Abstract
Cyclodextrin glucanotransferases (CGTases) have attracted major interest from industry due to their unique capacity of forming large quantities of cyclic α-(1,4)-linked oligosaccharides (cyclodextrins) from starch. CGTases produce a mixture of cyclodextrins from starch consisting of 6 (α), 7 (β) and 8 (γ) glucose units. In an effort to identify the structural factors contributing to the evolutionary diversification of product specificity amongst this group of enzymes, we selected nine CGTases from both mesophilic, thermophilic and hyperthermophilic organisms for comparative product analysis. These enzymes displayed considerable variation regarding thermostability, initial rates, percentage of substrate conversion and ratio of α-, β- and γ-cyclodextrins formed from starch. Sequence comparison of these CGTases revealed that specific incorporation and/or substitution of amino acids at the substrate binding sites, during the evolutionary progression of these enzymes, resulted in diversification of cyclodextrin product specificity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-009-1988-6) contains supplementary material, which is available to authorized users.
Keywords: Protein evolution, CGTase, α-Amylase, Reaction specificity, Protein stability
Introduction
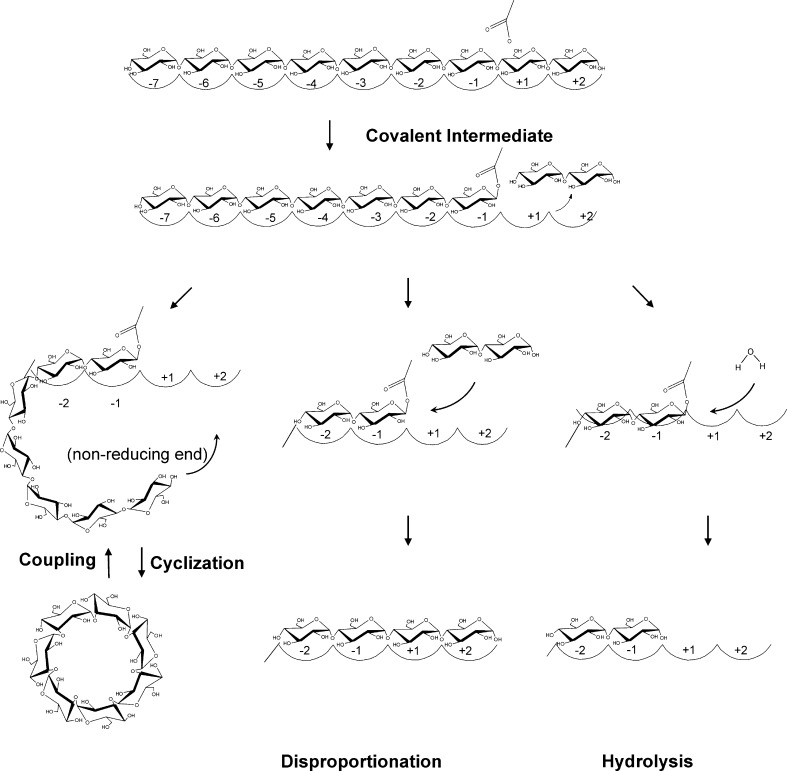
Cyclodextrin glucanotransferases (CGTases) are members of the largest family of glycoside hydrolases acting on starch and related α-glucans, glycoside hydrolyase family 13 (GH13; Stam et al. 2006; Svensson 1994; Buchholz and Seibel 2008). Family members share a conserved active site architecture along with four short conserved sequence regions embedded in a TIM (β/α)8 structural fold (Janecek 1997). All family members either hydrolyse and/or transglycosylate α-glucosidic linkages, with catalysis proceeding via a double displacement mechanism (McCarter and Withers 1994). CGTases are composed of five domains (A–E; Lawson et al. 1994; Klein and Schulz 1991). The A and B domains form the substrate binding groove and contain the catalytic residues. The additional domains C and E assist in starch binding (Penninga et al. 1996), while the function of the D domain is unknown. CGTases are unique members of the GH13 family: They use the non-reducing end of a bound oligosaccharide as acceptor molecule for circular α-(1,4)-linked oligosaccharide formation (cyclization, Fig. 1). The cyclization reaction starts with binding of a linear saccharide chain across the nine sugar binding subsites labelled −7 to +2 (Fig. 1; Strokopytov et al. 1996), with glycosidic bond cleavage occurring between the −1 and +1 subsites, to yield a stable covalent glycosyl–enzyme intermediate (Uitdehaag et al. 1999b). The sugar at the acceptor subsites, labelled +1/+2, is then exchanged for a new acceptor, which is the non-reducing end of the covalently bound oligosaccharide, to yield a cyclodextrin molecule (Fig. 1). CGTases can also transfer the covalently bound oligosaccharide to a water molecule (hydrolysis) or to a second sugar molecule (disproportionation). In addition, CGTases can breakdown cyclodextrins in the coupling reaction, which involves cyclodextrin binding and ring cleavage at the active site before being transferred to an acceptor molecule, to yield linear products (Fig. 1). These enzymes primarily produce three types of cyclodextrin, differing in their degree of polymerization α (6), β (7), and γ (8; Li et al. 2007; Qi and Zimmermann 2005). The quantity and cyclodextrin size specificity vary greatly with CGTases from various organisms, ranging from predominantly α-cyclodextrin to β- or γ-cyclodextrin. There is also considerable variation in the thermostability properties of CGTases, largely depending on the habitat of the host organism, e.g. meso- up to hyperthermophilic.
Fig. 1.
Schematic representation of the reactions catalysed by CGTase. A linear glucan chain binds to the substrate binding subsites of CGTase followed by bond cleavage to yield a covalent glycosyl–enzyme intermediate. The nature of the acceptor molecule in the second step of the reaction, to which the covalently bound oligosaccharide is transferred, determines the enzyme reaction specificity
To identify and understand the structural and molecular factors contributing to the evolutionary diversification amongst this group of enzymes, we have selected nine CGTases from both meso-, thermo- and hyperthermophilic organisms for comparative product analysis. Amino acid sequence analysis revealed a clear trend in cyclodextrin product specificity based on the nature of various amino acids at the donor substrate binding subsites, close to the active site. Thus, CGTases most likely evolved from a common ancestor to diversify in product specificity by acquisition of specific mutations at the substrate binding sites.
Experimental procedures
Phylogenetic analysis
All amino acid sequences of functional CGTases were selected from the CAZY web server (Coutinho and Henrissat 1999). These sequences were then blasted against the protein database at Swissprot (http://www.ncbi.nlm.nih.gov/BLAST/) for identification of a number of extra putative CGTase proteins (indicated by “p” in Fig. 2). Sequences were aligned using the ClustalW interface in the Mega 4 programme (Tamura et al. 2007). The phylogenetic tree of CGTases was created with Mega 4 (Tamura et al. 2007), using the neighbour-joining method (Saitou and Nei 1987), and default settings for gap and extension penalties. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. Accession numbers of the proteins are listed in Table S1 in Electronic supplementary material.
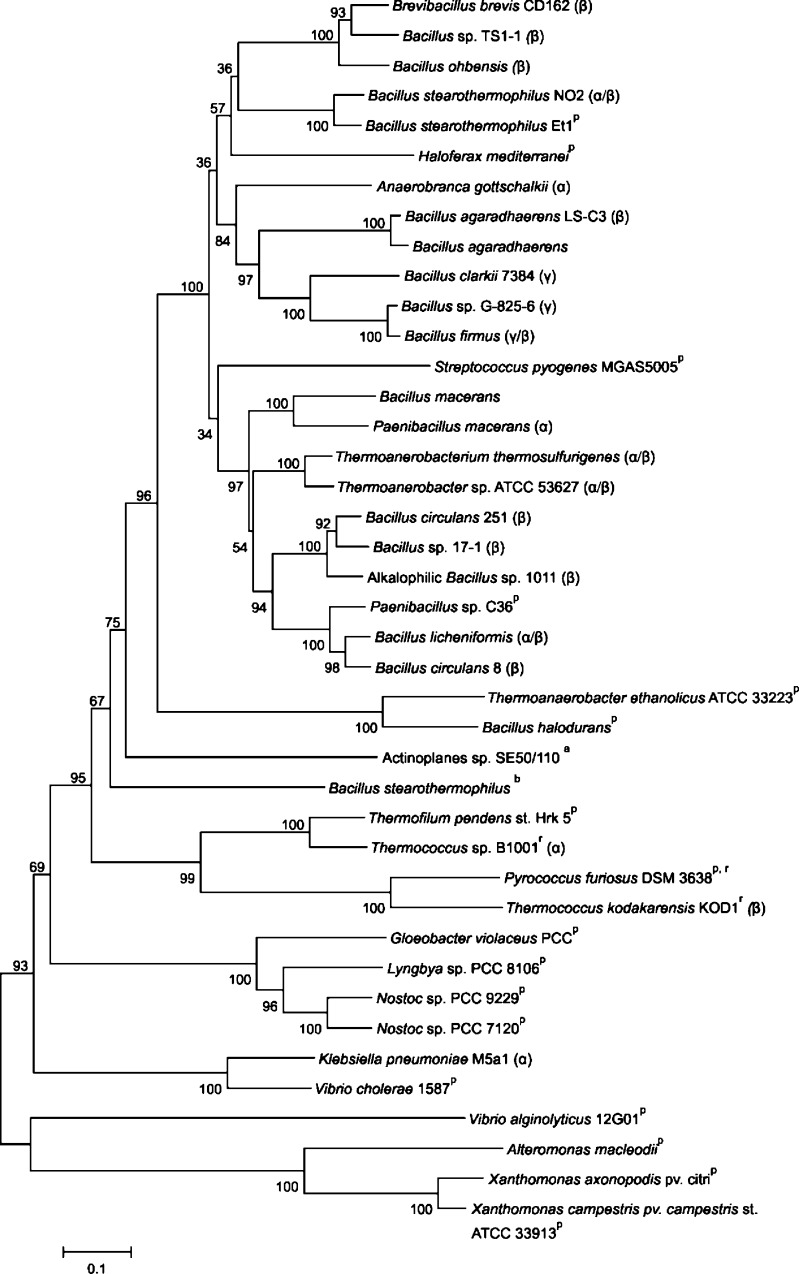
Fig. 2.
