Abstract
Cell migration is a dynamic process that requires temporal and spatial regulation of integrin activation and focal adhesion assembly-disassembly1. Talin, an actin and β integrin tail-binding protein, is essential for integrin activation and focal adhesion formation2,3. Calpain-mediated cleavage of talin plays a key role in focal adhesion turnover3; however, the talin head (TH) domain, one of the two cleavage products, stimulates integrin activation, localizes to focal adhesions, and maintains cell edge protrusions2,4,5, suggesting that additional steps, downstream of talin proteolysis, are required for focal adhesion disassembly. Here we show that TH binds Smurf1, an E3 ubiquitin ligase involved in cell polarity and migration6,7, more tightly than full length talin and that this interaction leads to TH ubiquitination and degradation. TH was a substrate for Cdk5, a regulator of cell migration and cancer metastasis8–11. Cdk5 phosphorylated TH at Ser425, inhibiting its binding to Smurf1, thus preventing TH ubiquitination and degradation. Expression of talS425A, which resists Cdk5 phosphorylation thereby increasing its susceptibility to Smurf1-mediated ubiqitination, resulted in extensive focal adhesion turnover and inhibited cell migration. Thus, TH produced by calpain cleavage of talin, is degraded via Smurf1-mediated ubiquitination; moreover, phosphorylation by Cdk5 regulates Smurf1 binding to TH and, in this way, controls TH turnover and adhesion stability and, ultimately, cell migration.
Talin is cleaved into an N-terminal ~47-kDa globular head domain and a ~190-kDa, C-terminal rod domain, by calpain12. The talin head domain contains a FERM (band four-point-one, ezrin, radixin, moesin homology domain) domain, which is comprised of three subdomains, named F1, F2, and F3, and is responsible for talin-binding to the β integrin tail13, PIP kinase (Type I)14,15, FAK16 and layilin17,18. The rod domain has several vinculin binding sites, a second integrin-binding site and two actin-binding sites19. Reducing talin expression by siRNA knockdown or genetic deletion inhibit integrin activation20,21, whereas transfecting the cells that have low talin expression with talin restores integrin activation22. Also, talin(−/−) ES cells are deficient in focal adhesion formation23, whereas talin knockdown or microinjection of anti-talin antibody inhibits cell migration4,24. Thus, talin is essential for integrin activation, focal adhesion formation, and mesenchymal cell migration.
Talin Ser425 is a potential phosphorylation site for Cdk525, a cyclin-dependent protein kinase that is essential for cell migration, synaptic transmission, and cancer metastasis8,11. Although Cdk5 phosphorylates a number of proteins, including microtubule-associated proteins, signaling molecules and synapse-related proteins8, the molecular mechanisms by which Cdk5 influences cell migration remains to be elucidated. We hypothesized that phosphorylation of talin by Cdk5 could regulate its stability and, in turn, cell migration.
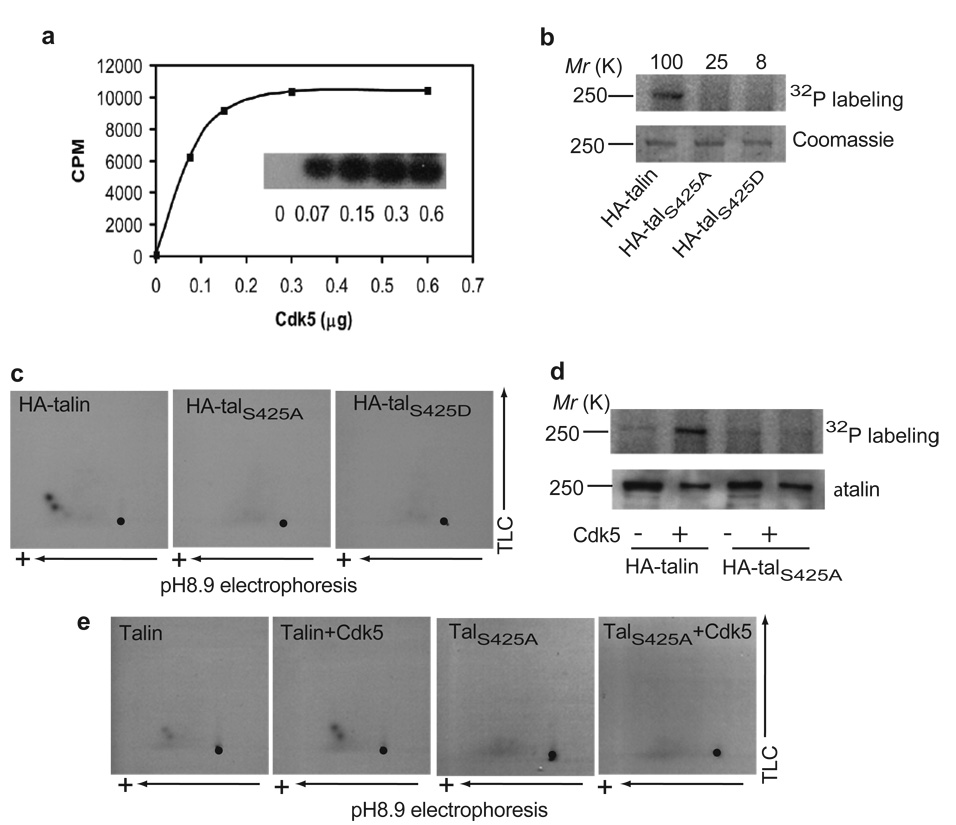
To learn whether talin is a substrate for Cdk5, recombinant His-tagged TH was incubated with active Cdk5/p25 in the presence of [γ-32P]ATP, and phosphorylation was measured by autoradiography and Cerenkov counting. Cdk5 efficiently phosphorylated TH in vitro (Fig. 1a). To assess whether Ser425 on talin is phosphorylated by Cdk5, HA-talin, -talS425A and –talS425D were transfected into CHO-K1 cells, immunoprecipitated with anti-HA antibody and phosphorylated with purified Cdk5/p35. Cdk5 phosphorylated wild-type (wt) talin but not talS425A or talS425D (Fig. 1b). These proteins were digested with trypsin and analyzed by two-dimensional (2-D) phospho-peptide mapping. There were two phosphopeptides in the map of wt talin, but not in the map of talS425A or talS425D (Fig. 1c). The two phosphopeptides were probably due to alternative cleavage of talin at Lys427 or Lys428 by trypsin because both were abolished by point mutations at Ser425. Therefore, talin is a substrate for Cdk5 in vitro and Ser425 of talin is the site of phosphorylation by Cdk5.
