Abstract
Current hypotheses regarding vertebrate left-right asymmetry patterns are based on the presumption that genetic regulatory networks specify sidedness via extracellular morphogens and/or ciliary activity. We show empirical time-lapse evidence for an asymmetric rotation of epiblastic nodal tissue in avian embryos. This rotation spans the interval when initial symmetric expression of Shh and Fgf8 becomes asymmetrical with respect to the midline.
Keywords: Hensen's node, node, asymmetry, time-lapse imaging, cellular motion, tissue motion, Fgf8, Shh
INTRODUCTION
In chicken embryos Shh and Fgf8 genes are initially expressed symmetrically around the node (HH stage 4); a few hours later Shh and Fgf8 mRNAs are distributed asymmetrically (Levin et al., 1995; Boettger et al., 1999; Dathe et al., 2002; Raya and Izpisua Belmonte, 2008) — however, the mechanism is unclear. Left-right asymmetry in mice is reported to emerge from extracellular “nodal-flow” (Hirokawa et al., 2006). Molecules characteristic of “nodal-flow” function are expressed in nodal cilia of chicken embryos and other vertebrate organizer regions; thus, leading to the suggestion that the “…activity of nodal cilia is probably a universal mechanism for specifying the vertebrate L-R axis” (Essner et al., 2002). However, the importance of “nodal flow” in avians is in dispute (Dathe et al., 2002), as a parallel mechanism, mediated via N-cadherin, might also exist (Garcia-Castro et al., 2000).
During early vertebrate embryogenesis new cellular patterns arise within constantly moving tissue fields (Yang et al., 2002; Cui et al., 2005; Chuai et al., 2006; Zamir et al., 2006; Zamir et al., 2008). Despite this fact, the possibility that relative motion occurs between nodal tissue and the surrounding epiblastic epithelium has been largely overlooked. Dathe and colleagues addressed the link between the biomechanics of left-right patterning and gene expression using chicken embryos fixed at HH stages 4-7 (Dathe et al., 2002). Scanning electron microscopy, semi-thin sections and whole-mount in situ mRNA hybridization was used to examine Hensen's node and the cranial primitive streak of chicken embryos. The images showed that “the asymmetric expression of two laterality genes, Shh and Fgf8, is preceded by an asymmetric morphology of the avian organizer.” Specifically, Hensen's node and the right lip of the primitive streak extends further dorsally compared to the left lip. Further, Dathe and colleagues show that the right nodal area contains a cylindrical cell condensation, which is connected with the head process. These workers argued against a “nodal flow” mechanism by noting that “…densely packed cells in Hensen's node and in the cranial part of the primitive streak connect the epiblast with the endoderm…” (Dathe et al., 2002).
Another study in avian embryos, which is compatible with a biomechanical basis for asymmetry, showed that N-cadherin is involved in the establishment of L-R asymmetry (Garcia-Castro et al., 2000). Blocking N-cadherin function increased the frequency of improper heart looping, which suggests that N-cadherin based cell-cell adhesions are required for proper establishment of L-R asymmetry.
In the course of conducting a time-lapse study of the positional fate of heart forming cells (Cui et al., unpublished data, Ms submitted, 2008), we observed that there was considerable non-uniform motion centered around Hensen's node. To pursue this unexpected observation, presumptive nodal epiblastic cells were selectively electroporated with a nuclear-directed fluorescent marker protein — either Histone 2B-Green Fluorescent Protein/H2B-GFP (a gift from Dr. R. Lansford, Caltech) or H2B-RFP (a gift from Dr. O. Pourquie, Stowers Institute for Medical Research). Electroporation of ten individual chicken embryos was performed as described (Cui et al., 2006) with the platinum wire electrode tip positioned either directly on Hensen's node, or approximately 100 μm lateral (right/left) to the node. Embryos were cultured on a microscope stage using standard methods (Zamir et al., 2006; 2008). Time-lapse recording and cellular tracking were performed as described elsewhere (Czirok et al., 2002; Zamir et al., 2006; Zamir et al., 2008 and Cui et al., Ms Submitted). The instrumentation captures cellular and tissue motion throughout an embryo at one micrometer resolution across millimeter-to-centimeter length scales (Filla et al., 2004).