Bootstrapped phylogenetic tree of CGTases and homologous proteins. The scale bar corresponds to a genetic distance of 0.1 substitution per position. CGTases displaying >95% similarity were not included. The main type of cyclodextrin formed by the functional CGTases is indicated by α, β, or γ. a ATase (Leemhuis et al. 2004b), b Novamyl (Dauter et al. 1999), p putative CGTase, r CGTase from Archaea. (See Table S1 of the Electronic supplementary material for protein accession numbers.)
Bacterial strains and plasmids
Bacillus circulans 251 (BC251) and Thermoanaerobacterium thermosulfurigenes EM1 (Tabium) CGTase proteins were produced from plasmids pDP66k- and pCScgt-tt, respectively, using Bacillus subtilis strain DB104A as host (Leemhuis et al. 2003b). The B. circulans 8 (BC8) CGTase protein was produced using plasmid pTZ19R in Escherichia coli JM103 containing the cgt gene together with endogenous regulatory sequence, as described (Nitschke et al. 1990). B. subtilis NA-1 was used as host for the pKB1 plasmid for the production of Bacillus stearothermophilus NO2 (BNO2) CGTase protein (Fujiwara et al. 1992). Production of Anaerobranca gottschalkii CGTase protein was carried out using the expression plasmid pET24a with E. coli BL21(DE3) as host (Thiemann et al. 2004). Thermococcus sp. B1001 CGTase protein was produced in E. coli BL21 (DE3) cells harbouring the expression plasmid pCGTA. The Thermococcus sp. B1001 CGTase was purified from inclusion bodies by solubilization in 6 M urea, refolded by dialysis and heated to 80°C for 20 min, as described (Yamamoto et al. 1999). Purity and molecular weight of all CGTase proteins were checked by SDS-PAGE. Protein concentrations were determined using the Bradford reagent from Bio-Rad (München, Germany) and bovine serum albumin as standard. Alkalophilic Bacillus sp. A2-5a (BA2-5a) and Klebsiella pneumoniae M5A1 CGTase proteins were kindly provided by Dr. T. Kuriki of Ezaki Glico (Japan) and Dr. M. Moick of Wacker Chemie AG (Germany), respectively. Dr. C. Andersen of Novozyme (Denmark) supplied us with the Thermoanaerobacter sp. ATCC 53627 (Toruzyme) CGTase protein.
Enzyme assays
Initial rates of BC251, Tabium, Toruzyme and BNO2 CGTases were measured in sodium citrate buffer (10 mM, pH 6.0) at 50, 60, 60 and 65°C, respectively. BC8 and K. pneumoniae CGTases were assayed in Tris–HCL buffer (10 mM, pH 6.8) at 30°C, A. gottschalkii CGTase was assayed in Tris–HCL (100 mM, pH 8.5), BA2-5a CGTase was assayed in sodium acetate buffer (10 mM, pH 5.5), and Thermococcus sp. B1001 CGTase was assayed in sodium acetate buffer (50 mM, pH 5.5). The β-cyclodextrin forming activity was determined by incubating 1.3–260 nM of enzyme with a 2.5% (w/v) solution of partially hydrolysed potato starch with an average degree of polymerisation of 50 (Paselli SA2; AVEBE, Foxhol, The Netherlands) in the presence of 3 mM CaCl2. The amount of β-cyclodextrin produced was quantified with phenolphthalein (Vikmon 1982). Starch hydrolysing activity was measured by following the increase in reducing power with dinitrosalicylic acid, using 1% (w/v) soluble starch (Sigma-Aldrich) and 0.66 nM–0.66 mM of enzyme.
HPLC product analysis
Formation of cyclodextrins from starch was analysed by incubating 10% (w/w) starch (Paselli SA2; AVEBE) for 72 h with 13 nM of CGTase enzyme. Samples (1 mL) were taken at regular time intervals, and the reactions were stopped by the addition of 100 µl 1.2 N HCl. Prior to HPLC analysis, 100 µl of 1.2 N NaOH was added to the samples. Products formed were analysed on an Econosphere NH2 5 U column (250 × 4.6 mm; Alltech, Breda, the Netherlands) linked to a refractive index detector. A mobile phase of acetonitrile/water (60/40, v/v) at a flow rate of 0.5 mL/min was used.
Differential scanning calorimetry
Thermal unfolding of CGTases and the maltogenic amylase from B. stearothermophilus (Novamyl) was measured using a MicroCal VP-differential scanning calorimetry (DSC) microcalorimeter (MicroCal, Northhampton, MA, USA). The cell volume was 0.52 mL, and experiments were performed at a scan rate of 1°C/min at a constant pressure of 2.75 bar. Samples were degassed prior to scanning. The enzyme concentrations used were 9.3 µM for Novamyl, BC8, BC251, Tabium CGTases and 4.6 µM for BCA2-5a CGTase, in 10 mM sodium acetate buffer with 3 mM CaCl2 (pH 5.5). Ca2+ ions were added as they have been shown to raise the unfolding temperature of CGTase (Leemhuis et al. 2004a). Unfolding of A. gottschalkii CGTase was measured in 20 mM potassium phosphate buffer supplemented with 3 mM CaCl2 (pH 7.5), at an enzyme concentration of 9.3 µM.
Stability assays
Resistance to thermal inactivation was determined by incubating CGTase protein at various temperatures in a water bath for 30 min. Samples were then cooled in an ice bath for 3 min. Residual activity was measured by the β-cyclization assay. The buffer and protein conditions used for assaying residual CGTase activity were identical to those used for DSC analysis and the β-cyclization activity assay, respectively. Both BNO2 CGTase and Toruzyme were assayed in 10 mM sodium acetate buffer with 3 mM CaCl2 (pH 5.5). K. pneumoniae CGTase activity was assayed in 20 mM potassium phosphate buffer supplemented with 3 mM CaCl2 (pH 7). We defined T 50 as the temperature at which 50% of the initial β-cyclization enzyme activity remained after incubation for 30 min.
Protein interactions calculator
Protein interactions calculator server was used to compute the ionic interactions and disulphide bonds of Tabium (1CIU), BC8 (1CGT), BC251 (1CDG), Alkalophilic Bacillus sp. 1011 (1PAM), BNO2 (1CYG) CGTase structures and Novamyl (1QHO; Tina et al. 2007). The structural PDB file accession numbers are given between brackets. Interactions between ionic pairs (Arg, Lys, His, Asp, Glu) were calculated within a distance of 6 Å.
Results
Phylogenetic analysis
CGTases differ greatly in their cyclodextrin product specificity and thermostability. In an effort to identify residues and/or sequence regions which may control their enzymatic properties, we compared the CGTase sequences. The acarviosyl transferase (ATase) from Actinoplanes sp. SE50/110 and the maltogenic amylase Novamyl were also included in the alignment (see Fig. S1 of the Electronic supplementary material for the full alignment) as these two GH13 enzymes have about 50% sequence identity with CGTases, and they acquire the ability to form cyclodextrins upon introduction of a few mutations (Beier et al. 2000; Leemhuis et al. 2004b). A phylogenetic tree generated from the CGTase sequences alignment (Fig. 2) showed that β-CGTase sequences are rather diverse, with these enzymes scattered throughout the phylogenetic tree. The three γ-CGTases, in contrast, are clustered together, indicating common specific (stretches of) amino acid sequence favouring γ-cyclodextrin production (Fig. 2). Despite their diverse product specificity, the CGTases from Archaea are also grouped together (indicated in Fig. 2 by “r”). The α-CGTase from K. pneumoniae is the only known CGTase from a gram-negative bacterium, which may explain its relatively low sequence similarity with other functional CGTases and its grouping with putative CGTases from various bacteria. These putative CGTases display large variations in the conserved sequence regions 1 and 2 of GH13 enzymes compared to the consensus sequence of functional CGTases (Fig. 3). Residues of regions 1 and 2 determine the orientation of the substrate within the active site and play crucial roles in glycosidic bond cleavage (Leemhuis et al. 2003c; Nakamura et al. 1993; Table 1).
