Summary
Sirtuins, also designated class III histone deacetylases, are implicated in the regulation of cell division, apoptosis, DNA damage repair, genomic silencing and longevity. The nucleolar Sirtuin7 (SIRT7) was reported to be involved in the regulation of ribosomal gene (rDNA) transcription, but there are no data concerning the regulation of SIRT7 during the cell cycle. Here we have analyzed the behavior of endogenous SIRT7 during mitosis, while rDNA transcription is repressed. SIRT7 remains associated with nucleolar organizer regions, as does the RNA polymerase I machinery. SIRT7 directly interacts with the rDNA transcription factor UBF. Moreover, SIRT7 is phosphorylated via the CDK1-cyclin B pathway during mitosis and dephosphorylated by a phosphatase sensitive to okadaic acid at the exit from mitosis before onset of rDNA transcription. Interestingly, dephosphorylation events induce a conformational modification of the carboxy-terminal region of SIRT7 before the release of mitotic repression of rDNA transcription. As SIRT7 activity is required to resume rDNA transcription in telophase, we propose that this conformational modification regulates onset of rDNA transcription.
Keywords: Sirtuins, Nucleolus, Cell cycle, NOR, CDK1
Introduction
The nucleolus is the nuclear compartment in which ribosome biogenesis takes place, i.e. ribosomal gene (rDNA) transcription, precursor ribosomal RNA (rRNA) processing and assembly of mature rRNAs with ribosomal proteins (Boisvert et al., 2007; Carmo-Fonseca et al., 2000; Hadjiolov, 1985). rDNA transcription is subject to extensive modulation during cell growth and/or cell cycle. In mammalian cells the correlation between cell cycle and rRNA synthesis is well established. Up- or downregulation of rRNA synthesis, which can be reversed in appropriate cases, correlates with situations such as nutrient starvation, hormone or drug treatment, aging, tumor and viral infections (Grummt, 2003; Jacob and Ghosh, 1999; Ruggero and Pandolfi, 2003). It is now becoming clear that several signaling pathways are involved in the regulation of rDNA transcription (Kim et al., 2003; Russell and Zomerdijk, 2005; Schmelzle and Hall, 2000; Stefanovsky et al., 2001).
A crucial event in nucleolar activity is the resumption of rDNA transcription at the exit from mitosis. In higher eukaryotic cells, rDNA transcription is repressed from late prophase to telophase (Prescott and Bender, 1962; Roussel et al., 1996), and consequently nucleoli are no longer maintained. Even if recent analyses performed in living cells showed that some subunits of RNA polymerase I (Pol I) are absent at nucleolar organizer regions (NORs) from metaphase to anaphase (Chen et al., 2005; Leung et al., 2004), the RNA Pol I machinery, as observed for RNA Pol I (Gilbert et al., 1995; Scheer and Rose, 1984), the promoter selectivity factor (SL1), the upstream binding factor (UBF) (Jordan et al., 1996; Roussel et al., 1996) and the transcription termination factor (TTF-1) (Sirri et al., 1999), remains mainly associated in an inactive state at NORs, i.e. in chromosomal sites where rDNAs are clustered. Phosphorylation of components of the RNA Pol I machinery by the cyclin-dependent kinase (CDK) 1-cyclin B pathway is responsible for the repression of rDNA transcription during mitosis, and dephosphorylation of these components resumes rDNA transcription at the exit from mitosis (Heix et al., 1998; Kuhn et al., 1998; Sirri et al., 2000). However, other players most probably participate in restoring rDNA transcription in late telophase.
Mammalian sirtuins (SIRT1-7), homologs of the yeast Sir2, have recently been proposed to be involved in the control of critical metabolic pathways as well as apoptosis, stress responses, DNA repair, cell cycle, genomic stability and gene expression. Sirtuins, also designated class III histone deacetylases, are protein deacetylases/ADP ribosyltransferases (Blander and Guarente, 2004). These enzymes are highly conserved from prokaryotes to eukaryotes. They all share a conserved NAD-dependent catalytic core domain, and exhibit variable N-terminal and C-terminal extensions that contribute to their unique subcellular localization and may also regulate their catalytic activity. The subcellular distribution, substrate specificity and cellular function of sirtuins are quite diverse (for reviews, see Denu, 2005; Guarente, 2000; North and Verdin, 2004). SIRT2 is a predominantly cytoplasmic protein (Dryden et al., 2003; Michishita et al., 2005; North et al., 2003), SIRT3-5 are mitochondrial (Michishita et al., 2005) and SIRT1, -6 and -7 are localized in the nucleus. SIRT1, the most closely related to yeast Sir2 and the best characterized sirtuin, possesses a large number of substrates, including p53, Ku70, NF-κB and forkhead transcription factors, that regulate cellular oxidative and genotoxic stresses (Brunet et al., 2004; Cohen et al., 2004; Luo et al., 2001; Motta et al., 2004; Vaziri et al., 2001; Yeung et al., 2004). SIRT6 is involved in important functions in preserving cells from genomic instability and progeroid phenotype (Mostoslavsky et al., 2006). Moreover, SIRT6 is the only sirtuin to exhibit a robust auto-ADP-ribosyltransferase activity (Liszt et al., 2005). SIRT7 is the only sirtuin localized in nucleoli (Ford et al., 2006; Michishita et al., 2005). It was shown to exhibit no deacetylase or ADP-ribosyltransferase activity when tested on acetylated histones and various acetylated components of the RNA Pol I machinery (Ford et al., 2006). Concerning the nucleolar function of SIRT7, Ford et al. (Ford et al., 2006) proposed that SIRT7 could be a positive regulator of rDNA transcription via its association with RNA Pol I. Its overexpression enhances rDNA transcription, whereas its inhibition reduces rDNA transcription. Interestingly, expression of SIRT7 is positively correlated with cell growth: SIRT7 is abundant in metabolically active tissues such as liver, spleen and testes (Ford et al., 2006; Michishita et al., 2005). To date, there are no data concerning the cell cycle regulation of SIRT7 and its fate during mitosis when rDNA transcription is repressed.
In the present study, we investigated the behavior of endogenous SIRT7 during mitosis and the involvement of SIRT7 at the onset of rDNA transcription as cells exit mitosis. By confocal microscopy analyses we showed that SIRT7 remains at NORs during mitosis, and consequently, as previously established for several RNA Pol I subunits, UBF, SL1 and TTF-1, SIRT7 is not released from rDNAs during mitotic repression of rDNA transcription. Glutathione S-transferase (GST) pull-down experiments showed that SIRT7 directly interacts with the rDNA transcription factor UBF. Moreover, we demonstrate that SIRT7 is phosphorylated during mitosis and that its phosphorylation is governed by the CDK1-cyclin B pathway. Interestingly, mitotic phosphorylation of SIRT7 decreases the reactivity of antibodies directed against its C-terminal region from prophase to anaphase. Conversely, inhibition of the CDK1-cyclin B pathway in late mitosis and consequently dephosphorylation of SIRT7 via an okadaic acid sensitive phosphatase increase the reactivity of antibodies to its C-terminal region. Moreover, inhibition of SIRT7 by sirtinol in late mitosis demonstrates that SIRT7 activity is required for onset of rDNA transcription at the exit from mitosis. We propose a model in which SIRT7 remains at NORs during mitosis in a phosphorylated inactive form and undergoes a dephosphorylation-dependent conformational rearrangement in telophase that precedes the resumption of rDNA transcription.
