Abstract
Context
The risk of breast cancer in BRCA1 and BRCA2 mutation carriers has been examined in many studies, but relatively little attention has been paid to the degree to which the risk may vary among carriers.
Objectives
To determine the extent to which risks for BRCA1 and BRCA2 carriers vary with respect to observable and unobservable characteristics.
Design, Setting and Participants
Probands were identified from a population-based case-control study of asynchronous contralateral breast cancer. Participants diagnosed with contralateral breast cancer (CBC) and with unilateral breast cancer (UBC) were genotyped for mutations in BRCA1 and BRCA2. All participants had their initial breast cancer diagnosed before age 55.
Main Outcome Measure
Incidence of breast cancer in first degree female relatives of the probands was examined and compared on the basis of proband characteristics and on the basis of variation between families.
Results
Seventy-three carriers of deleterious mutations (42 BRCA1, 31 BRCA2) were identified among the 1394 participants with UBC and 108 carriers (67 BRCA1, 41 BRCA2) were identified among the 704 participants with CBC. Among relatives of carriers, risk was significantly associated with younger age at diagnosis in the proband (p=0.04), and there was a trend towards higher risk for relatives of CBC versus UBC participants (OR=1.4, p=0.28). In addition there were significant differences in risk between carrier families after adjusting for these observed characteristics.
Conclusions
There exists broad variation in breast cancer risk among carriers of BRCA1 and BRCA2 mutations.
The magnitude of the risk of breast cancer in carriers of mutations in BRCA1 or BRCA2 is critical for guiding decisions concerning cancer prevention options. Many previous studies have reported on the cumulative risk to various ages (penetrance) of breast cancer in carriers. The recent literature has involved primarily studies of breast cancer incidence in the relatives of probands identified without consideration of family history. This literature has included studies of self-selected volunteers,1 but there appears to be some degree of consensus that the most reliable approach is to use “population-based” ascertainment .2-8 Most of this literature has been focused on the magnitude of the risk, with relatively little attention being paid to the degree by which risk may vary among carriers.
Population-based studies to date have used as probands incident cases from existing case-control investigations. Estimates of risk based on studies of incident cases are inevitably inflated if there exists risk variation among carriers, caused by additional, possibly unknown genetic variants that influence risk.9-11 The thesis that there exists substantial variation in the risk of breast cancer due to unknown genetic factors has become well established,12 and this has been supported by statistical modelling of disease aggregation,13 and theoretical models to explain the aggregation.14 However, although there is little doubt that other genes influence the risk of breast cancer, such as relatively rare mutations in TP53,15 and possibly CHEK216 and ATM,17 as well as common low penetrance variants in genes that remain unidentified at this point, there is little direct evidence that variation in risk exists among BRCA1 and BRCA2 mutation carriers specifically.
In this article, we report the results of an investigation that provides direct evidence about risk variation among carriers. We use the information on lifetime risk of breast cancer in first degree relatives of breast cancer patients (probands) who were identified as BRCA1 or BRCA2 carriers in the population-based case-control WECARE Study (Women’s Environmental Cancer and Radiation Epidemiology).18 The WECARE Study is novel in that it involved recruitment of cases of asynchronous contralateral breast cancer (CBC) and matched controls who had experienced a prior unilateral breast cancer (UBC). If there is no appreciable risk variation among BRCA1 or BRCA2 mutation carriers, then we would expect the risk estimates in relatives of carriers with CBC to be similar to the estimates from relatives of carriers with UBC. Likewise, evidence of risk variation may be demonstrated by significant differences in risk in groups of carrier families distinguished by any factors that are plausibly associated with risk, such as age at diagnosis of the proband.
METHODS
The WECARE Study is a population-based, cancer registry based, nested case-control study of CBC. Participant recruitment was completed in 2004. The design has been described in detail in a previous article,18 but the essential features are as follows. Participants were identified and interviewed through five population-based cancer registries, four in the USA (covering Iowa, the Orange County and San Diego regions of California, Los Angeles County in California, and three counties in the Seattle, Washington area) and one covering all of Denmark. All participants had to have had a diagnosis of a first invasive breast cancer between January 1985 and December 2000. The cancer had to have occurred prior to the age of 55 years without evidence of spread beyond the regional lymph nodes at diagnosis. Participants also had to have had no cancer other than breast cancer prior to the diagnosis of the second primary, for CBC participants, and to the corresponding matching date for UBC controls, to be alive at the time of contact, and to be able to complete the interview and provide a blood sample. Participants were eligible if they had an in situ or invasive diagnosis of a contralateral breast cancer at least one year after the first primary breast cancer diagnosis, and if they resided in the same reporting area for both diagnoses. Control participants were selected randomly from the pool of available breast cancer patients in the cohort, after matching individually on the basis of year of birth (5-year strata), year of diagnosis (4-year strata), registry and race. Controls were also “counter-matched” in a ratio of two to each CBC case on the basis of whether or not they had received radiotherapy treatment as recorded in the cancer registry.19 The study was reviewed and approved by local Institutional Review Boards at each of these registry sites, and all biological samples and data were obtained after the participants provided informed consent.
