Abstract
Background
AGO1 associates with microRNAs (miRNAs) and regulates mRNAs through cleavage and translational repression. AGO1 homeostasis entails DCL1-dependent production of miR168 from MIR168a and MIR168b transcripts, post-transcriptional stabilization of miR168 by AGO1, and AGO1-catalyzed miR168-guided cleavage of AGO1 mRNA.
Principal Findings
This study reveals that MIR168a is highly expressed and predominantly produces a 21-nt miR168 species. By contrast, MIR168b is expressed at low levels and produces an equal amount of 21- and 22-nt miR168 species. Only the 21-nt miR168 is preferentially stabilized by AGO1, and consequently, the accumulation of the 22-nt but not the 21-nt miR168 is reduced when DCL1 activity is impaired. mir168a mutants with strongly reduced levels of 21-nt miR168 are viable but exhibit developmental defects, particularly during environmentally challenging conditions.
Conclusions/Significance
These results suggest that 22-nt miR168 ensures basal cleavage of AGO1 mRNA whereas 21-nt miR168 permits an effective response to endogenous or environmental fluctuations owing to its preferential stabilization by AGO1.
Introduction
20- to 30-nt long small RNAs regulate gene expression by guiding transcriptional silencing, mRNA cleavage and translation repression of homologous target genes [1], [2]. MicroRNAs (miRNAs) are produced by the precise excision of an approximately 21-nt miRNA/miRNA* duplex from the stem of a single-stranded, stem-loop RNA precursor [3]. Short interfering RNAs (siRNAs) derive from long perfect double-stranded RNAs (dsRNAs) that result from convergent transcription or transformation of single-stranded RNA into dsRNA by RNA-dependent RNA polymerases [4]. PIWI-associated RNAs (piRNAs) are produced by the shortening of a long single-stranded precursor RNA [5]. A common characteristic of all types of small RNAs is their association with proteins of the ARGONAUTE family, which is comprised of both AGO and PIWI members [6], [7]. The identity of the associated AGO protein determines the functional output of the associated small RNA (transcriptional silencing, mRNA cleavage or translation repression). Plants have miRNAs and siRNAs but no piRNAs. Consistently, plants encode AGO proteins but no PIWI proteins. The model plant species Arabidopsis thaliana potentially encodes 10 AGO proteins [7]. AGO1 catalyzes broad miRNA- and siRNA-guided mRNA cleavage and translation repression [8], [9], [10], [11]. AGO10 appears to be limited to miRNA- and siRNA-guided translation repression [9]. AGO7 specifically associates with miR390 and cleaves TRANS-ACTING siRNA3 (TAS3) RNA to set the nucleotide frame for tasiRNA production, but the basis of this specific association is unknown [12]. AGO4 and AGO6 associate with endogenous siRNAs that are 24-nt long and guide transcriptional silencing [13], [14], [15], [16]. The functions of the five other Arabidopsis AGO proteins remain unknown.
AGO1-catalyzed miRNA-directed mRNA cleavage tolerates some mismatches, particularly near the miRNA 3′ end of the miRNA/mRNA duplex [17], [18]. Consequently, a given miRNA can target mRNAs that differ slightly in their targeted sequences [19]. However, these targets generally are members of a multigene family. Reciprocally, a given mRNA can be targeted by multiple miRNAs that differ only slightly in their sequences and, as such, derive from members of a MIR multigene family. Indeed, while most non-conserved miRNAs derive from a single locus, conserved miRNAs commonly are produced from multiple MIR loci [20], [21]. Therefore assigning the contribution of each MIR locus to the total miRNA pool and to target regulation often is difficult. So far, only a handful of phenotypic mir single mutants have been described, including Petunia hybrida bl/mir169, Antirrhinum majus fis/mir169, Zea mays ts4/mir172e, and Arabidopsis thaliana mir164a, miR164b and mir164c [22], [23], [24], [25], [26].
AGO1 is both an essential actor and a target of miRNA and siRNA pathways. Indeed, the level of AGO1 mRNA is regulated by both the miRNA miR168 and by siRNAs generated from the AGO1 mRNA after miR168-mediated cleavage [11], [27]. In addition, we previously reported that AGO1 and MIR168a are transcriptionally co-regulated, which together with the coordinated action of miR168-guided AGO1 mRNA cleavage and AGO1-mediated stabilization of miR168, keeps AGO1 levels in check [11], [28]. However, miR168 is produced from two loci, MIR168a and MIR168b, evoking questions about the respective contributions of these two loci. Here, we examine the roles of the MIR168a and MIR168b loci and show that they produce both 21-nt and 22-nt miR168 species. While both the 21-nt and 22-nt miR168 contribute to basal AGO1 mRNA cleavage, the preferential stabilization of 21-nt miR168 by AGO1 protein likely provides a layer of adaptive response to endogenous or environmental fluctuations.