Results
Endogenous SIRT7 localizes in rDNA transcription sites during interphase and remains associated with NORs during mitosis via interaction with UBF
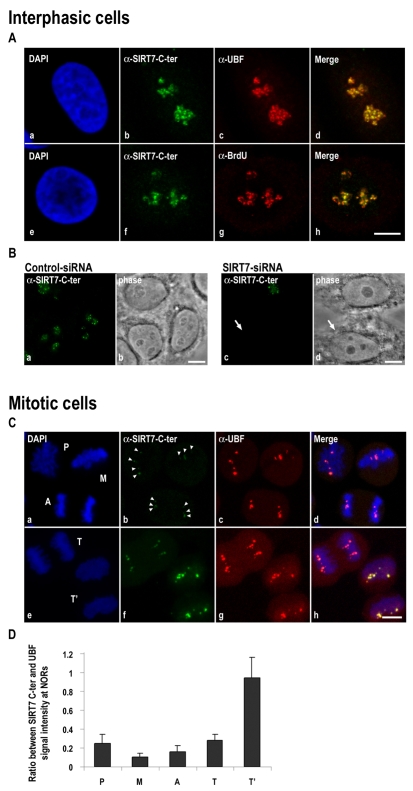
To begin assessing the biological function of SIRT7, we investigated its cellular localization by immunofluorescence confocal microscopy. We first analyzed in interphase HeLa cells the localization of endogenous SIRT7 together with that of UBF or with the rDNA transcription sites revealed by bromouridine 5′-triphosphate (BrUTP) incorporation. The superimposition of labelings revealed co-localization of SIRT7 and UBF (Fig. 1Aa-d) at the level of the rDNA transcription sites (Fig. 1Ae-h). Knockdown of SIRT7 by siRNAs eliminated the nucleolar signal (Fig. 1Bc) demonstrating the specificity of the anti-SIRT7-C-terminal antibodies raised against a peptide located in the C-terminal region (amino acids 384-400) of SIRT7. SIRT7 is the only sirtuin to present an arginine-rich domain (amino acids 1-74) that is involved in its nucleolar localization (supplementary material Fig. S1). In contrast to endogenous SIRT7, the exogenous C-terminal GFP-tagged SIRT7 (GFP-SIRT7) showed a more diffuse nucleolar localization (supplementary material Fig. S2Aa-d). Moreover, in HeLa cells treated with actinomycin D (AMD), which inhibits rDNA transcription and consequently induces nucleolar reorganization (Hadjiolov, 1985), the GFP-SIRT7 fusion was largely excluded from the caps containing UBF (supplementary material Fig. S2Ae-h), whereas the endogenous SIRT7 remained co-localized with UBF in these caps (supplementary material Fig. S2Ai-l).
Fig. 1.
Endogenous SIRT7 co-localizes with UBF in interphase and mitotic HeLa cells. (A) Immunodetection of SIRT7, UBF and rDNA transcription sites was obtained in fixed interphase HeLa cells using anti-SIRT7-C-terminal (b,f), anti-UBF (c) and anti-BrdU (g) antibodies. Images were obtained using confocal laser microscopy. In merged images (d,h), yellow labeling indicates co-localization of green (SIRT7) and red (UBF or BrdU) signals. (B) SIRT7 signal obtained by immunofluorescence in control HeLa cells (a,b) is no longer detected after SIRT7 knockdown (c,d; arrow). (C) HeLa cells were synchronized in mitosis. Mitotic stages were determined on Z projections of focal planes by DAPI staining (a,e). SIRT7 (arrowheads b,f) and UBF (c,g) were immunodetected in mitotic HeLa cells. Immunofluorescence labelings were merged (d,h). (D) SIRT7 and UBF signal intensity was quantified in 30 NORs for each stage of mitosis. The ratios of the mean intensity value were quantified (P, 0.24±0.10; M, 0.11±0.03; A, 0.16±0.06; T, 0.28±0.06; T', 0.94±0.22). Bars, 10 μm. A, anaphase; M, metaphase; P, prophase; T, early telophase; T', late telophase.
It was previously reported that GFP-SIRT7 remains associated with condensed chromosomes in mitotic living human cells (Ford et al., 2006; Michishita et al., 2005). The association of GFP-SIRT7 with metaphase chromosomes appears diffuse rather than NOR-specific. Because in interphase cells endogenous and exogenous SIRT7 did not exhibit the same localization, we verified the location of endogenous SIRT7 during mitosis. Using anti-SIRT7-C-terminal antibodies for immunofluorescence confocal microscopy, we compared endogenous SIRT7 to UBF known to remain associated with NORs throughout mitosis (Roussel et al., 1996). In mitotic cells, SIRT7 detected as dots during prophase, metaphase, anaphase and telophase co-localized with UBF (Fig. 1Ca-d,e-h). Therefore in contrast to GFP-SIRT7, endogenous SIRT7 remains associated with NORs during mitosis, as do components of the RNA Pol I machinery. Interestingly, the comparison between the UBF and SIRT7 signals showed that the intensity of the UBF signal was similar whatever the mitotic stage, whereas that of SIRT7 was weak in early mitosis (Fig. 1Ca-d) and high in late mitosis (Fig. 1Ce-h). The yellow signals in merge images represent co-localization of equivalent red and green labelings (compare Fig. 1Cd,h). The quantification of UBF and SIRT7 signals indicated that the SIRT7 signal was about almost 25% that of UBF during early stages of mitosis (Fig. 1D). The SIRT7 signal increased during telophase and became comparable to that of UBF at late telophase. Thus, endogenous SIRT7 remains associated with the RNA Pol I machinery during mitosis, yet its labeling signal increases at the exit from mitosis.
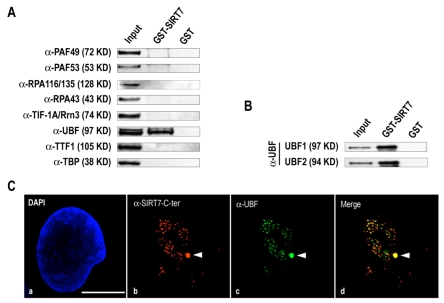
To identify with which component(s) of the RNA Pol I machinery SIRT7 interacts, pull-down experiments using GST-SIRT7 on whole HeLa cell extracts were performed. Several RNA Pol I subunits (RPA116/135, RPA43, PAF49, PAF53), as well as TIF-IA/Rrn3, TTF-1, TBP and UBF were tested. Only UBF interacted with SIRT7 (Fig. 2A). To determine if this was a direct interaction, GST pull-down experiments were performed with purified recombinant UBF produced using a baculovirus expression system. The results of these experiments (Fig. 2B) indicate that SIRT7 interacts directly with both isoforms UBF1 and UBF2 and suggest that SIRT7 localization to rDNAs might be due to this direct interaction. To confirm this possibility, recruitment at pseudo-NORs was investigated. Pseudo-NORs are structures that form around UBF-binding site arrays at ectopic sites on human chromosomes (Mais et al., 2005). They are bound by UBF throughout the cell cycle. During metaphase they appear morphologically similar to endogenous NORs. In interphase cells, they can be visualized as novel sub-nuclear structures distinct from nucleoli. Here we analyzed the pseudo-NOR containing cell line 3D-1, which harbors a 1.4 Mb UBF-binding site array on chromosome 10. Antibody staining clearly demonstrates that endogenous SIRT7 is efficiently recruited to pseudo-NORs along with UBF (Fig. 2Ca-d). Conversely, in 3D-1 cells the SIRT7-YFP fusion protein does not target pseudo-NORs (supplementary material Fig. S2Ba-d), thus confirming mislocalization of ectopically expressed SIRT7. Taken together, these results show that endogenous SIRT7 remains at NORs during mitosis, most probably via direct interaction with UBF.
Fig. 2.
SIRT7 is recruited to pseudo-NORs via direct interaction with UBF. (A) GST and GST-SIRT7 pull-down assays were performed using whole HeLa cell extracts. 1% of extract used for each pull-down assay (Input), 5% of the proteins eluted after GST-SIRT7 pull-down assays and 5% of the proteins eluted after GST pull-down assays were submitted to 10% SDS-PAGE. Immunoblotting was performed using anti-PAF49, anti-PAF53, anti-RPA116/135, anti-RPA43, anti-TIF-1A/Rrn3, anti-UBF, anti-TTF-1 and anti-TBP antibodies. (B) GST and GST-SIRT7 pull-down assays were performed using 1 μg purified human UBF1 or UBF2. (C) The pseudo-NOR containing cell line 3D-1 was stained with anti-SIRT7-C-terminal and anti-UBF antibodies (a-d). Red and green labelings were merged (d). Pseudo-NORs are identified by arrowheads. Bar, 10 μm.
