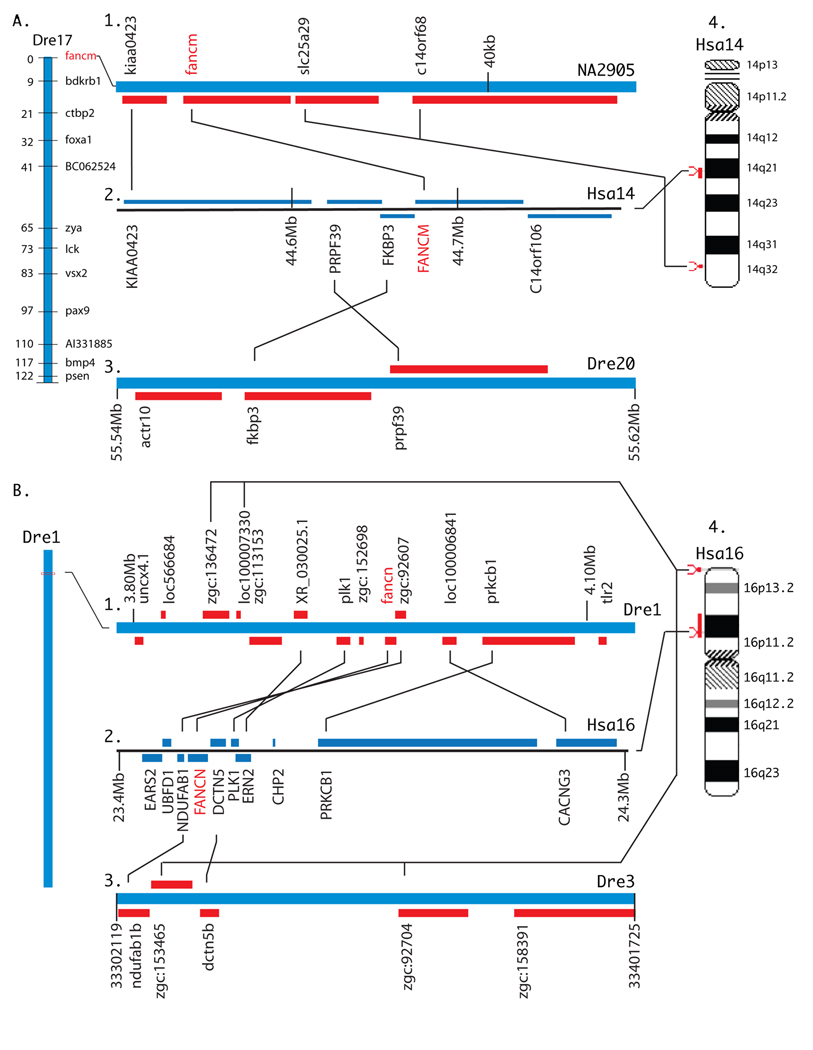
A tBLASTn search using human FANCM as query produced significant hits to just four exons on unassembled contig Zv7_NA1567. Because sequencing of this region in the zebrafish genome project is currently incomplete, we queried another sequenced teleost genome, the pufferfish Fugu rubripes (http://fugu.biology.qmul.ac.uk/blast/). Using the human FANCM sequence, we found significant matches to 21 exons on Fugu scaffold N000010. We then used the translated Fugu exons to query the zebrafish genome and EST databases for other fancm sequences. These searches identified matches to the Fugu sequences on several zebrafish sequences, including contig NA465, the computational prediction XM_689089, and EST CK703773. Using these additional sequences together with sequences from the original NA1567, we designed overlapping primer sets for 3’ RACE and standard PCR amplifications, from which we cloned and sequenced the entire zebrafish fancm cDNA (GenBank submission EF088195). Our cDNA sequence is 97 to 100% identical to sequences on four zebrafish unassembled contigs (nucleotides (nt) 1–862 on NA1567, nt 874–1313 on NA2384, nt 1693–4207 on NA2905, and nt 4202–4601 on NA1217). Finally, the 3’ end of the transcript (nt 5194–5504) appears on assembled chromosome Dre17 in Zv7. The predicted fancm translation product is 1,761 amino acids with 36% overall identity to human (Fig. 3A). Zebrafish Fancm contains the characteristic N-terminal helicase domain [13], which is 62% identical to human FANCM. No FANCM missense mutations have been reported.
Figure 3.
Fancm and Fancn. A.) Fancm. A1.) Part of Dre17 containing fancm. We mapped fancm using mapping primers TGGCTAGTGAAAATGGCGAGTGG and TACGGCTGAGTGGAGGAACATTACA (NW_001881046 nucleotides 6059–6298). A2.) Part of Hsa14 containing FANCM. A3.) Much of Dre20 is a duplicate of Dre17, but it has no fancm ortholog. A4.) Hsa14 showing the location of FANCM. B. Fancn. B1.) Part of Dre1 containing fancn. B2.) Six of eight genes surrounding FANCN have orthologs surrounding fancn. B3.) The two genes flanking FANCN in Hsa16 have orthologs on Dre3, but there is no fancn gene between them. NDUFAB1 is duplicated on Dre3 and Dre1 (ndufab1b and zgc:92607). B4.) Hsa16 showing the location of FANCN.
A portion of our cDNA sequence for fancm hits Scaffold Zv7_NA2905, which contains zebrafish orthologs of human FANCM exons 11, 12, 13, and 19. Because this scaffold has not yet been integrated into the zebrafish genome, we used the heat shock doubled haploid meiotic mapping panel [41] to locate fancm on the upper end of LG17 (Fig. 3A1). In a BLASTX search of the human genome, scaffold Zv7_NA2905 returned FANCM and three other genes: KIAA0423, which resides three genes from FANCM in Hsa14q21), and SLC25A29 and C14orf68, which lie near each other in Hsa14q32 (Fig. 3A2, 4). We conclude that the KIAA0423/FANCM conserved synteny was in place before the divergence of the zebrafish and human lineages; this independent evidence from genomic neighborhood analysis is as predicted from the hypothesis that fancm and FANCM are orthologs. Orthologs of FANCM’s three immediate neighbors (FKBP3, PRPF39, C14orf106, Fig. 3A2) were not on contig NA2905, so we searched for their orthologs in the zebrafish genome. Results showed that an ortholog of C14orf106 resides on Dre17 (sequence si:busm1-142b24.1 beginning at location 33,249,930) quite a distance from fancm (not diagramed) and orthologs of the other two genes (FKBP3 and PRPF39) are adjacent on chromosome Dre20, although fkbp3 (at location Dre20:55564447- 55588365) is not annotated in Zv7 (Fig. 3A3). Large portions of Dre17 and Dre20 are known to be duplicated portions of Hsa14 [52,54,55]; thus, the missing duplicate of fancm would likely have originally resided on Dre20 but has since been lost.
3.1.4. FANCN
A search of the zebrafish genome by tBLASTn using the human FANCN sequence returned many hits with low levels of identity between 20 and 25%. Two of the marginally best hits were on Linkage Group 1 (NW_001878018) and corresponded to exons 3 and 13 in human FANCN. These two short zebrafish genomic sequences were used in reciprocal BLASTX queries to human refseq proteins and recovered FANCN. We used Genomescan (http://genes.mit.edu/genomescan.html) to construct a translational model for zebrafish Fancn by comparing the human FANCN protein (NP_078951) to the zebrafish genomic contig (NW_001878018). Although the coding sequence for our protein model had 13 exons, the same as human [25,26], the sequence contained 1,381 amino acids, substantially longer than its human counterpart with 1,186 amino acids. A BLASTx search of our translational model against zebrafish ESTs confirmed approximately 50% of the coding sequence by EST hits. Large gaps in some EST/protein local alignments, however, indicated inaccuracies in the model. Using primers designed from our gene model, we cloned and sequenced a portion of fancn from zebrafish embryonic cDNA and deposited the sequence in Genbank (accession FJ032295).
Given the low level of sequence identity between human FANCN and the zebrafish predicted Fancn protein (31% in our cloned cDNA fragment but much less in the rest of the predicted sequence), independent evidence of orthology is essential. Conserved synteny analysis showed that zebrafish fancn is embedded in the middle of seven genes, six of which are orthologous with six of eight genes immediately surrounding human FANCN (Fig. 3B1 and 2). Human chromosome Hsa16p12 conserves the order of four of five genes in a row, including fancn/FANCN (Fig. 3B4), and the next two genes are inverted and separated from the group of four by a single gene (Fig. 3B1 and 2). Despite the low percent identity between the zebrafish and human Fancn proteins, the location of the zebrafish gene in an extensive region of conserved synteny leave no doubt that this is the location of the zebrafish ortholog of FANCN. Cloning the rest of the zebrafish fancn cDNA is necessary to fully understand the structure of this gene.