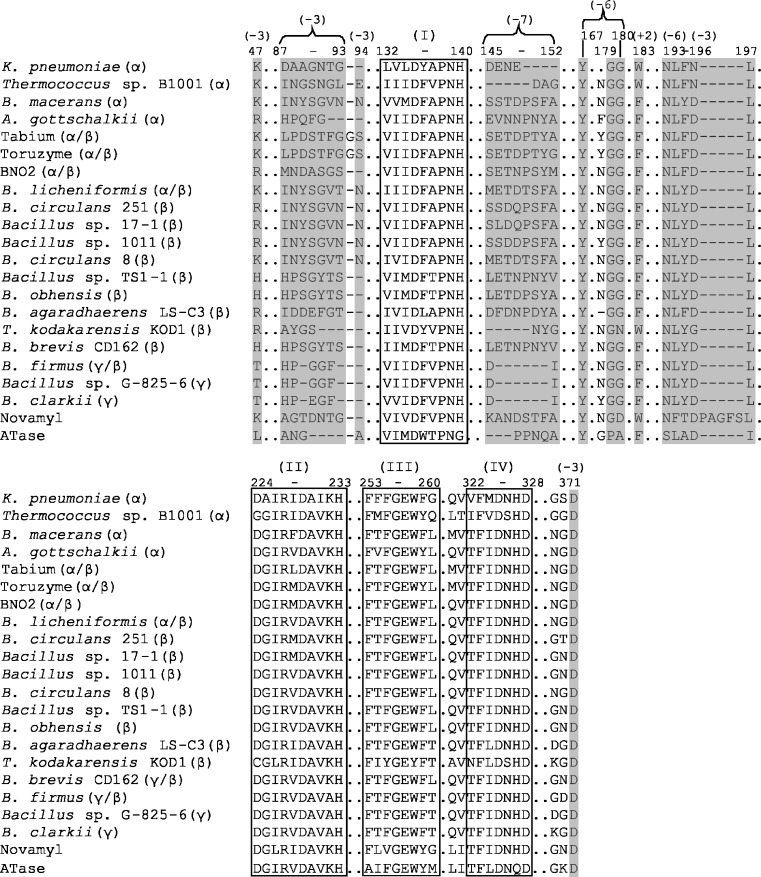
Fig. 3.
Alignment of functional CGTase regions. The four conserved GH13 sequence regions are boxed and marked I–IV. The numbers in brackets indicate the residues involved in substrate binding at subsites +2, −3, −6 and −7 and are coloured in grey. Numbering of residues according to BC251 CGTase. (See Fig. S1 of the Electronic supplementary material for full sequence alignment)
Table 1.
The function of residues in CGTase, according to B. circulans strain 251 numbering
| Positions | Residue | Mutations | Function/effect |
|---|---|---|---|
| Acceptor subsite +2 | |||
| 183 | Phe, Trp | Asn, Ser, Leu | Cyclization, reduction of hydrolysisa,b,c,d |
| 232 | Lys, Ala | Leu, Glu | General activityc,e |
| 259 | Phe, Tyr | Leu, Asn, Ile, Glu, His, Arg | Cyclization, reduction of hydrolysisa,b,c,d,f,g |
| Acceptor subsite +1 | |||
| 194 | Leu | Thr | Cyclizationh |
| 195 | Tyr, Phe | All possibilities | Cyclizationa,b,f,h,i,j |
| 230 | Ala | Val | Transglycosylation specificityd,e,k |
| 233 | His | Asp, Asn, Arg | General activityl,m |
| Catalytic subsite −1 | |||
| 100 | Tyr | Phe, Ser | General activityn,o |
| 140 | His | Asp, Asn, Arg | General activityl,m |
| 227 | Arg | Ala, Lys | General activityp |
| 229 | Asp | Ala, Asn | Catalytic nucleophileq,r,s,t |
| 257 | Glu | Ala, Gln | Acid–base catalystq,s,t |
| 327 | His | Asp, Asn, Arg | General activityl,m |
| 328 | Asp | Asn, Ala | General activity, transition state stabilisers,t |
| Donor subsite –2 | |||
| 98 | His | Asp | General activitym |
| Donor subsite −3 | |||
| 47 | Arg, Lys, His, Thr | Trp, Leu, Gln | Cyclizationk,u |
| 89 | Tyr, Asp, Ser, Gln, Ala, Gly | Phe, Ser, Asp, Gly | General activity, product specificityo,v |
| 94 | Asn, Ser, Tyr, Glu | Ser, Gln | Slightly decreased activitieso,k |
| 196 | Asp, Asn, Gly | His | Reduced activity, more α-cyclodextrinw |
| 371 | Asp | Gly, Asn, Arg | General activity, less α-cyclodextrinw |
| Donor subsites −4/−5 | No side-chain interactions | ||
| Donor subsite –6 | |||
| 167 | Tyr | Phe | Selection of long substratesx |
| 179 | Gly | Leu | Selection of long substratesx |
| 180 | Gly, Asn | Leu | Selection of long substratesx |
| 193 | Asn | Leu, Gly | Selection of long substrates x |
| Donor subsite –7 | |||
| 145 | Ser, Met, Leu, Asp, Glu | Ala, Glu | Cyclodextrin specificity |
| 146 | Ser, Glu, Val, Phe, Leu | Pro | Cyclodextrin specificityv |
| 147 | Thr, Asp, Asn | Trp | Cyclodextrin specificity |
| Loop 145–151 | See Fig. 3 | Asp | Cyclodextrin specificityh |
| Other important residues | |||
| 21 | Phe, Val | Leu | Limitation of hydrolysisk |
| 76 | Ser, Thr | Pro | Hydrolysisn |
| 135 | Asp | Asn, Ala | Influences conformation of Glu257 and Arg227p |
| 326 | Asp, Ser | Asp, Gln, Leu | Cyclization and general activityw |
a(Leemhuis et al. 2002a)
b(Nakamura et al. 1994)
c(van der Veen et al. 2001)
d(Kelly et al. 2007)
e(Kelly et al. 2008a)
f(Fujiwara et al. 1992a)
g(Shim et al. 2004)
h(Parsiegla et al. 1998)
i(Penninga et al. 1995)
j(Sin et al. 1994)
k(Leemhuis et al. 2003d)
l(Nakamura et al. 1993)
m(Mattsson et al. 1995)
n(Kelly et al. 2008b)
o(Kim et al. 1997)
p(Leemhuis et al. 2003c)
q(Uitdehaag et al. 1999b)
r(Mosi et al. 1997)
s(Knegtel et al. 1995)
t(Klein et al. 1992)
u(van der Veen et al. 2000a)
v(van der Veen et al. 2000b)
w(Wind et al. 1998)
x(Leemhuis et al. 2002b)
Enzymatic properties of CGTases
The β-cyclization and hydrolysis rates of nine CGTases were determined (Table 2). The BC251, BNO2 and Tabium CGTases displayed the highest initial rates for β-cyclodextrin formation. Toruzyme and Tabium CGTases exhibited elevated hydrolytic rates on starch, while in contrast the BA2-5a, BC8 and K. pneumoniae CGTases displayed the lowest hydrolytic rates. As a general trend, Bacillus species CGTases displayed much higher cyclization/hydrolysis ratios compared to other functional CGTases (Table 2).
Table 2.