Figure 1. Cdk5 phosphorylates talin at Ser425.
a, Direct phosphorylation of TH by Cdk5 in vitro. Recombinant His-tagged TH (2.8 µg) was incubated with active Cdk5/p25 in 40 µl of kinase buffer containing 40 µM [γ-32P]ATP (10 µCi). A 1:8 stoichiometry of phosphorylation of the TH was achieved in the experiments. b, Phosphorylation of Ser425 on talin by Cdk5 in vitro. HA-talin, -talS425A or -talS425D were immunoprecipitated from transfected CHO-K1 cells, and incubated with active Cdk5/p35 in 50 µl of kinase buffer containing 20µM [γ-32P]ATP (10 µCi). The numbers on the top indicates phosphorylation levels (% of wt talin) quantified as described in “Methods”. c, 2-D phospho-peptide mapping analysis of wt talin, talS425A, and talS425D. d, Talin is phosphorylated at Ser425 by Cdk5 in vivo. Cells were transfected with HA-talin or –talS425A in the presence or absence of Cdk5/p35. After 18 hr of transfection, cells were incubated with [32P]orthophosphoric acid (0.8 mCi/ml) for 12 hr. Cells were then harvested and analyzed as described in “Methods”. Over-expression of Cdk5 promotes phosphorylation of talin but not of talS425A. e, 2-D phospho-peptide mapping analysis of talin and talS425Ain the absence and presence of recombinant Cdk5.
To ascertain whether talin is a substrate for Cdk5 in vivo, CHO-K1 cells were transfected with HA-talin or –talS425A with or without Cdk5/p35 co-transfection, and labeled with [32P]orthophosphoric acid. HA-talin and –talS425A were immunoprecipitated with an anti-HA antibody and phosphorylation was detected by autoradiography. Phosphorylation of wt talin was enhanced by Cdk5/p35 co-transfection, whereas that of mutant talS425A was not. The talin and talS425A bands from Fig. 1d were excised, digested with trypsin and subjected to 2-D phosphopeptide mapping analysis. Two major phosphopeptides were observed on the 2-D maps of wt talin, and co-transfection of Cdk5/p35 caused an increase in the intensity of the phosphorylation. However, the phosphopeptides were absent on the 2-D maps of talS425A (Fig. 1e). To test whether endogenous talin is a substrate for Cdk5 under physiological condition, endogenous talin phosphorylation and phosphopeptide maps were analyzed in rat phaeochromocytoma PC-12 cells. Talin phosphorylation was stimulated by nerve growth factor (NGF), a known activator of Cdk526, whereas the stimulation was inhibited by roscovitine, a specific inhibitor of Cdk5 (Supplementary Fig. 1). The tryptic maps of the endogenous talin also showed the two major phosphopeptides and these were abolished by roscovitine. Therefore, talin is also a substrate for Cdk5 in vivo, and the same site is phosphorylated by Cdk5 in vitro and in vivo.
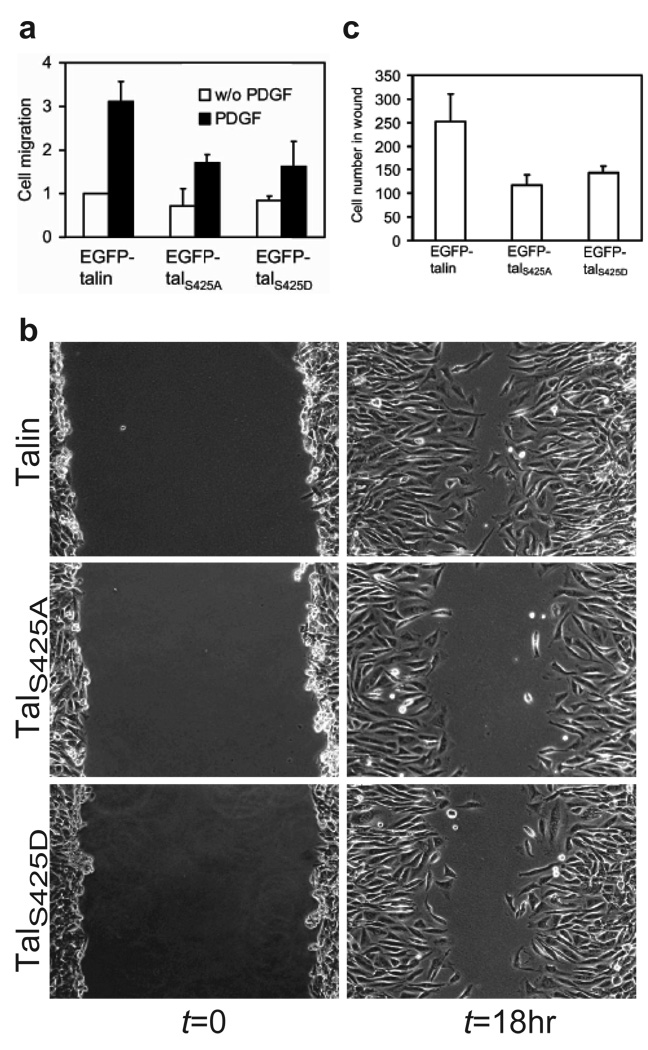
Since Cdk5 is essential for neuronal migration9,10, the physiological role of Cdk5-mediated talin phosphorylation was assessed in neuroblastoma SH-SY5Y cells. We transfected these cells with EGFP-talin, –talS425A or –talS425D, and cell migration was measured using transwell assays. Whereas the migration of cells expressing EGFP- talin was stimulated by PDGF, expression of EGFP-talS425A or EGFP-talS425D markedly retarded PDGF-induced cell migration (Fig. 2a; Supplementary Information, Fig. S2a). Moreover, roscovitine inhibited PDGF-induced migration of SH-SY5Y cells in a dose-dependent manner (Supplementary Information, Fig. S2b), and reducing Cdk5 expression by shRNA knockdown also significantly inhibited neuroblastoma cell migration (Supplementary Information, Fig. S2c).