The resulting time-lapse movies demonstrated a reproducible, left-side biased, counterclockwise rotation of fluorescent cells (nuclei) within Hensen's node, when viewed en face from the dorsal surface. Right side cellular displacements are more complex. To confirm that these robust physical displacements of nodal cells take place during the interval that L-R asymmetry is established at the molecular level, we conducted in situ mRNA hybridization analysis for Shh and Fgf8 expression. In situ mRNA hybridization of whole-mounted embryos was conducted using a method described previously (Benazeraf et al., 2006).
RESULTS AND DISCUSSION
Time-lapse recordings allowed motion tracking as epiblastic cells move within and near the node. It must be kept in mind that in avian embryos, Hensen's node is constantly moving “down” the primitive streak (caudally). For the purposes of this report, software is employed to register the image files to the node itself; thus, the QuickTime movies show Hensen's node being held “stationary” in the center of the visual field (Czirok et al., 2002). Embryos were electroporated at HH stage 3+ to 4-, and time-lapse sequences were recorded during HH stages 4-6. The viewer's perspective is from the dorsal side in all whole-mounted specimen images.
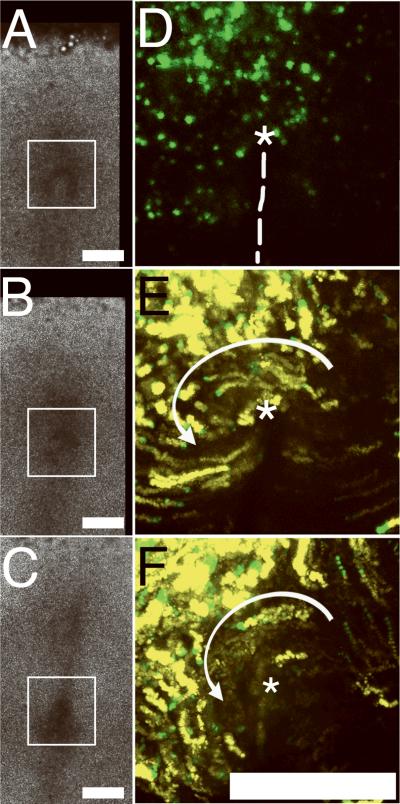
An embryo in which predominantly left-side nodal cells were tagged is shown in Figure 1 and the corresponding Movie 1. Our wide-field imaging approach allows recording of entire tissue fields (Fig. 1A-C) with no loss of resolution. The white box indicates the region of interest (ROI) shown in the epifluorescence time-lapse movies. Panel 1D depicts H2B-GFP cells early in the time-lapse sequences with the presumptive axis and position of Hensen's node indicated. Panels 1E and 1F summarize the motion tracks of fluorescent cells (nuclei) within Hensen's node. A sector encompassing approximately 9 o'clock to 1 o'clock (centered on the node) appears to rotate counterclockwise (see Movie 1), especially cells near the center of the node (asterisk).
Figure 1. Whole-mounted chicken embryos subjected to time-lapse imaging after H2B-GFP electroporation.
A-C. Three brightfield image frames extracted from a 360 min time-lapse sequence (Movie 1) demonstrate the extensive degree of tissue-level motion and expansion around a region of interest (ROI; white box), which is centered on Hensen's node, at HH stages 4-6. The brightfield Movie 1 is a collage composed of six image frames (2×3) blended into a single wide-field view using TiLa-KUMC software.
D. A corresponding H2B-GFP epifluorescence image within the ROI. The node (asterisk) and the presumptive anterior-posterior axis (dashed line) are indicated. The epifluorescence image depicts one 10X image field.
E and F. Epifluorescence cellular tracking within the same ROI as Panels A-C demonstrate the counter-clockwise vortical motion of some nodal tissue indicated by the white arrow (see corresponding Movie 1). The cellular (nuclear) tracks from 20 consecutive frames are colored yellow and superimposed with the last 5 frames colored green. Simple inspection confirms that the motion pattern within the node is not bilaterally equivalent with respect to the anterior-posterior axis (dashed line in Panel D; see Movies 1 and 2). Magnification bars=250 μm.
