Abstract
Environmental fungi, in particular primary pathogens and Cryptococcus spp. can be responsible for skin lesions mimicking sporotrichosis. In this paper, we report a case of subcutaneous cryptococcosis in an apparently healthy young male patient due to a non-C. neoformans Cryptococcus species, C. diffluens. The isolate showed in vitro phenotypic switching that may affect virulence and host inflammatory and immune responses, and in vitro resistance to amphotericin B and 5-flucytosin. This species shares several phenotypic traits with C. neoformans, and, therefore, decisive diagnosis should be based on biopsy and culturing results followed by molecular identification.
Keywords: Cryptococcus diffluens, primary subcutaneous cryptococcosis, phenotypical switching, cryptococcal antigens, antifungals
Introduction
Cryptococcoses are most frequently caused by Cryptococcus neoformans and C. gattii, more rarely by other Cryptococcus species [1]. Other Cryptococcus species, however, have been reported from clinical sources as well. Cryptococcus albidus and C. laurentii have been isolated from lung, cerebrospinal fluid, and blood samples [1–5]. Cryptococcus flavescens, reported as C. nodaensis or C. laurentii, has been isolated from cerebrospinal fluid [6–8]. Cryptococcus adeliensis has been recently reported to be involved in meningitis, and has been isolated from a lung biopsy of a male adult suffering from a progressive lung disease and from the oral cavity from an HIV-infected 8-year-old-human [9,10]. Finally, Cryptococcus curvatus has been reported from cerebrospinal fluid of a 30-year-old HIV-infected male patient [11]. Clinical isolates of this latter species from the CBS culture collection originated from sputum, urine, and feces [12].
Here we report a subcutaneous cryptococcosis case in an apparently healthy, young male patient due to a non-C. neoformans Cryptococcus species, C. diffluens. This is a recently re-established species that is only known from a few strains present in culture collections [13].
Case report
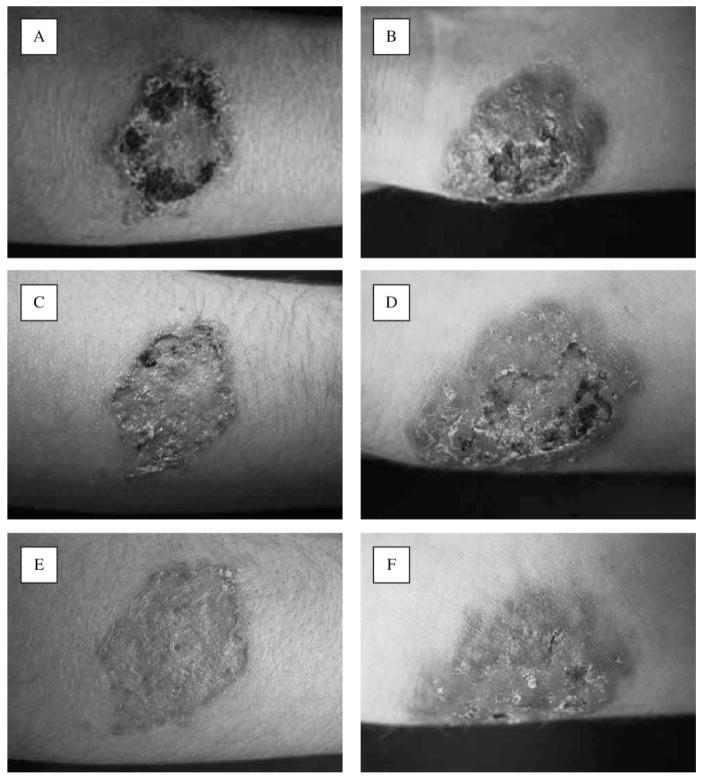
The patient was an otherwise healthy, 17-year-old white, male university student in Istanbul, Turkey, who originated from Turkmenistan. He was admitted to a students’ medical unit in Istanbul, having a two-month history of nontender nodules that first occurred on the right wrist, followed by involvement of the right arm. This condition was diagnosed as pyoderma and treated with topical and systemic antibiotics, which did not result in improvement of his condition. He was transferred to the Dermatology Department of Sisli Etfal Training and Research Hospital (SETRH) in Istanbul for further examinations and treatment. Dermatological examination on his admission revealed two, nontender, erythematous, crusted, noduler lesions of 5–6 cm diameter on both the right wrist and arm. Three subcutaneous lesions with linear arrangement were palpeted on the right forearm (Fig. 1A,B). The regional lymph nodes in the right axilla were not enlarged. Routine clinical laboratory data were unremarkable. Pathological examination of punch biopsy material was performed. Pseudoepitheliomatous hyperplasia, lymphocytic exocytosis and formation of micro-abscess containing mixed type inflammatory cells were seen in the epidermis. A dense infiltrate of lymphocytes, plasma cells, histiocytes and occasionally eosinophyls invading epithels were present in the dermis. Multi-nuclear histiocytes and PAS-positive round to oval fungal cells in various dimensions were also observed. The fungus did not grow on culture media. A second punch biopsy sample was obtained and sent to the Cerrahpasa Medical Faculty (CMF) Deep Mycoses Laboratory for further mycological study. Gram-stained smears from both first and second biopsy specimens revealed no bacteria. No bacterial organisms were cultured from the specimens. Stains for acid-fast bacteria and mycobacterial cultures also were negative. Giemsa-stained tissue was negative for Leishmania amastigotes. The second skin biopsy showed again the presence of round to oval yeast cells and culture of the biopsy material yielded pure growth of Cryptococcus sp. The patient was found HIV-seronegative. The patient had travelled to deserts in Turkmenistan to work two months before the lesions appeared. He reported frequent skin injuries while working in the desert. Itraconazole therapy, 100 mg/day, was started. Twenty-five days after starting this treatment, there was a marked flattening of the lesions (Fig. 1C–F). Therapy was continued for two more months and at the end of the therapy complete resolution was reported.