Recruitment of participants took place during the period from January 2000 to July 2004. A total of 998 women with CBC were eligible and were approached for inclusion in the study, and 708 of these women (71%) agreed to participate. Of the 2112 women who were selected as potential UBC participants 1399 (66%) agreed to participate. The non-participants were similar to the participants with respect to age and calendar year of diagnosis and radiotherapy treatment of the initial primary breast cancer. Successful genotyping was accomplished in 704 CBC and 1394 UBC participants, and these 2098 individuals represent the “probands” for the analyses in this article.
All participants were interviewed by telephone using a structured questionnaire. They were questioned about the breast cancer incidence in each of their first and second degree relatives. For each relative the interviewer ascertained the age at diagnosis of breast cancer, and the vital status and dates of death (if relevant) of the relatives. For the purposes of this article we use only the information on female first degree relatives, in order to restrict the analysis to relatives for which the data are most likely to have high accuracy.20,21
Mutation screening
Coding and flanking intronic regions were screened for mutations or polymorphic variants by denaturing high-performance liquid chromatography (DHPLC). BRCA1 was covered by 30 PCR amplicons, while 41 amplicons were used for BRCA2. The majority of fragments were run at more than one DHPLC elution temperature condition for increased sensitivity. A few fragments were screened by direct sequencing because of complex melting profiles unsuitable for DHPLC or because of the presence of multiple common variants and combinations thereof that made interpretation of chromatograms difficult. With the exception of the very prevalent polymorphic variants (occurring in >10% of samples) with clearly distinguishable chromatograms, all variant DHPLC results (extra, shoulder, widened/shifted peaks) were followed up by direct sequencing of the appropriate amplicons. Three laboratories performed the screening using fixed sets of primers and DHPLC protocols. Consistency in screening between and within laboratories was assured via a laboratory quality control plan including: (1) blinded screening of an initial set of 21 positive controls by all laboratories; (2) initial screening of the same randomly selected 21 samples by all laboratories; (3) re-screening by one laboratory of a randomly selected 10% sample of all cases screened at each of the participating laboratories; and (4) blinded re-screening of a random 10% sample of each laboratory’s own sample by that same laboratory.22
We focus our analyses exclusively on those sequence variants that are considered to have a clearly deleterious effect based on current evidence. Specifically, the following sequence variant categories were classified as deleterious: (1) changes known or predicted to truncate protein production including all frameshift and nonsense variants with the exception of BRCA2 K3326X and other variants located 3′ thereof; (2) splice site mutations occurring within 2 bp of an intron/exon boundary or shown to result in aberrant splicing; and (3) missense changes that have been demonstrated to have a deleterious effect on, for example, the function of the BRCA1 RING finger and BRCT domains. The classification of missense changes of unknown clinical significance is an on-going challenge in the field and we recognize that a small portion of the numerous missense changes identified and scored as unclassified variants may actually be deleterious. Our approach to classifying mutations as deleterious is comparable to that used in the clinical care sector and it is compatible with classifications employed by the Breast Cancer Information Core (http://research.nhgri.nih.gov/projects/bic/). In the present study, we made no attempt to screen for larger genomic deletions or duplications. Thus, we anticipate that some deleterious mutations may have escaped detection due to technical reasons or location in a region not covered by the current methodological approach.
Statistical Analysis
All data analyses involve the incidence rates of breast cancer in the identified first degree biological relatives of the probands (parents, full siblings, and children). These rates exclude the proband, although analyses involve subgroups defined by characteristics of the proband. Person years at risk of breast cancer were determined for each relative up to the age at diagnosis of breast cancer, if diagnosed, age at death, or current age at the time the proband was interviewed.