Figure 2. Expression of phosphorylation-deficient talin mutants inhibits cell migration.
a, Inhibition of PDGF-stimulated migration of SH-SY5Y neuroblastoma cells by expressing EGFP-talS425A or –talS425D. SH-SY5Y cells were transfected with EGFP-talin, -talS425A or –talS425D, using Nucleofactor Kit V (Amaxa). At 24 hr post-transfection, cells were trypsinized for transwell migration assays employing collagen I (10µg/ml)-coated filters and 20 nM PDGF in lower charmber. c and d, Inhibition of wound closure in CHO cells expressing EGFP-talS425A or –talS425D. Confluent monolayers of CHO cells stably expressing EGFP-talin, talS245A or –talS425D were wounded and images were taken when the wounds were made (left panels) and after incubating for 18 hr (right panels). Data are expressed as mean ± SEM of three independent experiments.
Besides neurons, Cdk5 is also expressed in many other tissues27–29. To test the role of Cdk5-mediated talin phosphorylation in the migration of cells of non-neuronal-origin, we investigated the effects of talS425A and talS425D on the migration of CHO-K1 cells, which express Cdk5 (unpublished observation). In wound healing assays, talin mutants EGFP-talS425A or -talS425D inhibited the migration of CHO-K1 cells by approximately 55% and 42%, respectively (Fig. 2b, c and Supplementary Information, Fig. S2d). Roscovitine also reduced the migration of CHO-K1 cells (Supplementary Information, Fig. S2e), Thus, phosphorylation of talin at Ser425 is required for the migration of SH-SY5Y neuroblastoma and CHO-K1 cells.
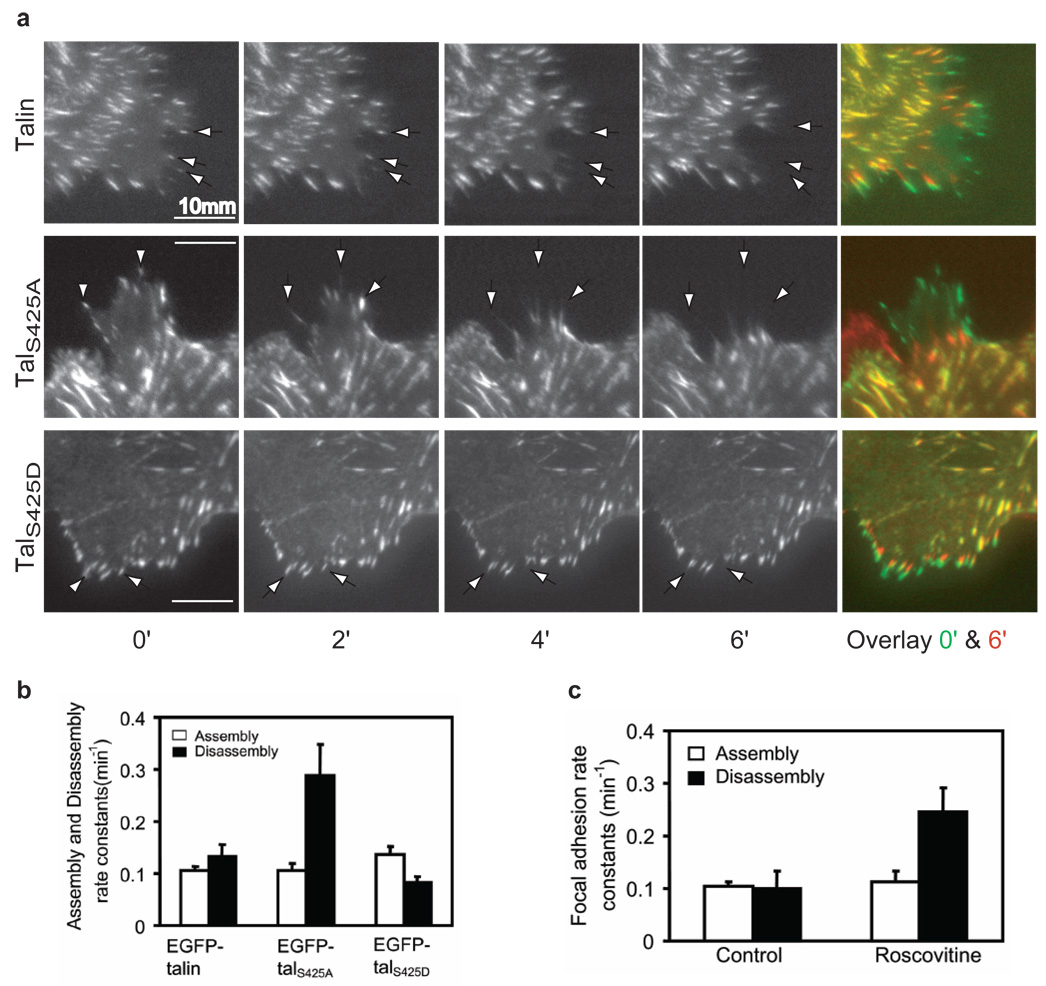
Talin regulates integrin activation and focal adhesion dynamics. In migratory cells, talin is found in both focal complexes--small, very dynamic adhesive structures at leading edges--and in focal adhesions30. Thus, it is possible that serine phosphorylation of talin might regulate focal adhesion turnover. This prompted us to examine the turnover of EGFP-talin, –talS425A and –talS425D at protrusive structures in spreading cells. Total internal reflection microscopy (TIRF) revealed that structures containing EGFP-talin disassembled much more slowly than those containing EGFP-talS425A (P=0.02), but somewhat faster than those containing EGFP-talS425D (P=0.06, Fig. 3a,b). In addition, roscovitine, the Cdk5 inhibitor, caused an increase in the disassembly rate (P=0.02, Fig. 3c). By contrast, there was a very little difference in the assembly rate constants between talin and the Ser425 mutants (Fig. 3b), and roscovitine also had no effect on the assembly rate constants (Fig. 3c). It is instructive to examine the ratio of the disassembly to assembly rate constants; these are significantly different: 1.27, 2.74, and 0.60 for talin, talS425A and talS425D, respectively (P≤0.02). Since constant focal adhesion disassembly and assembly are required for cell migration, these differences could be related to the effects of talin mutants on migration in the following way: When phosphorylation of Ser425 is reduced with the S425A mutant or by roscovitine, focal adhesion disassembly is favored preventing stabilization of protrusions because firm adhesion to the substrate is lacking. On the other hand, in the phosphomimetic S425D mutant, adhesion assembly is favored relative to disassembly resulting in overly firm adhesion to the substrate in the protruding region. Either effect would result in lower rates of cell migration relative to wild type cells.