Fig. 1.
(A–F) Clinical appearance of the lesions, (A, B) before treatment, (C, D) at 10 days after treatment and (D, E) after 25 days of therapy.
Mycology study
Materials and methods
Isolation and phenotypic characterization of the yeast
Specimens obtained at SETRH by a punch biopsy from subcutaneous tissue were submitted to CMF, Department of Microbiology and Clinical Microbiology, Deep Mycoses Laboratory, on 4 December 2002 with a clinical suspicion of sporotrichosis. Direct microscopical examination was performed using Gram, Ehrlich-Ziehl-Nielsen (EZN), Giemsa, and methylene blue stained slide preparations of the imprinted tissue specimens. Aseptically divided pieces of the specimens were inoculated by embedding onto Sabouraud dextrose agar (SDA), brain heart infusion agar (BHIA), Oatmeal agar (OA), potato dextrose agar (PDA), Czapeck dextrose agar (CDA) and malt extract agar (MEA) plates as primary culture media and incubated at 35, 30 and 25°C. All plates were sealed with parafilm to maintain adequate humidity. Microscopical morphology of the isolate was examined by staining with the same techniques and india ink preparations. Carbohydrate assimilation tests were performed using freshly prepared modified Wickerham medium with indicator [14] and fermentation tests were done for six sugars (viz. dextrose, galactose, sucrose, maltose, lactose and raffinose) using the techniques outlined by Kregervan Rij [15]. Christensen agar was used for urease test. Candida albicans ATCC 90028 (American Type Culture Collection, Manassas, VA, USA), ATCC 10231 and Cryptococcus neoformans ATCC 90112 were tested each time along with the case isolates as controls for biochemical tests. Additional tests were performed using API Candida kit (bioMerieux sa, Lyon, France). The strain was cultured on SDA and PDA and incubated at 37±1°C to detect growth at 37°C. The isolate was cultured, along with all molecularly identified C. diffluens strains, on Guizotia abyssinica agar [16] and PAL’s agar [17,18] and incubated at 30°C for two weeks and the color of the colony was examined every 24 h for phenol oxidase activity.
Molecular analyses
Genomic DNA was isolated from 2-day-old cultures according to Bolano et al. [19] with minor modifications. No ureum incubation was performed. Phenol-chloroform (1:1, pH 8.0) and lysis buffer (0.5% w/v SDS, 0.5% Sarkosyl in TE, pH 7.5) were added 1:1 to the pelleted yeast cells and the cells were bead beated for 3 min at 2500 rpm with sterile sand. After centrifugation the DNA fraction was ethanol precipitated and dissolved in TE buffer. For the sequencing of the ITS 1+2 regions and the D1/D2 domains of the large subunit ribosomal DNA (LSU rDNA) the conditions described in Gupta et al. [20] were used with the following modifications. The annealing in the initial PCR reaction with primers V9 and RLR3R took place at 52°C, and for sequencing of the ITS regions the forward primer ITS1 (5′-TCC GTA GGT GAA CCT GCG G) was used in combination with ITS4. GenBank accession numbers are ITS DQ242643 and LSU rDNA DQ242644.
Determination of antigenic formula
The strain was grown on modified Sabouraud dextrose agar containing 2% glucose, 1% polypeptone, 0.5% yeast extract, and 1.5% agar for 2 days at 27°C. The cells were harvested in physiological saline solution (PSS), heated at 100°C for 20 min, washed with PSS, and resuspended in 0.5% formalinized saline solution adjusted to McFarland no. 10 turbidity and subjected to cell slide agglutination test by using eight factor sera [3,21]. Factor sera were prepared based on the antigenic formulas of C. neoformans serotypes by adsorption of anti-C. neoformans serotypes A, B, C and D rabbit sera with killed cells [3,21]. For example, anti-C. neoformans serotype A serum adsorbed with serotype B cells was named factor 2, which contained antibodies reacted with serotype A, D and A–D of C. neoformans. Equal volumes of factor serum and heat-killed cell suspension were mixed on a glass slide and rotated for 5 min, and then the results of agglutination were observed.