SIRT7 signal increase is due to increased reactivity of antibodies to the SIRT7 C-terminal region
Increase of SIRT7 signal intensity during telophase could be due to either increased recruitment of SIRT7 at NORs or to increased reactivity of anti-SIRT7-C-terminal antibodies. To discriminate between these two possibilities, we first took advantage of roscovitine, a highly selective inhibitor of CDKs. In mitotic cells CDK1-cyclin B is the only known CDK inhibited by roscovitine treatment (De Azevedo et al., 1997; Meijer et al., 1997). We have previously demonstrated that in mitotic cells, roscovitine treatment induces events that normally occur in telophase, i.e. dephosphorylation of several components of RNA Pol I machinery and resumption of rDNA transcription (Sirri et al., 2000). Thus, it was first verified whether roscovitine treatment of nocodazole-arrested mitotic cells could reproduce the increase in SIRT7 signal detected in telophase. As shown in Fig. 3A, the signal of SIRT7 is increased in mitotic cells treated with roscovitine (Fig. 3Af) compared with control cells (Fig. 3Ac). The quantification of SIRT7 signals indicated that in roscovitine-treated cells the SIRT7 signal increased about 4-fold compared with untreated cells (Fig. 3B). The existence of a soluble pool of SIRT7 in the cytoplasm of mitotic cells treated or not with roscovitine was investigated. For this purpose mitotic cells treated or not with roscovitine were separated into chromosome and cytoplasmic fractions and the distribution of SIRT7 was compared to that of UBF and fibrillarin. Previous results had shown that UBF remains associated with NORs during mitosis (Roussel et al., 1993), whereas fibrillarin relocalizes to NORs during telophase or after roscovitine treatment (Sirri et al., 2002). As expected, UBF could only be detected in the chromosome fraction (Fig. 3C, lanes 2 and 4) and fibrillarin was mainly detected in the cytoplasmic fraction in mitotic cells (Fig. 3C, lane 1), whereas it was mainly detected in the chromosome fraction after roscovitine treatment (Fig. 3C, lane 4). SIRT7 was mainly detected in the chromosome fraction prepared from mitotic HeLa cells treated or not with roscovitine (Fig. 3C, lanes 2 and 4), suggesting that the increased SIRT7 signal is not due to increased recruitment of SIRT7 from a cytoplasmic pool at NORs during telophase.
Fig. 3.
Increase in SIRT7 signal corresponds to increased reactivity of antibodies to the SIRT7 C-terminal region. (A) In nocodazole-arrested HeLa cells treated (d-f) or not (a-c) with roscovitine (150 μM for 1 hour), SIRT7 and UBF were immunodetected using anti-SIRT7-C-terminal (c,f) and anti-UBF (b,e) antibodies. DNA was stained by DAPI and merged with red and green labelings (a,d). (B) SIRT7 signal intensity was quantified in 10 cells. The means of intensity value were quantified (Control: 0.25±0.06, Roscovitine: 0.83±0.15). (C) Cytoplasmic (lanes 1 and 3) and chromosome (lanes 2 and 4) fractions prepared from nocodazole-arrested HeLa cells treated (lanes 3 and 4) or not (lanes 1 and 2) with roscovitine were submitted to 10% SDS-PAGE. Immunoblotting was performed using anti-UBF, anti-SIRT7-N-terminal and anti-fibrillarin antibodies. (D) HeLa cells were synchronized in mitosis and mitotic stages were determined by DAPI staining (a,d,g). In immunofluorescence assays, SIRT7 and UBF were detected using anti-SIRT7-N-terminal (c,f,i) and anti-UBF (b,e,h) antibodies, respectively. Immunofluorescence labelings were merged with DAPI (a,d,g). Bars, 10 μm.
Thus, the reactivity of anti-SIRT7 antibodies to SIRT7 was tested during mitosis. For this purpose, the intensity of the SIRT7 signal obtained with polyclonal antibodies raised against a peptide located in the N-terminal region (amino acids 32-51) of SIRT7 was analyzed. Immunofluorescence analyses of mitotic HeLa cells using anti-SIRT7-N-terminal antibodies showed no variation in the intensity of the SIRT7 signal between early and late stages of mitosis (Fig. 3D). Moreover, comparison between UBF and SIRT7 signals showed equivalent red and green labelings throughout mitosis (Fig. 3D). Taken together, these results indicate that SIRT7 remains mainly localized at NORs during mitosis, but that the reactivity of antibodies to its C-terminal region varies, i.e. weak in early stages and high in late stages of mitosis.
Increased reactivity of antibodies to the SIRT7 C-terminal region occurs before, and independently of, onset of rDNA transcription
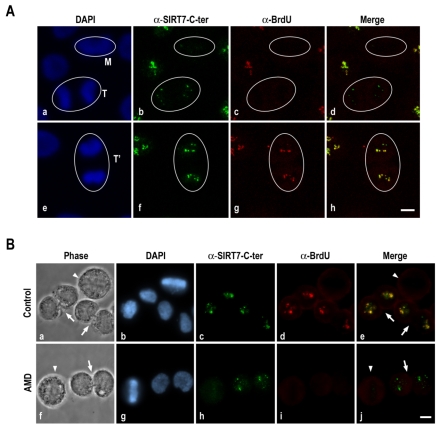
As in cycling cells rDNA transcription is arrested at the beginning of mitosis and restored at telophase (Roussel et al., 1996), we investigated whether the reactivity of antibodies to the SIRT7 C-terminal region at telophase is linked to the onset of rDNA transcription in HeLa cells. As expected, rDNA transcription, revealed by BrUTP incorporation, was detected in interphase cells and in late telophase (Fig. 4Ac,g) but not in metaphase and early telophase (Fig. 4Ac). As for SIRT7, its increased signal was already detected in early telophase (Fig. 4Ab) when rDNA transcription was not yet detectable (Fig. 4Ac). The SIRT7 signal was maximum in late telophase and co-localized with rDNA transcription sites (Fig. 4Ae-h). These results suggest that the reactivity of antibodies to the SIRT7 C-terminal region at NORs in late mitosis does not depend on the onset of rDNA transcription. To validate this hypothesis, we analyzed the reactivity of antibodies to the SIRT7 C-terminal region in mitotic synchronized cells inhibited or not for rDNA transcription. Nocodazole-arrested mitotic HeLa cells were treated or not with AMD for 60 minutes, washed to eliminate nocodazole and analyzed after 90 minutes, i.e. when the cells proceeded through and exited from mitosis, in the absence or presence of AMD. In the absence of any treatment the reactivity of antibodies to the SIRT7 C-terminal region was increased in early-G1 rDNA-transcribing cells compared with non-transcribing metaphase cells (Fig. 4Ba-e). It is noteworthy that the increase in reactivity of antibodies to the SIRT7 C-terminal region in early G1 cells compared with metaphase cells was also observed in the absence of rDNA transcription, i.e. after AMD treatment (Fig. 4Bf-j). These results demonstrate that increased reactivity of antibodies to the SIRT7 C-terminal region, which probably translates a change of SIRT7 conformation, occurs before, and independently of, onset of rDNA transcription.
Fig. 4.
Increased reactivity of antibodies to the SIRT7 C-terminal region occurs before, and independently of, onset of rDNA transcription. (A) Surrounded mitosis stages (M, T, T') of synchronized HeLa cells were determined on Z projections of focal planes by DAPI staining (a,e). SIRT7 and rDNA transcription sites were detected in fixed cells using respectively anti-SIRT7-C-terminal (b,f) and anti-BrdU (c,g) antibodies. Red and green labelings were merged (d,h). (B) Immunodetection of SIRT7 (c,h) and rDNA transcription sites (d,i) was performed in early G1 fixed cells in the absence (a-e) or presence (f-j) of AMD (0.1 μg/ml for 2 hours). Early G1 cells (arrows) were identified by phase-contrast (a,f) and DAPI staining (b,g). Arrowheads indicate metaphase cells. Immunofluorescence labelings were merged (e,j). Bars, 10 μm.