To look for the duplicated FANCN chromosome segment, we investigated genes that flank FANCN in human, and found that the two immediate neighbors (NDUFAB1 and DCTN5) are duplicated in zebrafish, one copy on Dre1 as described above and a duplicate copy on Dre3 (Fig. 3B3). This duplicated segment has a sequence called zgc:153465 instead of a FANCN ortholog located between ndufag1b and dctn5b (Fig. 3B3). This Dre3 sequence zgc:153465 is an ortholog of RPUSD1 located in Hsa16p13.3 immediately adjacent to CHTF18 and GNG13, which are orthologs of the Dre1 sequences zgc:136472 and loc100007330, respectively, the neighbors of rpusd1(zgc:153465)(Fig. 3B4). Note that, judging from the order of transcription of the NDUFAB1 and DCTN5 orthologs in human and zebrafish, an inversion must have occurred between these two genes in Dre3 after the genome duplication event that moved the Hsa16p13.3 ortholog zgc:153465 to a location between ndufab1b and dctn5b; this inversion may have simultaneously destroyed the duplicate fancn gene, or it may have decayed before or after the translocation. BLAST searches revealed no trace of fancn between ndufab1b and dctn5b on Dre3. We conclude that fancn is present on Dre1 in a well-conserved chromosome region and that the region is duplicated on Dre3 except for the loss of the second fancn copy.
In partial summary, the isolation of the last four zebrafish orthologs of human FANC genes supports the conclusion that zebrafish has a complete FA system. Furthermore, this work shows that all of the 13 zebrafish fanc genes were reduced from two copies to one copy after the teleost genome duplication. The genomic stage is set to exploit this knowledge to further understand the mechanisms of FA disease.
3.2. When and where are zebrafish FA genes expressed during development?
Abnormal blood cell development, either bone marrow failure or leukemia, is the main cause of morbidity and mortality in FA [56,57]. But FA is often accompanied by characteristic congenital abnormalities including slow growth, short stature, microcephaly, and microphthalmia [58]. The most common congenital anomaly in FA is an abnormal or missing thumb and radius, but kidney and reproductive organs are also frequently affected [59]. A gap in our knowledge is the mechanism by which FA leads to developmental anomalies in skeleton, blood, eyes, and other organs. One hypothesis is that FA genes are expressed preferentially in tissues that appear frequently in clinical manifestations of FA. Furthermore, the hypothesis that fanc genes consistently function as a unit in a single DNA-repair pathway predicts that FANC genes should have identical expression patterns during development. To test these hypotheses, we investigated the expression patterns of all thirteen FA genes in zebrafish embryonic development.
3.2.1. Expression of fanc genes during embryonic development: RT-PCR
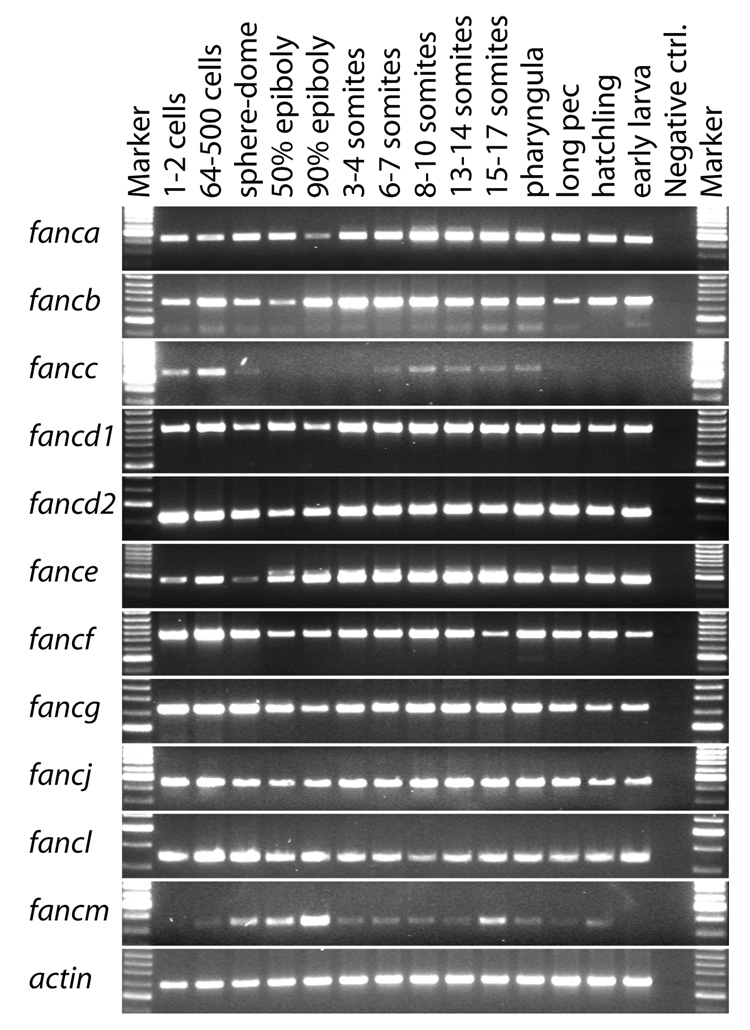
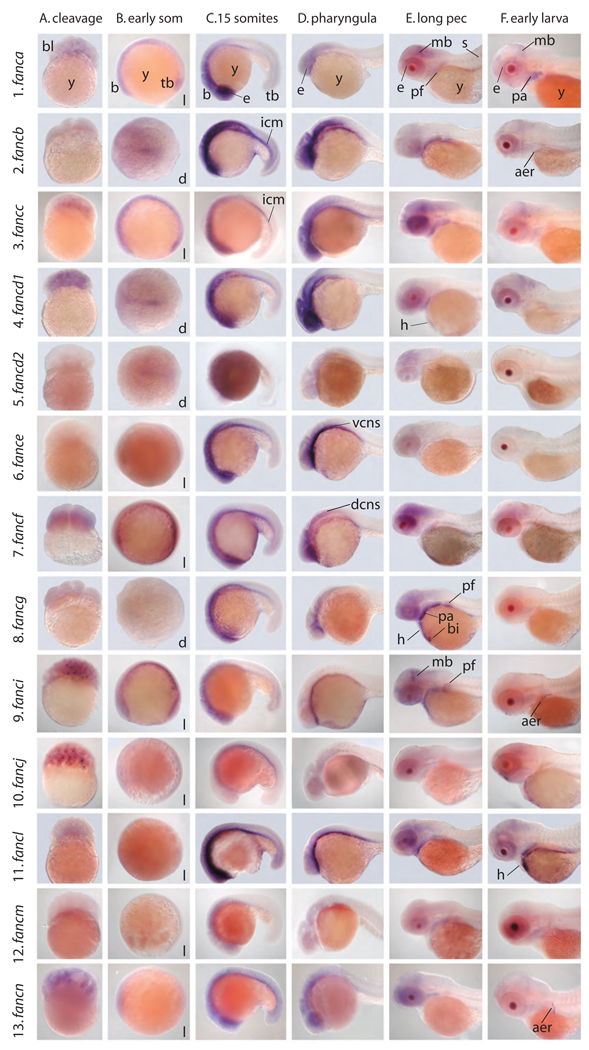
As an initial assay for the expression of fanc genes during development, we used RT-PCR to amplify 11 fanc genes at 14 different developmental stages from shortly after oviposition to 5 dpf larvae (Fig. 4). PCR conditions were designed to give a qualitative rather than quantitative view of fanc gene expression, and actin served as a loading control. Transcripts were detected from all developmental stages examined for nine of the 11 fanc genes tested (fanca, fancb, fancd1, fancd2, fance, fancf, fancg, fancj, and fancl) (Fig. 4). For this group of genes, bands were generally strong in early cleavage, decreased somewhat in intensity through epiboly as the embryo encompsses the yolk, and then increased during segmentation stages, remaining strong until at least 5 dpf, which is two or three days after hatching. Like most fanc genes, fancc showed maternal transcripts during early cleavage stages that diminished during epiboly. This was followed by reappearance of zygotic transcript during early somitogenesis to 1 dpf, which is the developmental phase during which primitive erythropoiesis occurs [60–63]. In contrast to most fanc genes, however, fancc showed decreasing band strength in 2 to 5 dpf embryos (Fig. 4). The fancm gene showed the most distinctive expression pattern: fancm appeared to have little maternal transcript followed by increasing band intensity as the number of zygotic cells increased, and then showed decreasing band intensity after the end of epiboly (Fig. 4). We conclude that transcripts of most fanc genes are maternally loaded into oocytes during oogenesis; that maternal transcripts gradually disappear during cleavage, and that zygotic transcription begins early. The distinctive expression of some fanc genes, however, may signal some specialized functions.
Figure 4.