Antifungal susceptibility tests
In vitro susceptibility of molecularly identified C. diffluens strains (n= 11) including our case isolate against seven antifungal agents were examined according to the reference NCCLS M27-A broth macrodilution method [22]. The tested antifungal agents were AMB (Bristol-Meyers Squibb, Wallingford, Conn.) (0.03–16 μg/ml), fluconazole (FLZ; Pfizer, Istanbul, Turkey) (0.125–64 μg/ml), itraconazole (ITZ; Janssen Pharmaceuticals, Beerse, Belgium) (0.03–16 μg/ml), ketoconazole (KTZ; Milen, Istanbul, Turkey) (0.03–16 μg/ml), miconazole (MCZ; Selectchemie AG, Zürich, Switzerland) (0.03–16 μg/ml), 5-Flucytosine (5-FC, Sigma Chemical Co., St. Louis, Missouri) (0.125–64 μg/ml), and terbinafine (TRB, Novartis, Istanbul, Turkey) (0.03–128 μg/ml). Antifungal susceptibility testing for seven antifungal agents was performed simultaneously with two different assay media, recommended in M27-A standard. Morpholinepropanesulfonic acid (0.05 mol/liter) (MOPS, Sigma) buffered yeast nitrogen base (YNB) medium (Difco) (pH 7.0 for AMB, azoles and TRB, 5.4 for 5-FC) and standard RPMI 1640 medium, with glutamine and without bicarbonate, both supplemented with 2% glucose were used as test media [22–24]. The strains were also simultaneously tested against AMB using Antibiotic medium 3 (M-3, Oxoid) (pH 7.0), recommended for the detection of C. neoformans resistance to AMB [25]. Inoculum suspensions were each prepared from five colonies of purified 5 day-old cultures grown on SDA at 35°C. Cell suspensions prepared in 5 ml of sterile (0.85%) saline, were vortexed for 15 sec and the turbidity of the suspensions adjusted to match that of a 0.5 McFarland turbidity standard at 530 nm wavelength and further diluted 1:200 in each test medium to obtain the final inoculum concentration of approximately 5.0×102− 2.5×103 CFU ml−1. Preliminary experiments were performed with 1 ml of each final inoculum suspension incubated at 30° and 35°C and growth was monitored from 24 h to four days to determine the optimal incubation temperature and time of reading. CLSI recommended strains Candida parapsilosis ATCC 22019, Candida krusei ATCC 6258, C. neoformans ATCC 90112 and Candida albicans ATCC 90028 were used for quality control. Drug-free and yeast-free controls were included for each isolate tested. Test tubes were incubated at 35°C. MICs were determined at the first 24-h interval when growth was observed in the drug-free control tube. The MIC endpoints were determined according to CLSI specifications of 100% reduction in growth for AMB and 80% reduction in turbidity for the other antifungals. Experiments were repeated twice on different days.
Although interpretive giidelines and breakpoints for susceptibility testing of C. neoformans are not yet available from CLSI, it has been suggested that isolates for which MICs are ≥ 2 μg/ml should be regarded resistant to AMB [25] and MICs are ≥ 32 μg/ml resistant to 5-FC [22]. In this study, these definitions were adopted for AMB and 5-FC, and only MIC comparisons were performed for the remaining antifungals tested.
Results
Mycology
Direct microscopical examination of stained imprinted tissue slides revealed globose or elliptical, encapsulated yeast cells in various dimensions (Fig. 2). After five days’ incubation, cultures on PDA, SDA, MEA and BHIA at 30°C and 25°C showed glossy, creamy colonies just around the tissue biopsy specimens. Relatively poor growth was obtained from tissue biopsy specimens when SDA and CDA plates were incubated at 35°C, while the remaining agar cultures showed good growth at this temperature.
Fig. 2.
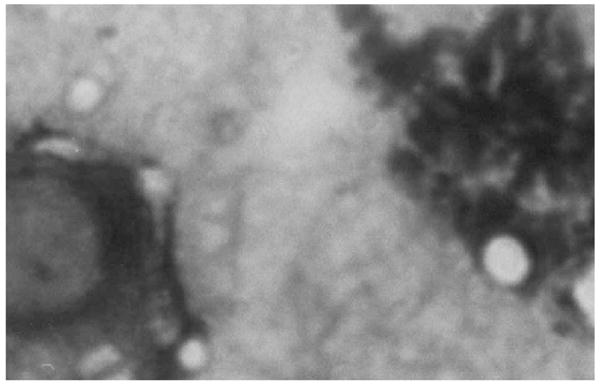
Imprinted tissue biopsy preparation showing encapsulated yeast cells (Giemsa stain, 100×).
The case isolate was persistantly mucoid. Stained and India ink preparations revealed encapsulated globose to subglobose yeast cells with elongated buds (Fig. 3). Fermentation tests were negative for the six sugars tested. Carbohydrate assimilation tests were run in triplicate and gave the following results: glucose +, maltose +, sucrose +, lactose −, galactose +, weak, trehalose +, dulcitol −, myo-inositol +, xylose +, cellobiose +, melibiose − and urease +. Candida albicans and C. neoformans control strains showed the expected assimilation profiles. Growth at 37°C was weak. The isolate did not produce pseudohyphae on cornmeal agar plates, and did not produce brown colonies on Guizzotia abyssinica agar nor on PAL’s agar like all tested C. diffluens strains. The overall results indicated that the isolate belonged to the genus Cryptococcus. Using the API Candida kit our isolate could not be identified.
Fig. 3.