SIRT7 is regulated by the CDK1-cyclin B pathway during mitosis
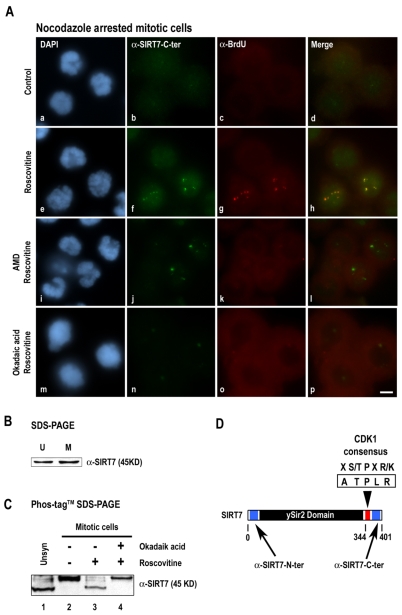
G2/M transition is triggered by activation of a protein kinase cascade, at the head of which is the CDK1-cyclin B (Dunphy and Newport, 1988; Gautier et al., 1988). Cyclin B degradation and consequently CDK1 inactivation starts at the end of metaphase and increases until exit from mitosis (Clute and Pines, 1999). Several components of the RNA Pol I machinery, including TBP (Heix et al., 1998), TAFI110, UBF (Klein and Grummt, 1999) and TTF-1 (Sirri et al., 1999), appear to be regulated during mitosis by the CDK1-cyclin B pathway. Furthermore, we previously demonstrated that inhibition of the CDK1-cyclin B pathway restores rDNA transcription in mitotic cells (Sirri et al., 2000). To verify whether inactivation of the CDK1-cyclin B pathway and reactivity of antibodies to the SIRT7 C-terminal region in late mitosis are correlated, BrUTP incorporation assays were carried out on nocodazole-arrested mitotic cells in the absence (Fig. 5Aa-d) or presence (Fig. 5Ae-h) of roscovitine. As expected, in nocodazole-arrested mitotic cells, rDNA transcription was not detected (Fig. 5Ac) and the SIRT7 signal was very weak (Fig. 5Ab). After roscovitine treatment, the SIRT7 signal was increased only in rDNA transcription-positive cells, i.e. responsive to roscovitine (Fig. 5Ae-h). To discriminate between a direct and indirect effect of inhibition of the CDK1-cyclin B pathway on the reactivity of antibodies to the SIRT7 C-terminal region, mitotic cells were treated with both roscovitine and AMD. The increased reactivity of antibodies to the SIRT7 C-terminal region was still detected even when the CDK1-cyclin B pathway was inhibited by roscovitine and the resumption of rDNA transcription was prevented by AMD (Fig. 5Ai-l), confirming that resumption of rDNA transcription is not necessary to increase the SIRT7 signal in mitotic cells. Thus, inhibition of the CDK1-cyclin B pathway is sufficient to increase reactivity of antibodies against the C-terminal region of SIRT7 at the level of the NORs. To analyze whether reactivity of antibodies to the SIRT7 C-terminal region in late mitosis is linked to dephosphorylation occurring when the CDK1-cyclin B pathway is inhibited, the reactivity of antibodies to the SIRT7 C-terminal region was analyzed in nocodazole-arrested mitotic cells pretreated with okadaic acid, a potent inhibitor of protein phosphatase 1 (PP1) and 2A (PP2A). As observed in Fig. 5A, in mitotic cells previously treated with okadaic acid, subsequent roscovitine treatment produced no effect on the reactivity of antibodies to the SIRT7 C-terminal region (Fig. 5Am-p). These results show that the reactivity of antibodies to the SIRT7 C-terminal region during mitosis, and probably the conformation of SIRT7, depends on phosphorylation/dephosphorylation events. To date, no post-translational modification of SIRT7 has been described. To analyze phosphorylation of SIRT7 during the cell cycle, the electrophoretic mobility of SIRT7 was compared using unsynchronized and mitotic cell extracts. No mobility shift of the 45 kDa SIRT7 was detected (Fig. 5B). This suggests either that SIRT7 is not phosphorylated during the cell cycle or that SIRT7 phosphorylation is not detectable by SDS-PAGE. Thus, the recently developed Phos-tag compound, which has specific phosphate-binding activity (Kinoshita et al., 2006; Kinoshita et al., 2004), was exploited to separate phosphorylated from unphosphorylated forms of SIRT7 by phospho-affinity gel electrophoresis. In these conditions, a specific mobility shift of SIRT7 in mitotic cells was observed. Indeed, in unsynchronized cells, a high-mobility form of SIRT7 corresponding to the unphosphorylated 45 kDa form of SIRT7 (Fig. 5C, lane 1) was detected. Conversely, in mitotic cells, a low-mobility band was detected, which corresponds to a phosphorylated form of SIRT7 (Fig. 5C, lane 2). As SIRT7 possesses a consensus CDK1 phosphorylation site (ATPLR) (Fig. 5D) in its C-terminal region (amino acids 344-348), the effect of CDK1-cyclin B inhibition by roscovitine treatment was analyzed on mitotic phosphorylation of SIRT7. As shown in Fig. 5C, in mitotic cells treated with roscovitine, the unphosphorylated form of SIRT7 visible in unsynchronized cells was detected (compare lane 3 to 1). These data indicate that in vivo CDK1-cyclin B activity is necessary to maintain phosphorylation of SIRT7 in nocodazole-arrested mitotic cells. Dephosphorylation of SIRT7 induced by inhibition of CDK1-cyclin B involved a phosphatase sensitive to okadaic acid. Indeed, when nocodazole-arrested mitotic cells were pretreated with okadaic acid and then treated with roscovitine, no unphosphorylated form of SIRT7 was detected (Fig. 5C, lane 4). Taken together, our results demonstrate that SIRT7 is directly or indirectly phosphorylated during mitosis by the CDK1-cyclin B pathway. The mitotic phosphorylation state of SIRT7 correlates with a decreased reactivity of antibodies to the SIRT7 C-terminal region that probably translates the mitotic conformation of SIRT7. Conversely dephosphorylation of SIRT7 increases reactivity of antibodies to the SIRT7 C-terminal region and most probably releases the transient mitotic conformation of SIRT7.
Fig. 5.
Inhibition of CDK1-cyclin B induces increased reactivity of antibodies to the SIRT7 C-terminal region by SIRT7 dephosphorylation. (A) SIRT7 and rDNA transcription sites were immunodetected using anti-SIRT7-C-terminal (b,f,j,n) and anti-BrdU (c,g,k,o) antibodies in prometaphase mitotic HeLa cells, in the absence (a-d) or presence (e-h) of roscovitine (150 μM for 1 hour). Mitotic HeLa cells were also treated with AMD (0.1 μg/ml, i-l) or okadaic acid (0.5 μM, m-p) for 1 hour before addition of 150 μM roscovitine for 1 hour. Red and green labelings were merged (d,h,l,p). (B) Whole cell extracts from unsynchronized (U) or nocodazole-arrested mitotic (M) HeLa cells were submitted to 10% SDS-PAGE. Immunodetection of SIRT7 was performed using anti-SIRT7-N-terminal antibodies. (C) Whole cell extracts prepared from unsynchronized HeLa cells (Unsyn, lane 1) and from nocodazole-arrested mitotic HeLa cells (lanes 2-4) treated with okadaic acid (0.5 μM for 1 hour) (lane 4) or not (lane 3) before addition of roscovitine (150 μM for 1 hour) (lanes 3 and 4) were submitted to 7.5% Phos-tag SDS-PAGE. Immunodetection of SIRT7 was performed using anti-SIRT7-N-terminal antibodies. (D) SIRT7 presents a consensus CDK1 phosphorylation site (ATPLR) at amino acids 344-348 (red box). Blue boxes indicate the peptides against which anti-SIRT7-N-terminal antibodies (amino acids 32-51) and anti-SIRT7-C-terminal antibodies (amino acids 384–400) are directed. Bar, 10 μm.