Expression of fanc genes during development as assayed by reverse transcriptase PCR. Columns are Marker (size standard), 1–2 cells (1hpf), 64–500 cells (2–3hpf), sphere-dome (4hpf), 50% epiboly (5hpf), 90% epiboly (9hpf), 3–4 somites (11hpf), 6–7 somites (12hpf), 8–10 somites (13hpf), 13–14 somites (15hpf), 15–17 somites (16hpf), pharyngula (24hpf), long pec (long pectoral fin, 48hpf), hatchling (72hpf), early larva (5dpf), Negative ctrl (control). Rows are fanc genes; actin was amplified as a loading control.
3.2.2. In situ hybridization in embryo whole mounts
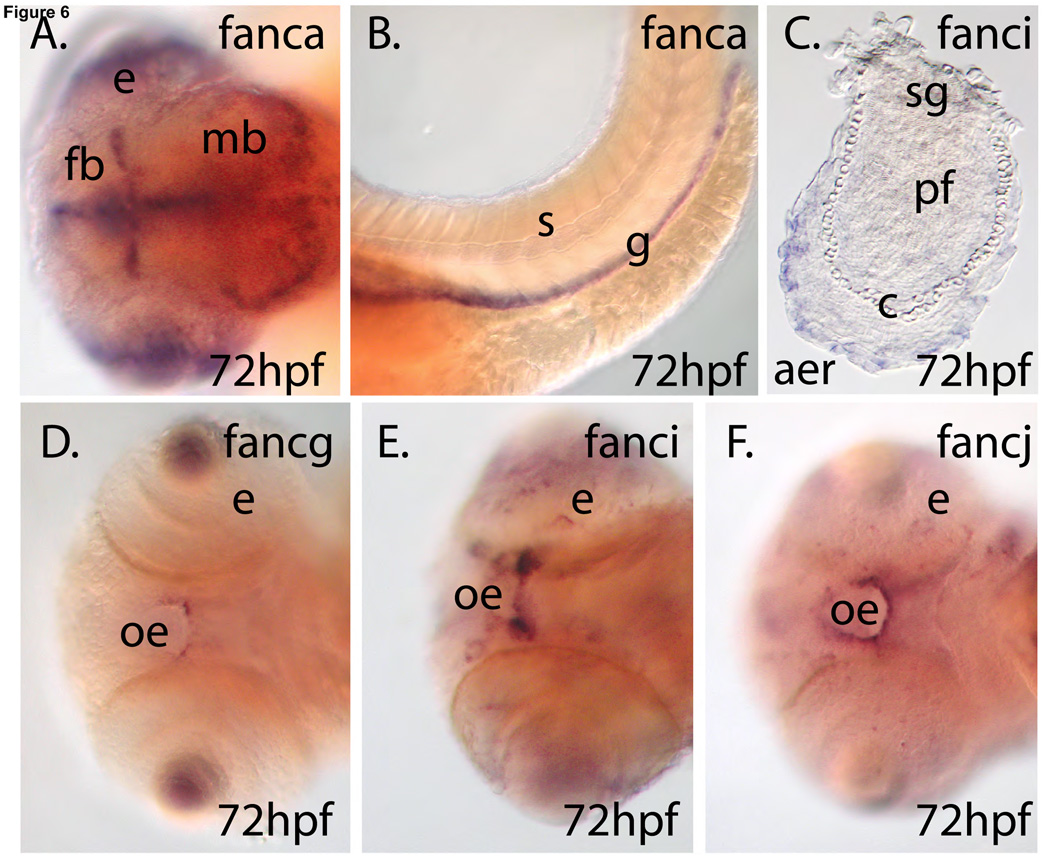
To understand tissue-specific gene expression patterns, we performed in situ hybridization experiments on embryo whole mounts. Consider first fanca, the zebrafish ortholog of the most frequently mutated gene in FA patients [64]. As expected from the RT-PCR results (Fig. 4), fanca transcript was already present in cleavage stage embryos at less than 2 hpf (Fig. 5A1 (column A, row 1)). At 10 hpf, as zebrafish embryos completed gastrulation and entered the segmentation stage (somite formation), fanca was expressed broadly throughout the embryo and most strongly in the rostral midline (Fig. 5B1). By 16 hpf, fanca expression appeared predominantly in the eye and in specific regions of the forebrain, midbrain, and hindbrain of the central nervous system (CNS)(Fig. 5.C.1). In the next few hours (Fig. 5.D.1), zebrafish embryos down-regulated the expression of fanca in the eye and refined expression to specific regions of the CNS. At 48 hpf (long pectoral fin) and 72 hpf (early larva), the strongest remaining expression domain was in the posterior midbrain (the tectum), with decreased intensity in the eye and branchial arches (Fig. 5E1 and 5F1). At 72 hpf, expression in the CNS was confined to a distinctive pattern in the ventricular zone of the forebrain and midbrain and the posterior of the tectum, sites of rapid neuroblast divisions (Fig. 6A). At this stage, expression of fanca was especially strong in the developing intestinal tract (Fig. 6B). In the pectoral fin bud, the homolog of the mammalian forelimb, fanca was expressed in the endochondral disc, which is fated to form the fin skeleton (Fig. 5E1). Although staining in the lens of the eye was strong, many genes are expressed in this domain and so it is unlikely that this indicates a special role for fanca in the lens.
Figure 5.
Developmental expression of fanc genes assayed by whole mount in situ hybridization. Columns represent developmental stages: A.) cleavage (<2hpf), B.) early som (early somitogenesis, 10–12hpf), C.) 15 somites (16hpf), D.) pharyngula (24hpf), E. long pec (long pectoral fin, 48hpf), F. early larva (72hpf). Rows represent fanc genes. Abbreviations: aer, apical ectodermal ridge; b, brain; bi, blood island; bl, blastomeres; d, dorsal view; dcns, dorsal central nervous system; e, eye; h, heart; icm, intermediate cell mass; l, lateral view; mb, midbrain; pa, pharyngeal arches; pf, pectoral fin bud; s, somites; tb, tailbud; vcns, ventral central nervous system; y, yolk.
Figure 6.
Expression of some fanc genes. A.) fanca is expressed in the ventricular zone of the brain and B.) in the gut. C.) fanci is expressed in the apical ectodermal ridge of the pectoral fin. D.–F.) fancg, fanci, and fancj are expressed in the oral epithelium. Abbreviations: aer, apical ectodermal ridge; c, chondrocytes; e, eye; fb, forebrain; g, gut; mb, midbrain; oe, oral epithelium; pf, pectoral fin; s, somites; sg, shoulder girdle.
Expression patterns of the zebrafish orthologs of other FA genes were largely similar to that for fanca, with some notable exceptions. Expression of fancb at 16 hpf (15 somite stage)(Fig. 5C2) extended more strongly into the intermediate cell mass (ICM), the site of primitive hematopoiesis, which begins to occur at about this time [60]. The 24 hpf pharyngula stage retained heavy fancb expression in the CNS but expression in the ICM diminished and was subsequently down-regulated in a manner similar to fanca. Like fanca, fancc maternal message was abundant (Fig. 5A3 and Fig. 6A). Embryos showed accumulation of fancc transcript at low levels in the CNS from 16 hpf onward (Fig. 5B,C,D). At 24 hpf, fancc expression was prominent in the eye, forebrain, midbrain-hindbrain border, and spinal cord (Fig. 5D, H). In addition, like fancb, fancc was expressed in the ICM at 16 through 24 hpf in the ICM.
fancd1 was expressed like fanca, but a bit stronger in the ICM and CNS at 16 hpf and more strongly in the heart at 48 hpf (Fig. 5C4,D4,E4). Like fanca, fancd1 was expressed at 72 hpf in the neural retina, brain ventricular zone, and tectum (Fig. 5E4). The expression pattern of fancd2 was similar to fanca in density and location, as would be expected if a major role of Fanca were to activate Fancd2 protein. fance expression was strong in the ventral CNS at 24 hpf (Fig. 5D6), while fancf, unique among fanc genes tested, was expressed dorsally in the CNS at 24 hpf (Fig. 5D7) and remained strong in the eye at least until 48 hpf. Expression of fancg was similar to fanca, except that fancg was expressed strongly in the branchial arches, heart, pectoral fin bud, and yolk blood islands at 48 hpf (Fig. 5E8). At 72 hpf, fancg was expressed strongly and specifically in the oral epithelium (Fig. 5F8), the site of squamous cell carcinomas in FA patients [65]. Like fancg, fanci was expressed at 48 hpf in the branchial arches, heart, and yolk blood islands, and oral epithelium (Fig. 5F), but in addition, was expressed strongly in the midbrain (Fig. 5E). At 48 hpf, fanci was expressed in the endochondral disc, but expression in this domain had faded by 72 hpf and was replaced by distinctively strong staining in a thin row of cells at the distal margin of the pectoral fin bud called the apical ectodermal ridge (AER) (Fig. 5F), a signaling center for outgrowth of fish fin buds and tetrapod limb buds [66,67]. The fancb, fancj, and fancn genes also showed strong expression in the AER. Like fancg, fanci transcript accumulated in the oral epithelium (Fig. 6D). In zebrafish embryos, fancj was expressed like fanca but generally weaker, except for maternal message (Fig. 5A10) and the oral epithelium (Fig. 6F). Zebrafish fancl exhibited the same CNS and ICM expression domains as fanca at 16 hpf, but lost CNS expression by 24 hpf. The heart expressed fancl at 72hpf. The fancm and fancn genes were expressed generally like fanca, except that fancn expression was detected more strongly in the apical ectodermal ridge of the pectoral fin bud (Fig. 5F13 and Fig. 6C).