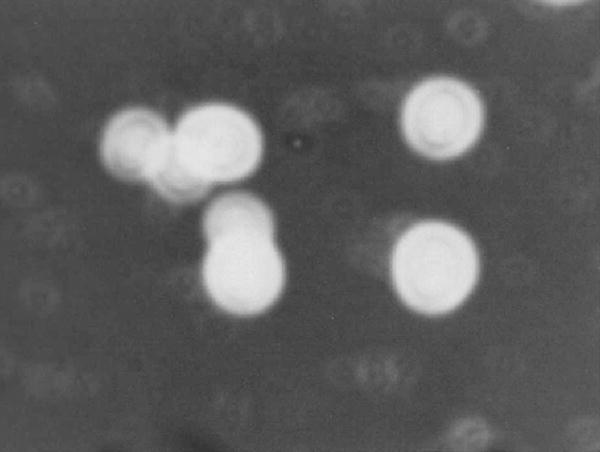
Microscopical morphology of the isolate: encapsulated yeast cells (India ink, 100×).
Colony and capsule phenotype
Colony types were evaluated for gross morphology after growth for 72 h on SDA. Two basic colony morphologies were identified as mucoid and wrinkled. The colony morphologies were however, unstable. In a switching experiment where a single wrinkled colony was plated many sectored and mixed colonies were detected (Fig. 4). Yeast cells were suspended in India ink (Becton Dickinson, NJ) and visualized at 1000× magnification with an Olympus AX70 microscope. Images were captured with a QImaging Retiga 1300 digital camera using the QCapture Suite V2.46 software (QImaging, Burnaby BC, Canada). The corresponding cellular phenotype also differed. The Wrinkled cell population was more heterogenous with respect to cell size and grew in clumps, whereas the Mucoid cell size was more homogenous and less clumps were observed (Fig. 5). Capsule measurements were made on 25 randomly chosen cells from each strain using Adobe Photoshop 7.0 for windows and capsule thickness was calculated using the conversion of 45 pixels per micron. The capsule of the mucoid colony phenotype was larger than the capsule of the Wrinkled yeast cells (1.06 μm ± 0.4 vs 0.74 μm ± 0.19, P= 0.003). Yeast cells of both colony types, were stained with monoclonal antibody 18-B7 (mAbs) to glucuronoxylomannan (GXM) and visualized with FITC-labeled sheep antibody to mouse IgG as previously described [21]. Both cellular phenotypes stained with mAb 2H-1 and exhibited similar annular staining patterns (Fig. 5).
Fig. 4.

Sectored colony with mucoid (MC) and wrinkled (WR) colony parts after growth on SDA for 72 h.
Fig. 5.
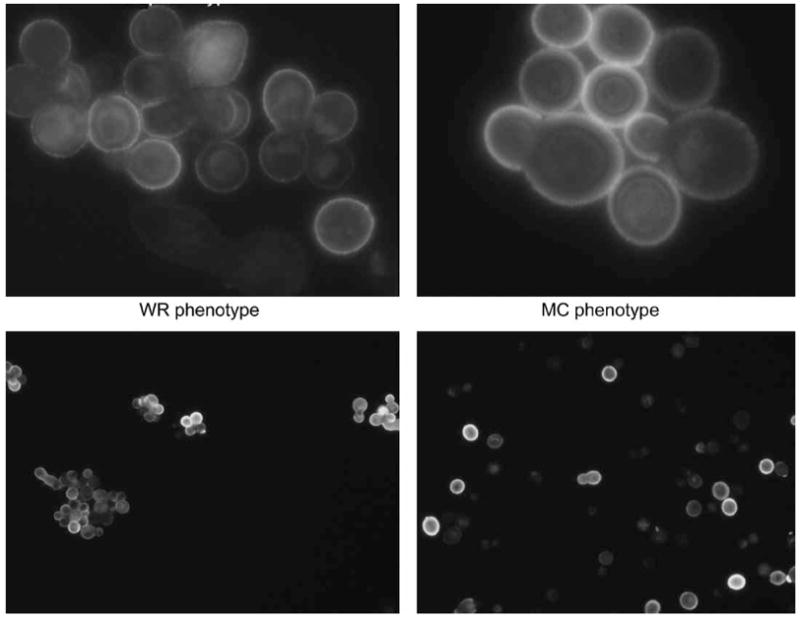
Indirect immunofluorescens with Mab 2H1 of wrinkled (WR) and mucoid (MC) cells respectively (upper panel: 100× and lower panel: 40×).
Molecular analysis
The identity of the isolate was revealed by sequencing the ITS 1+2 regions and the D1/D2 domains of the LSU rDNA of both the smooth and wrinkled variants of the strain. Both isolates investigated showed a 99% match with the ITS and LSU rDNA sequences of C. diffluens (i.e., Genebank accession numbers AB032669, AF145330, AJ510142, AF406909) and, hence, the isolates were identified as belonging to this species. The isolate is preserved as CBS 10266 in the collection of the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
Antigenic formula
The cells reacted with factor sera 1, 2, and 3 by slide agglutination test, and this pattern was the same as that of isolate CBS160 [3,13,26].