To examine risk variation among carriers on the basis of characteristics of the proband, and to construct formal statistical tests for its presence, we conducted Poisson regression analyses of the incidences of breast cancer in family members of carrier probands. In these analyses, the time periods at risk were grouped into 10-year age intervals and stratified on the basis of the relationship of the relative to the proband (mother sister, daughter). Various characteristics of the proband, such as CBC versus UBC status, age at diagnosis, and geographic site of recruitment (USA versus Denmark), were also included as covariates. The analyses also adjusted for the location of the individual mutation on the BRCA1 or BRCA2 genes. Mutations on BRCA1 were grouped into 3 regions: nucleotides 1-2400 (47 probands); nucleotides 2401-4184 (36 probands); and nucleotides 4185+ (26 probands). BRCA2 mutations were classified as within the ovarian cancer cluster region (nucleotides 3059-6629, 22 probands) or not (50 probands). These classifications are consistent with the meta-analysis of Antoniou et al.23 To account for residual variation in risk between carriers in these analyses, a random effect was included for each family, where the random effects were assumed to conform to a normal distribution. In this method each family is assumed to have a distinct risk. The estimated variance of these random effects was then evaluated for departure from zero to test for the presence of unexplained risk variation. This analysis was performed using Stata software.24
The cumulative incidences of breast cancer to various ages in relatives of carriers and in relatives of non-carriers were calculated using the Kaplan-Meier method, and the penetrance, the imputed cumulative risk in a defined population of mutation carriers, was calculated by the kin-cohort method proposed by Chatterjee and Wacholder.25 In our analyses we calculate the penetrance in several populations defined by the observed risk factors. Conceptually, the method calculates the penetrance as double the rate observed in the first degree relatives of carriers (since approximately half of these will be carriers), with an adjustment for the “baseline” incidence rate in relatives of non-carrier probands. As an approximate benchmark for evaluating the estimated penetrance curves, a population cumulative incidence curve was constructed to reflect the population incidence of breast cancer. We used reported SEER age-specific rates for this purpose, weighted to account for the calendar time periods in which the individual relatives were at risk for breast cancer. [We note that for calendar periods prior to 1975 we have used the 1975 rates, the earliest rates reported by SEER.]
RESULTS
Mutation screening of all coding exons and flanking intronic regions of BRCA1 and BRCA2 resulted in the identification of 470 unique sequence variants, among which a total of 113 unique deleterious mutations were identified, 57 located in BRCA1 and 56 in BRCA2. Of the 113 unique deleterious mutations, 73 consisted of small frameshift deletions or insertions predicted to cause protein truncation, 26 were nonsense mutations and 7 were splice site mutations. Seven missense mutations were defined as deleterious, including C44S and C61G in the BRCA1 RING domain, R1699W, A1708E, G1738E and M1775R in the BRCA1 BRCT domains, as well as M1I, disrupting the translation initiation codon of BRCA2.
There was a total of 73 carriers of deleterious mutations among the 1394 UBC participants (42 BRCA1, 31 BRCA2) and 108 carriers among the 704 CBC participants (67 BRCA1, 41 BRCA2). Data were reported for 598 first degree female relatives of these 181 carrier probands (350 in BRCA1 families, 248 in BRCA2 families), among whom 103 breast cancers were reported (61 in BRCA1 families, 42 in BRCA2 families). For the 1917 non-carrier probands there were 525 relatives with breast cancer. In total 628 breast cancers were reported among the 7156 first degree female relatives.
The crude familial aggregation that forms the basis for our analyses is displayed for descriptive purposes in Table 1. [Note that these frequencies do not reflect the varying numbers of relatives in each family and the ages of the relatives.] The preponderance (75%) of the probands had no evidence of breast cancer in their first degree relatives. This is true even for BRCA1 (58%) and BRCA2 (58%) carriers. Of the relatively few families that demonstrate very strong familial aggregation (≥ 3 first degree female relatives in addition to the proband), eight of ten occurred in probands with CBC, and three of these ten occurred in carriers.
Table 1.
Aggregation of Breast Cancer in Families of WECARE Study Probands
| Number of First Degree Female Relatives with Breast Cancer | ||||
|---|---|---|---|---|
| 3+ | 2 | 1 | 0 | |
| All Probands | 10 (0.5%) | 73 (4%) | 449 (21%) | 1565 (75%) |
| BRCA1 Carriers | 1 (1%) | 14 (13%) | 30 (28%) | 64 (58%) |
| BRCA2 Carriers | 2 (3%) | 5 (8%) | 23 (31%) | 41 (58%) |
| UBC Probands | 2 (0.1%) | 39 (3%) | 260 (19%) | 1093 (78%) |
| BRCA1 Carriers | 0 | 4 (10%) | 13 (31%) | 25 (60%) |
| BRCA2 Carriers | 0 | 2 (6%) | 10 (32%) | 19 (61%) |
| CBC Probands | 8 (1%) | 34 (5%) | 189 (27%) | 472 (67%) |
| BRCA1 Carriers | 1 (2%) | 10 (15%) | 17 (26%) | 39 (58%) |
| BRCA2 Carriers | 2 (5%) | 3 (10%) | 13 (30%) | 22 (55%) |
The results of our analyses of risk variation are presented in Table 2. Analyses are conducted initially in a joint analysis of all carrier families displayed in the first set of columns. and then separately for the relatives of BRCA1 carriers (second set of columns) and BRCA2 carriers (third set of columns). All three analyses are multivariate analyses that include all of the factors listed in the table. There is a statistically significant trend for higher risks in relatives of women carriers diagnosed with breast cancer at younger ages (p=0.04). Risks are also noticeably higher (OR=1.4) in relatives of CBC versus UBC probands, though this comparison is not statistically significant (p=0.28). The magnitudes of the trends are replicated broadly in the separate analyses of BRCA1 and BRCA2 carriers. For the analysis involving BRCA1, there is no evidence that risk is affected by location of the mutation on the gene (p=0.99), while for BRCA2, significantly higher breast cancer risks are evident for mutations outside of the ovarian cancer cluster region (p=0.03). For BRCA1, sisters (p=0.07) and daughters (p=0.03) appear to be at a higher risk than mothers. There is no apparent difference in overall risk for BRCA1 versus BRCA2 mutations (OR=1.1, 95% CI 0.6-1.8, data not shown). There is strong evidence of residual between-family variation in risk, even after adjusting for CBC versus UBC status, proband age at diagnosis, and mutation location. This is evidenced by the statistically significant tests of residual between-family variation in all three analyses (p=0.004 overall, p=0.04 for BRCA1, p=0.03 for BRCA2).