3.2.3. Gene-specific fanc expression domains in adult zebrafish
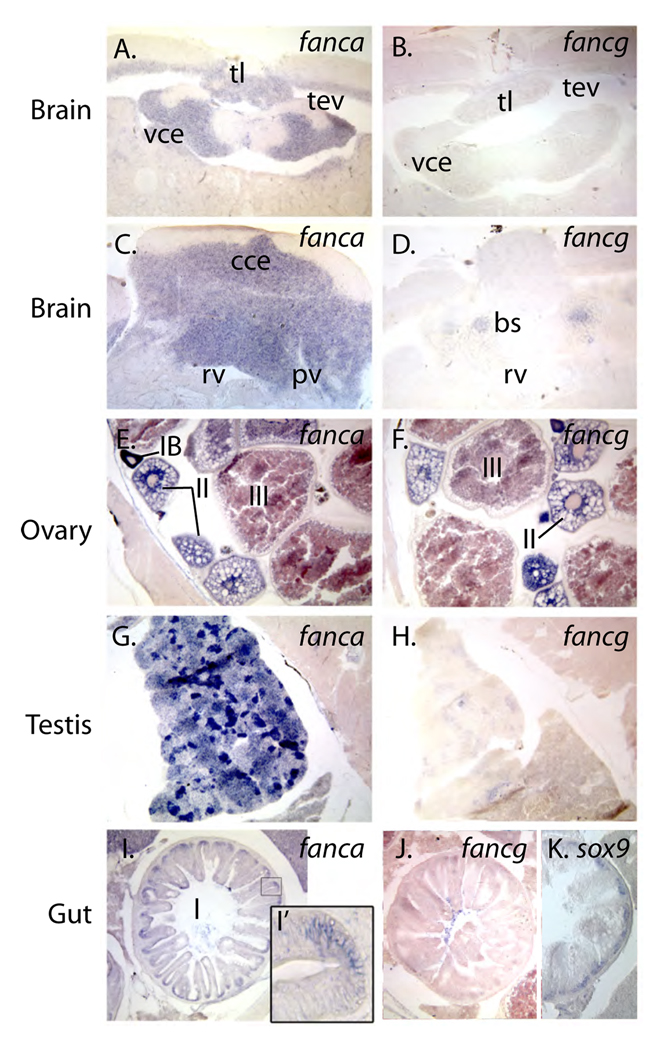
To learn whether expression patterns detected in embryos extended into the adult phase of the life cycle, we performed in situ hybridization experiments on histological sections of adult zebrafish using probe to fanca and fancg, the orthologs of the most frequently mutated genes in human FA patients. In hatching stage embryos, fanca and other fanc genes are expressed in the ventricular zones of the developing brain where cells are proliferating. Likewise, in the adult brain, fanca -- and to a lesser degree fancg -- are expressed in the periventricular gray zone (Fig.7A, B), a site of active proliferation in the adult zebrafish brain [68]. In the valvula cerebelli, however, fanca is expressed strongly in the granular layer, but the zone of most active proliferation is in the molecular layer [68]. In the corpus cerebelli, the most actively proliferating zone is the molecular layer [68], which does not express fanca (Fig. 7C). The granular zone of the corpus cerebelli expresses fanca most strongly while these cells are proliferating, but expression is reduced compared to cells in the molecular layer. Fig. 7D shows that fancg is expressed strongly in specific brain stem nuclei.
Figure 7.
Adult expression patterns. A., C., E., G., I.) fanca. B., D., F., H., J.), fancg. A.–D.) Brain, showing that fanca and fancg are expressed in different regions of the adult brain. E., F.) Ovary, showing that both fanc genes are expressed in stage I oocytes. G., H.) Testis, showing expression of fanca and fancg in young spermatocytes. I.–K.) Intestine, showing expression in the generative layer, co-expressed with sox9b, a marker of proliferative cells in the intestine. Abbreviations: IA, IB, II, III, stages of oocyte development; bs, brain stem nuclei; cce, corpus cerebelli; pv, periventricular gray zone; rv, rhombencephalic ventricle; sc, stratum compactum; tev, tectal ventricle; tl, torus longitudinalis; vce, valvula cerebelli.
Infertility is a common feature of the FA phenotype in human patients and in mutant mice [69–71]. We found that the gonads of adult zebrafish strongly express fanc genes. Both fanca and fancg are expressed in immature oocytes, most strongly in the earliest stages (Stage IA and IB), and the message was maintained through more mature stages (II and III) (Fig. 7E, F). This maternal message was still detected in cleavage stage embryos (Fig. 5). Testis expressed fanca and fancg strongly only at specific stages, and fanca was expressed more strongly than fancg (Fig. 7D, I).
In the intestine, fanca, but not fancg, is expressed at the base of intestinal folds in the stratum compactum (Fig. 7I, J). This is a region of rapid proliferation [72] that is marked by sox9b expression (Fig. 7K).
4. Discussion
4.1. Zebrafish has a complete complement of Fanc proteins
The data we report here complete the identification of zebrafish orthologs of human FA genes and reveal the dynamic expression patterns of these genes in embryonic and adult life stages. The isolation of zebrafish orthologs of all 13 human FANC genes is significant for several reasons. First, it shows that the full complement of FA genes had a more ancient origin than had originally been assumed. The presence of 13 FA genes in zebrafish shows that the genomic system had evolved before the divergence of ray fin and lobe fin fishes about 450 million years ago.
Second, knowing that zebrafish has all 13 FA genes supports the notion that zebrafish will be a suitable model for the investigation of FA. Other non-mammalian forward-genetic models, including the fruitfly Drosophila melanogaster and the nematode worm Caenorhabditis elegans, have parts of the FA system, but do not have the upstream, regulatory components of the core complex [73–78]. In addition to the FA network, zebrafish also shares with mouse and human a common organization of organs, organ systems, and physiology that validate connectivities among the systems.
Third, our finding that many zebrafish FA components show quite low levels of sequence conservation with the human Fanc proteins presents advantages for analysis. The 450 million years of phylogenetic divergence separating zebrafish and humans ensures that non-essential genome sequences exhibit considerable divergence. This situation allows functionally constrained sequences, including regulatory elements in genomes and functional motifs in proteins, to identify themselves by evolutionary conservation [79]. Thus, retained evolutionarily conserved domains highlight protein motifs and amino acid positions that provide a focus for targeted mutagenesis studies designed to link protein structure to function.
Finally, the demonstration that zebrafish has all 13 FA genes suggests that other teleost fishes probably also have a complete FA system. In teleosts such as stickleback and killifish, some populations of which inhabit waters polluted with DNA-damaging contaminants [80], may have evolved genetic modifiers of the FA system that allow them to repair DNA more efficiently. (It has been shown that these fish can evolve surprisingly rapidly over several decades to adapt to changing environments [80–84].) Work should be directed towards the identification of evolved genetic changes and physiological adaptations in vertebrates that enhance survival in the presence of DNA damaging agents. Such knowledge might help in the design of innovative therapies for human FA patients to survive in the presence of greater than normal DNA damage.
Curiously, we find no evidence that any of the fanc genes currently exist as duplicate copies in the zebrafish genome. Because about 30% of zebrafish genes remain duplicated from the teleost genome duplication event [51], it is unlikely that for 13 randomly chosen genes, none would be present in duplicate copy (1.6 × 10−7). Thus, the finding that all 13 fanc genes are present as singletons suggests that the evolutionary pressures on this gene set differ in some way from the selective forces acting on most genes in the genome. The conclusion that Fanc genes do not appear to tolerate duplication extends to other genome duplication events besides the pre-teleost genome duplication. Two rounds of genome duplication (called R1 and R2) preceded the explosion of vertebrate lineages after vertebrates diverged from their non-vertebrate chordate ancestors, the urochordates and cephalochordates [85,86]. In some cases, all gene duplicates from the R1 and R2 events were retained, providing four paralogs of many genes, such as Hoxa4, Hoxb4, Hoxc4, and Hoxd4 in the four Hox clusters. But most families reverted to two or three copies of the preduplication ancestral gene. In contrast, we conclude that all 13 Fanc genes reverted to single copy after R1 and R2 because none currently have paralogs within mammalian genomes. The only FA genes that appear as paralogs in mammalian genomes are Fanci and Fancd2 [20,22]. The gene duplication event that produced Fanci and Fancd2 occurred well before the R1 and R2 genome duplications because the genome of Drosophila, whose lineage diverged from chordates long before R1 and R2, contains orthologs of both Fanci and Fancd2 [20]. The overwhelming conclusion is that in three separate rounds of genome duplication, none of 13 Fanc genes were retained in duplicate copy. What is special about Fanc genes that makes natural selection drive them to single copy after genome duplication events?