Figure 3. Talin phosphorylation by Cdk5 is important for the disassembly of focal adhesions.
a, CHO cells stably expressing EGFP–talin, EGFP–talS425A, or EGFP-talS425D were plated on fibronectin (5µg/ml)-coated glass-bottom dishes (MatTek) and analyzed by time-lapse TIRF microscopy. Arrowheads point to disassembling focal adhesions. b, Quantification of the rate constants of EGFP–talin, EGFP–talS425A, or EGFP-talS425D assembly into and dissociation from focal adhesions. (c) Effects of roscovitine on the rate constants of EGFP–talin, assembly into and dissociation from focal adhesions. Quantifications are expressed as means ± SEM of 30 focal adhesions from 7 cells.
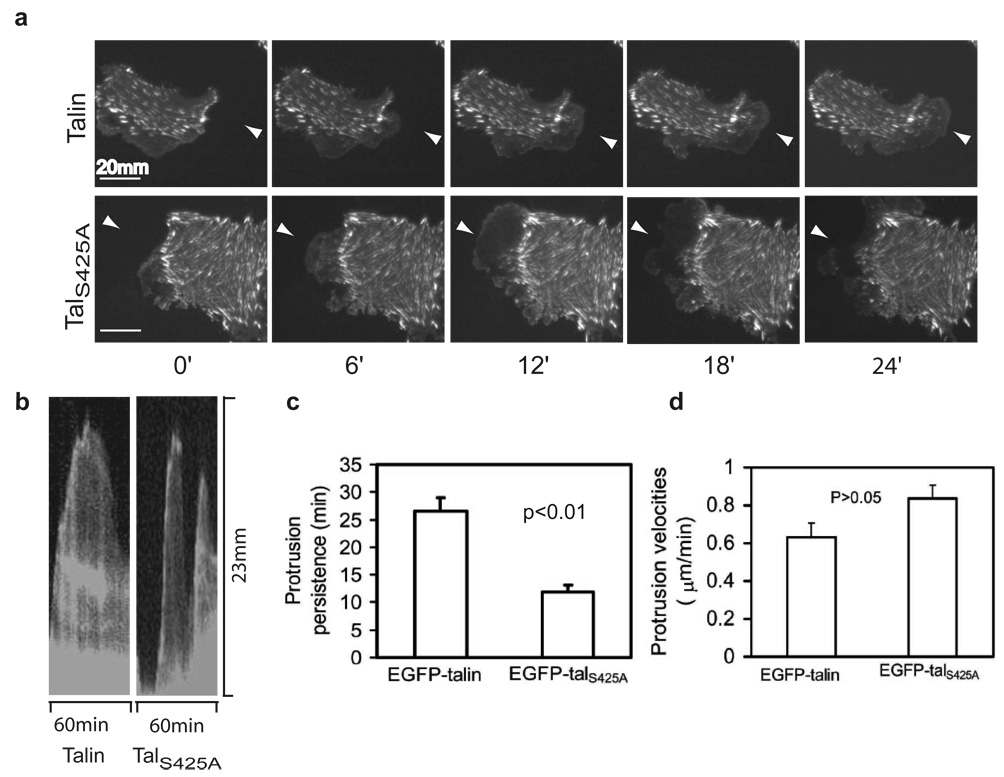
To test the idea that the effects of talin mutants on focal adhesion dynamics influenced cell protrusion, we examined the effects of talin and the Ser425 mutants on protrusion dynamics. Cells expressing EGFP-talS425A formed unstable protrusions, whereas cells expressing EGFP-talin generated more persistent protrusions and lamellipodia (Fig. 4a). Kymographic analysis showed that cells expressing EGFP-talS425A had significantly lower protrusion persistence than cells expressing wild-type talin (P<0.01, Fig. 4 b, c), whereas cells expressing EGFP-talS425D had prolonged protrusion persistence (Supplementary Information, Fig. S3a, b). There was less difference in protrusion velocities (Fig. 4d). Furthermore, Cdk5 localized to focal complexes in the lamellipodium (Supplementary Information, Fig. S3c), a site at which adhesion assembly/disassembly regulates protrusion dynamics. These results indicate that talin phosphorylation by Cdk5 is an important factor for stabilizing focal adhesions and lamellipodia during cell migration.
Figure 4. Talin phosphorylation by Cdk5 stabilizes lamellipodia.
a, CHO cells expressing EGFP–talin, or EGFP–talS425A were plated on 5 µg/ml fibronectin and analysed by time-lapse TIRF microscopy. Arrow heads points to lamellipodium formation and retraction. b, Kymograph of lamellipodium dynamics in CHO cells expressing EGFP–talin or EGFP–talS425A. c, The lamellipodial persistence in cells expressing wild-type Talin is significantly higher than that of mutant S425A. d, There is no significant difference in protrusion velocities between cells expressing the wild-type and mutant S425A. Quantifications are expressed as means ± SEM.