β-Cyclization and hydrolysis activities of CGTases on potato starch
| Enzyme | β-Cyclization (µmol min−1 mg−1) | Hydrolysis (µmol min−1 mg−1) | Cyclization/hydrolysis ratio |
|---|---|---|---|
| B. sp. strain A2-5a | 94 ± 2 | 0.3 ± 0.04 | 313 |
| B. circulans strain 251 | 265 ± 5 | 2.1 ± 0.1 | 126 |
| B. stearothermophilus NO2 | 255 ± 6 | 1.4 ± 0.1 | 182 |
| B. circulans strain no. 8 | 52 ± 2 | 0.3 ± 0.01 | 173 |
| Tabium | 255 ± 7 | 57 ± 1.8 | 4.5 |
| Toruzyme | 127 ± 5 | 78 ± 5 | 1.6 |
| K. pneumoniae M5a1 | 0.5 ± 0.02 | 0.3 ± 0.03 | 1.7 |
| A. gottschalkii | 28 ± 2 | 7.7 ± 0.5 | 3.6 |
| Thermococcus sp B1001 | 23 ± 1 | 5.6 ± 0.3 | 4.1 |
Cyclodextrin size specificity
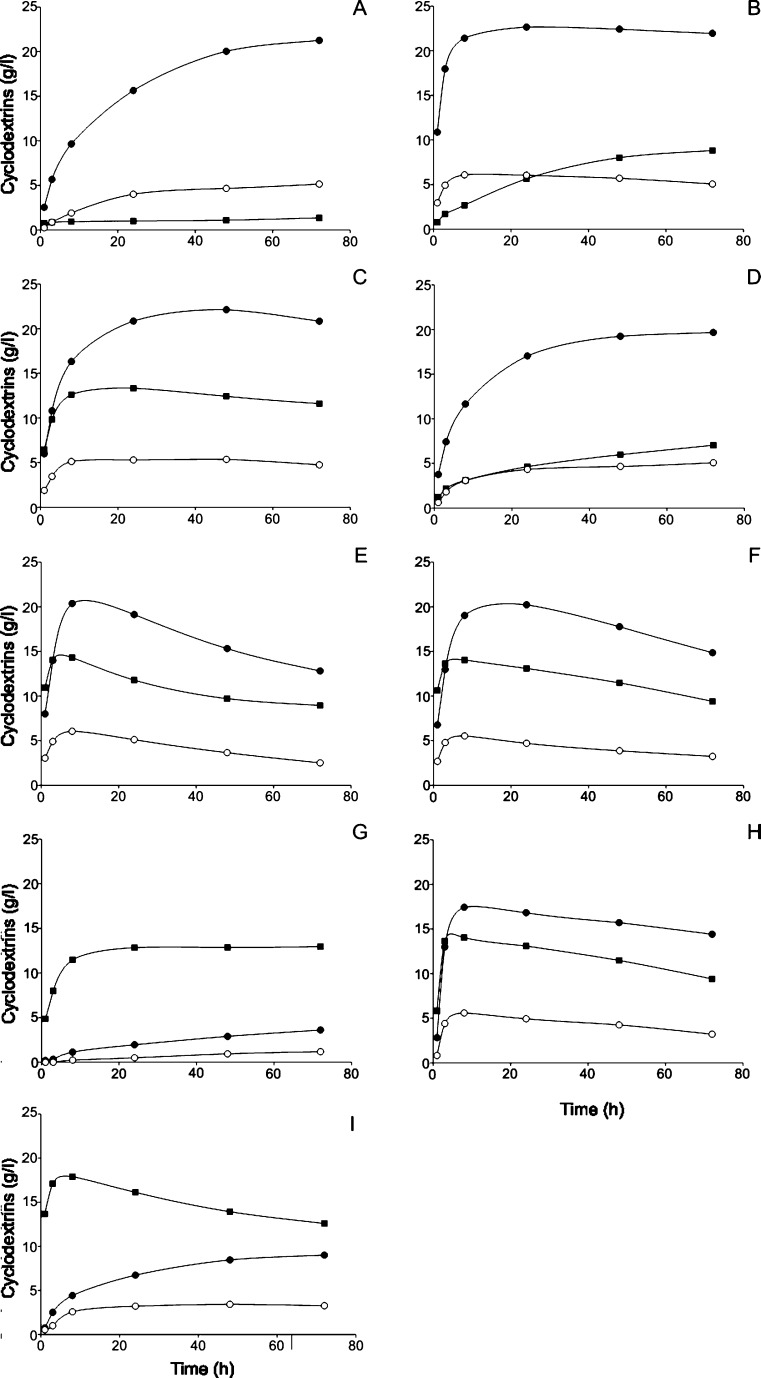
The amount of α-, β- and γ-cyclodextrins produced from starch by the CGTases was followed in time (Fig. 4). The four CGTases from Bacillus species were all primary β-cyclodextrin producers, yielding up to 22.6 g β-cyclodextrin per litre (Table 3). BC251 CGTase initially produced β-cyclodextrin at the fastest rate before levelling off after 24 h at 22.6 g/L (Fig. 4 and Table 3). BA2-5a CGTase was the most “specific” β-cyclodextrin producer with 77% β-cyclodextrin being formed (Table 3). Both Tabium and Toruzyme CGTases produced greater initial amounts of α-cyclodextrin compared to CGTases from Bacillus species. These enzymes also had the highest percentage conversion of starch into cyclodextrins along with BNO2 CGTase (Fig. 4 and Table 3). However, after reaching the maximum cyclodextrin yield, a continuous decrease in the amount of α-, β- and γ-cyclodextrins was noted for both Tabium and Toruzyme CGTases. A. gottschalkii CGTase also degraded cyclodextrins in the later stages of the starch incubations. This strong reduction in cyclodextrin yield may be attributed to the elevated hydrolytic rates of these enzymes on starch, providing short oligosaccharides for the breakdown of cyclodextrins in the coupling reaction (Fig. 1). The absolute yield of α-cyclodextrin was greatest for Thermococcus sp. B1001 CGTase (Fig. 4), even though the percentage of α-cyclodextrin was higher for the K. pneumoniae CGTase (Table 3).
Fig. 4.
Cyclodextrin production in time (g/L) from Paselli SA2 starch (100 g/L) by cyclodextrin glucanotransferases. aBacillus sp. A2-5a, bB. circulans 251, cBacillus stearothermophylus NO2, dB. circulans 8, eT. thermosulfurigenes EM1, f Toruzyme, gK. pneumoniae M5a1, hA. gottschalkii and iThermococcus sp. 1001. α, β and γ cyclodextrins are indicated by squares, filled circles and open circles, respectively
Table 3.
Cyclodextrin production from 100 g/L potato starch following incubation with various CGTases
| Enzyme | Main cyclodextrin formed | Cyclodextrin mass% ratio at 72 h | Total% starch into CDsb | |||
|---|---|---|---|---|---|---|
| Mass (g/L) | type | %a | Time (h) | (α:β:γ) | ||
| B. sp. strain A2-5a | 21.2 | β | 77 | 72 | 5:77:18 | 28 |
| B. circulans strain 251 | 22.6 | β | 66 | 24 | 25:61:14 | 36 |
| B. stearothermophilus NO2 | 22.1 | β | 56 | 48 | 31:56:13 | 40 |
| B. circulans strain no. 8 | 19.7/L | β | 62 | 72 | 22:62:16 | 32 |
| Tabium | 20.4/L | β | 50 | 8 | 37:53:10 | 41 |
| Toruzyme | 20.2 | β | 53 | 24 | 34:54:12 | 39 |
| K. pneumoniae M5a1 | 12.9 | α | 73 | 72 | 73:20:7 | 18 |
| A. gottschalkii | 17.4 | β | 47 | 8 | 35:53:12 | 37 |
| Thermococcus sp. B1001 | 17.9 | α | 72 | 8 | 51:36:13 | 26 |
aPercentage of the major cyclodextrin formed as mass percentage of all three cyclodextrins at the indicated time
bThe percentage Paselli starch converted into cyclodextrins over 72 h
Thermostability of CGTases
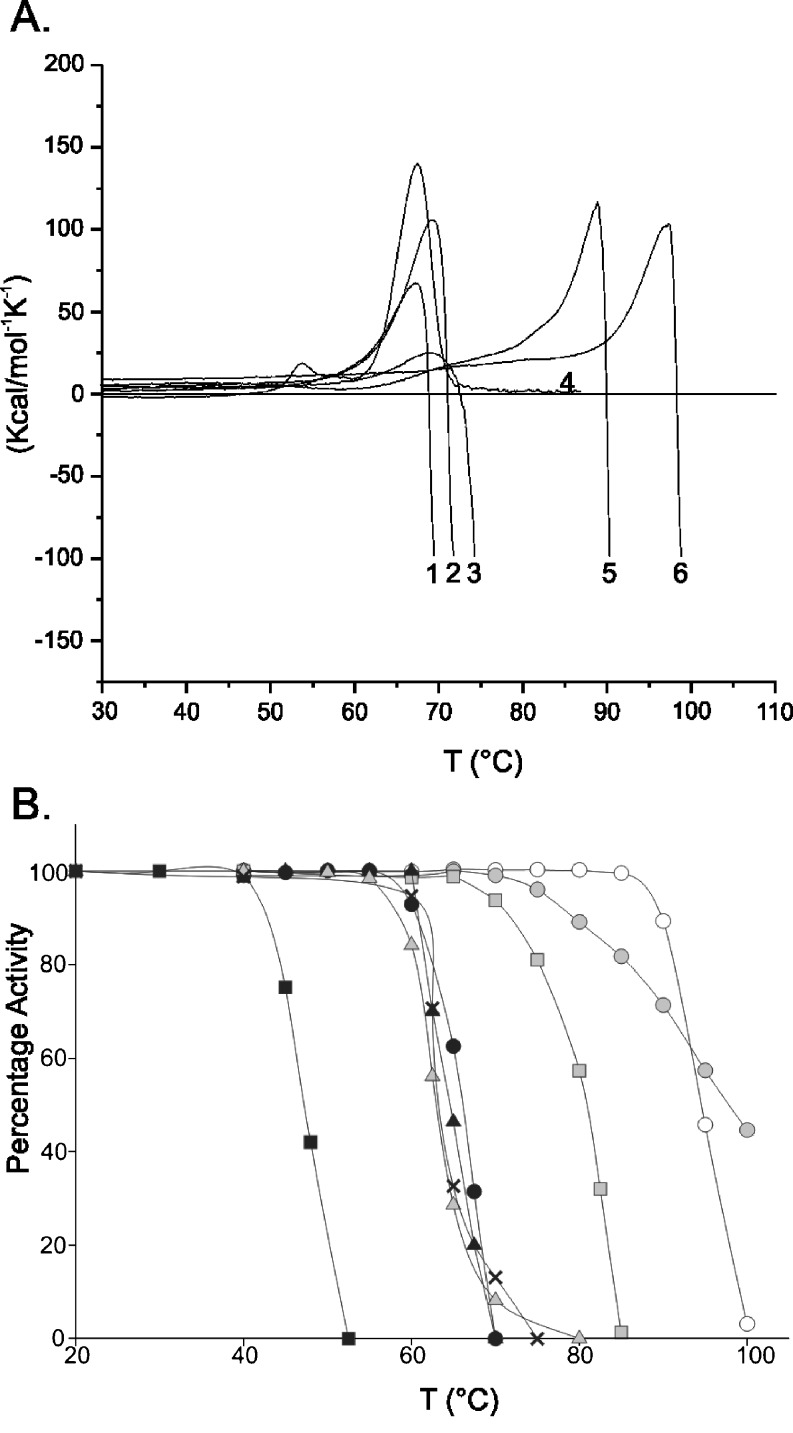
Thermostabilities of selected bacterial and archaeal CGTases along with Novamyl were determined by differential scanning calorimetry and thermal inactivation assays. Due to the irreversible unfolding nature of all CGTases analysed, apparent melting temperatures were estimated from the denaturation curves (Fig. 5). The resistance to thermal inactivation (T 50 temperature) was determined by incubating protein samples at various temperatures followed by measuring the remaining β-cyclization enzyme activity. The Bacillus species and A. gottschalkii CGTase showed similar apparent melting temperatures and T 50 temperatures (Fig. 5). Novamyl, Tabium and Thermococcus sp. B1001 CGTases were far more thermostable than their Bacillus CGTase counterparts (Table 4), whereas the K. pneumoniae CGTase was the least thermostable with a T 50 temperature of 47°C (Fig. 5). The CGTase of Thermococcus sp. B1001 was most resistant to thermal inactivation, with an apparent melting temperature of 106°C (Yamamoto et al. 1999).