Antifungal susceptibility test results
Isolates grew in the drug-free control tubes within 24–28 h in YNB and M-3 broth, and in 48 h in RPMI 1640 medium. Elevated MICs were found for all antifungal agents with YNB medium and against AMB with M-3 medium. In general, MICs of AMB and 5-FC were one twofold dilution higher in YNB and M-3 broth. Reduced MIC values were obtained against all antifungals tested with RPMI 1640 medium. In general the MIC values for each isolate duplicate were identical. Seven strains (7/10) had MICs of AMB ≥ 2 μg/ml, five (5/10) exhibited MICs of 5-FC ≥ 32 μg/ml, including the case isolate. Five strains (5/10) had elevated MICs against FLZ. Antifungal susceptibility results obtained by CLSI reference macrodilution method were shown in Table 1. Quality control strains showed that the MICs ranged within acceptable limits.
Table 1.
Susceptibility to antifungals of molecularly identified Cryptococcus diffluens strains in standard RPMI 1640, YNB and M-3 media.
| Isolate | Media | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| AMB | 5-FC | FLZ | ITZ | KTZ | MCZ | TRB | ||
| CBS 160 | RPMI 1640 | 8 | <0.125 | 32 | 1 | 8 | 8 | 16 |
| YNB | 16 | 0.5 | 64 | 16 | 8 | >16 | >64 | |
| M-3 | 16 (R) | |||||||
| CBS 926 | RPMI 1640 | 2 | <0.125 | 2 | 1 | 1 | 8 | 16 |
| YNB | 4 | 0.25 | 4 | 16 | 1 | >16 | >64 | |
| M-3 | 8 (R) | |||||||
| CBS 966 | RPMI 1640 | 1 | 0.5 | 1 | 0.5 | 2 | 8 | 32 |
| YNB | 2 | 1 | 4 | 4 | 4 | >16 | >64 | |
| M-3 | 2 (R) | |||||||
| CBS 6436 | RPMI 1640 | <0.03 | <0.125 | 16 | <0.3 | 8 | 8 | 2 |
| YNB | <.03 | 0.5 | >64 | 16 | 16 | 16 | 4 | |
| M-3 | <0.03 (S) | |||||||
| CBS 964 | RPMI 1640 | 4 | <0.125 | 16 | 16 | 4 | 4 | 2 |
| YNB | 8 | 0.5 | 4 | >16 | 8 | 8 | 4 | |
| M-3 | 16 (R) | |||||||
| CBS 965 | RPMI 1640 | <0.03 | <0.125 | 1 | <0.03 | 8 | 8 | 2 |
| YNB | <0.03 | 0.5 | 1 | 8 | 16 | >16 | >64 | |
| M-3 | <0.03 (S) | |||||||
| CBS 986 | RPMI 1640 | 0.5 | 0.5 | 0.25 | 8 | 0.5 | 2 | 2 |
| YNB | 1 | 32 (R) | 0.5 | 16 | 1 | 4 | 4 | |
| M-3 | 1 (S) | |||||||
| CBS 1925 | RPMI 1640 | 8 | 64 | <0.125 | 1 | 0.06 | 0.125 | 2 |
| YNB | 16 | 64 (R) | <0.125 | 16 | 0.25 | 0.25 | 4 | |
| M-3 | 16 (R) | |||||||
| CBS 1929 | RPMI 1640 | 8 | 32 (R) | 32 | 0.5 | 0.125 | 1 | <0.03 |
| YNB | 16 | 64 | 64 | 4 | 0.25 | 1 | 0.25 | |
| M-3 | 16 (R) | |||||||
| CBS 2824 | RPMI 1640 | 8 | 32 (R) | 8 | <0.03 | 1 | 1 | 0.5 |
| YNB | 16 | 64 | 64 | <0.03 | 2 | 2 | 1 | |
| M-3 | 16 (R) | 16 | ||||||
| CBS 10266 (this case) | RPMI 1640 | 8 | 16 | 1 | 0.25 | 0.5 | 0.5 | <0.03 |
| YNB | 16 | 32 (R) | 4 | 0.5 | 0.5 | 0.5 | 8 | |
| M-3 | 16 (R) | |||||||
Abbrevations: S: susceptible; R: resistant.
Discussion
Skin infections with a classic ‘sporotrichoid’ pattern in an otherwise healthy patient could be due to relatively limited aetiologies [27–29]. The major causes are Sporothrix schenckii, Nocardia brasiliensis or Mycobacterium marinum [27,29]. Careful examination of the patient’s history, and study of biopsy and appropriate cultures are essential in establishing the correct diagnosis and course of therapy. In the present case, differential diagnosis was carried out by histological stains and cultures of biopsied tissue samples and the findings were suggestive of a fungal infection. The primary pathogen dimorphic fungus Sporothrix schenckii is one of the causes of primary cutaneous fungal infection known as sporotrichosis. However, several other fungi have the ability to cause infections when they enter the body through the skin. Direct inoculation by infecting particles of Coccidioides immitis, Blastomyces dermatitidis, Histoplasma capsulatum, Pseudallescheria boydii, and C. neoformans through skin injuries can cause lesions similar to that seen with Sporothrix schenckii [28,30–34].
Here we report, for the first time, the involvement of C. diffluens in primary subcutaneous sporotrichioidal lesions occurring on the forearm of an otherwise healthy young Asian male patient who had an exposure-prone outside working history two months prior to the onset of the lesions. Histopathology from the lesions was compatible with that of cryptococcal infections. Decisive diagnosis was based on biopsy and culturing results followed by molecular identification.