Results
Both 4m-MIR168a and 4m-MIR168b constructs rescue the 4m-AGO1 phenotype
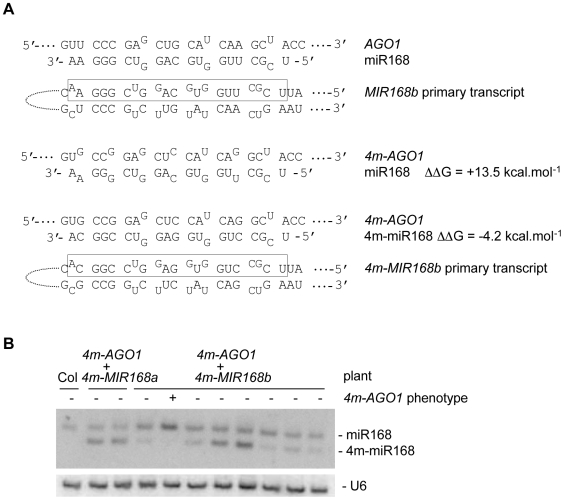
We previously reported that introducing an AGO1 mutant construct (4m-AGO1) carrying four silent mutations in the miR168 complementarity site in Arabidopsis results in a series of developmental defects that can be rescued by co-introducing a MIR168a mutant construct (4m-MIR168a) carrying compensatory mutations that produces a miRNA (4m-miR168) that can pair with the 4m-AGO1 mRNA [11]. These results indicate that the MIR168a gene is functional and sufficient for the proper regulation of its target gene AGO1. To determine whether the MIR168b gene, which also produces miR168, is functionally redundant with MIR168a or if MIR168a is the only active gene, the genomic region corresponding to the MIR168b gene, i.e. the segment of DNA comprised between the two adjacent genes, was mutagenized to introduce compensatory mutations (Figure 1A) and wildtype Arabidopsis plants were co-transformed with the 4m-AGO1 construct and either the 4m-MIR168a or 4m-MIR168b construct. Whereas 18 out of 27 4m-AGO1 transformants displayed the 4m-AGO1 phenotype, only five out of 39 4m-AGO1/4m-MIR168a co-transformants and six out of 46 4m-AGO1/4m-MIR168b co-transformants displayed this phenotype. Double transformants that exhibited a wildtype phenotype accumulated detectable levels of 4m-miR168, which migrated faster than miR168 (Figure 1B). In contrast, 4m-miR168 was undetectable in double 4m-AGO1/4m-MIR168b transformants that exhibited the 4m-AGO1phenotype, and the endogenous miR168 level was increased in these plants (Figure 1B lane 5), consistent with previous observations in phenotypic 4m-AGO1 plants [28]. Since both 4m-MIR168b and 4m-MIR168a constructs are expressed under the control of their own regulatory elements, these results suggest that both the MIR168a and MIR168b genes are capable of regulating AGO1 mRNA. Nevertheless, 4m-miR168 accumulation generally was lower than miR168 in 4m-AGO1/4m-MIR168b plants (Figure 1B) whereas there accumulation was similar in 4m-AGO1/4m-MIR168a plants [11]. This suggests that the MIR168b gene is expressed at a level lower than that of the MIR168a gene, consistent with miR168b* being less abundant than miR168a* in wildtype plants (Table 1 and [20]).
Figure 1. Compensatory mutations in the MIR168b gene rescue developmental defects induced by silent mutations in the miR168 complementary site of the AGO1 mRNA nearly as efficiently as compensatory mutations in the MIR168a gene.
(A) The MIR168b gene encodes a primary transcript that is partially paired (unpaired nucleotides are in superscript). The miRNA is boxed. Compensatory mutations in the 4m-MIR168b transgene conserved the structure of the primary transcript and restored pairing with the 4m-AGO1 mRNA. Original mismatches were kept. ΔΔG was calculated using mfold. (B) Accumulation of the compensatory miRNA (4m-miR168) in plants transformed with 4m-AGO1 and either 4m-MIR168a or 4m-MIR168b. RNA gel blot analysis was performed using 20 µg of total RNA extracted from non-transformed plants (Col) and independent double transformants. The blot was first hybridized with a probe complementary to 4m-miR168, and re-hybridized with a probe complementary to miR168. U6 serves as a loading control.