In other specimens the electroporation resulted in predominately right-side labeling (Movie 2). The latter embryos displayed more complex motion patterns, which differed from the left side. Cells between the 2 o'clock to 4 o'clock sector showed a bifurcated or forked pathway. Thus, epiblastic cells in this sector appeared to move toward the node until reaching a position approximately 60-100 μm distant — at which point the more anterior cells curved cranially and moved counterclockwise; whereas, the more posterior cohort curved caudally toward the primitive streak (Movie 2 and Figure 3).
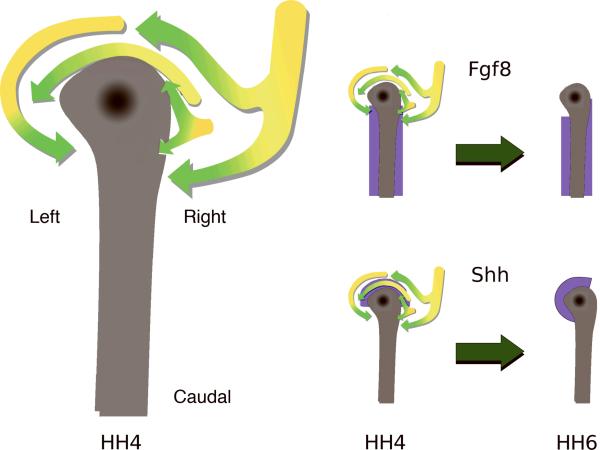
Figure 3. A model of motion in and near Hensen's Node.
An interpretative diagram of epiblastic cellular/tissue motion patterns in and near Hensen's node, based on empirical time-lapse data (n=10). Also depicted are two possible scenarios whereby gene products, Fgf8 and Shh, initially expressed in a symmetrical manner, could be moved into asymmetrical positions as a direct result of the empirically observed biophysical/mechanical tissue deformations. Purple color denotes the respective distribution of a gene product.
At HH stage 4 the avian node is a quasi-discoidal shaped tissue with a slight asymmetry toward the anatomical left (dorsal view). As embryogenesis progresses the empirical time-lapse data show that the cranial-most nodal/peri-nodal cells move counterclockwise. The arrows correspond to the color scheme of the Movies 1 and 2, with yellow depicting early image frames and green depicting later time points. On the anatomical right we envision a stream of cells approaching the node and then forking, with one stream moving in a cranially or counterclockwise fashion, while the other cellular stream flows caudally or somewhat clockwise — this latter motion pattern can be observed in Movie 2.
Let the morphogen Fgf8 be symmetrically expressed (purple) at HH stage 4. After being subjected to the motion patterns indicated by the arrows, for six hours — the morphogen producing cells would be positioned asymmetrically as shown at HH stage 6. Similarly, let Shh expressing cells be distributed symmetrically at HH stage 4 (purple), then be moved to an asymmetrical position at HH stage 6. Note that the molecular distribution patterns proposed in this model correspond to the cognate mRNA expression patterns shown in Figure 2 and by Dathe and colleagues (2002).
It must be kept in mind that the tissue patterns we depict in this diagram occur within a continuously deforming biophysical framework in vivo.
In the case of specimens electroporated in such a way that there was little right/left positional bias (i.e., at the center of the node) the motion pattern was a combination of the patterns described above (not shown). In control movies recorded from the ventral side, newly ingressed cells do not move in the vortical pattern depicted in Figure 3 (data not shown).