SIRT7 activity is essential for the resumption of rDNA transcription at the exit from mitosis
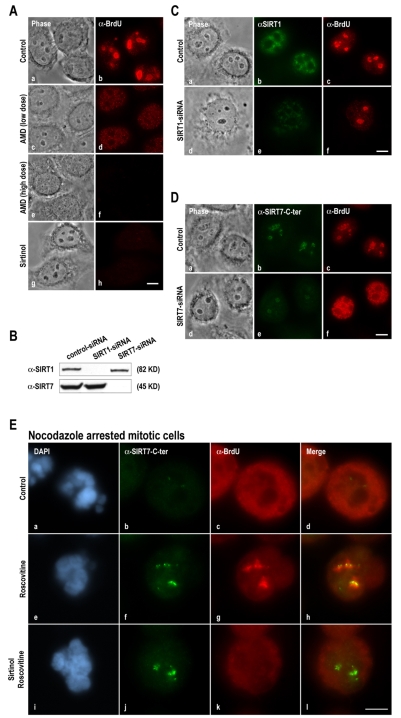
To study the effect of SIRT7 inhibition on the resumption of rDNA transcription, the specific sirtuin inhibitor sirtinol (Grozinger et al., 2001) was used. As expected, because at least 70% of the total RNA in a cell corresponds to rRNA (Jacob and Ghosh, 1999), in the absence of any treatment BrUTP incorporation in vivo was predominantly detected within nucleoli (Fig. 6Aa,b). After inhibition of rDNA transcription with a low dose of AMD, only non-nucleolar transcription activity was detected (Fig. 6Ac,d). In addition, when all RNA transcription activity was inhibited using a high dose of AMD, neither nuclear nor nucleolar transcription was detected (Fig. 6Ae,f). Consequently, this assay was appropriate to analyze the effect of sirtuins on RNA transcription activity in the cell. To inhibit sirtuins, HeLa cells were treated with sirtinol, which affects the catalytic activity of all sirtuins, including SIRT1, known to regulate several RNA Pol II transcription factors and to repress RNA Pol I transcription during caloric restriction (Murayama et al., 2008). After sirtinol treatment, both nuclear and nucleolar transcription decreased dramatically (Fig. 6Ag,h). To further examine the specific involvement of SIRT7 and/or SIRT1 on rDNA transcription, the effect of knockdown of SIRT7 and SIRT1 was examined using SIRT7 and SIRT1 siRNAs (Fig. 6B). In SIRT1-depleted HeLa cells, nucleolar BrUTP incorporation was not, or poorly, affected compared with control cells (Fig. 6Cc,f). In SIRT7-depleted HeLa cells, nucleolar BrUTP incorporation was very weak or absent compared with control cells, whereas nuclear BrUTP incorporation was not affected (Fig. 6Dc,f). The increase in nuclear BrUTP incorporation might be a consequence of the absence of nucleolar BrUTP incorporation. Taken together, these results indicate that sirtinol affects rDNA transcription in vivo predominantly through SIRT7 inhibition.
Fig. 6.
SIRT7 is involved in resumption of rDNA transcription at the exit from mitosis. (A) BrUTP incorporation in living cells was detected using anti-BrdU antibodies in interphase HeLa cells in the absence (a-b) or presence of AMD (0.1 μg/ml, c-d, or 4 μg/ml, e-f, for 2 hours) or sirtinol (100 μM for 6 hours, g-h). (B) Knockdown of SIRT1 and SIRT7 was analyzed by 10% SDS-PAGE and immunoblotting using anti-SIRT1 and anti-SIRT7-N-terminal antibodies. (C) SIRT1 and BrUTP incorporation were detected using anti-SIRT1 (b,e) and anti-BrdU (c,f) antibodies, respectively, in control (a-c) or SIRT1-depleted (d-f) HeLa cells. (D) SIRT7 and BrUTP incorporation were detected using anti-SIRT7 (b,e) and anti-BrdU (c,f) antibodies, respectively, in control (a-c) or SIRT7-depleted (d-f) HeLa cells. (E) SIRT7 and rDNA transcription sites were immunodetected using anti-SIRT7-C-terminal (b,f,j) and anti-BrdU (c,g,k) antibodies in prometaphase mitotic HeLa cells, in the absence (a-d) or presence (e-h) of roscovitine (150 μM for 1 hour). Mitotic HeLa cells were also treated with 100 μM sirtinol for 2 hours before addition of 150 μM roscovitine for 1 hour (i-l). Red and green labelings were merged (d,h,l). Bars, 10 μm.
To study SIRT7 implication in the resumption of rDNA transcription at the exit from mitosis, we analyzed the effect of sirtinol on BrUTP incorporation in nocodazole-arrested mitotic cells treated with roscovitine. As expected, in mitotic cells treated with roscovitine, the increased reactivity of antibodies to the SIRT7 C-terminal region at NORs and resumption of rDNA transcription were induced (Fig. 6Ee-h), compared with mitotic cells not treated with roscovitine (Fig. 6Ea-d). In mitotic cells pretreated for 2 hours with sirtinol, roscovitine treatment induced an increased reactivity of antibodies to the SIRT7 C-terminal region but not onset of rDNA transcription (Fig. 6Ei-l), indicating that inhibition of SIRT7 activity via sirtinol prevents resumption of rDNA transcription. These results demonstrate that SIRT7 is necessary to restore rDNA transcription at the exit from mitosis.
Discussion
The nucleolar sirtuin, SIRT7, has been described as a positive regulator of rDNA transcription and its localization to rDNAs reported to depend on ongoing rDNA transcription (Ford et al., 2006). In the present study, we localized endogenous SIRT7 during mitosis, i.e. when rDNA transcription is repressed. As previously established for the RNA Pol I machinery, endogenous SIRT7 co-localizes with UBF in nucleoli during interphase and remains localized at NORs during mitosis. In addition, contrary to previous results obtained with U2OS cells treated with AMD (Ford et al., 2006), our results obtained in AMD-treated HeLa cells indicate that endogenous SIRT7 remains associated with the RNA Pol I machinery independently of ongoing rDNA transcription. These results are confirmed by the recruitment of SIRT7 to transcriptionally silent pseudo-NORs (Mais et al., 2005). The localization of endogenous SIRT7 during the cell cycle appears different from that reported for the GFP-SIRT7 fusion (Ford et al., 2006; Michishita et al., 2005) (this study). In the present study we show that SIRT7 interacts with UBF but not with TTF-1, TBP and several components of RNA Pol I (RPA116/135, RAP43, PAF53, PAF49), nor with TIF-IA/Rrn3. In addition, in a cellular context UBF is able to recruit SIRT7 at pseudo-NORs. Therefore, as SIRT7 directly interacts with the purified UBF1 and UBF2 isoforms, we propose that SIRT7 localization to rDNAs in vivo occurs most probably via direct interaction between SIRT7 and UBF.
SIRT7 immunofluorescence analyses during mitosis using antibodies directed against the SIRT7 C-terminal region revealed a weak SIRT7 signal during the early mitotic stages (from prophase to anaphase) and a high SIRT7 signal in late telophase, unlike UBF, which presents a constant level of immunofluorescence labeling during mitosis. An equivalent increase of SIRT7 signal is also induced by treatment using the CDK inhibitor roscovitine. Interestingly, the comparison between chromosomal and cytoplasmic proteins prepared from mitotic HeLa cells treated or not with roscovitine shows that SIRT7 remains fully associated with NORs during mitosis, as does UBF, revealing that increase of SIRT7 signal during telophase is not due to increased recruitment of SIRT7 at NORs. Immunofluorescence analyses using antibodies directed against either the N-terminal or C-terminal region of SIRT7 showed that only the reactivity of antibodies to the SIRT7 C-terminal region changes during mitosis. Conversely, the reactivity of antibodies to the SIRT7 N-terminal region is high and unchanged. These results indicate that SIRT7 most probably undergoes a conformational rearrangement during mitosis.