Table 2.
Relative Risks of Breast Cancer for Relatives BRCA1 and BRCA2 Mutation Carriers
| All Carrier Probands | BRCA1 Probands | BRCA2 Probands | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Factor1 | #2 | PY2 | Odds Ratio3 | # | PY | Odds Ratio | # | PY | Odds Ratio |
| UBC Proband | 35 | 12,027 | 1.0 | 21 | 6516 | 1.0 | 14 | 5511 | 1.0 |
| CBC Proband | 68 | 16,021 | 1.4 (0.8 - 2.4) | 40 | 9661 | 1.3 (0.7-2.7) | 28 | 6360 | 1.4 (0.6 - 3.4) |
| p=0.28 | p=0.43 | p=0.46 | |||||||
| Dx. Age 45-542 | 28 | 9,407 | 1.0 | 10 | 3397 | 1.0 | 18 | 6010 | 1.0 |
| Dx. Age 35-44 | 53 | 13,703 | 1.8 (0.9 - 3.3) | 35 | 8806 | 1.9 (0.8-4.7) | 18 | 4897 | 1.5 (0.6 - 3.9) |
| Dx. Age <35 | 22 | 4,938 | 2.2 (1.0 - 4.7) | 16 | 3974 | 1.9 (0.7-5.2) | 6 | 964 | 3.6 (1.0 - 13.3) |
| p=0.04 | p=0.25 | p=0.07 | |||||||
| Mothers | 52 | 11,778 | 1.0 | 27 | 7094 | 1.0 | 25 | 4684 | 1.0 |
| Sisters | 47 | 12,015 | 1.5 (1.0 - 2.4) | 30 | 6924 | 1.7 (1.0-3.2) | 17 | 5091 | 1.1 (0.5 - 2.4) |
| Daughters | 4 | 4,255 | 1.5 (0.5 - 4.9) | 4 | 2159 | 4.6 (1.3-16.0) | 0 | 2096 | NE |
| p=0.18 | p=0.03 | p=0.75 | |||||||
| USA | 80 | 21,048 | 1.0 | 51 | 12841 | 1.0 | 29 | 8207 | 1.0 |
| Denmark | 23 | 7,000 | 0.7 (0.4 - 1.3) | 10 | 3336 | 0.6 (0.3-1.5) | 13 | 3664 | 0.8 (0.3 - 1.9) |
| p=0.27 | p=0.27 | p=0.55 | |||||||
| BRCA1 (1-2400) | 27 | 6,943 | 1.0 | 27 | 6,943 | 1.0 | -- | ||
| BRCA1 (2401-4184) | 20 | 5,488 | 1.0 (0.5 - 2.1) | 20 | 5,488 | 1.0 (0.5 - 2.1) | -- | ||
| BRCA1 (4185+) | 14 | 3,746 | 1.0 (0.4 - 2.1) | 14 | 3,746 | 1.1 (0.5 - 2.5) | -- | ||
| p=0.99 | p=0.99 | ||||||||
| BRCA2 (3059-6629) | 6 | 3,331 | 1.0 | -- | 6 | 3331 | 1.0 | ||
| BRCA2 (<3059, 6630+) | 36 | 8,540 | 3.2 (1.1 - 9.4) | -- | 36 | 8540 | 3.6 (1.1 - 11.6) | ||
| p=0.03 | p=0.03 | ||||||||
| RE Variance4 | 0.90 (p=0.004) | 0.79 (p=0.04) | 0.98 (p=0.03) | ||||||
All factors are characteristics of the probands, with the exception of mothers, sisters, and daughters, which are characteristics of the relatives in the analyses.