One hypothesis to explain this finding relates to the mechanisms that preserve gene duplicates. According to the Duplication, Degeneration, Complementation model for the resolution of gene duplicates, the complementary partitioning of gene subfunctions between two gene duplicates initially preserves most duplicates (subfunctions are independently mutable gene functions, such as tissue or time specific regulatory elements or protein domains) [87,88]. Theory predicts that the greater the number of gene subfunctions, the greater the likelihood that both duplicates would be preserved [87]. Consistent with this idea, many developmental gene regulators have complicated gene expression patterns and both duplicates remain preserved. An example is the lhx1a/lhx1b and tbx2a/tbx2b duplicates in Fig. 2 for the teleost genome duplication (R3) and the human paralogs LHX1 and LHX5 [89]and TBX2 and TBX3 [90]for R1 and R2. In contrast to the complicated, dynamic, and highly cell-specific expression patterns of genes of the HOX, LHX, TBX and many other families, we observed rather widespread expression of Fanconi anemia genes, suggesting fewer subfunctions regulating transcript quantity. With fewer transcriptional subfunctions, FA genes would have been less likely to experience subfunction partitioning and hence less likely to be preserved as duplicate copies following genome duplication. An alternate hypothesis to explain the retention of Fanc genes in single copy would be that Fanc proteins might be required in precise stoichiometric amounts to perform the FA network’s function. The retention of multiple gene copies after genome duplication might cause problems in the protein interactions needed in the nuclear core complex or other protein-protein interactions.
4.2. Zebrafish fanc genes are expressed in tissues of clinical relevance to FA
To learn about the complexity of fanc gene expression during development, we examined in situ hybridization patterns in embryos at several stages and in adults. We found transcripts for most fanc genes already present in early cleavage stage embryos. Our RT-PCR experiments ruled out the possibility that the broad expression detected by whole-mount in situ hybridization during early cleavage was due to non-specific background staining. Because embryos at this stage have not yet passed the mid-blastula transition, the time at which zygotic genes commence expression [91], early cleavage transcripts represent maternal message molecules produced during oogenesis and stored in oocytes. Examination of adult ovaries confirmed this conclusion -- fanca, fancc, and other fanc transcripts accumulated in developing oocytes during oogenesis. Maternal fanc message is thus available for translation into Fanc protein before zygotic genes begin to be expressed at the mid-blastula transition (the tenth cleavage division at 3 hpf). These Fanc protein molecules translated from maternal message would be available for DNA repair during the rapid replications that occur in swiftly dividing cleavage stage embryos. (In zebrafish embryos, from the second to the tenth cleavage division, each cell cycle lasts just 15 minutes at 28.5C [43]. During cleavage divisions, each cell replicates the entire zebrafish genome, which is more than half the size of the human genome [92]. In contrast, in human and mouse embryos, cleavage cell cycles last an average of about 20 hours (80 times longer than zebrafish) at 37C [93,94].) Given the requirement of the FA network for the efficient resolution of interstrand cross link-induced S-phase arrest of the cell cycle [95], it is not surprising that fanc transcripts are loaded into eggs and stored as maternal message, which is thus available to help resolve DNA damage associated with the normal, but rapid, cleavage cell divisions.
Observations of adult ovaries confirmed the accumulation of fanc transcripts in developing oocytes, which is the source of the maternal message that we observed in freshly oviposited oocytes. Transcripts from both fanca and fancg accumulated in greatest concentration in stage I oocytes, the stage during which meiotic recombination occurs [96]. Expression of fanc genes in germ cells is expected under the hypothesis that these genes are employed in homologous recombination during meiosis [16,97]. In humans, FANCD2 protein accumulates in fetal oocytes, which are still undergoing meiosis, consistent with a role in meiotic recombination [97]. In addition, Fanca, Fancd1, or Fancd2 knockout mice show a defect in homologous pairing and aberrant meioses [70,98,99].
Fetal human oocytes accumulate FANCD2 protein, but adult human oocytes do not [97]. This contrasts with adult zebrafish oocytes, which accumulate transcripts for fanc genes that are maintained in mature oocytes and early cleavage embryos. This difference between zebrafish and mammals in the quantity of maternal fanc message may relate to differences in the biology of cleavage stage embryos. In mammalian embryos, activation of zygotic genes begins by the two-cell stage (first cleavage division), as evidenced by the synthesis of paternally-derived protein variants and of new mRNAs [100–102]. Thus, in mammalian embryos, machinery for the transcription of Fanc genes is already available after the first cleavage division. In contrast, zebrafish embryos begin to transcribe zygotic genes only after the tenth cleavage division [91]. Therefore, if zebrafish embryos are to have protective Fanc proteins available during cleavage, they must rely on maternal components.
Bone marrow failure and high risks of acute myelogenous leukemia are serious threats to FA patients. In zebrafish as in mammals, hematopoiesis occurs in two major phases, primitive and definitive [63,103,104]. In primitive hematopoiesis, primitive macrophages develop from the cephalic mesoderm (rostral blood islands) and move onto the yolk before migrating throughout the embryo [62] and primitive erythrocytes differentiate in the intermediate cell mass (ICM) before entering the circulation at about 24 hours post fertilization [60,61]. In the definitive phase of hematopoiesis, definitive hematopoietic progenitors arise between 28 and 48 hpf. This is when erythromyeloid progenitors develop in the posterior blood island (the posterior ICM) before 36hpf and form the definitive myeloid and erythroid cells [105,106]. Multipotent hematopoietic stem cells arise between axial blood vessels in the zebrafish equivalent of the mammalian aorta, gonad and mesonephros region (AGM), and colonize the thymus and kidney marrow, the major definitive hematopoietic organs in zebrafish [107,108]. Our expression analysis showed that at 16hpf (15 somites), nearly all fanc genes were expressed in the ICM. Functional analyses knocking down the activity of fanc genes are necessary to explore the functions of fanc genes in this domain. Expression in hematopoietic domains would be predicted under the hypothesis that fanc genes play similar roles during hematopoiesis in both zebrafish and humans.
Microcephaly and microphthalmia are typical symptoms of FA for several complementation groups. In zebrafish embryos, the brain and eyes increase rapidly in cell number during somitogenesis stages and are major domains of fanc gene expression. At about the time of hatching, expression of many fanc genes, especially fanca, fancb, fancd1, fancf, fanci, and fancn, becomes localized to the ventricular zone of the CNS. In general, cells that strongly express fanc genes in the CNS also express PCNA (proliferating cell nuclear antigen) [109,110], a marker of rapidly proliferating cells, and ascl1 (Mash1 in mouse), which controls the transition of cortical progenitor cells from proliferation to neurogenesis [111,112]. We conclude that fanc genes tend to be expressed in rapidly proliferating cells of the embryonic CNS. The strong expression of fanc genes in proliferating neural cells presumably repairs DNA damage associated with rapid cell divisions. This surmise is supported by the finding that xrcc6, a component of the nonhomologous end-joining (NHEJ) pathway of DNA repair, is also strongly expressed in the ventricular zone of the brain in hatching stage zebrafish embryos [113]. In the absence of Fanc protein action, the growth of eyes and brain is apparently slowed, leading to the observed phenotypic effects in FA patients.
The most common skeletal anomaly among FA patients is a hypoplastic thumb and radius of the pectoral appendage [56]. The homolog of the human arm in teleost fishes is the pectoral fin, but the fish fin does not have a homolog of a radius, a thumb, or other fingers [114,115]. We found strong expression of several fanc genes initially in the mesenchymal component of the fin bud, which gives rise to the skeleton, followed by strong and specific expression in the apical ectodermal ridge, a regulatory center that uses FGF (fibroblast growth factor) signaling to promote continued outgrowth and elongation of the limb bud in tetrapods and the fin bud of teleost fish [66,67,116]. Expression of FA genes in pectoral appendage buds in the patterns we observed would be predicted if Fanc proteins are important for normal appendage development in fish and humans. The specific essential role that fanc genes play in pectoral appendage development is unknown for any species, and the mechanisms that make these genes especially important for thumb and radius development remain a mystery.
After leukemias, the most frequent tumors in FA patients are oropharyngeal squamous cell carcinomas [117]. This tumor spectrum in FA patients may be related to the strong accumulation of transcripts from some fanc genes in the zebrafish mouth, assuming this expression domain is conserved with human embryos. The second most common non-hematopoietic tumors in FA patients are in the gastrointestinal tract [117]. Correlated with this observation is the specific expression of fanc genes in the rapidly proliferating intestinal stem cell domain of zebrafish larvae and adults, a domain also observed in mouse FANCD2 [97]. This expression domain would be predicted if Fanc proteins are necessary for DNA repair during the rapid cell divisions that maintain gastrointestinal integrity.