Fig. 5.
Thermostability properties of CGTases. a Thermal denaturation curves of CGTases as measured with differential scanning calorimetry. (1) B. circulans 8, (2) B. circulans 251, (3) Bacillus sp. A2-5a, (4) A. gottschalkii, (5) Novamyl, (6) Tabium. b Resistance to thermal inactivation (T 50) of CGTases. Black squares K. pneumoniae, grey triangles Bacillus 8, multiplication signs A. gottschalkii, black triangles BA2-5a, black circles BC251, grey squares BNO2, white circles Tabium, grey circles Toruzyme
Table 4.
Resistance to thermal inactivation (T 50) and apparent melting temperatures of various CGTases
| Enzyme | Interactions | T 50 for β-cyclization activity (°C) | Apparent melting | |
|---|---|---|---|---|
| Ionic | Disulfide bonds | DSC (°C) | ||
| Tabium | 57 | 0 | 94.5 | 96.9 |
| B. circulans strain no. 8 | 53 | 1 | 62.9 | 67.1 |
| B. circulans strain 251 | 58 | 1 | 66.1 | 69.1 |
| B. stearothermophilus NO2 | 67 | 1 | 80.9 | nd |
| Novamyl | 66 | 0 | 88a | 88.8 |
| Alkalophilic Bacillus sp. 1011b | 55 | 1 | –b | –b |
| Toruzyme | nc | nc | 97.8 | nc |
| Bacillus sp. strain A2-5a | nc | nc | 64.6 | 69 |
| Anaerobranca gottschalkii | nc | nc | 63.3 | 67.2 |
| Klebsiella pneumoniae | nc | nc | 47.2 | nc |
| Thermococcus sp. B1001 | nc | nc | nd | 106c |
Also structural factors possibly contributing to thermostability is given for CGTases with known 3D structure
nc 3D structure has not been solved, nd not determined
a(Beier et al. 2000). The T 50 value of Novamyl was determined by measuring residual starch hydrolyzing activity
bThis CGTase was not investigated in this study
c(Yamamoto et al. 1999)
Discussion
Evolution of product specificity
The type of cyclodextrin (α, β or γ) formed by CGTases is dependent upon the number of glucose units bound at the donor substrate binding subsites prior to glycosidic bond cleavage in the cyclization reaction (Fig. 1 and Table 3). It is therefore likely that the substitution of residues at the donor subsites or mutations that indirectly affect the properties of the donor subsites during the evolutionary progression of CGTases has led to diversification of product specificity amongst this group of enzymes. Structural studies of CGTases complemented by mutational analysis have helped to identify several of such residues (Table 1; Leemhuis et al. 2002b; van der Veen et al. 2000b).
Several conserved CGTase residues are found at the −1 and −2 subsites, which are crucial in glycosidic bond cleavage. These residues are also conserved in α-amylases (Janecek 1997). The −3 subsite is unique to CGTases in that it provides specific interactions with substrate, assisting in the circularization process of the oligosaccharide chain. This subsite shows variation in amino acid composition between primary α, α/β, β and γ producing CGTases. During the circularization process, the sugar at the −3 subsite becomes stabilised by R47 and D371 (Figs. 3 and 6). In the primarily α-cyclodextrin producing CGTases from Thermococcus sp. B1001 and K. pneumoniae (Table 3), a lysine is found at position 47, in β-CGTases a lysine, histidine or arginine, whereas in β/γ and γ-CGTases, a threonine is present. Indeed, mutations (R47L and R47Q) at this position in BC251 CGTase altered the cyclodextrin product specificity (van der Veen et al. 2000a). The H43T (position 47 in BC251 CGTase) mutation at −3 subsite of the Bacillus sp. G1 β-CGTase increased γ-cyclodextrin production from 10% to approximately 39% using tapioca starch (Goh et al. 2008).
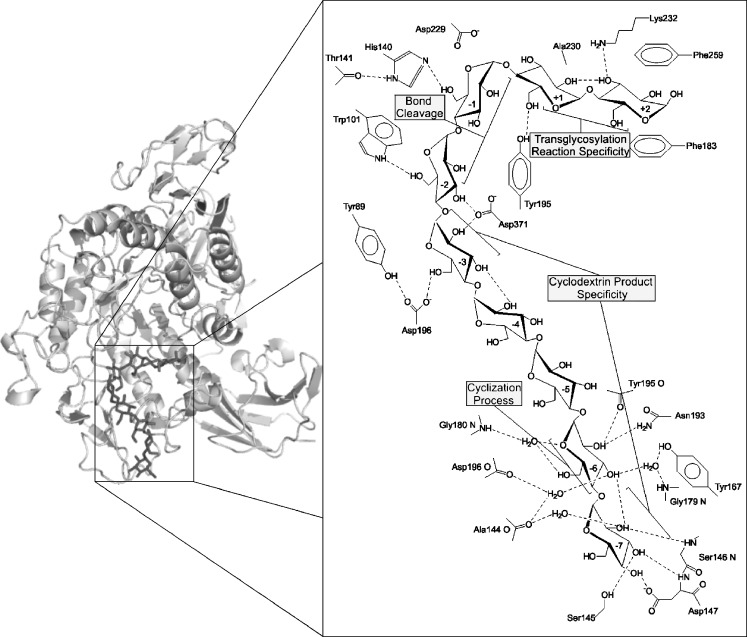
Fig. 6.
Structural representation of BC251 CGTase protein, with a maltononaose substrate bound at the –7 to +2 subsites (Protein Data Bank 1CXK). The side panel displays a close-up view of the catalytic core region with schematic representation of the interactions between residues of the nine substrate binding sites of BC251 CGTase and the maltononaose substrate. The major contributions of each subsite for the formation of cyclodextrins have been highlighted. This side panel has been adapted from Uitdehaag et al. (1999a) and was created using PyMOL (http://www.pymol.org)
The highly variable loop region 87–93 also provides substrate interaction at the −3 subsite. A Y89D mutation shifted BC251 CGTase in the direction of α-cyclodextrin production (van der Veen et al. 2000b). In the γ/β- and γ-CGTases, the 87–93 loop is shorter (Fig. 3), which is thought to allow for more space for larger cyclodextrin formation during the circularization process. The N94 residue at the −3 subsite is present only in certain α- and β-CGTases but is absent in all γ/β- and γ-CGTases (Fig. 3). The importance of this residue in product specificity determination was shown by the Bacillus 1–5 CGTase mutant, N94S, which resulted in enhanced α-cyclodextrin production with increased conversion of starch into cyclodextrins (Kim et al. 1997). Clearly, the loss, gain and substitution of amino acid residues at the −3 donor subsite during the evolutionary divergence of various CGTases has played an important role in the variation in cyclodextrin product specificity. There are, however, exceptions to the rule regarding amino acid composition of the −3 subsite and cyclodextrin size specificity. A. gottschalkii CGTase for example is primarily an α-cyclodextrin producer with an arginine at position 47 and a shortened 87–93 loop similar to that of β- and γ-CGTases, respectively. Other structural features thus appear to be involved here.