Primary cryptococcosis cases due to direct skin inoculation were recently reviewed [33,35]. A history of skin injury and participation in outdoor activities predisposing to wounds were found to be important for direct inoculation of the fungus, even in immunocompentent persons. Our patient had a travel history of working in a desert in Turkmenistan, and wound and skin injuries occurred regularly while working. Although the geographic distribution of C. diffluens is not yet known due to the small number of isolates available (Table 2), other Cryptococcus species including C. neoformans, C. gattii and C. albidus have been known to be associated with plants and trees [36–38].
Table 2.
| Strain | Source | Origin |
|---|---|---|
| CBS 160 | Diseased fingernail of 48-year-old milliner | Austria |
| CBS 926 | Atmosphere | Unknown |
| CBS 964 | Skin lesion | Estonia |
| CBS 965 | Sputum | Netherlands |
| CBS 966 | Skin | Netherlands |
| CBS 986 | Sputum | Netherlands |
| CBS 1925 | Washed beer bottle | Sweden |
| CBS 1929 | Biopsy of 12th thoracic vertebra of a 9-year-old boy | Netherlands |
| CBS 2824 | (ATCC Unknown 32037) | Unknown |
| CBS 6436 | Purification tank | Uruguay |
| CBS 10266 (this study) | Biopsy of sporotrichoidal lesions on arm of a young male patient | Turkmenistan |
Recently, Fonseca et al. [39] reinstated C. diffluens as a distinct species based on analysis of the D1/D2 domains of the LSU rDNA and the ITS 1+2 regions of the C. albidus complex. Following this, Sugita et al. [13] reidentified 19 strains of C. albidus, present in culture collections, including nine clinical strains, as C. albidus or C. diffluens.
Since this species shares several phenotypic traits with C. neoformans, the most common cause of cryptococcosis, in routine practice phenol oxidase negativity may be a clue to suspect C. diffluens in clinical laboratories. Molecularly C. diffluens strains shared three antigens 1, 2, and 3 when tested with eight factor sera derived from C. neoformans serotypes [13]. Therefore, to correctly identify clinical C. diffluens isolates molecular analysis of the D1/D2 domains of the LSU rDNA and the ITS 1+2 are required as proposed before [39].
The antifungal susceptibility of known C. diffluens strains was analysed by the M27-A susceptibility method. Modifications of the standard procedure, due to poor growth of C. neoformans and other non-fermentative yeasts with the standard RPMI 1640 medium as recommended by CLSI, have been evaluated [24,40,41]. Previously, YNB buffered with MOPS was shown to enhance growth of C. neoformans and yielded more reliable MICs for AMB, FLZ and 5-FC than those obtained by the RPMI 1640 medium [25,42,43]. Currently, glucose supplemented RPMI 1640 and YNB media are possible alternative media for Cryptococcus neoformans strains in the CLSI M27-A procedure. The YNB medium was previously found not to be capable to distinguish AMB-susceptible and resistant isolates of C. neoformans. The use of M-3 medium has been shown to be useful in identifying AMB-resistant isolates of C. neoformans [25]. Regarding these previous reports, we tried different assay media. We observed that both YNB and M-3 allowed satisfactory growth of the non-C. neoformans Cryptococcus case isolate, and all strains belong to C. diffluens, permitting the performance of susceptibility testing. In our experiments, all three media generated reproducible in vitro susceptibility data, however, an effect of the medium was observed, and YNB and M-3 were found comparable to distinguish AMB-resistant isolates.
Sugita et al. [13] analysed in vitro susceptibilities of isolates to various antifungals with a broth microdilution method using a commercial kit. Both C. albidus and C. diffluens strains were found to be resistant to 5-FC (>64 μg/ml), but were sensitive to AMB with MICs ranging from 0.125–0.5 μg/ml. Of five tested clinical isolates of C. diffluens, only one was found to be resistant to FLZ (>64 μg/ml) and showed a high MIC value against ITZ (>8 μg/ml), but the remaining were susceptible to FLZ, ITZ and MCZ. The discrepancies found between our susceptibility results obtained by CLSI M27-A method and those reported by Sugita et al. [13] may be attributable to methodological differences. In our study, the C. diffluens case isolate showed relatively low MICs against the azoles tested and to TRB as well. The patient was treated with ITZ according to the susceptibility test results and responded to this drug.
Environmentally occurring fungi may be responsible for unexplained skin lesions that are unresponsive to antibiotics, even in apparently healthy persons [44]. The occurrence of infections by such environmental fungal pathogens is usually associated with outdoor activities. Diagnosis of such infections should be based on biopsy, culturing results and molecular identifications.
Acknowledgments
The present work was partly supported by the Research Fund of Istanbul University, Project No: UDP-761/23052006.