Table 1. Cloning frequency of miR168 species.
| miRNA species | Locus origin | Sequence (5′to 3′) | Cloning frequency |
| 22-nt miR168 | MIR168a/b | TCGCTTGGTGCAGGTCGGGAAC | 254 |
| 21-nt miR168 | MIR168a/b | TCGCTTGGTGCAGGTCGGGAA | 5039 |
| 22-nt miR168a* | MIR168a | TCCCGCCTTGCATCAACTGAAT | 59 |
| 21-nt miR168a* | MIR168a | CCCGCCTTGCATCAACTGAAT | 1912 |
| 22-nt miR168b* | MIR168b | TCCCGTCTTGTATCAACTGAAT | 15 |
| 21-nt miR168b* | MIR168b | CCCGTCTTGTATCAACTGAAT | 63 |
Cloning frequency indicates the number of times each small RNA sequence was cloned among a population of 887,000 reads [20].
The MIR168b promoter has more limited expression than the MIR168a promoter
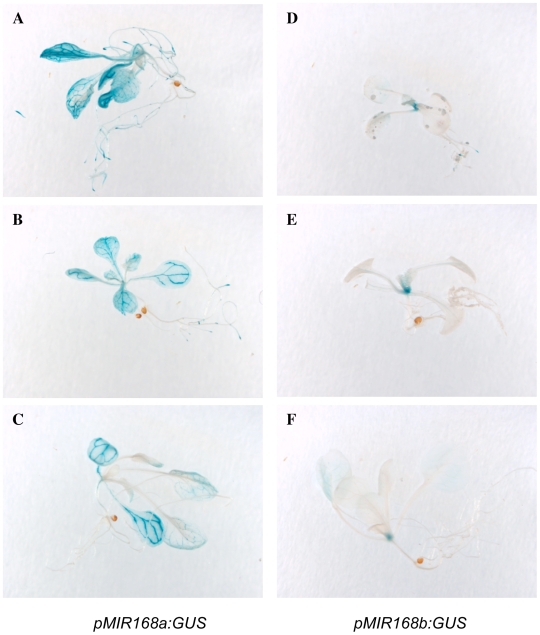
To determine MIR168a and MIR168b promoter strengths and expression patterns, the 5′ sequences of the MIR168a and MIR168b genes, i.e. the genomic region comprised between the miR168 hairpins and the upstream adjacent genes, were fused upstream of the uidA reporter gene encoding GUS, and the resulting transgenes were introduced into wildtype Arabidopsis plants. The MIR168a promoter was subcloned as a 1339 bp fragment (position −1358 to −19 relative to the beginning of the hairpin), to remove an ATG located at position −15 that could compromise the use of the GUS reporter initiation codon and to keep the entire promoter region previously used to express the compensatory miRNA 4m-miR168 from the 4m-MIR168a transgene [11], [28]. Similarly, the MIR168b promoter was cloned as a 479 bp fragment (position −516–37 relative to the beginning of the hairpin) to remove an ATG located at position −29 and to keep the promoter region that is sufficient to express the compensatory miRNA 4m-miR168 from the 4m-MIR168b transgene (Figure 1A). Among 94 pMIR168a∶GUS transformants, 91 exhibited detectable GUS expression, ranging from faint to strong GUS staining as expected from T-DNA insertion positions affecting transgene expression. In contrast, only 24 out of 83 pMIR168b∶GUS transformants had detectable GUS expression, suggesting that the strength of the pMIR168b promoter is much lower than that of the pMIR168a promoter, consistent with the lower cloning frequency of miR168b* versus miR168a* (Table 1 and [20]). GUS expression was detected in shoot and root apical meristems of both pMIR168a∶GUS and pMIR168b∶GUS transformants whereas GUS expression in leaves was more restricted from pMIR168b∶GUS compared with pMIR168a∶GUS (Figure 2). It is unlikely that the lower and restricted expression of the pMIR168b∶GUS construct is due to a lack of regulatory elements of the MIR168b promoter because constructs carrying longer versions of the promoter, i.e. up to position −737 or −1520 relative to the beginning of the hairpin, exhibited similar patterns of GUS expression [29]. Together, these results indicate that expression from the MIR168a promoter is stronger and broader than that from the MIR168b promoter.
Figure 2. Expression from the MIR168a promoter is stronger and broader than that of the MIR168b promoter.
The promoters of the MIR168a and MIR168b genes were fused upstream of the uidA reporter gene encoding GUS, and the resulting transgenes were introduced into wildtype Arabidopsis plants. Over-night X-gluc staining of three representative pMIR168a∶GUS (A–C) and pMIR168b∶GUS (D–F) transformants is shown (blue color indicates the presence of GUS).