The donor subsites −4 and −5 display low affinity for substrate (Uitdehaag et al. 1999a), which may be a selective strategy applied during evolution to prevent the binding of oligosaccharides too short for cyclodextrin formation. At the −6 subsite, interactions between substrate and residues, including G179, G180, Y167 and N193, are strongly conserved in functional CGTases (Fig. 3 and Table 1). Only the archaeal Thermococcus kodakarensis CGTase shows variation at subsite −6 with an N180 instead of the typical G180 (BC251 numbering). A computational model of the cyclization reaction indicated that circularization begins with the extended conformation of the linear oligosaccharide chain changing into a conformation that more resembles that of amylose (spiral-like shape). The hydrogen bonds in the O2 and O3 atoms of adjacent glucoses are formed, which are absent in the starting conformation. During the reorientation of the sugar chain to optimise its internal hydrogen bond network, the sixth, seventh and eighth sugar residues glide along G179 and G180 and successively bind to N193 and Y167 at subsite −6 (Fig. 6; Uitdehaag et al. 2001). The conservation of the residues at subsite −6 and the large effect of mutations at this remote subsite on the catalytic rate confirm their essential role in the cyclization process (Leemhuis et al. 2002b; Fig. 3 and Table 1). The conservation of subsite −6 indicates, in addition, that this subsite is less important in determining cyclodxtrin size specificity. Several of these typical −6 subsite residues are, however, replaced in the putative CGTases (Fig. S1). This may indicate a distinctive evolutionary cluster of CGTases with alternative interactions with substrate at the −6 subsite for cyclodextrin formation. Alternatively, it may be that these putative CGTases are not functional CGTases but closely related enzymes, like Novamyl and ATase, which also show variation in these amino acids (Fig. 3 and Fig. S1).
Subsite −7, unlike subsite −6, shows considerable variation in amino acid sequence amongst CGTases, which has been linked to cyclodextrin size specificity (Fig. 6). Residues of the 145–152 loops form the −7 subsite in β-CGTases (Fig. 3). The α/β-CGTases from Tabium, Toruzyme and BNO2 share the similar loop sequence of SETXPXY (X denotes variation in amino acid sequence). Both γ/β- and γ-CGTases completely lack the six amino acids in this loop region. The shortened loop structure, like that of the −3 subsite, is thought to create extra space for longer linear oligosaccharide binding required for the production of larger cyclodextrins (Takada et al. 2003). Support for this hypothesis comes from a mutagenesis study, which demonstrated enhanced γ-cyclodextrin production for BC8 CGTases upon replacing the 145–151 loop by a single aspartate residue in anology with the γ-CGTases (Parsiegla et al. 1998).
Despite not being part of the donor subsite architecture, residue 195 at the active site cleft of CGTase, in the vicinity of subsite +1, also has a considerable influence in controlling cyclodextrin size specificity. This 195 residue guides the non-reducing end of the covalent intermediate into the acceptor subsites in the cyclization process (Uitdehaag et al. 1999a). A Tyr is present at position 195 in all β- and γ-CGTase, whereas it is a Phe in CGTases producing larger quantities of α-cyclodextrin (Fig. 3 and Fig. 4). Mutagenesis studies indicate that larger residues at this position favour the formation of larger cyclodextrins (γ; Penninga et al. 1995; Nakamura et al. 1994; Parsiegla et al. 1998).
The CGTase sequence analysis and subsite–substrate interactions, together with mutagenesis studies, clearly show that the accumulative effects of specific amino acid residues of the −3/−7 donor subsites and neighbouring active site cleft are essential for efficient production of specific cyclodextrin size.
Acceptor substrate specificity
In addition to the formation of cyclodextrins, CGTase enzymes possess hydrolytic activity as a minor side reaction (Table 2). To minimise the number of hydrolytic events, the active site architecture of CGTases deters water from effectively attacking the glycosyl–enzyme intermediate during cyclodextrin ring formation (Fig. 1). During the final stages of the cyclization process, F183 and F259 (subsite +2) form stacking interactions with the incoming acceptor oligosaccharide, while residues A230, L194 and L197 (subsite +1) form a hydrophobic cavity that traps the non-reducing end of sugar chain prior to nucleophilic attack on the covalent intermediate (Uitdehaag et al. 2001). Directed evolution and site-directed mutagenesis studies have revealed that the +1 and +2 acceptor subsites determine CGTase preference for transglycosylation reactions (Table 1), as mutations in these residues drastically lower the rate of cyclization and usually enhance the hydrolysis activity (van der Veen et al. 2001; Leemhuis et al. 2002a; Leemhuis et al. 2003d; Kelly et al. 2008a). A triple mutant A231V/F184Q/F260W at the +1/+2 subsites virtually abolished cyclization activity and enhanced the hydrolytic activity of Tabium CGTase close to that of α-amylases (Kelly et al. 2007). Recent directed evolution studies of Tabium CGTase showed that only a single mutation (S77P) located far from the substrate binding sites improved the cyclization/hydrolysis ratio of the enzyme by lowering the rate of the hydrolytic side reaction, while cyclization activity was only marginally lowered (Kelly et al. 2008b). In contrast, directed evolution of acceptor subsite residues yielded no mutants with a higher cyclization/hydrolysis ratio (Kelly et al. 2008b). This proves that the residues at the acceptor substrate binding sites of CGTases have optimally evolved for selective transfer of the covalent glycosyl intermediate to sugar acceptors instead of water. Recent structural studies of the 4-α-glucanotransferase from Thermus thermophilus revealed that this enzyme minimises the competing hydrolysis reaction efficiently (e.g. transgycosylation versus hydrolysis ratio of 5,000:1) by ensuring that the acid/base catalyst E340 only assumes its catalytic competent conformation upon binding of an efficacious oligosaccharide acceptor (Barends et al. 2007; Kaper et al. 2007). Such a mechanism has not been identified in CGTases, and even if present in CGTases, it is clearly less effective as CGTases have much lower transgycosylation versus hydrolysis ratios than 4-α-glucanotransferases.
CGTase stability
The factors contributing to the stability of thermophilic proteins has been the focus of numerous experimental and computational investigations (Kumar et al. 2000; Fitter and Haber-Pohlmeier 2004; Tokuriki et al. 2007; Vieille and Zeikus 2001; Sterner and Liebl 2001; Eijsink et al. 2004), while the general consensus from these investigations is that numerous structural factors are thought to partially contribute to increased protein stability (Zhao and Arnold 1999; Szilagyi and Zavodszky 2000; Tokuriki et al. 2007). Berezovsky et al. suggested that bacterial hyperthermostable proteins and their mesophilic homologues are structurally similar and differ only in a small number of strong interactions such as salt bridges (Berezovsky and Shakhnovich 2005). In an attempt to identify these factors contributing to the variation of thermostabilities in CGTases, the number of ionic interactions and disulfide bonds was examined for five CGTases and Novamyl with known 3D structure, using the Protein Interactions Calculator (PIC) programme (Tina et al. 2007). All CGTase proteins were found to have one or no disulfide bonds, providing no clear correlation between enhanced thermostability and the number of disulfide bonds. The only variation found was in the number of ionic interactions, which appeared to be higher in the more thermostable enzymes, with the exception of Tabium CGTase (Table 4). The increased number of ionic interactions in the more thermostable proteins is thought to be due to selective incorporation of specific point mutations during the evolutionary adaptation of the host organism to hotter environments (Duy and Fitter 2005). This hypothesis may help explain why CGTases from Klebsiella and Bacillus species display such a large variation in apparent melting temperatures (Fig. 5) compared to homologues from anaerobic bacteria and archaea, which inhabit extremely hot environments.
In the biotechnological processes of starch based industries, which often operate at extreme conditions, various methods are employed to increase the poor stability of enzymes such as K. pneumoniae CGTase, including the addition of sugars and polyols (glycerol, xylitol and sorbitol; Kelly et al. 2009; Fagain 1995). The addition of polyols to the aqueous medium has been shown to enhance the stability of carbohydrate modifying enzymes by increasing the degree of structural organisation of water molecules around the protein surface (Gangadhara and Prakash 2008; Combes et al. 1987; Martins and Hatti-Kaul 2003).