The frequencies and PY follow-up refer to the occurrences of breast cancer, and years at risk, in the first degree relatives of the 181 carrier probands in the study.
All p-values reported are likelihood ratio tests that examine the significance of the indicated factor in affecting risk, after adjusting for the other listed factors in a multivariate model . In the case of diagnosis age and BRCA1 mutation location the tests are linear trend tests.
Estimated variance of the between-family random effects. The magnitude of the estimated variance represents the variance of the distinct relative risks estimated for each of the families on a logarithmic scale.
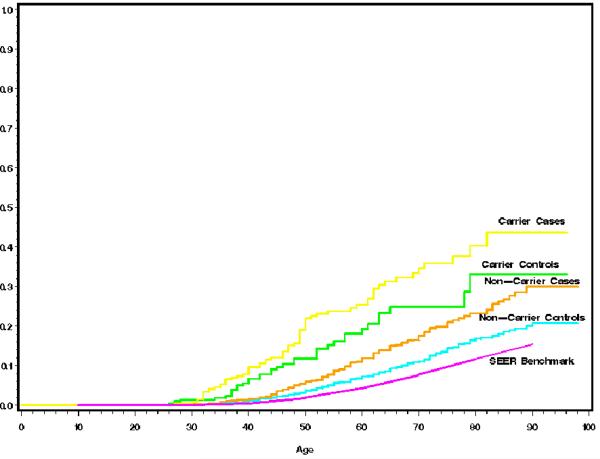
The estimated cumulative risks of breast cancer are displayed in Figure 1. The plots indicate that relatives of BRCA1 or BRCA2 mutation carriers have a substantially greater risk than relatives of non-carriers, and that relatives of case (CBC) probands have higher risk than relatives of control (UBC) probands, regardless of carrier status. Both of these differences are highly statistically significant in a Poisson regression analysis, similar in structure to Table 2, but which includes the families of all non-carrier and carrier probands (BRCA1 vs. non-carriers: OR=2.4, 95% CI 1.7-3.5; BRCA2 vs. non-carriers: OR=2.6, 95% CI 1.7-4.0; CBC vs. UBC OR=1.7, 95% CI 1.4-2.0). Penetrance estimates in mutation carriers are imputed from these curves (Table 3). From relatives of UBC probands, the penetrance is estimated to be 20% by age 50, rising to 40% by age 70 and 50% by age 80. The corresponding penetrance estimates from relatives of CBC probands are 32% by age 50, 51% by age 70, and 57% by age 80. Table 3 also displays the penetrance estimates obtained separately from relatives of carriers diagnosed in distinctive age ranges. The quantitative impact on the penetrance of the observed between-family residual risk variation can be interpreted as follows. Table 3 shows that the “average” risk to age 70 in a first degree relative of a UBC proband is 40%. Our random effects analysis demonstrates that the actual risks in individual carrier families may be much higher or much lower than this average value. In fact, assuming a constant risk of breast cancer from age 30 to age 70 in carriers, our random effects variance of 0.90 (Table 2) implies that carriers in carrier families at the upper 95th percentile of the risk distribution have a risk to age 70 of 92% rather than 40%, while carriers at the lower 5th percentile have risks similar to the population risk of breast cancer.
Figure 1. Cumulative Breast Cancer Incidence in Relatives of Probands.
The graphs show the estimated cumulative risks of breast cancer in first degree female relatives of different categories of proband. The benchmark is the cumulative risk of breast cancer reported by the SEER registries (see text). Pink - SEER 1990-1994; Blue - Cumulative risks in relatives of non-carrier UBC probands; Orange - Cumulative risks in relatives of non-carrier CBC probands; Green - Cumulative risks in relatives of BRCA1/2 carrier UBC probands; Yellow - Cumulative risks in relatives of BRCA1/2 carrier CBC probands.
Table 3.