MIR168a and MIR168b loci produce distinct amounts of 21- and 22-nt miRNA
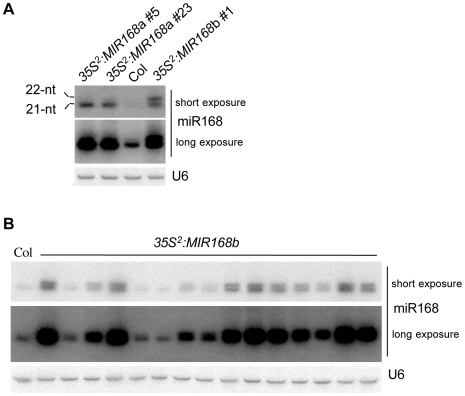
To further examine the contributions of the MIR168a and MIR168b genes, the MIR168a and MIR168b stem-loops were expressed under the control of the 35S promoter in wildtype Arabidopsis plants. As expected from the T-DNA insertion position affecting transgene expression and as previously reported for the 35S2∶MIR168a construct [28], 35S2∶MIR168b transformants accumulated miR168 at varying levels, with those plants accumulating high levels of miR168 exhibiting large and serrated leaves and flowering late compared with wildtype plants (data not shown), indicating that both 35S2∶MIR168a and 35S2∶MIR168b constructs are functional. However, in contrast to 35S2∶MIR168a transformants that over-accumulated a miR168 species that co-migrated with the endogenous 21-nt miR168 [28], 35S2∶MIR168b transformants over-accumulated equal amounts of 21-nt and 22-nt miR168 species (Figure 3A and data not shown). To rule out a construct artifact, the 35S2∶MIR168b construct was rebuilt into a different binary vector and introduced into wildtype Arabidopsis. Analysis of a series of 15 transformants confirmed in plants accumulating miR168 at levels higher than the Col wildtype control an equal and consistent production of both 21-nt and 22-nt miR168 species from the MIR168b stem-loop (Figure 3B).
Figure 3. A 22-nt miR168 species accumulates in 35S2∶MIR168b plants but not in 35S2∶MIR168a plants.
(A) miR168 accumulation in untransformed Col and individual 35S2∶MIR168a and 35S2∶MIR168b transformants generated using the binary vector pBin+. RNA gel blot analysis of 10 µg of RNA with a probe complementary to miR168. U6 serves as a loading control. (B) miR168 accumulation in untransformed Col and 15 individual 35S2∶MIR168b transformants generated using the binary vector pCambia1200. RNA gel blot analysis of 10 µg of RNA hybridized with a probe complementary to miR168. U6 serves as a loading control.
In wildtype plants, the 22-nt miR168 species often was masked by or merged with the signal of the abundant 21-nt species. However, high resolution northern blots allowed detection of the 22-nt miR168 species in wildtype plants (Figure 4A). Pyrosequencing revealed that the 22-nt TCGCTTGGTGCAGGTCGGGAAC miR168 species represents 5% of the overall cloned miR168 sequences from wildtype plants [20] and differs from the 21-nt species by the addition of a C at its 3′ end (Table 1). This 3′ C is present in both the MIR168a and MIR168b sequences, suggesting that the 22-nt species likely is a bona fide miRNA deriving from the two MIR168 loci and does not result from the addition of a C after processing of the 21-nt species. Supporting this hypothesis, both 21- and 22-nt miR168a* and miR168b* sequences have been cloned [20]. The 21-nt miR168a* species was 40 times more abundant than the 22-nt miR168a* species whereas the 21-nt miR168b* species was only 4 times more abundant than the 22-nt miR168b* species (Table 1). Although different miRNA* molecules may have different stabilities, their cloning frequencies and the analysis of 35S2∶MIR168a and 35S2∶MIR168b transformants support the hypothesis that the MIR168b stem-loop is more prone to produce 22-nt miR168 species than the MIR168a stem-loop.
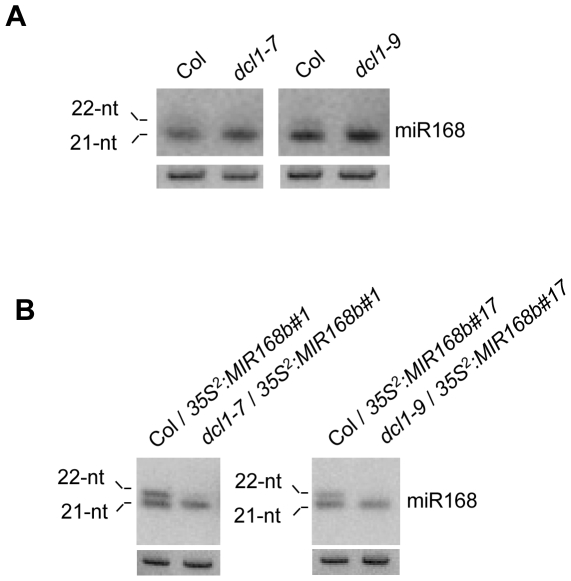
Figure 4. The 22-nt miR168 species is sensitive to dcl1 mutations.