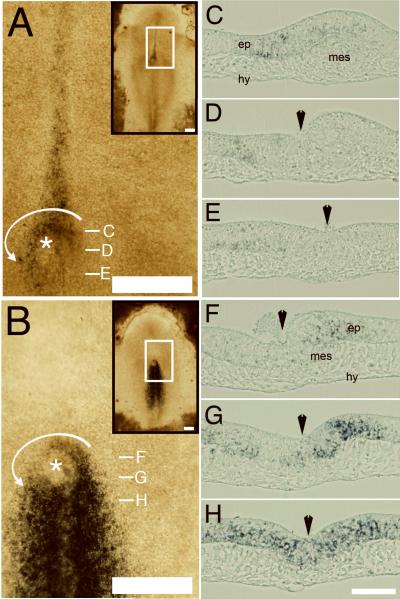
These unexpected epiblastic/nodal motion patterns prompted us to conduct in situ mRNA hybridization analysis for two genes, Shh and Fgf8, reported to play a key role in establishing L-R asymmetry. Embryos at HH stage 5+ were selected for analyses because this is a time when nodal rotation/displacement is proceeding robustly. As others have shown, Shh (Panel 2A) is expressed near the node predominantly on the left; whereas, Fgf8 (Panel 2B) is expressed with a right-side bias (Dathe et al., 2002). Expression of both markers occurs within the epiblastic layer and by some mesodermal cells, but not in the hypoblastic layer (Panels 2C-H) at HH stage 5+.
The practical result of these intricate cellular and tissue motion patterns is that nodal epiblastic cells within the 12 o'clock-to-1 o'clock sector are moved to the anatomical left, while epiblastic cells on the anatomical right near the node (1 o'clock-to-2 o'clock sector) are first moved toward, then pass across, the midline (arrows, Panels 1E and F; Movie 1; Figure 3). Close inspection of all the time-lapse data show that relative motion is occurring between nodal tissue and the surrounding epiblastic tissue (Movies 1 and 2). For example, the brightfield time-lapse sequences in Movie 1 (left panel) demonstrate that tissues bordering on, and outside, the white box (ROI) do not display the complex vortical and streaming motion patterns described above for nodal/peri-nodal tissue. Prompted by these observations, we inspected dozens of earlier time-lapse movies prepared for other purposes. In every specimen for which we have data there is evidence for local physical displacement of epiblastic nodal cells when compared to surrounding epiblastic tissue (data not shown).
These empirical data represent dynamic recordings at the tissue level of resolution—i.e., nuclear fluorescence (1-10 μm-sized objects) examined over an approximately 0.5 mm × 0.5 mm area. The focal plane depth is ±10 μm. These initial observations beg a host of questions, all of which will require higher spatial and temporal resolution to address, including confocal/deconvolution analysis at multiple focal planes. Nevertheless, our findings are noteworthy as they document the complex motion patterns of cells around the node at a critical time during the establishment of LR asymmetry in bird embryos.
As reported earlier (Dathe et al., 2002), the mRNA hybridization data show that at HH stage 5+, Shh mRNA expression in the epiblast is distributed asymmetrically toward the anatomical left side of the node (Panel 2A). Moreover, Fgf8 expression in the epiblast near the midline extends further anteriorly on the right side (Panel 2B). Both these distinct HH stage 5 mRNA expression patterns are manifested during the period covered by the time-lapse movies (HH stages 4-6). Based on these empirical, and highly reproducible observations, we suggest that Shh and Fgf8 expressing epiblastic cells are shifted to asymmetrical positions by mechanical rotation of nodal tissue. An arguably weak analogy to the motion pattern can be made to a “Lazy Susan” on a dining table, where a 180 degree rotation of a tray allows positioning of a food item on one side of the table versus the other (see Figure 3).
The time-lapse data and the in situ mRNA hybridization data suggest that the motion patterns delineated above occur in the uppermost (epiblastic) image plane. The evidence strongly suggests that some molecular markers — reported to be activated or repressed on one side of the embryo or “switched-on/off” by means of L-R regulatory mechanisms — are distributed asymmetrically due to physical rotation of the node. This statement is based on the fact that our software permits acquisition of time-resolved optical data at the tissue-level of organization in the X-Y plane, without sacrificing the cellular level detail. Thus, our data show that long-range tissue deformations provide a novel mechanical basis for conferring asymmetrical cellular position-fates in early amniote embryos. These data do not preclude regulatory circuits that impose L-R asymmetry.