Estimates of Risk (Penetrance) of Breast Cancer for BRCA1/2 Carriers1 Classified by Proband Characteristics
| Age | |||
|---|---|---|---|
| Population | 50 | 70 | 80 |
| UBC Probands | 20% (11%-32%) | 40% (26%-58%) | 50% (31%-71%) |
| CBC Probands | 32% (20%-44%) | 51% (36%-69%) | 57% (42%-78%) |
| BRCA1 UBC Probands | 30% (16%, 46%) | 36% (21%, 58%) | 58% (30%, 80%) |
| BRCA1 CBC Probands | 38% (23%, 54%) | 48% (30%, 67%) | 58% (38%, 81%) |
| BRCA2 UBC Probands | 9% (2%-22%) | 47% (25%-100%) | 47% (25%-100%) |
| BRCA2 CBC Probands | 22% (6%-40%) | 59% (30%-84%) | 60% (34%-92%) |
| Proband Age < 35 | 34% (16%-56%) | 52% (29%-100%) | 95% (39%-100%) |
| Proband Age 35 – 44 | 32% (20%-44% | 50% (35%-66%) | 54% (38%-75%) |
| Proband Age ≥ 45 | 14% (5%-24%) | 36% (18%-56%) | 44% (24%-68%) |
Estimated cumulative risk to given age in carriers (95% confidence interval)
COMMENT
Our study is one of the largest individual population-based family studies to date to address the breast cancer risks in BRCA1 and BRCA2 carriers, comprising 181 carrier probands, with a total of 103 breast cancers reported in the 598 first degree female relatives of these probands. We examined variation of risk between carrier families by determining whether distinct risk profiles can be identified when carrier families are sorted by observed characteristics of the probands. We observed a statistically significant trend of increasing risk with decreasing age at diagnosis of the proband (p=0.04). Furthermore, there is strong evidence of residual variation in risk between carrier families due to unobserved risk factors on the basis of a statistically significant random effects variance, even after accounting for observable proband characteristics (p=0.004). We observe that risks in relatives of CBC probands are higher than risks in relatives of UBC probands (p<0.001), although this comparison is not statistically significant when conducted solely in the carrier families (p=0.28).
These trends are consistent with the hypothesis that risks to BRCA1 or BRCA2 mutation carriers vary substantially due to the presence of additional unknown risk factors for breast cancer which are more prevalent in the families of women diagnosed at a younger age, and in the families of women with CBC. These unknown factors, which could include variants in candidate genes such as ATM or CHEK2 or other unknown genes, may ultimately explain the strong familial clustering in the families exhibiting multiple cases of breast cancer, the preponderance of which are not linked to either BRCA1 or BRCA2 mutations. Our results complement recent studies that examined risk variation in carriers on the basis of factors such as parity,26-28 age at first live birth,26-28 breast feeding,26 and mammographic density.29 Although the results from those studies are not fully consistent, they suggest that the relative risks conferred by these risk factors in carriers may be similar to the relative risks in non-carriers. In a recent study Chen and Parmigiani30 have examined between-study heterogeneity of BRCA1 and BRCA2 risks, but we emphasize that our analyses address between-family risk variation. Our results underscore the conclusion that there is no single risk associated with BRCA1 or BRCA2 carrier status. On the contrary, risks for carriers vary substantially based on observable factors, such as the characteristics of the affected relatives (probands in our case) examined in this study, host factors such as the preceding ones, and other as yet undetermined factors.
An alternative explanation for the observed risk heterogeneity is the possibility that individual variants in the BRCA1 and BRCA2 genes lead to substantially different breast cancer risks.31,32 In our study we adjusted for potential within-gene effects of this nature by classifying the variants broadly using their position on the gene. We observed no trend for the location of BRCA1 mutations, but mutations in BRCA2 outside the ovarian cancer cluster region were shown to have substantially elevated breast cancer risk compared to mutations within it. However, our sensitivity for exploring variations at the level of the individual mutation is low due to the low frequencies of occurrence of individual variants. Regardless of whether risk variation within BRCA1 and BRCA2 contributes meaningfully to the overall risk variation observed, it seems likely that other genetic factors play a major role. This conclusion is supported by a recent detailed review of studies that addressed this issue.33 Also, recently published genome-wide association studies suggest elevated breast cancer risk at several candidate loci.34,35 Furthermore, the fact that the preponderance of familial clustering occurs in families in which the proband is not a BRCA1 or BRCA2 carrier, a phenomenon discussed in depth in an earlier investigation by Cui and Hopper,36 also point to the existence of unexplained risk variation in the entire sample. It is also possible that some of the risk variation is due to environmental or life-style factors that aggregate in families, such as for example age at first birth, and that also act as modifiers of risk in carriers, although a genetic explanation is more plausible.37
The overall penetrance estimates for BRCA1 or BRCA2 mutation carriers from our study are consistent with the literature on this topic from other population-based studies, but are at the low end of a very broad range that has been reported. The most comprehensive study of this type is the pooled analysis of 22 studies by Antoniou et al..23 These authors derive a risk to age 70 of 65% in BRCA1 carriers and 45% in BRCA2 carriers. Their analysis included hospital-based as well as population-based studies, many of which used probands with early onset breast cancer (similar to our study), and some with ovarian cancer or male breast cancer probands. Although our study is population-based, the sampling of probands was unusual, and this could affect the results through unforeseen selection effects. Probands were selected if they were alive and eligible during the recruitment period between 2000 and 2004, if they had suffered a diagnosis of breast cancer from 1985 onwards (two diagnoses for CBC probands), and if they had no prior cancer other than breast cancer. This corresponds to a single ascertainment family-based design,38 and it could lead to overestimates of risk if families were inadvertently ascertained twice. We attempted to compare in an algorithmic fashion the family information of all pairs of probands, but this search revealed only one pair of sisters among the probands, confirmed on follow-up. Thus we believe that double counting of members is not a concern requiring statistical adjustment.39 The ascertainment of probands who have survived sufficiently long to be eligible for the study could lead to a selection bias if some of the heritable factors affecting cancer risk also affect prognosis, but we have no way to test this assumption. Our recruitment of women with a relatively young age at diagnosis is likely to have led to generally higher risk estimates from their relatives than would be expected in a study involving women of unrestricted age at diagnosis. Interestingly, we observed a significantly higher risk in sisters of probands than in mothers. There is no obvious explanation for this finding, though it is consistent with a previous meta-analysis.40 This trend was also observed when we analyzed non-carrier probands (data not shown). Finally, our analysis is based on first degree relatives, and so information on risk factors of these relatives is unavailable, except for age.