(A) miR168 accumulation in Col and dcl1-7 and dcl1-9 mutants. RNA gel blot analysis of 10 µg of RNA with a probe complementary to miR168. U6 serves as a loading control. (B) miR168 accumulation in Col and dcl1-7 or dcl1-9 siblings deriving from the transformation of dcl1-7/DCL1 or dcl1-9/DCL1 heterozygotes by the 35S2∶MIR168b construct in the binary vector pCambia1200. RNA gel blot analysis of 10 µg of RNA with a probe complementary to miR168. U6 serves as a loading control.
22-nt but not 21-nt miR168 is sensitive to dcl1 mutations
Two features that distinguish 21-nt miR168 from other conserved miRNAs have been described previously: i) miR168 over-accumulates in 4m-AGO1 plants, and ii) miR168 appears insensitive to mutations in the miRNA-processing enzyme DCL1. Indeed, the level of miR168 is similar in wildtype Arabidopsis and dcl1-7, dcl1-8 and dcl1-9 hypomorphic mutants whereas these mutations negatively affect the accumulation of the other tested conserved miRNAs (Figure 3 and [28], [30]). The level of miR168 is unchanged in dcl2, dcl3 and dcl4 single mutants and in double, triple and quadruple dcl mutant combinations [30], indicating that miR168 is not processed by the other DCLs. Rather, these results suggest that AGO1 is limiting in wildtype plants and that miR168 competes with other miRNAs for incorporation into AGO1 [28]. In cases where AGO1 levels are not limiting, such as in a dcl1 hypomorphic mutants in which the processing of many MIRNA precursors is impaired or in 4m-AGO1 plants where AGO1 levels are increased, more miR168 successfully associates with AGO1. As a result, the preferential stabilization of miR168 by the excess AGO1 in dcl1 mutants likely masks the reduced processing of MIR168 precursors.
Having revealed the production of a 22-nt miR168 species, the sensitivity of the 21-nt and 22-nt miR168 species to hypomorphic dcl1 mutations was compared by RNA gel blot analysis. Whereas dcl1 mutations do not impact the accumulation of 21-nt miR168, the 22-nt miR168 is below detectable levels in dcl1-7 and dcl1-9 mutants (Figure 4A). To confirm this distinct behavior of the two miR168 species, the 35S2∶MIR168b construct was introduced into plants heterozygous for either the dcl1-7 or dcl1-9 mutation. In each mutant background, one dcl1/DCL1 transformant showing detectable accumulation of the 22-nt miR168 species and insertion of the 35S2::MIR168b construct at a single locus was analyzed further. After self-fertilization, isogenic dcl1 and DCL1 siblings were harvested by bulk, allowing a comparative analysis of miR168 accumulation in wildtype and mutant backgrounds. The accumulation of the 22-nt miR168 species was strongly reduced in dcl1-7 and dcl1-9 mutants, similar to the vast majority of conserved miRNAs, whereas the level of the 21-nt miR168 species remained unchanged (Figure 4B), suggesting that these two miR168 species are distinctly regulated.
21-nt but not 22-nt miR168 species are preferentially stabilized by AGO1
To further characterize the behavior of the 21- and 22-nt miR168 species, mir168a and mir168b mutants were analyzed. Likely due to the presence of a 35S promoter within the T-DNA inserted in the MIR168a and MIR168b promoters, mir168a-1d (SALK_113514) and mir168b-1d (SALK_066855) mutants accumulated miR168 to levels slightly higher than wildtype plants and did not exhibit obvious developmental defects ([28]; data not shown). Loss-of-function mir168b mutants were not available in the public databases. In contrast, the mir168a-2 mutant (CSHL_GT305) carries a Ds element inserted between the transcription start and the miRNA stem-loop and miR168 accumulation was reduced to 15% of the wildtype level (Figure 5A). This result is consistent with the pMIR168a∶GUS fusion being more strongly expressed than the pMIR168b∶GUS fusion (Figure 2) and with miR168a* being more frequently cloned than miR168b* (Table 1) and indicates that the majority of miR168 derives from the MIR168a locus. As a consequence of the decrease in miR168 production in the mir168a-2 mutant, AGO1 mRNA levels were increased by three-fold, revealing that the MIR168b locus per se is insufficient to maintain AGO1 homeostasis, at least at the mRNA level (Figure 5B).