Next, we explored the mechanism whereby Cdk5-mediated talin phosphorylation regulated adhesion disassembly starting with the observation that calpain cleavage of talin is essential for disassembly3. Since the calpain cleavage site on talin is close to Ser425, the Cdk5 phosphorylation site, we tested whether wt EGFP-talin or EGFP-talS425A behave differently with respect to calpain cleavage. Mutation at Ser425 had no effect on the cleavage by calpain (Supplementary Information, Fig. S4a) nor did phosphorylation of talin by Cdk5 have any effect on the cleavage of talin by calpain (Supplementary Information, Fig. S4b). We therefore explored other potential events regulated by talin phosphorylation that could lead to focal adhesion disassembly.
Smurf1 is an E3 ubiquitin ligase that ubiquitinates Rho A and causes Rho A degradation at the leading edge of migrating cells, promoting lamellipodium formation6,7. Since Smurf1 possesses a NPXY motif, a motif responsible for talin-integrin tail interation31, and a small portion of Smurf1 localized to focal adhesions (Supplementary Information, Fig. S5a), we suspected that Smurf1 was associated with talin and played a role in talin-regulated focal adhesion dynamics.
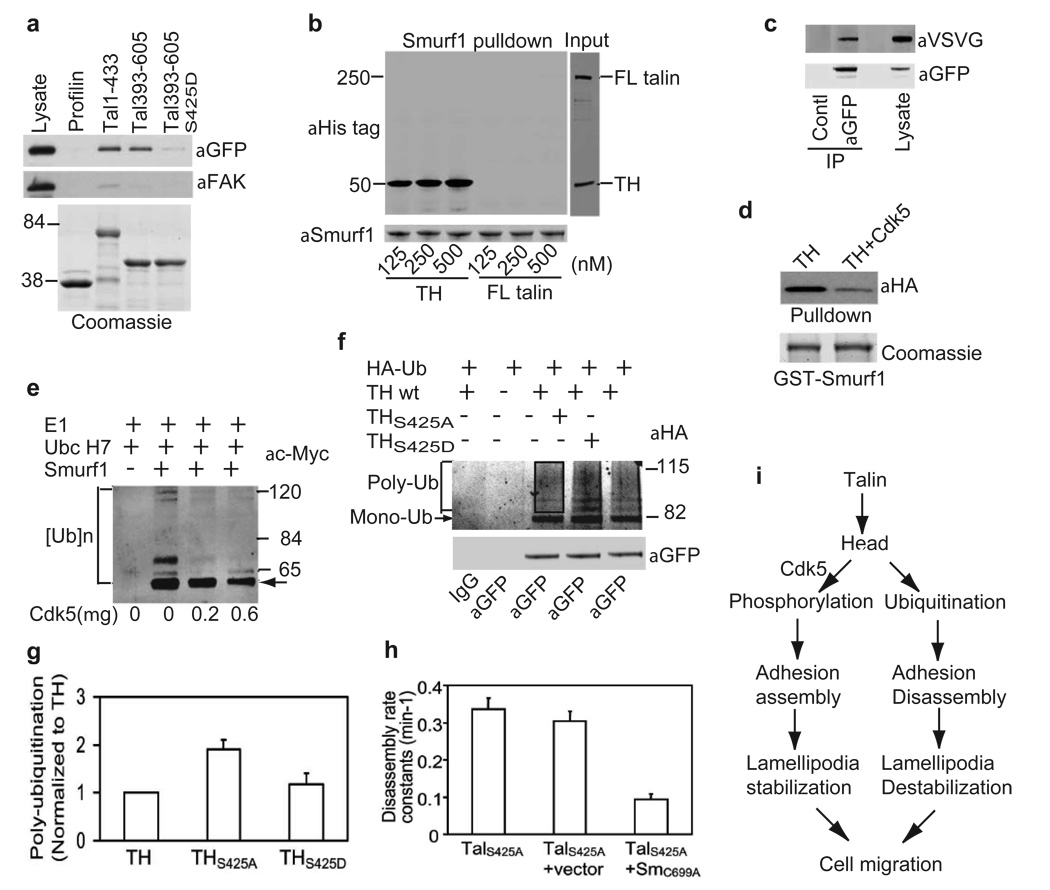
To test whether Smurf1 interacts with talin, we examined the binding of recombinant Smurf1, expressed in CHO cells, to GST talin fragments and mutants. Both TH (Tal1-433) and Tal393-605 bound Smurf1, whereas profilin had no detectable interaction with Smurf1 (Fig. 5a). Furthermore, purified TH strongly interacted with purified GST-Smurf1 but not with GST-profilin in vitro, whereas full-length talin did not show significant interaction (Fig. 5b and Supplementary Information, Fig. S5b). However, the NPXY motif on Smurf1 is not essential for TH binding, since mutation at this motif had little effect on the Smurf1-TH interaction (Supplementary Information, Fig. S5c). Endogenous Smurf1 immunoprecipitated from MDA-MB-231 human breast cancer cells interacted with EGFP-TH in CHO cell lysates (Supplementary Information, Fig. S5d). To examine whether TH is associated with Smurf1 in cells, EGFP-TH and VSVG-Smurf1C699A, a catalytically inactive Smurf1 mutant, were co-transfected into CHO-K1 cells in combination with H-RasG12V, a small GTPase that amplifies the expression of Smurf1 or its mutants (unpublished data). Smurf1C699A co-precipitated with TH (Fig. 5c). These data indicate that Smurf1 binds to TH, and imply that TH associates with Smurf1 through a short sequence (residues 393–433) immediately after the F3 domain.