References
- 1.Boekhout T, Gueho E. Basidiomycetous yeasts. In: Howard Dexter H., editor. Pathogenic Fungi in Humans and Animals. 2. New York: Marcel Dekker; 2003. pp. 535–564. [Google Scholar]
- 2.De Hoog GS, Guarro J, Gené JL, et al. Atlas of Clinical Fungi. 2. Utrecht/Reus: Centraalbureau voor Schimmelcultures, Universitat Rovira i Virgili; 2000. pp. 132–133. [Google Scholar]
- 3.Ikeda R, Sugita T, Shinoda T. Serological relationship of Cryptococcus spp: distribution of antigenic fators in Cryptococcus and intrapsecies diversity. J Clin Microbiol. 2000;38:4021–4025. doi: 10.1128/jcm.38.11.4021-4025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauters TGM, Swinne D, Boekhout T, et al. Repeated isolation of Cryptococcus laurentii in the oropharynx despite treatment with fluconazole. Mycopathologia. 2002;153:133–135. doi: 10.1023/a:1014551200043. [DOI] [PubMed] [Google Scholar]
- 5.Averbuch D, Boekhout T, Falk R, et al. Fungemia in a cancer patient caused by fluconazole resistant Cryptococcus laurentii. Med Mycol. 2002;40:479–484. doi: 10.1080/mmy.40.5.479.484. [DOI] [PubMed] [Google Scholar]
- 6.Kordossis T, Avlami A, Velegraki A, et al. First report of Cryptococcus laurentii meningitis and a fatal case of Cryptococcus albidus cryptococcaemia in AIDS patients. Med Mycol. 1998;36:335–339. [PubMed] [Google Scholar]
- 7.Sugita T, Takashima M, Ikeda R, et al. Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J Clin Microbiol. 2000;38:1468–1471. doi: 10.1128/jcm.38.4.1468-1471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takashima M, Sugita T, Shinoda T, et al. Three new combinations from the Cryptococcus laurentii complex: Cryptococcus aureus, Cryptococcus carnescens and Cryptococcus peneaus. Int J Syst Evol Microbiol. 2003;53:1187–1194. doi: 10.1099/ijs.0.02498-0. [DOI] [PubMed] [Google Scholar]
- 9.Rimek D. First report of a case of meningitis caused by Cryptococcus adeliensis in a patient with acute myeloid leukemia. J Clin Microbiol. 2004;42:481–483. doi: 10.1128/JCM.42.1.481-483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tintelnot K, Lostert H. Isolation of Cryptococcus adeliensis from clinical samples and the environment in Germany. J Clin Microbiol. 2005;43:1007. doi: 10.1128/JCM.43.2.1007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dromer F, Moulignier A, Dupont B, et al. Myeloradiculitis due to Cryptococcus curvatus in AIDS. AIDS. 1995;9:395–396. [PubMed] [Google Scholar]
- 12.Available from <www.cbs.knaw.nl>
- 13.Sugita T, Takashima M, Ikeda R, et al. Intraspecies diversity of Cryptococcus albidus isolated from humans as revealed by sequences of the internal transcribed spacer regions. Microbiol Immunol. 2001;45:291–297. doi: 10.1111/j.1348-0421.2001.tb02621.x. [DOI] [PubMed] [Google Scholar]
- 14.Adams ED, Cooper BH. Evaluation of a modified Wickerham medium for identifying medically important yeasts. Am J Med Technol. 1974;40:377–388. [PubMed] [Google Scholar]
- 15.Kreger-van Rij NJW, editor. The Yeasts: A Taxonomic Study. 3. Amsterdam: Elsevier Science Publishers BV; 1984. [Google Scholar]
- 16.Kwon Chung KJ, Bennett JE. Medical Mycology. Philadelphia: Lea and Febinger; 1992. p. 819. [Google Scholar]
- 17.Pal M, Onda C, Hasegawa A. Isolation of saprophytic Cryptococcus neoformans. Jpn J Vet Sci. 1990;52:1171–1174. doi: 10.1292/jvms1939.52.1171. [DOI] [PubMed] [Google Scholar]
- 18.Pal M. Pulmonary mycosis in a pigeon handler due to Cryptococcus neoformans var. neoformans 2nd International Conference on Cryptococcus and cryptococcosis; Milan (Italy). 1993. Abstracts, 119. [Google Scholar]
- 19.Bolano A, Stinchi S, Preziosi R, et al. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 2001;1:221–224. doi: 10.1111/j.1567-1364.2001.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 20.Gupta AK, Boekhout T, Theelen B, et al. Identification and typing of Malassezia species by amplified fragment length polymorphism (AFLP) and sequence analysis of the internal transcribed spacer (ITS) and large subunit (LSU) regions of ribosomal DNA. J Clin Microbiol. 2004;42:4253–4260. doi: 10.1128/JCM.42.9.4253-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda R, Shinoda T, Fukazawa Y, et al. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping of clinical isolates. J Clin Microbiol. 1982;16:22–29. doi: 10.1128/jcm.16.1.22-29.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards Reference Method for Broth Dilution Antifungal Susceptibility Testing for Yeasts. Approved Standard; Document M27-A. 2. Wayne, PA: National Committee for Laboratory Standards; 2002. [Google Scholar]