Our results imply that the risk of breast cancer in carriers who might be identified at random in the population without evidence of familial breast cancer may be even lower than the 40% at age 70 that we estimate from families of probands with unilateral breast cancer. It is reasonable to infer that the reduction in risk from the estimates in CBC probands to the estimates in UBC probands may be mirrored in a corresponding reduction if we were able to measure risk in carriers identified from unselected population disease-free controls. Although population-based screening for these mutations is not recommended at this time, it is entirely possible that in the future, as technology advances and genotyping costs are reduced, widespread genetic screening for important risk factors for breast cancer and other diseases may become routine, and will likely serve as the foundation for tailored risk reduction interventions. For this reason, accurate estimation of the risks conferred in the population and identification of important sources of variation in these risks constitute important scientific goals with significant implications for the clinical management of female carriers of BRCA1 or BRCA2 mutations.
Acknowledgments
Funding/Support: The study was supported by the National Cancer Institute, awards CA097397 and CA098438.
Footnotes
Financial Disclosures: None.
REFERENCES
- (1).Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–8. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- (2).Anglian Breast Cancer Study Group Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83:1301–8. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Antoniou AC, Gayther SA, Stratton JF, Ponder BA, Easton DF. Risk models for familial ovarian and breast cancer. Genet Epidemiol. 2000;18:173–90. doi: 10.1002/(SICI)1098-2272(200002)18:2<173::AID-GEPI6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- (4).Hopper JL, Southey MC, Dite GS, Jolley DJ, Giles GG, McCredie MR, et al. Population-based estimate of the average age-specific cumulative risk of breast cancer for a defined set of protein-truncating mutations in BRCA1 and BRCA2. Cancer Epidemiol Biomarkers Prev. 1999;8:741–7. [PubMed] [Google Scholar]
- (5).Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–10. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Risch HA, McLaughlin JR, Cole DEC, Rosen B, Bradley L, Fan I, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Nat Cancer Inst. 2006;98:1694–706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- (7).Thorlacius S, Struewing JP, Hartge P, Olafsdottir GH, Sigvaldason H, Tryggvadottir L, et al. Population-based study of risk of breast cancer in carriers of BRCA2 mutation. Lancet. 1998;352:1337–9. doi: 10.1016/s0140-6736(98)03300-5. [DOI] [PubMed] [Google Scholar]
- (8).Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–7. doi: 10.1093/jnci/91.14.1241. [DOI] [PubMed] [Google Scholar]
- (9).Begg CB. On the use of familial aggregation in population-based case probands for calculating penetrance. J Nat Cancer Inst. 2002;94:1221–6. doi: 10.1093/jnci/94.16.1221. [DOI] [PubMed] [Google Scholar]
- (10).Gong G, Whittemore AS. Optimal designs for estimating penetrance of rare mutations of disease-susceptibility genes. Genet Epidemiol. 2003 doi: 10.1002/gepi.10219. [DOI] [PubMed] [Google Scholar]
- (11).Whittemore AS, Gong G. Re: On the use of familial aggregation in population-based case probands for calculating penetrance. J Nat Cancer Inst. 2003;95:76–77. doi: 10.1093/jnci/95.1.76. [DOI] [PubMed] [Google Scholar]
- (12).Ponder BAJ, Antoniou A, Dunning A, Easton DF, Pharoah PDP. Polygenic inherited predisposition to breast cancer. Cold Spring Harbor Symposia on Quantitative Biology. 2005;LXX:35–41. doi: 10.1101/sqb.2005.70.029. [DOI] [PubMed] [Google Scholar]
- (13).Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BAJ. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- (15).Garber JE, Goldstein AM, Kantor AF, Dreyfus MG, Fraumeni JF, Li FP. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51:6094–7. [PubMed] [Google Scholar]
- (16).Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in non-carriers of BRCA1 and BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- (17).Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene. 2006;25:5906–11. doi: 10.1038/sj.onc.1209873. [DOI] [PubMed] [Google Scholar]