Figure 5. Cdk5 phosphorylation inhibits Smurf1-mediated TH ubiquitination.
a, Talin-Smurf1 interaction. Binding of EGFP-Smurf1 to immobilized GST-profilin, -tal1-433, -tal393-605, or -tal393-605S425D. Smurf1 was detected by immunoblotting with anti-GFP. b, TH but not full-length talin binds Smurf1 tightly,. Binding of purified His-tagged TH or FL talin to immobilized GST-Smurf1 was detected by immunoblotting with anti-His tag mAb. c, TH and catalytically-inactive Smurf1(SmC699A) interact in cells. EGFP-TH and VSVG-SmC699A were co-transfected in combination with H-RasG12V into CHO-K1 cells. Anti-GFP immunoprecipitated EGFP-TH and bound Smurf1C699A was detected with anti-VSVG. d, Inhibition of TH-Smurf1 interaction by Cdk5-mediated phosphorylation. HA-TH was purified from CHO-K1 cell lysates and phosphorylated in vitro with Cdk5/p35 in vitro, and binding to GST-Smurf1 was analyzed as in panel b. e, Inhibition of Smurf1-mediated ubiquitination Cdk5-mediated phosphorylation. Purified His-tagged TH was incubated 120 min with the indicated quantity of Cdk5/p25 and then with Myc-ubiquitin, E1, and UbcH7 in the presence or absence of GST-Smurf1. f, Effect of TH phosphorylation on ubiquitination in vivo. CHO-K1 cells were co-transfected with EGFP-TH, -THS452A, or -THS425D and HA-ubiquitini. Twenty-four hr post-transfection, cells were re-plated on fibronection for 5 hr and then were treated with MG132 (40µM) for an additional 4 hr before ati-HA immunoblotting. g, The poly-ubiquitination of EGFP-TH, -THS452A and -THS425D was quantified by densitometry of the rectangular area (see lane 3). Depicted are the mean and range of two independent experiments. h, CHO cells expressing EGFP–talS425A, were transfected with vector or DsRed-Smurf1C699A and plated on fibronectin for 4 hr. The rate constants of EGFP–talS425A dissociation from focal adhesions were quantified by time-lapse microscopy (mean ± SEM, n=30). i, Proposed mechanism whereby talin phosphorylation by Cdk5 regulates TH ubiquitination, focal adhesion disassembly, lamellipodial dynamics, and cell migration. Full scans of western blots shown in b, c, e, and f are available in Supplementary Information, Fig. S7.
Since Ser425 is within TH (393–433), Cdk5-mediated phosphorylation may regulate the binding of TH to Smurf1. To examine this hypothesis, the His-tagged TH was expressed in CHO-K1 cells and purified by Ni2+ chelate affinity chromatography. We then assessed the effects of in vitro phosphorylation with Cdk5 on binding to GST-Smurf1. Phosphorylation by Cdk5 markedly inhibited the direct binding of purified TH to Smurf1, as did the phosphomimetic mutation Tal393-605S425D (Fig. 5a, d). Furthermore, substituting Ser425 with Ala in Tal210-605, expressed in CHO cells, markedly increased its binding to Smurf1 (Supplementary Information, Fig. S5e), suggesting that phosphorylation of Ser425 inhibits binding of Tal210-605 to Smurf1. If this inference is correct, inhibition of Cdk5 in cells should enhance Tal210-605 binding to Smurf1. Indeed, transfection of Cdk5-144N, a dominant negative mutant of Cdk5, promoted the interaction of Tal210-605 with Smurf1 (Supplementary Information, Fig. S5f). These results suggest that talin phosphorylation by Cdk5 on Ser425 inhibits its interaction with Smurf1.
Smurf1 is an E3 ubiquitin ligase that specifies targets for ubiquitination and proteasomal degradation32. To learn whether the binding of Smurf1 leads to degradation of TH, we co-transfected TH with Smurf1 and observed a reduction in TH steady-state levels. This reduction in TH abundance was due to proteasomal degradation because it was reversed by MG132, a proteasome inhibitor. In contrast, co-transfection of either Sm1-267 (a Smurf1 mutant without the ligase domain) or VASP had no effect on TH abundance (Supplementary Information, Fig. S6a). Transfection of Smurf1 did not cause a reduction in the level of full-length talin (Supplementary Information, Fig. S6b), indicating that full-length talin is not a major target for Smurf1. Smurf1 did not cause a reduction in the level of VASP, FAK and paxillin (Supplementary Information, Fig. 6c, d). These results suggest that Smurf1-mediated TH ubiquitination leads to its degradation.
To learn whether Smurf1 could mediate TH ubiquitination and to examine the role of talin phosphorylation in regulating its ubiquitination, recombinant His-tagged TH was phosphorylated with Cdk5/p25 and its capacity to serve as substrate for Smurf1-directed ubiquitination was assayed in vitro. TH was ubiquitinated by Smurf1 in vitro and ubiquitination was inhibited by Cdk5-mediated phosphorylation (Fig. 5e; Supplementary Information, Fig. S6e). To examine the ubiquitination of TH and its mutants in cells, HA-ubiquitin was co-transfected with EGFP-TH, -THS425A, or -THS425D into CHO-K1 cells. The cells were plated on fibronectin and treated with MG132 to block proteasomal degradation and ubiquitination was assessed by Western blotting to the immunoprecipitated talin fragments. Ubiquitination of TH was enhanced by substituting Ser425 with Ala but not with Asp (Fig. 5f, g). In addition, Smurf1 stimulated the ubiquitination of EGFP-tal210-605, whereas Smurf1C699A, a ligase-dead mutant of Smurf1, retarded its ubiquitination (Supplementary Information, Fig. S6f), indicating the role of Smurf1 in this ubiquitination. Furthermore, expressing Cdk5-144N, a dominant negative mutant of Cdk5, stimulated the ubiquitination of EGFP-tal210-605 (Supplementary Information, Fig. S6g). Thus, we conclude that talin phosphorylation by Cdk5 negatively regulates TH ubiquitination by modulating the TH-Smurf1 interaction.
If TH ubiquitination by Smurf1 is important for focal adhesion disassembly, inhibiting Smurf1 should block focal adhesion disassembly. Indeed, SmC699A, a catalytically inactive Smurf1 mutant, markedly reduced focal adhesion disassembly rate in cells expressing EGFP-talS425A (P<0.01, Fig. 5h). Furthermore, the effects of talin Ser425 mutants on Smurf1-mediated ubiquitination (Fig. 5f, g) correlated with the rates of focal adhesion disassembly (Fig. 3b). Thus, Cdk5 regulates focal adhesion disassembly by controlling Smurf1-mediated ubiquitination and degradation of TH.