4.3. Zebrafish as a model for FA research
The use of model organisms for human disease research depends upon evolutionary constraints that result in genetic, developmental, and functional similarities between humans and other organisms. Close phylogenetic relationships among mammals, including mouse, rat, dog, and human, have made these models tremendously attractive and beneficial for the study of human disease. Phylogenetically more distant organisms, however, like Drosophila and C. elegans have also yielded important insights into the understanding and treatment of human disease. Teleost fish like zebrafish, provide especially convenient genomic, developmental, and physiological models for human health and disease because of their unique evolutionary distance from humans [118–124]. Zebrafish, as vertebrates, are close enough to humans to share important fundamental features of organ structure, physiological function, and developmental genetic regulatory mechanisms. Similarities involve organ systems important for FA, including the hematopoietic system, immune system, oral epithelia, endocrine system, central nervous system, and skeletal and muscular systems. Other useful models, like flies and worms, do not share vertebrate-specific features. On the other hand, zebrafish is sufficiently distant from humans that genetic sequences that are unconstrained by selective forces have had substantial opportunities to diverge; in contrast, sequence identity among mammals may merely reflect sites that have had insufficient evolutionary time for genetic drift to change sequence.
Zebrafish is highly tractable for disease-related studies because of their small size and rapid growth and the ability to culture large numbers of animals inexpensively. Zebrafish embryos are optically clear and develop outside the mother unencumbered by the uterus. This arrangement provides experimental access to zebrafish cells and organs for cell and tissue transplants or for the introduction of various substances, including mRNAs, proteins, gene constructs, or antisense oligonucleotides. Moreover, embryos can be exposed to environmental toxicants or libraries of drug-like molecules for chemical screens [125–130]. Importantly, zebrafish are amenable to forward and reverse genetic screens by induced mutagenesis [131–134]. Phenotypic screens for mutations have provided many models for human disease, including hematopoietic failure and carcinogenesis [123,135,136].
Tools are available to make zebrafish models for FA. Morpholino antisense oligonucleotides (MOs) inhibit splicing or translation of target genes [137,138], and have been used to effectively knockdown the expression of fancd2 [44]. The sequence information in this and other papers [46]; [139]; [44] provides data for the design of MOs targeted to any zebrafish fanc gene. MOs are commercially available for any sequenced gene, but because MOs are typically injected into early cleavage stage embryos and their concentration per cell decreases as the number of cells increases during development. RNase protection assays showed that splice-inhibiting MOs are effective only until about day five (e.g., see [42]). In some cases MO phenotypes include nonspecific toxic effects, some of which mimic expected FA phenotypes, so well designed controls are essential (see [140]). Mutations provide an alternative method to reduce the activity of zebrafish genes. Mutations can be identified in sequenced zebrafish genes either by resequencing the target gene from mutagenized chromosomes or by using TILLING [141–143]. Mutations in specific genes are more difficult to obtain than MO knockdown phenotypes, but have the advantage that an individual carries the mutation throughout its lifetime. Therefore, the phenotypic outcome of fanc gene abrogation can be studied across all ontogenetic stages rather than the first few days afforded by MOs.
Zebrafish provides an exceptional opportunity to screen for therapeutic compounds to help FA patients. For the first week of life, zebrafish can be cultured in 96 well microtiter plates. Animals hatch by 3dpf and fry possess organ systems that function like those of a human. People and zebrafish larvae have epithelia, through which substances must pass to enter the body, a liver, which can metabolize small molecules to more active or less active or toxic compounds, a nephric system that can rapidly excrete drugs, and a central nervous system sensitive to neurotoxins. Thus, developing zebrafish fry provide a model that is more similar in many ways to a human patient than is a human tissue culture cell line. Furthermore, because zebrafish are aquatic, test compounds can be placed directly into the medium, from which animals can absorb most hydrophilic and lipophilic agents [126–129,144]. The small size of zebrafish fry makes the testing of expensive compounds more cost effective than similar testing in mammals such as mouse. A whole animal model for FA disease provides a less biased approach than targeting a specific protein like FANCD2 already known to be involved in the process because whole animal tests simultaneously screen all known Fanc proteins as well as proteins that are involved but are not yet known to investigators. Elucidation of the mode of action of a small molecule that rescues a zebrafish FA model can help identify undiscovered players in a pathway, can facilitate an understanding of epistatic relationships of various players, and can provide lead compounds for therapeutic molecules to improve outcomes for FA patients.
Exploiting the advantages zebrafish provides for the investigation of FA should improve our understanding of the basic molecular genetic and developmental mechanisms of the disease and the screening of small molecules for compounds that ameliorate the symptoms of zebrafish models of FA should provide lead compounds for the development of therapeutics to help FA patients.
Acknowledgements
We thank grant sponsors: The Fanconi Anemia Research Foundation and the National Center for Research Resources (NCRR) grant numbers R01 RR10715 and R01 RR020833, National Institute of Child Health and Human Development grant number P01 HD22486. The paper’s contents are solely the responsibility of the authors and do not necessarily represent the official views of grant sponsors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
"The authors declare that there are no conflicts of interest. "
References
- 1.de la Fuente J, Reiss S, McCloy M, Vulliamy T, Roberts IA, Rahemtulla A, Dokal I. Non-TBI stem cell transplantation protocol for Fanconi anaemia using HLA-compatible sibling and unrelated donors. Bone Marrow Transplant. 2003;32:653–656. doi: 10.1038/sj.bmt.1704219. [DOI] [PubMed] [Google Scholar]
- 2.Dufour C, Svahn J. Fanconi anaemia: new strategies. Bone Marrow Transplant. 2008;41 Suppl 2:S90–S95. doi: 10.1038/bmt.2008.63. [DOI] [PubMed] [Google Scholar]
- 3.Huck K, Hanenberg H, Nurnberger W, Dilloo D, Burdach S, Gobel U, Laws HJ. Favourable long-term outcome after matched sibling transplantation for Fanconi-anemia (FA) and in vivo T-cell depletion. Klin Padiatr. 2008;220:147–152. doi: 10.1055/s-2008-1065326. [DOI] [PubMed] [Google Scholar]
- 4.Motwani J, Lawson SE, Darbyshire PJ. Successful HSCT using nonradiotherapy-based conditioning regimens and alternative donors in patients with Fanconi anaemia--experience in a single UK centre. Bone Marrow Transplant. 2005;36:405–410. doi: 10.1038/sj.bmt.1705071. [DOI] [PubMed] [Google Scholar]
- 5.Muller LU, Milsom MD, Kim MO, Schambach A, Schuesler T, Williams DA. Rapid lentiviral transduction preserves the engraftment potential of Fanca(−/−) hematopoietic stem cells. Mol Ther. 2008;16:1154–1160. doi: 10.1038/mt.2008.67. [DOI] [PubMed] [Google Scholar]
- 6.Wagner JE, Eapen M, MacMillan ML, Harris RE, Pasquini R, Boulad F, Zhang MJ, Auerbach AD. Unrelated donor bone marrow transplantation for the treatment of Fanconi anemia. Blood. 2007;109:2256–2262. doi: 10.1182/blood-2006-07-036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93:511–517. doi: 10.3324/haematol.12234. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- 9.Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Carreau M, Callen DF, Savoia A, Cheng NC, van Berkel CG, Strunk MH, Gille JJ, Pals G, Kruyt FA, Pronk JC, Arwert F, Buchwald M, Joenje H. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 10.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, Hoatlin ME, Joenje H, Wang W. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 11.Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G, de Winter JP, Wang W, Joenje H. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 12.Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- 13.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature. 1992;358:434. doi: 10.1038/358434a0. [DOI] [PubMed] [Google Scholar]