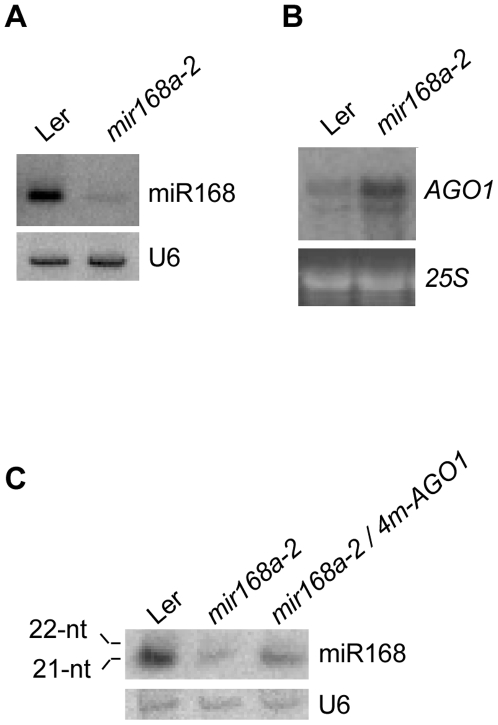
Figure 5. A mir168a loss-of-function mutant has reduced miR168 accumulation and increased AGO1 levels.
(A) miR168 accumulation in wildtype Ler and the mir168a-2 mutant. RNA gel blot analysis of 10 µg of RNA with a probe complementary to miR168. U6 serves as a loading control. (B) AGO1 mRNA accumulation in wildtype Ler and the mir168a-2 mutant. RNA gel blot analysis of 10 µg of RNA with a T7-transcribed RNA probe complementary to AGO1. Ethidium bromide staining of 25S rRNA is shown as a loading control. (C) miR168 accumulation in wildtype Ler, the mir168a-2 mutant and the mir168a-2 transformed with the 4m-AGO1 construct. RNA gel blot analysis of 10 µg of RNA with a probe complementary to miR168. U6 serves as a loading control.
Because an equal amount of 21-nt and 22-nt miR168 species is observed in 35S2∶MIR168b transformants in which miR168 is predominantly produced by the 35S2∶MIR168b transgene (Figure 3A), the mir168a-2 mutant, which produces miR168 only from the MIR168b locus, was expected to produce an equal amount of 21-nt and 22-nt miR168 species. However, only the 21-nt miR168 was detected in the mir168a-2 mutant (Figure 5A). The fact that 35S2∶MIR168b transformants with lower than wildtype AGO1 levels exhibit a lower 21/22 ratio than wildtype plants whereas the mir168a-2 mutant with higher than wildtype AGO1 levels exhibits a higher 21/22 ratio than wildtype plants suggests that preferential stabilization of miR168 by AGO1 only affects the 21-nt miR168 species but not the 22-nt species. Supporting this hypothesis, expression of the 4mAGO1 transgene in the mir168a-2 mutant led to increased levels of the 21-nt miR168 species compared to the mir168a-2 mutant but did not affect 22-nt miR168 accumulation (Figure 5C).
mir168a-2 mutants exhibit developmental defects
When grown in vitro on vertically-oriented plates, the mir168a-2 mutant consistently exhibited an increased number of lateral roots compared with wildtype plants grown on the same plates (Figure 6A), consistent with a previous report showing that ago1 null alleles have a decreased number of lateral roots [31]. Aerial parts of the mir168a-2 mutant did not exhibit obvious developmental defects when grown in vitro or when sown directly on soil and grown in a growth chamber under standard controlled conditions (Figure 6B). However, high temperature promoted early flowering of mir168a-2 compared with wildtype plants (Table 2). Moreover, when grown in a glasshouse where plants are exposed to abiotic stresses such as temperature, light and water fluctuations, mir168a-2 mutants were less vigorous and consistently exhibited narrow, twisted leaves and flowered earlier than wildtype plants (Figure 6C). Altogether, these results indicate that the MIR168b locus is not sufficient for proper development, particularly during environmentally challenging conditions.
Figure 6. A mir168a loss-of-function mutant exhibits developmental defects.
(A) Ten-day-old wildtype Ler and the mir168a-2 mutant sown in vitro on vertical plates and grown in a controlled growth chamber at 18°C. (B) Twenty-two-day-old wildtype Ler and the mir168a-2 mutant sown directly on soil and grown in a controlled growth chamber at 22°C. (C) Twenty-five-day-old wildtype Ler and the mir168a-2 mutant sown directly on soil and grown in a glasshouse where temperature varies between a minimum of 13°C at night and a maximum of 30°C during the day.
Table 2. Flowering characteristics of the mir168a-2 mutant.
| Growth conditions | Ler (DAG) | mir168a-2 (DAG) |
| In vitro 18°C | 28 | 29 |
| In vitro 24°C | 23 | 19 |
| Soil growth chamber 22°C | 27 | 24 |
| Soil glasshouse 13–30°C | 22 | 19 |
| 15 days in vitro 18°C ->soil growth chamber | 28 | 27 |
| 15 days in vitro 18°C ->soil glasshouse | 26 | 25 |
DAG indicates the number of days after germination at which 50% of the plants had open flowers.