In migratory cells, talin is cleaved into head and rod domains by calpain. The head domain in turn binds to Smurf1 and is consequently ubiquitinated and degraded. This step, downstream of calpain, provides a more complete explanation for calpain-mediated focal adhesion turnover. We find that talin phosphorylation by Cdk5 inhibits talin head ubiquitination and degradation, thus limiting focal adhesion turnover and stabilizing lamellipodia at the leading edges that, in turn, leads to optimal rates of cell migration (Fig. 5i). Thus, our studies explain the capacity of talin proteolysis to promote focal adhesion disassembly, identify talin as an important target of Smurf1-induced ubiquitination and degradation, and establish talin as a relevant substrate of Cdk5.
Methods
Cell culture and transfection
CHO-K1 cells and SH-SY5Y cells were from ATCC and maintained in DMEM/F-12 medium (Gibco) containing 10% FBS, 100 units/ml penicillin and 100µg/ml streptomycin. CHO K1 cells were transfected with Lipofectamine Plus transfection reagent (Invitrogen) according to the manufacturer’s protocol. Cells stably expressing EGFP-talin, -talS425A, or –talS425D were obtained by sorting EGFP positive cells after G418 selection in the UNC Flow Cytometry Facility.
In vitro phosphorylation
CHO-K1 cells were transfected with HA-talin, -talS425A and -talS425D, respectively. At 24 hr after transfection, cells were lysed in Lysis Buffer A (50 mM Tris-HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail), and talin and its mutants were reacted with anti-HA polyclonal antibodies, and the immunoprecipitates were captured on Protein A Sepharose beads. The beads were washed 3 times with lysis buffer and once with a kinase buffer33 and then were incubated with purified Cdk5 (~0.1 µg) in 50 µl of the kinase buffer containing 20 µM ATP (10 µCi [γ-32P]ATP) for 30 min at room temperature. The reactions were terminated by adding SDS-sample buffer. The samples were fractionated by SDS-PAGE and transferred to nitrocellulose membranes for autoradiography.
In vivo phosphorylation
Cells were incubated with [32P]orthophosphoric acid in sodium phosphate-deficient DMEM (Gibco). After 10 hr, cells were stimulated with PDGF where indicated and harvested and lysed with a RIPA buffer33 including protease inhibitor cocktail and 500 nM Okadaic acid. The lysates were immunoprecipitated with anti-HA or anti-GFP polyclonal antibodies. The immune complexs were analyzed by SDS-PAGE and transferred to nitrocellulose membranes for detection of phosphorylation by autoradiography.
Peptide mapping
Peptide mapping was performed as described previously33. Briefly, protein bands were cut from nitrocellulose membrane and digested with trypsin (sequencing grade, Promega) in NH4HCO3, pH7.8 with shaking at 37°C for 8 hr. The samples were dried and washed in a speed-vac, and then spotted onto cellulose plate for 2-D phosphopeptide mapping, using pH8.9 buffer for electrophoresis and phospho chromatography buffer for TLC.
GFP TIRF imaging
CHO-K1 cells stably expressing EGFP-talin, –talS425A or –talS425D were plated on MatTek dishes (with a glass coverslip at the bottom) precoated with fibronectin (5 µg/ml) and grown for 3 hr. TIRF images were taken using an Olympus IX81 microscope equipped with a ×60, 1.45NA objective, an auto-focusing system and a Sensicam camera (Cooke Corp.). The temperature was maintained at 37°C using a DH-35i dish culture incubator (Warner Instruments). Images were recorded at 1-min interval for a 60min period. Focal adhesion assembly and disassembly rate constants were calculated as described by Webb et al.34 Protrusion persistence and velocities were analyzed as described by Bear et al.35
Protein interaction
Cells were lysed with Lysis Buffer A, and the lysates were cleared and incubated with glutathione Sepharose beads loaded with GST fusion proteins at 4°C for 2hr. The beads were washed with the lysis buffer for three times and were resuspended in SDS sample buffer. Samples were analyzed by SDS-PAGE and transferred to nitrocellulose membrane to detect interacting proteins.
Protein ubiquitination
In vitro ubiquitination
Purified recombinant His-tagged TH (3 µg) was incubated in the absence or presence of Cdk5/p25 in 12 µl of a kinase buffer (25 mM Hepes, pH 7.5, 6 mM MgCl2, 3 mM MnCl2, 10 µM Na3VO4, 1 mM DTT, 200 µg/ml poly(ethylence glycol) 20,000) for 2 hrs. The mixtures were incubated with E1, UbcH7, GST-Smurf1 and Myc-Ub in 40 µl of the Ubiquitination buffer (50 mM Tris-HCl, pH7.4, 10 mM MgCl2, 10 µM DTT, 4 mM ATP) for 120 min. The reactions were terminated by adding 800 µl of 8M urea containing 100 mM NaH2PO4, 10 mM Tris, 0.1% Tween 20, pH 8.0. Additional 13 µg His-tagged TH was added to each reaction, and the TH was purified with Ni-NTA agarose chromatography, resolved by SDS-PAGE, and transferred to nitrocellulose membrane for detection of ubiquitination by anti-c-Myc immunoblotting.
In vivo ubiquitination
CHO-K1 cells were co-transfected with HA-ubiquitin and EGFP-talin mutants. After 19 hr, cells were treated with MG132 (40µM) for 4 hr and lysed with RIPA buffer containing protease inhibitor cocktail and 40 µM MG132. The lysates were immunoprecipitated with anti-GFP polyclonal antibodies. The immune complexes were analyzed by SDS-PAGE and transferred to nitrocellulose membrane for detection of ubiquitination by anti-HA immunoblotting.
Cell migration
Wound healing assays were performed as described previously33. Mitomycin C (2 µg/ml) was included to inhibit cell proliferation.