- 15.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 16.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea AD, Moses R, Grompe M. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 17.de Winter JP, Waisfisz Q, Rooimans MA, van Berkel CG, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, Pronk JC, Arwert F, Hoehn H, Digweed M, Buchwald M, Joenje H. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 18.de Winter JP, Rooimans MA, van Der Weel L, van Berkel CG, Alon N, Bosnoyan-Collins L, de Groot J, Zhi Y, Waisfisz Q, Pronk JC, Arwert F, Mathew CG, Scheper RJ, Hoatlin ME, Buchwald M, Joenje H. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 19.de Winter JP, Leveille F, van Berkel CG, Rooimans MA, van Der Weel L, Steltenpool J, Demuth I, Morgan NV, Alon N, Bosnoyan-Collins L, Lightfoot J, Leegwater PA, Waisfisz Q, Komatsu K, Arwert F, Pronk JC, Mathew CG, Digweed M, Buchwald M, Joenje H. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am J Hum Genet. 2000;67:1306–1308. doi: 10.1016/s0002-9297(07)62959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D, Mohan S, Schindler D, Arwert F, Pals G, Mathew CG, Waisfisz Q, de Winter JP, Joenje H. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 23.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, Arwert F, Mathew CG, Zdzienicka MZ, Hiom K, De Winter JP, Joenje H. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 24.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, Ott J, Petrini J, Schindler D, Hanenberg H, Auerbach AD. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 25.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 26.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish SD, Lach FP, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew CG, Auerbach AD, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 27.Gurtan AM, Stuckert P, D'Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281:10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 28.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
- 32.Shimamura A, de Oca RM, Svenson JL, Haining N, Moreau LA, Nathan DG, D'Andrea AD. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood. 2002;100:4649–4654. doi: 10.1182/blood-2002-05-1399. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, Mok SC, D'Andrea AD. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 34.Chirnomas D, Taniguchi T, de la Vega M, Vaidya AP, Vasserman M, Hartman AR, Kennedy R, Foster R, Mahoney J, Seiden MV, D'Andrea AD. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther. 2006;5:952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Sejas DP, Zhang X, Qiu Y, Nattamai KJ, Rani R, Rathbun KR, Geiger H, Williams DA, Bagby GC, Pang Q. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briot D, Mace-Aime G, Subra F, Rosselli F. Aberrant activation of stress-response pathways leads to TNF-alpha oversecretion in Fanconi anemia. Blood. 2008;111:1913–1923. doi: 10.1182/blood-2007-07-099218. [DOI] [PubMed] [Google Scholar]
- 37.Sejas DP, Rani R, Qiu Y, Zhang X, Fagerlie SR, Nakano H, Williams DA, Pang Q. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178:5277–5287. doi: 10.4049/jimmunol.178.8.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uziel O, Reshef H, Ravid A, Fabian I, Halperin D, Ram R, Bakhanashvili M, Nordenberg J, Lahav M. Oxidative stress causes telomere damage in Fanconi anaemia cells - a possible predisposition for malignant transformation. Br J Haematol. 2008;142:82–93. doi: 10.1111/j.1365-2141.2008.07137.x. [DOI] [PubMed] [Google Scholar]
- 39.Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 40.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 41.Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan Y-L, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 42.Yan YL, Miller CT, Nissen R, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- 43.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 44.Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, Kanki JP, D'Andrea AD, Look AT. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5:903–914. doi: 10.1016/s1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- 45.Blom E, van de Vrugt HJ, de Vries Y, de Winter JP, Arwert F, Joenje H. Multiple TPR motifs characterize the Fanconi anemia FANCG protein. DNA Repair (Amst) 2004;3:77–84. doi: 10.1016/j.dnarep.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Titus TA, Selvig DR, Qin B, Wilson C, Starks AM, Roe BA, Postlethwait JH. The Fanconi anemia gene network is conserved from zebrafish to human. Gene. 2006;371:211–223. doi: 10.1016/j.gene.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 47.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 48.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischkowitz M, Xia B, Sabbaghian N, Reis-Filho JS, Hamel N, Li G, van Beers EH, Li L, Khalil T, Quenneville LA, Omeroglu A, Poll A, Lepage P, Wong N, Nederlof PM, Ashworth A, Tonin PN, Narod SA, Livingston DM, Foulkes WD. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 51.Postlethwait JH, Yan Y-L, Gates M, Horne S, Amores A, Brownlie A, Donovan A, Egan E, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly J-S, Larhammar D, Talbot WS, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 52.Taylor J, Braasch I, Frickey T, Meyer A, Van De Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest H. Crollius Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 54.Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 2004;14:820–828. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starback P, Lundell I, Fredriksson R, Berglund MM, Yan YL, Wraith A, Soderberg C, Postlethwait JH, Larhammar D. Neuropeptide Y receptor subtype with unique properties cloned in the zebrafish: the zYa receptor. Brain Res Mol Brain Res. 1999;70:242–252. doi: 10.1016/s0169-328x(99)00152-7. [DOI] [PubMed] [Google Scholar]
- 56.Tischkowitz MD, Hodgson SV. Fanconi anaemia. J Med Genet. 2003;40:1–10. doi: 10.1136/jmg.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bagby GC, Lipton JM, Sloand EM, Schiffer CA. Marrow failure. Hematology (Am Soc Hematol Educ Program) 2004:318–336. doi: 10.1182/asheducation-2004.1.318. [DOI] [PubMed] [Google Scholar]
- 58.D'Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17:1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 59.De Kerviler E, Guermazi A, Zagdanski AM, Gluckman E, Frija J. The clinical and radiological features of Fanconi's anaemia. Clin Radiol. 2000;55:340–345. doi: 10.1053/crad.2000.0445. [DOI] [PubMed] [Google Scholar]
- 60.Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- 62.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 63.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 65.Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP, Singh B. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003;129:106–112. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- 66.Fallon JF, Lopez A, Ros MA, Savage MP, Olwin BB, Simandl BK. FGF-2: apical ectodermal ridge growth signal for chick limb development. Science. 1994;264:104–107. doi: 10.1126/science.7908145. [DOI] [PubMed] [Google Scholar]
- 67.Grandel H, Schulte-Merker S. The development of the paired fins in the zebrafish (Danio rerio) Mech. Dev. 1998;79:99–120. doi: 10.1016/s0925-4773(98)00176-2. [DOI] [PubMed] [Google Scholar]
- 68.Zupanc GK, Hinsch K, Gage FH. Proliferation, migration, neuronal differentiation, and long-term survival of new cells in the adult zebrafish brain. J Comp Neurol. 2005;488:290–319. doi: 10.1002/cne.20571. [DOI] [PubMed] [Google Scholar]
- 69.Whitney MA, Royle G, Low MJ, Kelly MA, Axthelm MK, Reifsteck C, Olson S, Braun RE, Heinrich MC, Rathbun RK, Bagby GC, Grompe M. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 70.Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 71.Koomen M, Cheng NC, van de Vrugt HJ, Godthelp BC, van der Valk MA, Oostra AB, Zdzienicka MZ, Joenje H, Arwert F. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum Mol Genet. 2002;11:273–281. doi: 10.1093/hmg/11.3.273. [DOI] [PubMed] [Google Scholar]
- 72.Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 73.Dequen F, St-Laurent JF, Gagnon SN, Carreau M, Desnoyers S. The Caenorhabditis elegans FancD2 ortholog is required for survival following DNA damage. Comp Biochem Physiol B Biochem Mol Biol. 2005;141:453–460. doi: 10.1016/j.cbpc.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 74.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2217. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collis SJ, Barber LJ, Ward JD, Martin JS, Boulton SJ. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair (Amst) 2006;5:1398–1406. doi: 10.1016/j.dnarep.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 76.Youds JL, Barber LJ, Ward JD, Collis SJ, O'Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fei P, Yin J, Wang W. New advances in the DNA damage response network of Fanconi anemia and BRCA proteins. FAAP95 replaces BRCA2 as the true FANCB protein. Cell Cycle. 2005;4:80–86. doi: 10.4161/cc.4.1.1358. [DOI] [PubMed] [Google Scholar]
- 78.Marek LR, Bale AE. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst) 2006;5:1317–1326. doi: 10.1016/j.dnarep.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 79.Allende ML, Manzanares M, Tena JJ, Feijoo CG, Gomez-Skarmeta JL. Cracking the genome's second code: Enhancer detection by combined phylogenetic footprinting and transgenic fish and frog embryos. Methods. 2006 doi: 10.1016/j.ymeth.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 80.Meyer JN, Smith JD, Winston GW, Di Giulio RT. Antioxidant defenses in killifish (Fundulus heteroclitus) exposed to contaminated sediments and model prooxidants: short-term and heritable responses. Aquat Toxicol. 2003;65:377–395. doi: 10.1016/j.aquatox.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 81.Kitano J, Bolnick DI, Beauchamp DA, Mazur MM, Mori S, Nakano T, Peichel CL. Reverse evolution of armor plates in the threespine stickleback. Curr Biol. 2008;18:769–774. doi: 10.1016/j.cub.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 82.Mulvey M, Newman MC, Vogelbein WK, Unger MA, Ownby DR. Genetic structure and mtDNA diversity of Fundulus heteroclitus populations from polycyclic aromatic hydrocarbon-contaminated sites. Environ Toxicol Chem. 2003;22:671–677. [PubMed] [Google Scholar]