It is well established that local and/or global conformational changes can be induced by modifying protein phosphorylation (Johnson and Lewis, 2001). Thus, the mitotic rearrangement of the N-terminal domain of histone H3 induced by phosphorylation modifies its accessibility to antibodies (Sauvé et al., 1999). Inhibition of the CDK1-cyclin B pathway at the exit from mitosis concomitantly induces dephosphorylation of several components of the RNA Pol I machinery (Heix et al., 1998; Kuhn et al., 1998; Sirri et al., 2000) and onset of rDNA transcription. The concomitant inhibition of the CDK1-cyclin B pathway by roscovitine and rDNA transcription by AMD in mitotic cells led us to conclude that inhibition of CDK1-cyclin B is sufficient to increase reactivity of antibodies to the SIRT7 C-terminal region independently of rDNA transcription. Moreover, experiments performed on mitotic cells pretreated with okadaic acid led us to propose that the increased reactivity of antibodies to the SIRT7 C-terminal region at the exit from mitosis is linked to dephosphorylation. Indeed, our results show that SIRT7 is undoubtedly phosphorylated during mitosis by the CDK1-cyclin B pathway. At present, we cannot conclude whether CDK1-cyclin B kinase directly phosphorylates SIRT7 or whether it activates another kinase responsible for SIRT7 phosphorylation. Interestingly, since SIRT7 presents a consensus CDK1 phosphorylation site at amino acids 344-348 in its C-terminal region, we can speculate that SIRT7 could be directly phosphorylated by CDK1-cyclin B. To verify if SIRT7 is indeed phosphorylated in vivo by CDK1-cyclin B during mitosis, ectopic expression of a SIRT7 form mutated at the CDK1 phosphorylation site would have been pertinent; however, the mislocalization of exogenous SIRT7 does not make this possible. Interestingly, SIRT2 has been reported to be phosphorylated by CDK1-cyclin B in its C-terminal region, and this phosphorylation is required for the mitotic function of SIRT2 (North and Verdin, 2007). Thus it is tempting to suggest that SIRT7 might itself be phosphorylated by CDK1-cyclin B in its C-terminal region and that this phosphorylation could induce a conformational change of SIRT7, impairing the reactivity of antibodies to its C-terminal region during mitosis. Indeed, as anti-SIRT7-C-terminal antibodies are directed against a peptide downstream of the CDK1 phosphorylation site, we can exclude that anti-SIRT7-C-terminal antibodies preferentially recognize the dephosphorylated form of SIRT7. The mitotic regulation of SIRT7 by the CDK1-cyclin B pathway is in agreement with previous results concerning the mitotic repression of the RNA Pol I machinery. Thus, the phosphorylation of SL1, TTF-1 and SIRT7 modify, respectively, the interaction between SL1 and UBF (Heix et al., 1998; Kuhn et al., 1998), the chromatin-binding affinity of TTF-1 (Sirri et al., 1999), and the conformation of SIRT7.
Our results indicate that the conformational rearrangement of SIRT7 occurs independently of and before the onset of rDNA transcription. Thus, we investigated the potential role of SIRT7 in the resumption of RNA Pol I transcription at the exit from mitosis, and first analyzed the effect of sirtinol on rDNA transcription: in interphasic HeLa cells inhibition of sirtuins by sirtinol affects both nuclear and nucleolar BrUTP incorporation. To date, only SIRT7 was reported to function as activator in the in vivo regulation of rDNA transcription (Ford et al., 2006) whereas SIRT1 was reported to repress rDNA transcription during caloric restriction (Murayama et al., 2008). Repression of nuclear transcription may be due to sirtinol inhibition of SIRT1, which is reported to be a regulator of several nuclear transcription factors (Brunet et al., 2004; Cohen et al., 2004; Luo et al., 2001; Motta et al., 2004; Vaziri et al., 2001; Yeung et al., 2004), but other sirtuins could be implicated. As for nucleolar transcription (i.e. rDNA transcription), the absence of BrUTP incorporation demonstrates that nucleolar SIRT7 is inhibited in vivo by sirtinol treatment. Inhibition of SIRT1 should have increased nucleolar transcription. The fact that such is not the case demonstrates either that SIRT1 does not function as a major repressor, or that SIRT7 plays a predominant role as activator on rDNA transcription. To verify this hypothesis, SIRT7 and SIRT1 were knocked down by specific siRNAs. As expected, SIRT7 knockdown significantly decreases nucleolar transcription, whereas unexpectedly SIRT1 knockdown does not clearly increase nucleolar transcription. The role of SIRT1 as repressor is probably not of major importance in HeLa cells during interphase even if it could be important at the onset of mitosis when rDNA transcription is switched off, as previously suggested (Ford et al., 2006). Taken together, these results indicate that inhibition of rDNA transcription by sirtinol treatment is most probably due to inhibition of SIRT7, even if involvement of other sirtuins cannot be completely ruled out. To verify the role of SIRT7 on the resumption of rDNA transcription, mitotic cells were concomitantly treated with roscovitine, which induces onset of rDNA transcription, and with sirtinol, which inhibits SIRT7. Sirtinol treatment impairs resumption of rDNA transcription without affecting reactivity of antibodies to the SIRT7 C-terminal region. Consequently, these results not only confirm the positive regulation role of SIRT7 on rDNA transcription (Ford et al., 2006) but also reveal that the activity of SIRT7 plays an indispensable role in restoring rDNA transcription at the exit from mitosis.
Our results lead us to propose the following model. During mitosis, rDNA transcription is repressed and SIRT7, as well as several components of the RNA Pol I machinery, is phosphorylated by the CDK1-cyclin B pathway. SIRT7 phosphorylation in its C-terminal region could change the conformation of SIRT7, making antibodies weakly reactive to the C-terminal region of SIRT7. Inhibition of the CDK1-cyclin B pathway increases the reactivity of antibodies against the C-terminal region of SIRT7 before the onset of rDNA transcription. Nevertheless, we cannot conclude that the increased reactivity of the SIRT7 antibodies is due only to a conformational change of SIRT7, resulting from its phosphorylation/dephosphorylation. Indeed, dephosphorylation of several components of the RNA Pol I machinery at the exit from mitosis could induce a conformational change of the RNA Pol I machinery, which could modify the position of SIRT7 inside the RNA Pol I machinery, increasing the reactivity of antibodies to SIRT7. Interestingly, the N-terminal and C-terminal regions of yeast sirtuin Hst2 and mammalian SIRT2 play a role in the regulation of their enzymatic activity (Zhao et al., 2003), suggesting that rearrangement of SIRT7 could regulate SIRT7 activity at the exit from mitosis. Thus, during mitosis SIRT7 would be in an inactive form, whereas at the exit from mitosis SIRT7 would be induced into a more active form by a conformational rearrangement that would be essential to restore rDNA transcription.
Materials and Methods
Cell culture and drugs
HeLa cells were cultured in minimum essential medium (MEM) with GlutaMAX (Invitrogen) supplemented with 10% fetal calf serum (FCS). Stable GFP-SIRT7 HeLa cells were obtained by co-transfection of GFP-SIRT7 and pBABE-PURO (Addagene) at a 10:1 ratio, using effectene (Qiagen) according to the manufacturer's instructions, and were selected with 10 μg/ml puromycin. GFP-SIRT7 HeLa cells were cultured as HeLa cells. Clone 3D-1 was maintained and transfected as previously described (Prieto and McStay, 2007). For synchronization in mitosis, HeLa cells were accumulated in prometaphase by nocodazole treatment (0.04 μg/ml for 4 hours), selectively harvested by mechanical shock, washed and resuspended in nocodazole-free medium for 60 or 90 minutes. The drugs used were AMD (Sigma) at 0.1 or 4 μg/ml for 2 hours, roscovitine (Calbiochem) at 150 μM for 1 hour, okadaic acid (Calbiochem) at 0.5 μM for 2 hours and sirtinol (Calbiochem) at 100 μM for 2 or 6 hours. For immunofluorescence labeling and BrUTP incorporation assays, cells were grown as monolayers on glass slides or coverslips and transfected or treated 24 hours later. Synchronized mitotic cells were transferred onto glass slides coated with poly-L-lysine.
Plasmid constructs and antibodies
Human SIRT7 (hSIRT7) cDNA (clone IMAGE 5087554) was PCR-amplified and subcloned into the EcoRI/SalI sites of the pMIGR1 retroviral vector provided by S.A. Leibovitch (INRA, Montpellier, France) to generate the GFP-SIRT7 construct and into the EcoRI/NotI sites of the pGEX4T1 (Amersham) to obtain the GST-SIRT7 construct. SIRT7-YFP was obtained by cloning hSIRT7 cDNA into the Gateway entry vector pENTR-D-TOPO (Invitrogen), and recombined into pdEYFP (Simpson et al., 2000).