Many genes are expressed in the node. A partial list includes: Gli transcriptional factors (Granata and Quaderi, 2005), cMid1 (Granata and Quaderi, 2003), Ptc1/2 (Pearse et al., 2001), HNF3-β (Levin et al., 1995), and cWnt-8c (Levin, 1997). During chicken embryonic HH stages 6/7, each of these genes is first expressed symmetrically then later asymmetrically. Some gene expression patterns are consistent with our model. For example, Gli1 and Gli2 expression in nodal epiblast shifted from a symmetrical expression pattern at HH 4 to an asymmetrical pattern at later stages (Granata and Quaderi, 2005). However, cMid1, does not conform to our model. It is expressed symmetrically at HH stage 4, and then later shows stronger expression on the embryonic-right side of the nodal epiblast; thus, suggesting cMid1 is upregulated in epiblastic cells, which are newly recruited from more peripheral regions. Unfortunately in the case of cMid1, and many other genes, there are no definitive data regarding in which cellular layer(s) the gene expression pattern occurs. Therefore, to fully understand establishment of L-R asymmetry there is a need for more thorough examination of which cellular layers exhibit asymmetrically expressed genes at HH stages 4 to 6. In this regard it is important to note that we document motion in the epiblastic layer, but not the newly ingressed cells.
Our data show that cells expressing L/R sidedness genes in the region of Hensen's node are moving; resulting in tissue-level displacements. However, it is not known what “drives” the rotational movements of the epiblastic cells surrounding the node. The differential expression of N-cadherin and resulting differences in adhesive activity reported by Garcia-Castro et al., (2000) could explain or partially explain the rotational movements. Another potential and related mechanism is differential cell-ECM interactions. The authors have shown in previous work that cells and the ECM move as a collective during ingression and gastrulation (Zamir et al., 2006; 2008), but it is not known if the ECM associated with Hensen's node is moving in concert with the rotating epiblastic cells. Likewise, differential expression or crosslinking of ECM molecules on the left and right sides of the midline axis could affect cell movements relative to the ECM (cellular autonomous motility).
The authors would like to note that Dr. Clifford Tabin's group (Harvard Medical School) has independently used time-lapse imaging and observed that cellular rearrangements around Hensen's node lead to a leftward movement of cells and establishment of asymmetric gene expression patterns in chicken embryos (C. Tabin, personal communication; Gros et al., MS submitted).
In summary, we conclude that vortical motion and tissue convection occur in and near the epiblastic “organizer” tissue layer of amniotes and that the resulting biophysical displacements might drive asymmetrical L-R expression of some genes.
Supplementary Material
Figure 2. In situ mRNA hybridizations of whole-mounted embryos and corresponding cross-sections.
A and B. In situ mRNA hybridization for Shh and Fgf8, respectively, at HH stage 5+ shows that gene expression occurs during the time and at the place where epiblastic nodal cells are vigorously moving. The insert in the upper left shows a ROI denoted by a white box. Magnification bars=250μm. Asterisks denote the position of Hensen's node.
C-H. Serial cross-sections of the HH stage 5+ specimens in Panels A and B reveal that Shh and Fgf8 mRNAs are asymmetrically expressed in the epiblastic layer (ep) of the node and in some mesodermal cells (mes), but not in the hypoblastic layer (hy). The white dashes in A and B denote the axial position of the tissue sections in C-H. Black arrowheads denote the midline. Magnification bars = 50μm.
ACKNOWLEGEMENTS
The authors would like to thank Dr. Rusty Lansford for H2B-GFP plasmid constructs (California Institute of Technology, Pasadena, CA). We thank Dr. Olivier Pourquie and Dr. Bertrand Benazeraf for kind assistance with the mRNA hybridization procedures and the H2B-RFP plasmid (Stowers Institute for Medical Research, Kansas City, MO). The authors are grateful to Dr. Clifford Tabin (Harvard Medical School) for shared, helpful discussions.