Transwell assays were performed using polycarbonate transwell filters (Corning, 8 µm pore size, 6.5 mm diameter) coated with 10 µg/ml of type I collagen and placed over a bottom chamber containing 20 ng/ml PDGF. The SH-SY5Y cells suspended in RPMI medium containing 0.1% bovine serum albumin were added to the upper chamber at a density of 5 × 104 cells/well. After 8 hours of incubation at 37 °C, the cells that had migrated to the lower side of the filter were collected by trypsinizing and counted by FACS.
Data quantification
Gel data were quantified by analyzing inverted images using ImageJ. Briefly, densities of protein bands (or areas) were measured and subtracted a mean background from 5 different locations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Keith Burridge for critical reading of this manuscript, Dr. Michael Schaller for critical reading of this manuscript and supporting of phosphorylation assays, Dr. Anna Huttenlocher for pEGFP-talin plasmid, Dr. Li-Huei Tsai for Cdk5 and p35 plasmids, Dr. Douglas Cyr for HA-ubiquitin plasmid, E1 and Ubc5α, and Michael Kerber for assisting in TIRF imaging. Supported by National Institutes of Health grant to M.H.G. and the Cell Migration Consortium grant (NIH GM64346) to to M.H.G. and K.J. and Ruth L. Kirschstein National Research Service Award (1F32 HL083215) to C.H..
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells [mdash] over and over and over again. Nature Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 2.Goksoy E, et al. Structural Basis for the Autoinhibition of Talin in Regulating Integrin Activation. Molecular Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco SJ, et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nature Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 4.Nuckolls GH, Romer LH, Burridge K. Microinjection of Antibodies against Talin Inhibits the Spreading and Migration of Fibroblasts. J. Cell Sci. 1992;102:753–762. doi: 10.1242/jcs.102.4.753. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nature Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H-R, et al. Regulation of Cell Polarity and Protrusion Formation by Targeting RhoA for Degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 7.Sahai E, Garcia-Medina R, Pouyssegur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J. Cell Biol. 2007;176:35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz JC, Tsai LH. A Jekyll and Hyde disease: roles for Cdk5 in brain development and disease. Curr. Opin. Neurobiol. 2004;14:390–394. doi: 10.1016/j.conb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Xie ZG, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- 10.Tsai LH. Regulation of neuronal migration by cdk5. FASEB J. 2001;15:A2–A2. [Google Scholar]
- 11.Strock CJ, et al. Cyclin-Dependent Kinase 5 Activity Controls Cell Motility and Metastatic Potential of Prostate Cancer Cells. Cancer Res. 2006;66:7509–7515. doi: 10.1158/0008-5472.CAN-05-3048. [DOI] [PubMed] [Google Scholar]
- 12.Nuckolls GH, Turner CE, Burridge K. Functional-Studies of the Domains of Talin. J. Cell Biol. 1990;110:1635–1644. doi: 10.1083/jcb.110.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood DA, et al. The talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 14.Di Paolo G, et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- 15.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I[gamma] phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 16.Chen HC, et al. Interaction of Focal Adhesion Kinase with Cytoskeletal Protein Talin. J. Biol. Chem. 1995;270:16995–16999. doi: 10.1074/jbc.270.28.16995. [DOI] [PubMed] [Google Scholar]
- 17.Borowsky ML, Hynes RO. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 1998;143:429–442. doi: 10.1083/jcb.143.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegener KL, et al. Structural Basis for the Interaction between the Cytoplasmic Domain of the Hyaluronate Receptor Layilin and the Talin F3 Subdomain. Journal of Molecular Biology. 2008;382:112–126. doi: 10.1016/j.jmb.2008.06.087. [DOI] [PubMed] [Google Scholar]
- 19.Critchley DR. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 2004;32:831–836. doi: 10.1042/BST0320831. [DOI] [PubMed] [Google Scholar]
- 20.Tadokoro S, et al. Talin Binding to Integrin {beta} Tails: A Final Common Step in Integrin Activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 21.Petrich BG, et al. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J. Exp. Med. 2007;204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han JW, et al. Reconstructing and deconstructing agonist-induced activation of integrin alpha IIb beta 3. Curr. Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Priddle H, et al. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol. 1998;142:1121–1133. doi: 10.1083/jcb.142.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C-elegans. J. Cell Sci. 2003;116:3871–3878. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- 25.Ratnikov B, et al. Talin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 2005;118:4921–4923. doi: 10.1242/jcs.02682. [DOI] [PubMed] [Google Scholar]
- 26.Harada T, Morooka T, Ogawa S, Nishida E. ERK induces p35, a neuron-specific activator of Cdk5, through induction of Egr1. Nat Cell Biol. 2001;3:453–459. doi: 10.1038/35074516. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Studzinski GP. Expression of the neuronal cyclin-dependent kinase 5 activator p35Nck5a in human monocytic cells is associated with differentiation. Blood. 2001;97:3763–3767. doi: 10.1182/blood.v97.12.3763. [DOI] [PubMed] [Google Scholar]
- 28.Lazaro J, et al. Cyclin dependent kinase 5, cdk5, is a positive regulator of myogenesis in mouse C2 cells. J Cell Sci. 1997;110:1251–1260. doi: 10.1242/jcs.110.10.1251. [DOI] [PubMed] [Google Scholar]
- 29.Negash S, Wang H-S, Gao C, Ledee D, Zelenka P. Cdk5 regulates cell-matrix and cell-cell adhesion in lens epithelial cells. J Cell Sci. 2002;115:2109–2117. doi: 10.1242/jcs.115.10.2109. [DOI] [PubMed] [Google Scholar]
- 30.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 31.Calderwood DA, et al. The phosphotyrosine binding-like domain of talin activates Integrins. J. Biol. Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 32.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 23:1972–1984. doi: 10.1038/sj.onc.1207436. 0000. [DOI] [PubMed] [Google Scholar]
- 33.Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- 34.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 35.Bear JE, et al. Antagonism between Ena/VASP Proteins and Actin Filament Capping Regulates Fibroblast Motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.