Discussion
It was recently shown that the association of small RNAs with different AGO proteins partly depends on the nature of their 5′ nucleotide. Indeed, miRNAs that have a 5′U, 5′A or 5′C associate with AGO1, AGO2 and AGO5, respectively, whereas 24-nt siRNAs, which predominantly have a 5′A, associate with AGO4 [12], [13], [32]. However, there are other associations that remain unexplained. For example, AGO7 exclusively associates with miR390, which has a 5′A [12]. In addition, some miRNAs likely are able to associate with other AGO in the absence of AGO1. Indeed, while the accumulation of most miRNAs is strongly reduced in a null ago1 mutant, the levels of miR156/157 and miR167 are unchanged [11]. It is likely that additional proteins influence the incorporation and/or stabilization of miRNA into AGO proteins. For example, 4m-AGO1 plants that have increased AGO1 levels exhibit unchanged accumulation of most miRNAs but strongly increased accumulation of miR168 and to some extent miR159 and miR165/166 [28]. Such a preferential stabilization of miR168 by AGO1, in conjunction with miR168-guided AGO1 mRNA cleavage and transcriptional co-regulation of AGO1 and MIR168 genes, maintains AGO1 homeostasis [28].
Pyrosequencing-based counting of miR168, miR168a* and miR168b* molecules in wildtype plants [20] and northern analysis of the null mir168a-2 mutant revealed that MIR168a contributes the majority of miR168 molecules and likely is sufficient for plants to develop normally. Supporting this hypothesis, a mutagenized 4m-MIR168a gene expressing a compensatory miRNA rescues developmental defects triggered by a 4m-AGO1 gene. By contrast, the MIR168b locus appears to contribute much less miR168 molecules than MIR168a. Nevertheless, a mutagenized 4m-MIR168b gene expressing the same compensatory miRNA also rescues developmental defects triggered by 4m-AGO1 expression, and the mir168a-2 mutant develops normally under non-stressful conditions. However, the mir168a-2 mutant, which carries only a MIR168b locus, does not develop normally when stressed in a glasshouse, suggesting a role for MIR168a during stress adaptation. Whether the MIR168b locus is dispensable for proper development awaits the identification of a null mir168b mutant.
Several characteristics of the MIR168b locus distinguish it from the MIR168a locus, suggesting that MIR168a and MIR168b have specialized functions. MIR168a predominantly produces a 21-nt miR168 species whereas MIR168b produces an equal amount of 21- and 22-nt miR168 species, which differ by one nucleotide at their 3′ end [20]. The alternative processing at the 3′ end of the MIR168b stem-loop RNA precursor could occur due to the different structural features of the MIR168a and MIR168b stem-loops. Whereas pairing within the MIR168a RNA precursor stem extends beyond the 3′ end of miR168 sequence, the MIR168b precursor has a large bulge at the 3′ end of the miR168 sequence [19]. The 22-nt miR168 species differs from the 21-nt miR168 species in that it is not preferentially stabilized by AGO1 and consequently is more sensitive to dcl1 mutations than the 21-nt species. This result suggests that the length or the 3′ nucleotide of miR168 influences its incorporation or stabilization by AGO1, raising the possibility that the length or 3′ nucleotide identity of other miRNAs could similarly affect their stability.
The existence of two MIR168 loci that produces different levels of 21- and 22-nt miR168 species suggests that the fine tuned regulation of AGO1 homeostasis requires the combinatory action of the dcl1-sensitive AGO1-insensitive 22-nt miR168, which is produced at low levels, primarily by MIR168b, and the dcl1-insensitive AGO1-sensitive 21-nt miR168, which is produced at high levels, primarily by MIR168a. Indeed, mir168a mutants that have a drastically reduced level of 21-nt miR168 exhibit developmental defects, particularly during environmentally challenging conditions, suggesting that the high level of 21-nt miR168 provided by the MIR168a locus is essential for proper development. In conclusion AGO1 homeostasis likely requires the production of a low level of 22-nt miR168 to ensure basal cleavage of AGO1 mRNA and a high level of 21-nt miR168 to allow an effective response to endogenous or environmental fluctuations owing to its post-transcriptional stabilization by AGO1.
Materials and Methods
Plant material
The dcl1-7 and dcl1-9 mutants [33] have been back-crossed four times to Col. The Salk Institute Genomic Analysis Laboratory [34] generated the sequence-indexed T-DNA insertion line SALK_066855 and the Cold Spring Harbor Laboratory [35] generated the sequence-indexed T-DNA insertion line CSHL_GT305.
Molecular cloning and plant transformation
The 4m-AGO1, 4m-MIR168a, 35S2∶MIR168a and pMIR168a∶GUS constructs have been described before [11], [28].