Grant sponsor: American Heart Association Postdoctoral Fellowship (CC) Grant number: 0620105Z
Grant sponsor: National Institutes of Health, INBRE Program, NCRR (BJR) Grant number: P20 RR016475
Grant sponsor: NIHLB R01 (CDL) Grant number: 068855
Grant sponsor: G. Harold and Lila Y. Mathers Charitable Foundation (CDL, BJR) Grant number: n/a
LITERATURE CITED
- Benazeraf B, Chen Q, Peco E, Lobjois V, Medevielle F, Ducommun B, Pituello F. Identification of an unexpected link between the Shh pathway and a G2/M regulator, the phosphatase CDC25B. Dev Biol. 2006;294:133–147. doi: 10.1016/j.ydbio.2006.02.035. [DOI] [PubMed] [Google Scholar]
- Boettger T, Wittler L, Kessel M. FGF8 functions in the specification of the right body side of the chick. Curr. Biol. 1999;9:277–280. doi: 10.1016/s0960-9822(99)80119-5. [DOI] [PubMed] [Google Scholar]
- Chuai M, Zeng W, Yang X, Boychenko V, Glazier JA, Weijer CJ. Cell movement during chick primitive streak formation. Dev Biol. 2006;296:137–149. doi: 10.1016/j.ydbio.2006.04.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Cheuvront TJ, Lansford RD, Moreno-Rodriguez RA, Schultheiss TM, Rongish BJ. Dynamic Positional Fate Map of the Primary Heart-Forming Region. 2008. Ms Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Lansford R, Filla MB, Little CD, Cheuvront TJ, Rongish BJ. Electroporation and EGFP labeling of gastrulating quail embryos. Dev Dyn. 2006;235:2802–2810. doi: 10.1002/dvdy.20895. [DOI] [PubMed] [Google Scholar]
- Cui C, Yang X, Chuai M, Glazier JA, Weijer CJ. Analysis of tissue flow patterns during primitive streak formation in the chick embryo. Dev Biol. 2005;284:37–47. doi: 10.1016/j.ydbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Czirok A, Rupp PA, Rongish BJ, Little CD. Multi-field 3D scanning light microscopy of early embryogenesis. J Microsc. 2002;206:209–217. doi: 10.1046/j.1365-2818.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- Dathe V, Gamel A, Manner J, Brand-Saberi B, Christ B. Morphological left-right asymmetry of Hensen's node precedes the asymmetric expression of Shh and Fgf8 in the chick embryo. Anat Embryol (Berl) 2002;205:343–354. doi: 10.1007/s00429-002-0269-2. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- Filla MB, Czirok A, Zamir EA, Little CD, Cheuvront TJ, Rongish BJ. Dynamic imaging of cell, extracellular matrix, and tissue movements during avian vertebral axis patterning. Birth Defects Res C Embryo Today. 2004;72:267–276. doi: 10.1002/bdrc.20020. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Vielmetter E, Bronner-Fraser M. N-Cadherin, a cell adhesion molecule involved in establishment of embryonic left-right asymmetry. Science. 2000;288:1047–1051. doi: 10.1126/science.288.5468.1047. [DOI] [PubMed] [Google Scholar]
- Granata A, Quaderi NA. The Opitz syndrome gene MID1 is essential for establishing asymmetric gene expression in Hensen's node. Dev Biol. 2003;258:397–405. doi: 10.1016/s0012-1606(03)00131-3. [DOI] [PubMed] [Google Scholar]
- Granata A, Quaderi NA. Asymmetric expression of Gli transcription factors in Hensen's node. Gene Expr Patterns. 2005;5:529–531. doi: 10.1016/j.modgep.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in vertebrate embryogenesis. Bioessays. 1997;19:287–296. doi: 10.1002/bies.950190406. [DOI] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- Pearse RV, 2nd, Vogan KJ, Tabin CJ. Ptc1 and Ptc2 transcripts provide distinct readouts of Hedgehog signaling activity during chick embryogenesis. Dev Biol. 2001;239:15–29. doi: 10.1006/dbio.2001.0430. [DOI] [PubMed] [Google Scholar]
- Raya A, Izpisua Belmonte JC. Insights into the establishment of left-right asymmetries in vertebrates. Birth. Defects. Res. C. Embryo. Today. 2008;84:81–94. doi: 10.1002/bdrc.20122. [DOI] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- Zamir EA, Czirok A, Cui C, Little CD, Rongish BJ. Mesodermal cell displacements during avian gastrulation are due to both individual cell-autonomous and convective tissue movements. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19806–19811. doi: 10.1073/pnas.0606100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir EA, Rongish BJ, Little CD. The ECM moves during primitive streak formation — computation of ECM versus cellular motion. Public Library of Science, Biology. 2008;6 doi: 10.1371/journal.pbio.0060247. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.