- 23.Viviani MA, Esposto MC, Cogliati M, et al. Flucytosine and cryptococcosis: which in vitro test is the best predictor of outcome? J Chemother. 2003;15:124–128. doi: 10.1179/joc.2003.15.2.124. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Tudela JL, Martin-Diez F, Cuenca-Estrella M, et al. Influence of shaking on antifungal susceptibility testing of Cryptococcus neoformans: a comparison of the NCCLS standard M27-A medium, buffered yeast nitrogen base, and RPMI-2% glucose. Antimicrob Agents Chemother. 2000;44:400–404. doi: 10.1128/aac.44.2.400-404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozano-Chiu M, Paetznick VL, Ghannoum MA, et al. Detection of resistance to amphotericin B among Cryptococcus neoformans clinical isolates: performances of three different media assessed by using E-test and National Committee for Clinical Laboratory Standards M27-A methodologies. J Clin Microbiol. 1998;36:2817–2822. doi: 10.1128/jcm.36.10.2817-2822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casadevall A, Mukherjee J, Devi SJ, et al. Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;165:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 27.Ellis HT, William WJ. Sporotrichoid lymphocutaneous infections: aetiology, diagnosis and therapy. Am Fam Physic. 2001;63:326–332. [PubMed] [Google Scholar]
- 28.Ryncarz RE, Heasley EC, Babinchak TJ. The clinical spectrum of nodulary lymphangitis. Hosp Physic. 1999;9:63–66. [Google Scholar]
- 29.Kostman JR, DiNubile MJ. Nodulary lymphangitis: a distinctive but often unrecognized syndrome. Ann Intern Med. 1993;118:883–888. doi: 10.7326/0003-4819-118-11-199306010-00009. [DOI] [PubMed] [Google Scholar]
- 30.Wilson JW. Cutaneous (chancriform) syndrome in deep mycoses. Arch Dermatol. 1963;87:121–125. doi: 10.1001/archderm.1963.01590130087013. [DOI] [PubMed] [Google Scholar]
- 31.Rippon JW. Medical Mycology. 3. Philadelphia: WB Saunders Company; 1988. p. 666. [Google Scholar]
- 32.Ng WF, Loo KT. Cutaneous cryptococcosis-primary versus secondary disease. Report of two cases with review of literature. Am J Dermatopathol. 1994;16:227. [PubMed] [Google Scholar]
- 33.Neuville S, Dromer F, Morin O, et al. Primary cutaneous cryptococcosis: a distinct clinical entity. Clin Infect Dis. 2003;36:337–347. doi: 10.1086/345956. [DOI] [PubMed] [Google Scholar]
- 34.Narayan S, Batta K, Colloby P, et al. Cutaneous cryptococcosis infection due to C. albidus associated with Sezary syndrome. Br J Dermatol. 2000;143:632–634. doi: 10.1111/j.1365-2133.2000.03724.x. [DOI] [PubMed] [Google Scholar]
- 35.Christianson JC, Engber W, Andes D. Primary cutaneous cryptococcosis in immunocompetent and immunocompromised hosts. Med Mycol. 2003;41:177–188. doi: 10.1080/1369378031000137224. [DOI] [PubMed] [Google Scholar]
- 36.Ellis DH, Pfeiffer TJ. Ecology, life cycle and infectious propagules of Cryptococcus neoformans. Lancet. 1990;336:923–925. doi: 10.1016/0140-6736(90)92283-n. [DOI] [PubMed] [Google Scholar]
- 37.Lazera MS, Pires FDA, Camillo-Coura L, et al. Natural habitat of Cryptococcus neoformans var. gattii in decaying wood forming hollows in living trees. J Vet Med Mycol. 1996;34:127–131. [PubMed] [Google Scholar]
- 38.Lazera MS, Salmito MA, Londero AT, et al. Possible primary ecological niche of Cryptococcus neoformans. Med Mycol. 2000;38:379–383. doi: 10.1080/mmy.38.5.379.383. [DOI] [PubMed] [Google Scholar]
- 39.Fonseca A, Scorzetti G, Fell JW. Diversity in the yeast Cryptococcus albidus and related species revealed by ribosomal DNA sequence analysis. Can J Microbiol. 2000;46:7–27. [PubMed] [Google Scholar]
- 40.Odds FC, De Backer T, Dams G, et al. Oxygen as limiting nutrient for growth of Cryptococcus neoformans. J Clin Microbiol. 1995;33:995–997. doi: 10.1128/jcm.33.4.995-997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuenca-Estrella M, Gomez-Lopez A, Mellado E, et al. In vitro activities of ravuconazole and four other antifungal agents against fluconazole-resistant or -susceptible clinical yeast isolates. Antimicrob Agents Chemother. 2004;48:3107–3111. doi: 10.1128/AAC.48.8.3107-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghannoum MA, Ibrahim AS, Fu Y, et al. Susceptibility testing of Cryptococcus neoformans: a microdilution technique. J Clin Microbiol. 1992;30:2881–2886. doi: 10.1128/jcm.30.11.2881-2886.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witt MD, Lewis RJ, Larsen RA, et al. Identification of patients with acute AIDS-associated cryptococcal meningitis who can be effectively treated with fluconazole: the role of antifungal susceptibility testing. Clin Infect Dis. 1996;22:322–328. doi: 10.1093/clinids/22.2.322. [DOI] [PubMed] [Google Scholar]
- 44.Krajden S, Summerbell RC, Kane J, et al. Normally saprobic cryptococci isolated from Cryptococcus neoformans infections. J Clin Microbiol. 1991;29:1883–1887. doi: 10.1128/jcm.29.9.1883-1887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]