Anti-SIRT7-C-terminal rabbit polyclonal antibodies were first provided by I. Horikawa (National Institute of Health, Bethesda, USA) and subsequently raised against the same peptide (GWFGRGCTKRTKRKKVT) by Eurogentec. Human RPA43, PAF49, PAF53 and TIF-1A/Rrn3 antibodies were raised in sheep immunized with full-length recombinant proteins produced using baculovirus or Escherichia coli expression systems. UBF antibodies used in Fig. 2C and supplementary material Fig. S2B were described in (Mais et al., 2005). The other antibodies were anti-SIRT7-N-terminal (S5947, Sigma-Aldrich), anti-UBF (F-9, Santa Cruz Biotechnology), anti-RPA116/135 (N-17, Santa Cruz Biotechnology), anti-SIRT1 (Abcam), anti-TBP (N-12, Santa Cruz Biotechnology), anti-TTF-1 (Sirri et al., 1999), anti-BrdU (Sigma-Aldrich) and Texas Red-, FITC- or horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories).
Immunofluorescence
The cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature (RT) and permeabilized with 0.5% Triton X-100 for 5 minutes at RT. They were then washed with PBS and incubated with primary antibodies at RT for 60 minutes. The antibodies were revealed with Texas Red- and/or FITC-conjugated secondary antibodies. Alternatively, the cells were fixed with methanol for 20 minutes at –20°C, air-dried for 5 minutes and rehydrated with PBS for 5 minutes before incubation with antibodies. They were then incubated with DAPI to visualize DNA and mounted with the antifading solution AF1 (Citifluor). Fluorescent microscopy was performed using a CCD camera Leitz DMRB. Optical sections (0.4 μm thick) were examined on a Leica SPD2 AOBS confocal microscope (Leica Microsystem) with a 63×, 1.32 NA PlanApo lens and acquired with a Micromax CCD camera (Princeton Instruments, France). An argon laser adjusted to 488 nm was used for the fluorescein signal, a krypton laser adjusted to 568 nm for Texas Red and a blue laser diode at 405 nm for DAPI. Images were assembled using Adobe Photoshop. SIRT7 and UBF signal intensity was quantified in Fig. 1C and Fig. 2B using ImageJ software. Cell staining and image capture in Fig. 2C and supplementary material Fig. S2B were as described in (Prieto and McStay, 2007).
siRNA-mediated mRNA knockdown
The siControl RISC-free siRNA and SIRT1 SMARTpool siRNA were purchased from Dharmacon. SIRT7 siRNA duplexes targeting 5′-GCCUGAAGGUUCUAAAGAA (sense) and 5′-UUCUUUAGAACCUUCAGGC (antisense) were synthetized by Eurogentec. siRNA duplexes (15 nM) were transfected using INTERFERin™ (Polyplus-transfection) according to the manufacturer's instructions 24 and 48 hours after cell seeding.
Pull-down experiments
For in vitro pull-down assays, GST-SIRT7 was overexpressed in E. coli BL21 DE3 in Staby™Switch auto-inducible medium (Eurogentec) according to the manufacturer's instructions. The GST protein was induced for 3 hours with 0.1 mM isopropylthiogalactoside in E. coli. Lysates were obtained by enzymatic reaction with lysozyme (100 μg/ml) and DNase I (20 ng/μl) and clarified by ultracentrifugation at 65,000 g for 30 minutes. The clarified lysates were incubated with glutathione-sepharose beads (Amersham) for 2 hours at 4°C. GST-fusion proteins bound to glutathione-sepharose beads were washed four times with wash buffer (50 mM Tris-HCl, pH 8, 1 M NaCl, 10% glycerol and complete) and once with binding buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.2% NP40, 10% glycerol and complete; complete corresponds to a cocktail of protease inhibitors (Roche)]. Proteins of HeLa cells were extracted using lysis buffer (50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 1 mM EDTA, 1% NP40, 10% glycerol and complete). After centrifugation at 16,000 g for 15 minutes, the supernatants were adjusted to the binding buffer conditions and these whole-cell lysates were incubated with GST and GST-SIRT7 bound to glutathione-sepharose beads. Otherwise, 1 μg of human UBF1 and UBF2, produced as previously described (McStay et al., 1997), was diluted in the binding buffer and incubated with GST and GST-SIRT7 bound to glutathione-sepharose beads. After gentle shaking overnight at 4°C, the beads were centrifuged at 500 g for 2 minutes and washed five times in wash buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 1 mM EDTA, 0.2% NP40, 10% glycerol and complete). Proteins corresponding to cell lysates and proteins bound to beads were resuspended in SDS loading buffer, boiled for 5 minutes, resolved by 10% SDS-PAGE and analyzed by immunoblotting.
Chromosome and cytoplasmic fractions
Mitotic HeLa cells were resuspended in 20% FCS for 20 minutes at RT. Cells were then mechanically disrupted by passage through a G22 needle until chromosomes appeared scattered, as assessed by phase microscopy. The pellet containing the chromosome fraction and the supernatant containing the cytoplasmic fraction were normalized to an equal volume of SDS loading buffer for quantitative comparison.
Phos-tag SDS-PAGE
Proteins were resolved by 7.5% SDS-PAGE containing 100 μM Phos-tag ligand (AAL-107, NARD Institute, Japan) and 100 μM MnCl2 as previously described (Kinoshita et al., 2006). Gels were run at 40 mA for 1.5 hours, rinsed once for 10 minutes in transfer buffer containing 1 mM EDTA to chelate MnCl2 and once in the same buffer without EDTA. Proteins were then submitted to immunoblotting.
Immunoblotting
Proteins resolved by SDS-PAGE were transferred to nitrocellulose membranes (Protran, Schleicher and Schuell) and incubated with the following antibodies: anti-SIRT1, anti-SIRT7-N-terminal, anti-UBF, anti-TBP, anti-TTF-1, anti-RPA116/135, anti-RPA43, anti-PAF49, anti-PAF53 or anti-TIF-1A/Rrn3. The membranes were then incubated with suitable horseradish peroxidase-conjugated secondary antibodies and immunoreactivity was detected by chemiluminescence (Pierce).
BrUTP incorporation
The assay described in (Valdez et al., 2004) was slightly modified as follows. Coverslips seeded with HeLa cells were briefly rinsed with hypotonic KH buffer (30 mM KCl and 10 mM Hepes, pH 7.4) and incubated with KH buffer containing 10 mM BrUTP for 10 minutes in a 5% CO2 incubator at 37°C. The cells were rinsed three times with culture medium and incubated for 30 minutes in MEM containing 20% FCS. They were rinsed with PBS, fixed in methanol for 20 minutes at –20°C, air-dried for 5 minutes and rehydrated with PBS for 5 minutes. BrUTP incorporation was detected using an anti-BrdU antibody. Alternatively, RNA Pol I activity was detected in fixed cells in conditions set up to preferentially reveal RNA Pol I as previously described (Roussel et al., 1996). Briefly, HeLa cells seeded as monolayers on glass slides were weakly fixed with ethanol/acetone for 5 minutes at 4°C and air dried. The cells were then incubated in assay solution (100 mM Tris-HCl, pH 7.9, 12 mM 2-mercaptoethanol, 150 mM sucrose, 12 mM MgCl2, 0.6 mM ATP, CTP and GTP each, and 0.12 mM BrUTP) at 37°C for 15 minutes. The cells were post-fixed in 2% paraformaldehyde for 20 minutes at RT. BrUTP incorporation was detected by immunofluorescence labeling using an anti-BrdU antibody.