Interchanging GH13 reaction specificities
Interchangeable reaction specificities of various GH13 members, as shown by mutagenesis studies, also suggest that CGTases evolved from “intermediate multi-specific” enzymes similar to that of neopullulanase or maltogenic amylase. These enzymes produce similar amounts of hydrolase and transglycosylase end products from activity on starch and maltodextrin (Kim et al. 1999; Takata et al. 1992). As mentioned, only three mutations were required to convert Tabium CGTase into an α-amylase like starch hydrolase, with virtually no cyclodextrin forming ability (Kelly et al. 2007). The hydrolytic activities of BC251 CGTase (mutant A230V) and alkalophilic Bacillus sp. I-5 CGTase (mutant M234T/F259I) were also strongly enhanced by mutations at the acceptor subsites, though these two variants retained significant cyclization activity (Leemhuis et al. 2003d; Shim et al. 2004). Deletion of a five amino acid residue loop along with creation of a double mutation, F188L/T189Y, in Novamyl, introduced β-cyclization activity into the maltogenic α-amylase enzyme (Beier et al. 2000). The opposite experiment, involving insertion of the five amino acid loop into Tabium CGTase, converted this enzyme into a starch hydrolase with a strong increase in exo-specificity, similar to Novamyl (Leemhuis et al. 2003a). The reaction specificities of acarviosyl transferase (ATase) and Tabium CGTase were interchanged upon substitution of a single residue at the −1 subsite (Leemhuis et al. 2004b). ATase acquired cyclodextrin forming activity on starch upon introduction of the G140H mutation, while deletion of the histidine 140 side chain in Tabium CGTase enabled the enzyme to transfer the acarviosyl moiety of acarbose to acceptor sugars (Leemhuis et al. 2004b).
To conclude, we have highlighted the considerable evolutionary diversification amongst CGTases regarding both cyclodextrin product specificity and thermostability properties. Sequence alignments and product profile analysis of CGTases reveal that substitution of residues and gain and loss of amino acid residues at the donor subsites −3/−7 are largely responsible for the diversity in product specificitity. Changes at the subsites −1/+1/+2, in contrast, are essential for interchanging reaction specificities with other types of GH13 familiy enzyme. These results provide a valuable insight for future engineering efforts to enhance the product specificity of CGTases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Accession numbers of CGTases, Novamyl and ATase used in the sequence alignment (Fig. 3 and Fig. S1) and the phylogenetic tree (Fig. 2). Figure S1 Alignment of functional and putative CGTases, Novamyl and acarviosyl transferase. The four conserved amino acid sequence regions of GH13 are highlighted in black along with conserved residues of the −6 subsite of functional CGTases. Numbering according to BC251 CGTase. Accession numbers are given in Table S1. (DOC 179 kb)
Acknowledgements
We thank Dr. T. Kuriki and K. Ohdan of Ezaki Glico Co., Ltd. for the alkalophilic Bacillus sp. A2-5a CGTase protein; Dr. M. Moick of Wacker Chemie AG for the K. pneumoniae M5A1 CGTase protein; Prof. Dr. R. Sterner for the expression plasmid, pET24a, containing the A. gottschalkii CGTase gene; Dr. C. Andersen of Novozyme for the Toruzyme CGTase protein; Prof. Dr. G. Schulz for the expression plasmid containing the B. circulans 8 CGTase gene; Prof. Dr. S. Fujiwara and Y. Yamada for the B. subtilis NA-1 harbouring the pKB1 plasmid containing the B. stearothermophilus NO2 cgt gene and E. coli BL21(DE3) harbouring the expression plasmid pCGTA containing the Thermococcus sp. B1001 cgt gene; and Gea Schuurman-Wolters for assistance with the DSC measurements.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Hans Leemhuis acknowledges financial support from the Netherlands Organization for Scientific Research (NWO).
References
- Barends TR, Bultema JB, Kaper T, van der Maarel MJ, Dijkhuizen L, Dijkstra BW. Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase. J Biol Chem. 2007;282:17242–17249. doi: 10.1074/jbc.M701444200. [DOI] [PubMed] [Google Scholar]
- Beier L, Svendsen A, Andersen C, Frandsen TP, Borchert TV, Cherry JR. Conversion of the maltogenic alpha-amylase Novamyl into a CGTase. Protein Eng. 2000;13:509–513. doi: 10.1093/protein/13.7.509. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Shakhnovich EI. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci U S A. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K, Seibel J. Industrial carbohydrate biotransformations. Carbohydr Res. 2008;343:1966–1979. doi: 10.1016/j.carres.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Combes D, Yoovidhya T, Girbal E, Willemot RM, Monsan P. Mechanism of enzyme stabilization. Ann N Y Acad Sci. 1987;501:59–62. doi: 10.1111/j.1749-6632.1987.tb45684.x. [DOI] [PubMed] [Google Scholar]
- Coutinho P M and Henrissat B (1999) Carbohydrate-active enzymes server. http://www.cazy.org/.
- Dauter Z, Dauter M, Brzozowski AM, Christensen S, Borchert TV, Beier L, Wilson KS, Davies GJ. X-ray structure of Novamyl, the five-domain “maltogenic” alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7 Å resolution. Biochemistry. 1999;38:8385–8392. doi: 10.1021/bi990256l. [DOI] [PubMed] [Google Scholar]
- Duy C, Fitter J. Thermostability of irreversible unfolding alpha-amylases analyzed by unfolding kinetics. J Biol Chem. 2005;280:37360–37365. doi: 10.1074/jbc.M507530200. [DOI] [PubMed] [Google Scholar]
- Eijsink VG, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, Vriend G. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Fagain CO. Understanding and increasing protein stability. Biochim Biophys Acta. 1995;1252:1–14. doi: 10.1016/0167-4838(95)00133-F. [DOI] [PubMed] [Google Scholar]
- Fitter J, Haber-Pohlmeier S. Structural stability and unfolding properties of thermostable bacterial alpha-amylases: a comparative study of homologous enzymes. Biochemistry. 2004;43:9589–9599. doi: 10.1021/bi0493362. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Kakihara H, Woo KB, Lejeune A, Kanemoto M, Sakaguchi K, Imanaka T. Cyclization characteristics of cyclodextrin glucanotransferase are conferred by the NH2-terminal region of the enzyme. Appl Environ Microbiol. 1992;58:4016–4025. doi: 10.1128/AEM.58.12.4016-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadhara RKP, Prakash V. Influence of polyols on the stability and kinetic parameters of invertase from Candida utilis: correlation with the conformational stability and activity. Protein J. 2008;27:440–449. doi: 10.1007/s10930-008-9154-z. [DOI] [PubMed] [Google Scholar]
- Goh KM, Mahadi NM, Hassan O, Rahman RNZRA, Illias RM (2008) A predominant ß-CGTase G1 engineered to elucidate the relationship between protein structure and product specificity. J Mol Catal B Enzym . doi:10.1016/j.molcatb.2008.09.016
- Janecek S. α-Amylase family: molecular biology and evolution. Prog Biophys Molec Biol. 1997;25:67–97. doi: 10.1016/S0079-6107(97)00015-1. [DOI] [PubMed] [Google Scholar]
- Kaper T, Leemhuis H, Uitdehaag JC, van der Veen BA, Dijkstra BW, van der Maarel MJ, Dijkhuizen L. Identification of acceptor substrate binding subsites +2 and +3 in the amylomaltase from Thermus thermophilus HB8. Biochemistry. 2007;46:5261–5269. doi: 10.1021/bi602408j. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Dijkhuizen L, Leemhuis H (2009) Starch and α-glucan acting enzymes, modulating their properties by directed evolution. J Biotechnol . doi:10.1016/j.jbiotec.2009.01.020 [DOI] [PubMed]
- Kelly RM, Leemhuis H, Dijkhuizen L. Conversion of a cyclodextrin glucanotransferase into an alpha-amylase: assessment of directed evolution strategies. Biochemistry. 2007;46:11216–11222. doi: 10.1021/bi701160h. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Leemhuis H, Gatjen L, Dijkhuizen L. Evolution toward small molecule inhibitor resistance affects native enzyme function and stability, generating acarbose-insensitive cyclodextrin glucanotransferase variants. J Biol Chem. 2008;283:10727–10734. doi: 10.1074/jbc.M709287200. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Leemhuis H, Rozeboom HJ, van Oosterwijk N, Dijkstra BW, Dijkhuizen L. Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution. Biochem J. 2008;413:517–525. doi: 10.1042/BJ20080353. [DOI] [PubMed] [Google Scholar]
- Kim TJ, Kim MJ, Kim BC, Kim JC, Cheong TK, Kim JW, Park KH. Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl Environ Microbiol. 1999;65:1644–1651. doi: 10.1128/AEM.65.4.1644-1651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Bae KH, Kim TJ, Park KH, Lee HS, Byun SM. Effect on product specificity of cyclodextrin glycosyltransferase by site-directed mutagenesis. Biochem Mol Biol Int. 1997;41:227–234. doi: 10.1080/15216549700201231. [DOI] [PubMed] [Google Scholar]