- 83.Ownby DR, Newman MC, Mulvey M, Vogelbein WK, Unger MA, Arzayus LF. Fish (Fundulus heteroclitus) populations with different exposure histories differ in tolerance of creosote-contaminated sediments. Environ Toxicol Chem. 2002;21:1897–1902. [PubMed] [Google Scholar]
- 84.Bell MA, Aguirre WE, Buck NJ. Twelve years of contemporary armor evolution in a threespine stickleback population. Evolution. 2004;58:814–824. doi: 10.1111/j.0014-3820.2004.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 85.Holland PWH, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994:125–133. [PubMed] [Google Scholar]
- 86.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Force A, Lynch M, Pickett FB, Amores A, Yan Y-L, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 89.Wada S, Tokuoka M, Shoguchi E, Kobayashi K, Di Gregorio A, Spagnuolo A, Branno M, Kohara Y, Rokhsar D, Levine M, Saiga H, Satoh N, Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- 90.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 91.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 92.Postlethwait J, Johnson S, Midson CN, Talbot WS, Gates M, Ballenger EW, Africa D, Andrews R, Carl T, Eisen JS, Horne S, Kimmel CB, Hutchinson M, Johnson M, Rodriguez A. A genetic linkage map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- 93.Shire JG, Whitten WK. Genetic variation in the timing of first cleavage in mice: effect of maternal genotype. Biol Reprod. 1980;23:369–376. doi: 10.1095/biolreprod23.2.369. [DOI] [PubMed] [Google Scholar]
- 94.Edwards RG, Purdy JM, Steptoe PC, Walters DE. The growth of human preimplantation embryos in vitro. Am J Obstet Gynecol. 1981;141:408–416. doi: 10.1016/0002-9378(81)90603-7. [DOI] [PubMed] [Google Scholar]
- 95.Akkari YM, Bateman RL, Reifsteck CA, D'Andrea AD, Olson SB, Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol Genet Metab. 2001;74:403–412. doi: 10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- 96.Selman K, Wallace RA, Sarka A, X Q. Stages of oocyte development in the zebrafish Brachydanio rerio. J. Morphol. 1993;218:203–224. doi: 10.1002/jmor.1052180209. [DOI] [PubMed] [Google Scholar]
- 97.Holzel M, van Diest PJ, Bier P, Wallisch M, Hoatlin ME, Joenje H, de Winter JP. FANCD2 protein is expressed in proliferating cells of human tissues that are cancer-prone in Fanconi anaemia. J Pathol. 2003;201:198–203. doi: 10.1002/path.1450. [DOI] [PubMed] [Google Scholar]
- 98.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, Flaws JA, Handel MA. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131:131–142. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- 100.Sawicki JA, Magnuson T, Epstein CJ. Evidence for expression of the paternal genome in the two-cell mouse embryo. Nature. 1981;294:450–451. doi: 10.1038/294450a0. [DOI] [PubMed] [Google Scholar]
- 101.Manejwala FM, Logan CY, Schultz RM. Regulation of hsp70 mRNA levels during oocyte maturation and zygotic gene activation in the mouse. Dev Biol. 1991;144:301–308. doi: 10.1016/0012-1606(91)90423-z. [DOI] [PubMed] [Google Scholar]
- 102.Schultz RM, Worrad DM. Role of chromatin structure in zygotic gene activation in the mammalian embryo. Semin Cell Biol. 1995;6:201–208. doi: 10.1006/scel.1995.0028. [DOI] [PubMed] [Google Scholar]
- 103.Palis J, Yoder MC. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp Hematol. 2001;29:927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 104.Cumano A, Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol. 2007;25:745–785. doi: 10.1146/annurev.immunol.25.022106.141538. [DOI] [PubMed] [Google Scholar]
- 105.Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- 107.Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 108.Jin H, Xu J, Wen Z. Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood. 2007;109:5208–5214. doi: 10.1182/blood-2007-01-069005. [DOI] [PubMed] [Google Scholar]
- 109.Wullimann MF, Knipp S. Proliferation pattern changes in the zebrafish brain from embryonic through early postembryonic stages. Anat Embryol (Berl) 2000;202:385–400. doi: 10.1007/s004290000115. [DOI] [PubMed] [Google Scholar]
- 110.Pendeville H, Peers B, Kas K, Voz ML. Cloning and embryonic expression of zebrafish PLAG genes. Gene Expr Patterns. 2006;6:267–276. doi: 10.1016/j.modgep.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 111.Sommer L, Shah N, Rao M, Anderson DJ. The cellular function of MASH1 in autonomic neurogenesis. Neuron. 1995;15:1245–1258. doi: 10.1016/0896-6273(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 112.Wullimann MT. MF Expression of Zash-1a in the postembryonic zebrafish brain allows comparison to mouse Mash1 domains. Brain Res Gene Expr Patterns. 2002;1:187–192. doi: 10.1016/s1567-133x(02)00016-9. [DOI] [PubMed] [Google Scholar]
- 113.Bladen CL, Navarre S, Dynan WS, Kozlowski DJ. Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci Lett. 2007;422:97–102. doi: 10.1016/j.neulet.2007.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clack JA. Paleontology. From fins to fingers. Science. 2004;304:57–58. doi: 10.1126/science.1096415. [DOI] [PubMed] [Google Scholar]
- 115.Cohn MJ, Lovejoy CO, Wolpert L, Coates MI. Branching, segmentation and the metapterygial axis: pattern versus process in the vertebrate limb. Bioessays. 2002;24:460–465. doi: 10.1002/bies.10088. [DOI] [PubMed] [Google Scholar]
- 116.Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- 117.Alter BP. Fanconi's anemia and malignancies. Am J Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 118.Chico TJ, Ingham PW, Crossman DC. Modeling cardiovascular disease in the zebrafish. Trends Cardiovasc Med. 2008;18:150–155. doi: 10.1016/j.tcm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 119.Stoletov K, Klemke R. Catch of the day: zebrafish as a human cancer model. Oncogene. 2008;27:4509–4520. doi: 10.1038/onc.2008.95. [DOI] [PubMed] [Google Scholar]
- 120.Skromne I, Prince VE. Current perspectives in zebrafish reverse genetics: moving forward. Dev Dyn. 2008;237:861–882. doi: 10.1002/dvdy.21484. [DOI] [PubMed] [Google Scholar]
- 121.Ekker SC. Zinc finger-based knockout punches for zebrafish genes. Zebrafish. 2008;5:121–123. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008;25:341–350. doi: 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 123.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goessling W, North TE, Zon LI. New waves of discovery: modeling cancer in zebrafish. J Clin Oncol. 2007;25:2473–2479. doi: 10.1200/JCO.2006.08.9821. [DOI] [PubMed] [Google Scholar]
- 125.Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parng C, Seng WL, Semino C, McGrath P. Zebrafish: a preclinical model for drug screening. Assay Drug Dev Technol. 2002;1:41–48. doi: 10.1089/154065802761001293. [DOI] [PubMed] [Google Scholar]
- 127.Rubinstein AL. Zebrafish: from disease modeling to drug discovery. Curr Opin Drug Discov Devel. 2003;6:218–223. [PubMed] [Google Scholar]
- 128.Stern HM, Zon LI. Cancer genetics and drug discovery in the zebrafish. Nat Rev Cancer. 2003;3:533–539. doi: 10.1038/nrc1126. [DOI] [PubMed] [Google Scholar]
- 129.Ton C, Parng C. The use of zebrafish for assessing ototoxic and otoprotective agents. Hear Res. 2005 doi: 10.1016/j.heares.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 130.Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 131.Grunwald DJ, Eisen JS. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 132.Scata KA, El-Deiry WS. Zebrafish: swimming towards a role for fanconi genes in DNA repair. Cancer Biol Ther. 2004;3:501–502. doi: 10.4161/cbt.3.6.946. [DOI] [PubMed] [Google Scholar]
- 133.Traver Dea. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nature Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 134.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SCF, Malicki J, Stemple DL, Stainier DYR, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 135.Feitsma H, Kuiper RV, Korving J, Nijman IJ, Cuppen E. Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. Cancer Res. 2008;68:5059–5066. doi: 10.1158/0008-5472.CAN-08-0019. [DOI] [PubMed] [Google Scholar]
- 136.Trede NS, Ota T, Kawasaki H, Paw BH, Katz T, Demarest B, Hutchinson S, Zhou Y, Hersey C, Zapata A, Amemiya CT, Zon LI. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn. 2008;237:2575–2584. doi: 10.1002/dvdy.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nasevicius A, Ekker SC. Effective targeted gene "knockdown" in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 138.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 139.Leveille F, Ferrer M, Medhurst AL, Laghmani el H, Rooimans MA, Bier P, Steltenpool J, Titus TA, Postlethwait JH, Hoatlin ME, Joenje H, de Winter JP. The nuclear accumulation of the Fanconi anemia protein FANCE depends on FANCC. DNA Repair (Amst) 2006;5:556–565. doi: 10.1016/j.dnarep.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 140.Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 141.Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 142.Comai L, Henikoff S. TILLING: practical single-nucleotide mutation discovery. Plant J. 2006;45:684–694. doi: 10.1111/j.1365-313X.2006.02670.x. [DOI] [PubMed] [Google Scholar]
- 143.Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parng C, Anderson N, Ton C, McGrath P. Zebrafish apoptosis assays for drug discovery. Methods Cell Biol. 2004;76:75–85. doi: 10.1016/s0091-679x(04)76005-7. [DOI] [PubMed] [Google Scholar]