- (18).Bernstein JL, Langholz B, Haile RW, Bernstein L, Thomas DC, Stovall M, et al. Study design: evaluating gene-environment interactions in the etiology of breast cancer. Breast Cancer Res Trt. 2004;6:R199–R214. doi: 10.1186/bcr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Langholz B, Goldstein L. Risk set sampling in epidemiologic cohort studies. Statist Science. 1996;11:35–53. [Google Scholar]
- (20).Ziogas A, Anton-Culver H. Validation of family history data in cancer family registries. Am J Prev Med. 2003;24:190–8. doi: 10.1016/s0749-3797(02)00593-7. [DOI] [PubMed] [Google Scholar]
- (21).Bondy ML, Strom SS, Colopy MW, Brown BW, Strong LC. Accuracy of family history of cancer obtained through interview with relatives of patients with childhood sarcoma. J Clin Epidemiol. 1994;47:89–96. doi: 10.1016/0895-4356(94)90037-x. [DOI] [PubMed] [Google Scholar]
- (22).Bernstein JL, Teraoka S, Haile RW, Borresen-Dale AL, Rosenstein BS, Gatti RA, et al. Designing and implementing quality control for multi-center screening of mutations in the ATM gene among women with breast cancer. Hum Mutat. 2003;21:542–550. doi: 10.1002/humu.10206. [DOI] [PubMed] [Google Scholar]
- (23).Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).StataCorp . Stata Statistical Software: Release 7.0. Stata Corporation; College Station, TX: 2001. [Google Scholar]
- (25).Chatterjee N, Wacholder S. A marginal likelihood approach for estimating penetrance from kin-cohort designs. Biometrics. 2001;57:245–52. doi: 10.1111/j.0006-341x.2001.00245.x. [DOI] [PubMed] [Google Scholar]
- (26).Andrieu N, Goldgar DE, Easton DF, Rookus M, Brohet R, Antoniou AC, et al. Pregnancies, breast feeding, and breast cancer risk in the International BRCA1/2 Carrier Cohort Study. J Natl Cancer Inst. 2006;98:535–44. doi: 10.1093/jnci/djj132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Antoniou AC, Shenton A, Maher ER, Watson E, Woodward E, Lalloo F, et al. Parity and breast cancer risk among BRCA1 And BRCA2 mutation carriers. Breast Cancer Res. 2006;8:R72. doi: 10.1186/bcr1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cullinane CA, Lubinski J, Neuhausen SL, Ghadirian P, Lynch HT, Isaacs C, et al. Effect of pregnancy as a risk factor for breast cancer in BRCA1/BRCA2 mutation carriers. Int J Cancer. 2005;117:988–91. doi: 10.1002/ijc.21273. [DOI] [PubMed] [Google Scholar]
- (29).Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, et al. Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Res. 2006;66:1866–72. doi: 10.1158/0008-5472.CAN-05-3368. [DOI] [PubMed] [Google Scholar]
- (30).Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–33. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Thompson D, Easton DF. Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomark Prev. 2002;11:329–37. [PubMed] [Google Scholar]
- (32).Risch HA, McLaughlin JR, Cole DEC, Rosen B, Bradley L, Kwan E, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–10. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–906. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- (34).Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic post-menopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cui J, Hopper JL. Why are the majority of hereditary cases of early-onset breast cancer sporadic? A simulation study. Cancer Epidemiol Biomark Prev. 2000;9:805–12. [PubMed] [Google Scholar]
- (37).Khoury MJ, Beaty TH, Liang KY. Can familial aggregation of disease be explained by familial aggregation of environmental risk factors? Am J Epidemiol. 1988;127:674–83. doi: 10.1093/oxfordjournals.aje.a114842. [DOI] [PubMed] [Google Scholar]
- (38).Thomas DC. Statistical Methods in Genetic Epidemiology. Oxford University Press; New York: 2004. pp. 137–8. [Google Scholar]
- (39).Langholz B, Ziogas A, Thomas DC, Faucett C, Huberman M, Goldstein L. Ascertainment bias in rate ratio estimation from case-sibling control studies of variable age-at-onset diseases. Biometrics. 1999;55:1129–36. doi: 10.1111/j.0006-341x.1999.01129.x. [DOI] [PubMed] [Google Scholar]
- (40).Pharoah PDP, Day NE, Duffy S, Easton DF, Ponder BAJ. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int J Cancer. 1997;71:800–9. doi: 10.1002/(sici)1097-0215(19970529)71:5<800::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]