- Klein C, Schulz GE. Structure of cyclodextrin glycosyltransferase refined at 2.0 Å resolution. J Mol Biol. 1991;217:737–750. doi: 10.1016/0022-2836(91)90530-J. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- Lawson CL, van Montfort R, Strokopytov B, Rozeboom HJ, Kalk KH, de Vries GE, Penninga D, Dijkhuizen L, Dijkstra BW. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J Mol Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Dijkstra BW, Dijkhuizen L. Mutations converting cyclodextrin glycosyltransferase from a transglycosylase into a starch hydrolase. FEBS Lett. 2002;514:189–192. doi: 10.1016/S0014-5793(02)02362-1. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Kragh KM, Dijkstra BW, Dijkhuizen L. Engineering cyclodextrin glycosyltransferase into a starch hydrolase with a high exo-specificity. J Biotechnol. 2003;103:203–212. doi: 10.1016/S0168-1656(03)00126-3. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Dijkstra BW, Dijkhuizen L. Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase: mechanism and kinetics of inhibition by acarbose and cyclodextrins. Eur J Biochem. 2003;270:155–162. doi: 10.1046/j.1432-1033.2003.03376.x. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. The fully conserved Asp residue in conserved sequence region I of the α-amylase family is crucial for the catalytic site architecture and activity. FEBS Lett. 2003;541:47–51. doi: 10.1016/S0014-5793(03)00286-2. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. Improved thermostability of Bacillus circulans cyclodextrin glycosyltransferase by the introduction of a salt bridge. Proteins. 2004;54:128–134. doi: 10.1002/prot.10516. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Wilbrink M, Euverink G-JW, Dijkstra BW, Dijkhuizen L. Conversion of cyclodextrin glycosyltransferase into a starch hydrolase by directed evolution: the role of Ala230 in acceptor subsite +1. Biochemistry. 2003;42:7518–7526. doi: 10.1021/bi034439q. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Uitdehaag JCM, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. The remote substrate binding subsite -6 in cyclodextrin glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism. J Biol Chem. 2002;277:1113–1119. doi: 10.1074/jbc.M106667200. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Wehmeier UF, Dijkhuizen L. Single amino acid mutations interchange the reaction specificities of cyclodextrin glycosyltransferase and the acarbose-modifying enzyme acarviosyl transferase. Biochemistry. 2004;43:13204–13213. doi: 10.1021/bi049015q. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang M, Wang F, Gu Z, Du G, Wu J, Chen J. Gamma-cyclodextrin: a review on enzymatic production and applications. Appl Microbiol Biotechnol. 2007;77:245–255. doi: 10.1007/s00253-007-1166-7. [DOI] [PubMed] [Google Scholar]
- Martins RF, Hatti-Kaul R (2003) Bacillus agaradhaerens LS-3C cyclodextrin glycosyltransferase: activity and stability features. Enzyme Microb Technol . doi:10.1016/S0141-0229(03)00215-1
- McCarter JD, Withers SG. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440X(94)90271-2. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Haga K, Yamane K. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry. 1993;32:6624–6631. doi: 10.1021/bi00077a015. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Haga K, Yamane K. Four aromatic residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of replacements on substrate binding and cyclization characteristics. Biochemistry. 1994;33:9929–9936. doi: 10.1021/bi00199a015. [DOI] [PubMed] [Google Scholar]
- Nitschke L, Heeger K, Bender H, Schulz GE. Molecular cloning, nucleotide sequence and expression in Escherichia coli of the beta-cyclodextrin glycosyltransferase gene from Bacillus circulans strain no. 8. Appl Microbiol Biotechnol. 1990;33:542–546. doi: 10.1007/BF00172548. [DOI] [PubMed] [Google Scholar]
- Parsiegla G, Schmidt AK, Schulz GE. Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production. Eur J Biochem. 1998;255:710–717. doi: 10.1046/j.1432-1327.1998.2550710.x. [DOI] [PubMed] [Google Scholar]
- Penninga D, Strokopytov B, Rozeboom HJ, Lawson CL, Dijkstra BW, Bergsma J, Dijkhuizen L. Site-directed mutations in tyrosine 195 of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 affect activity and product specificity. Biochemistry. 1995;34:3368–3376. doi: 10.1021/bi00010a028. [DOI] [PubMed] [Google Scholar]
- Penninga D, van der Veen BA, Knegtel RM, van Hijum SAFT, Rozeboom HJ, Kalk KH, Dijkstra BW, Dijkhuizen L. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J Biol Chem. 1996;271:32777–32784. doi: 10.1074/jbc.271.51.32777. [DOI] [PubMed] [Google Scholar]
- Qi Q, Zimmermann W. Cyclodextrin glucanotransferase: from gene to applications. Appl Microbiol Biotechnol. 2005;66:475–485. doi: 10.1007/s00253-004-1781-5. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shim JH, Kim YW, Kim TJ, Chae HY, Park JH, Cha H, Kim JW, Kim YR, Schaefer T, Spendler T, Moon TW, Park KH. Improvement of cyclodextrin glucanotransferase as an antistaling enzyme by error-prone PCR. Protein Eng Des Sel. 2004;17:205–211. doi: 10.1093/protein/gzh035. [DOI] [PubMed] [Google Scholar]
- Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Sterner R, Liebl W. Thermophilic adaptation of proteins. Crit Rev Biochem Mol Biol. 2001;36:39–106. doi: 10.1080/20014091074174. [DOI] [PubMed] [Google Scholar]
- Strokopytov B, Knegtel RM, Penninga D, Rozeboom HJ, Kalk KH, Dijkhuizen L, Dijkstra BW. Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity. Biochemistry. 1996;35:4241–4249. doi: 10.1021/bi952339h. [DOI] [PubMed] [Google Scholar]
- Svensson B. Protein engineering in the alpha-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- Szilagyi A, Zavodszky P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure. 2000;8:493–504. doi: 10.1016/S0969-2126(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Takada M, Nakagawa Y, Yamamoto M. Biochemical and genetic analyses of a novel gamma-cyclodextrin glucanotransferase from an alkalophilic Bacillus clarkii 7364. J Biochem (Tokyo) 2003;133:317–324. doi: 10.1093/jb/mvg043. [DOI] [PubMed] [Google Scholar]
- Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at alpha-(1-4)- and alpha-(1-6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thiemann V, Donges C, Prowe SG, Sterner R, Antranikian G. Characterisation of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch Microbiol. 2004;182:226–235. doi: 10.1007/s00203-004-0717-x. [DOI] [PubMed] [Google Scholar]
- Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007;35:473–476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JCM, Kalk KH, van der Veen BA, Dijkhuizen L, Dijkstra BW. The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a gamma-cyclodextrin-CGTase complex at 1.8-Å resolution. J Biol Chem. 1999;274:34868–34876. doi: 10.1074/jbc.274.49.34868. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JCM, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. Nature Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JCM, van der Veen BA, Dijkhuizen L, Elber R, Dijkstra BW. Enzymatic circularization of a malto-octaose linear chain studied by stochastic reaction path calculations on cyclodextrin glycosyltransferase. Proteins. 2001;43:327–335. doi: 10.1002/prot.1044. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Leemhuis H, Kralj S, Uitdehaag JCM, Dijkstra BW, Dijkhuizen L. Hydrophobic amino acid residues in the acceptor binding site are main determinants for reaction mechanism and specificity of cyclodextrin glycosyltransferase. J Biol Chem. 2001;276:44557–44562. doi: 10.1074/jbc.M107533200. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Uitdehaag JCM, Dijkstra BW, Dijkhuizen L. The role of arginine 47 in the cyclization and coupling reactions of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 implications for product inhibition and product specificity. Eur J Biochem. 2000;267:3432–3441. doi: 10.1046/j.1432-1327.2000.01353.x. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Uitdehaag JCM, Penninga D, Van Alebeek GJ, Smith LM, Dijkstra BW, Dijkhuizen L. Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production. J Mol Biol. 2000;296:1027–1038. doi: 10.1006/jmbi.2000.3528. [DOI] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikmon M. Rapid and simple spectrophotometric method for determination of microamounts of cyclodextrins. In: Szejlti J, editor. Proceedings of the first international symposium on cyclodextrins. Dordrecht: Reidel; 1982. pp. 64–74. [Google Scholar]
- Yamamoto K, Shiraki K, Fujiwara S, Takagi M, Fukui K, Imanaka T. In vitro heat effect on functional and conformational changes of cyclodextrin glucanotransferase from hyperthermophilic archaea. Biochem Biophys Res Commun. 1999;265:57–61. doi: 10.1006/bbrc.1999.1629. [DOI] [PubMed] [Google Scholar]
- Zhao H, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 1999;12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers of CGTases, Novamyl and ATase used in the sequence alignment (Fig. 3 and Fig. S1) and the phylogenetic tree (Fig. 2). Figure S1 Alignment of functional and putative CGTases, Novamyl and acarviosyl transferase. The four conserved amino acid sequence regions of GH13 are highlighted in black along with conserved residues of the −6 subsite of functional CGTases. Numbering according to BC251 CGTase. Accession numbers are given in Table S1. (DOC 179 kb)