The 4m-MIR168b construct was made as follows: The MIR168b gene was sub-cloned from BAC K9E15 into the Bluescript vector pKS+ as a 930 bp fragment (position 39950–40880 on K9E15). Compensatory mutations that restore complementarity to the 4m-AGO1 mRNA were introduced into the MIR168b gene using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) . The miR168 sequence was first mutageneized using the following pair of primers: 5′-cgtgtcggtgtcagccaatcagttcccgacctgcaccaagcgaatccgagaccgccggtaac-3′ and 5′-cgtgtcggtgtcagccaatcagtgccggacctccaccaggcgaatccgagaccgccggtaac-3′. The miR168* sequence was subsequently mutageneized using the following pair of primers : 5′-cacctcggactccgattcagctgatagaagaccggcgctcacaaaccaaccatgacaagacacg-3′ and 5′-cgtgtcttgtcatggttggtttgtgagcgccggtcttctatcagctgaatcggagtccgaggtg-3′. The mutagenized fragment was sequenced to ensure that no other mutations have been introduced and transferred from pKS+ into the pCambia1200 binary vector.
The 35S2∶MIR168b construct was made as follows: The MIR168b region homologous to the MIR168a EST H77185 was amplified as a 435-nt fragment (position 40001–40435 on BAC K9E15) and cloned between the SalI and EcoRI sites in the pLBR19 vector. The p35S2∶MIR168b:t35S fragment was excised as a KpnI-XbaI and cloned between the KpnI and XbaI sites in the pBin+ or pCambia1200 binary vectors.
The construct pMIR168b∶GUS is a transcriptional fusion with a 479 bp fragment of the MIR168b gene (position −516–37 relative to the beginning of the hairpin). These nucleotide positions were chosen so as to exclude an ATG located at position −29 that could compromise the use of the GUS reporter initiation codon and to keep the entire promoter region previously used to express the compensatory miRNA 4m-miR168 from a 4m-MIR168b transgene, which rescued the 4m-AGO1 phenotype. To built pMIR168b∶GUS, the MIR168b promoter was amplified from the 4m-MIR168b construct in pKS+ as a 474-nt fragment (position 40407–40880 on BAC K9E15) and cloned in the pBI101.2 binary vector.
All constructs were transferred to Agrobacterium tumefaciens by triparental mating and Arabidopsis plants were transformed using the floral dip method [36]. Collected seeds were surface sterilized and transformants were selected by plating seeds on a medium supplemented with either 50 µg/ml kanamycin (pBin+) or 30 µg/ml hygromycin (pCambia1200). Plants were grown under cool-white light in long days (16 h of light, 8 h of dark) at 23°C.
RNA analysis
Total RNA was isolated from rosette leaves as described [11].
RNA gel blot analysis of low molecular weight RNAs was carried out on denaturing 15% polyacrylamide gels, followed by blotting to a nylon membrane (Genescreen Plus, PerkinElmer Inc) as described [11]. Blots were hybridized with gamma-ATP 32P end-labeled oligonucleotides. Blots were re-hybridized with an end-labeled oligonucleotide probe complementary to U6.
For mRNA gel blot analysis, RNA was separated on denaturing 1% agarose gels and trasferred to nylon membrane (Genescreen Plus, PerkinElmer Inc). Blots were hybridized with an alpha-UTP 32P labeled RNA probe complementary to AGO1 mRNA in ULTRAhyb buffer at 68°C (Ambion). AGO1 RNA probes were generated by PCR using primers 5′-CTACAGGGATGGAGTCAGTGAGGGAC-3′, 5′-GGCCGTAATACGACTCACTATAGGCTCCCACTAGCCATTGAGCCACTG-3′ followed by T7-mediated in vitro transcription.
Acknowledgments
I thank David Bartel for use of his lab facilities at the beginning of this project and for helpful discussions. I also thank Allison Mallory for critical reading of the manuscript.
Footnotes
Competing Interests: The author has declared that no competing interests exist.
Funding: This work was supported by the Agence Nationale de la Recherche (ANR-06-BLAN-0203 and ANR-06-POGM-007 to H.V.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Getting to the Root of miRNA-Mediated Gene Silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, et al. Criteria for Annotation of Plant MicroRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 5.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 6.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 7.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 10.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 13.Mi S, Cai T, Hu Y, Chen Y, Hodges E, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–652. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. Embo J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 17.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. Embo J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 26.Cartolano M, Castillo R, Efremova N, Kuckenberg M, Zethof J, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet. 2007;39:901–905. doi: 10.1038/ng2056. [DOI] [PubMed] [Google Scholar]
- 27.Mallory AC, Vaucheret H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009;10:521–526. doi: 10.1038/embor.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gazzani S, Li M, Maistri S, Scarponi E, Graziola M, et al. Evolution of MIR168 paralogs in Brassicaceae. BMC Evol Biol. 2009;9:62. doi: 10.1186/1471-2148-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. Embo J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 33.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 34.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 35.Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, et al. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 36.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]