Supplementary Material
The authors are grateful to I. Horikawa and S. A. Leibovitch for generously providing constructs or antibodies, to C. Chamot and A. Jobart-Malfait for technical support in confocal microscopy and to A. L. Haenni for critical reading of the manuscript. A.G. is the recipient of an undergraduate grant from Ministère de l'Education Nationale, de la Recherche et de la Technologie. This study was supported in part by grants from the Centre National de le Recherche Scientifique and the Association pour la Recherche sur le Cancer (contrat 3303). B.M. was supported by the MRC, UK. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/4/489/DC1
References
- Blander, G. and Guarente, L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417-435. [DOI] [PubMed] [Google Scholar]
- Boisvert, F. M., van Koningsbruggen, S., Navascues, J. and Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 8, 574-585. [DOI] [PubMed] [Google Scholar]
- Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y. et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011-2015. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Mendes-Soares, L. and Campos, I. (2000). To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107-E112. [DOI] [PubMed] [Google Scholar]
- Chen, D., Dundr, M., Wang, C., Leung, A., Lamond, A., Misteli, T. and Huang, S. (2005). Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168, 41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute, P. and Pines, J. (1999). Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1, 82-87. [DOI] [PubMed] [Google Scholar]
- Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., de Cabo, R. and Sinclair, D. A. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390-392. [DOI] [PubMed] [Google Scholar]
- De Azevedo, W. F., Leclerc, S., Meijer, L., Havlicek, L., Strnad, M. and Kim, S. H. (1997). Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur. J. Biochem. 243, 518-526. [DOI] [PubMed] [Google Scholar]
- Denu, J. M. (2005). The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 9, 431-440. [DOI] [PubMed] [Google Scholar]
- Dryden, S. C., Nahhas, F. A., Nowak, J. E., Goustin, A. S. and Tainsky, M. A. (2003). Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell. Biol. 23, 3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy, W. G. and Newport, J. W. (1988). Unraveling of mitotic control mechanisms. Cell 55, 925-928. [DOI] [PubMed] [Google Scholar]
- Ford, E., Voit, R., Liszt, G., Magin, C., Grummt, I. and Guarente, L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, J., Norbury, C., Lohka, M., Nurse, P. and Maller, J. (1988). Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54, 433-439. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., Lucas, L., Klein, C., Menager, M., Bonnet, N. and Ploton, D. (1995). Three-dimensional co-location of RNA polymerase I and DNA during interphase and mitosis by confocal microscopy. J. Cell Sci. 108, 115-125. [DOI] [PubMed] [Google Scholar]
- Grozinger, C. M., Chao, E. D., Blackwell, H. E., Moazed, D. and Schreiber, S. L. (2001). Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276, 38837-38843. [DOI] [PubMed] [Google Scholar]
- Grummt, I. (2003). Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691-1702. [DOI] [PubMed] [Google Scholar]
- Guarente, L. (2000). Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14, 1021-1026. [PubMed] [Google Scholar]
- Hadjiolov, A. A. (1985). The Nucleolus and Ribosome Biogenesis. New York: Springer-Verlag.
- Heix, J., Vente, A., Voit, R., Budde, A., Michaelidis, T. M. and Grummt, I. (1998). Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 17, 7373-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, S. T. and Ghosh, A. K. (1999). Control of RNA polymerase I-directed transcription: recent trends. J. Cell. Biochem. Suppl. 32-33, 41-50. [DOI] [PubMed]
- Johnson, L. N. and Lewis, R. J. (2001). Structural basis for control by phosphorylation. Chem. Rev. 101, 2209-2242. [DOI] [PubMed] [Google Scholar]
- Jordan, P., Mannervik, M., Tora, L. and Carmo-Fonseca, M. (1996). In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133, 225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. H., Sarbassov, D. D., Ali, S. M., Latek, R. R., Guntur, K. V., Erdjument-Bromage, H., Tempst, P. and Sabatini, D. M. (2003). GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11, 895-904. [DOI] [PubMed] [Google Scholar]
- Kinoshita, E., Takahashi, M., Takeda, H., Shiro, M. and Koike, T. (2004). Recognition of phosphate monoester dianion by an alkoxide-bridged dinuclear zinc(II) complex. Dalton Trans. 8, 1189-1193. [DOI] [PubMed] [Google Scholar]
- Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K. and Koike, T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749-757. [DOI] [PubMed] [Google Scholar]
- Klein, J. and Grummt, I. (1999). Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 96, 6096-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A., Vente, A., Doree, M. and Grummt, I. (1998). Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 284, 1-5. [DOI] [PubMed] [Google Scholar]
- Leung, A. K., Gerlich, D., Miller, G., Lyon, C., Lam, Y. W., Lleres, D., Daigle, N., Zomerdijk, J., Ellenberg, J. and Lamond, A. I. (2004). Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166, 787-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt, G., Ford, E., Kurtev, M. and Guarente, L. (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313-21320. [DOI] [PubMed] [Google Scholar]
- Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L. and Gu, W. (2001). Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137-148. [DOI] [PubMed] [Google Scholar]
- Mais, C., Wright, J. E., Prieto, J. L., Raggett, S. L. and McStay, B. (2005). UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 19, 50-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay, B., Sullivan, G. J. and Cairns, C. (1997). The Xenopus RNA polymerase I transcription factor, UBF, has a role in transcriptional enhancement distinct from that at the promoter. EMBO J. 16, 396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, L., Borgne, A., Mulner, O., Chong, J. P., Blow, J. J., Inagaki, N., Inagaki, M., Delcros, J. G. and Moulinoux, J. P. (1997). Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243, 527-536. [DOI] [PubMed] [Google Scholar]
- Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C. and Horikawa, I. (2005). Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky, R., Chua, K. F., Lombard, D. B., Pang, W. W., Fischer, M. R., Gellon, L., Liu, P., Mostoslavsky, G., Franco, S., Murphy, M. M. et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315-329. [DOI] [PubMed] [Google Scholar]
- Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M. and Guarente, L. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551-563. [DOI] [PubMed] [Google Scholar]
- Murayama, A., Ohmori, K., Fujimura, A., Minami, H., Yasuzawa-Tanaka, K., Kuroda, T., Oie, S., Daitoku, H., Okuwaki, M., Nagata, K. et al. (2008). Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133, 627-639. [DOI] [PubMed] [Google Scholar]
- North, B. J. and Verdin, E. (2004). Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North, B. J. and Verdin, E. (2007). Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J. Biol. Chem. 282, 19546-19555. [DOI] [PubMed] [Google Scholar]
- North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M. and Verdin, E. (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437-444. [DOI] [PubMed] [Google Scholar]
- Prescott, D. M. and Bender, M. A. (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell. Res. 26, 260-268. [DOI] [PubMed] [Google Scholar]
- Prieto, J. L. and McStay, B. (2007). Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21, 2041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, P., Andre, C., Masson, C., Geraud, G. and Hernandez-Verdun, D. (1993). Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104, 327-337. [DOI] [PubMed] [Google Scholar]
- Roussel, P., Andre, C., Comai, L. and Hernandez-Verdun, D. (1996). The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133, 235-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D. and Pandolfi, P. P. (2003). Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179-192. [DOI] [PubMed] [Google Scholar]
- Russell, J. and Zomerdijk, J. C. (2005). RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30, 87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvé, D. M., Anderson, H. J., Ray, J. M., James, W. M. and Roberge, M. (1999). Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J. Cell Biol. 145, 225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U. and Rose, K. M. (1984). Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc. Natl. Acad. Sci. USA 81, 1431-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T. and Hall, M. N. (2000). TOR, a central controller of cell growth. Cell 103, 253-262. [DOI] [PubMed] [Google Scholar]
- Simpson, J. C., Wellenreuther, R., Poustka, A., Pepperkok, R. and Wiemann, S. (2000). Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 1, 287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri, V., Roussel, P. and Hernandez-Verdun, D. (1999). The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J. Cell Sci. 112, 3259-3268. [DOI] [PubMed] [Google Scholar]
- Sirri, V., Roussel, P. and Hernandez-Verdun, D. (2000). In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 148, 259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri, V., Hernandez-Verdun, D. and Roussel, P. (2002). Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 156, 969-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky, V. Y., Pelletier, G., Hannan, R., Gagnon-Kugler, T., Rothblum, L. I. and Moss, T. (2001). An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8, 1063-1073. [DOI] [PubMed] [Google Scholar]
- Valdez, B. C., Henning, D., So, R. B., Dixon, J. and Dixon, M. J. (2004). The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 101, 10709-10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. and Weinberg, R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149-159. [DOI] [PubMed] [Google Scholar]
- Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A. and Mayo, M. W. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., Chai, X., Clements, A. and Marmorstein, R. (2003). Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 10, 864-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.