Abstract
Cytotoxic lymphocytes use the granule exocytosis pathway to kill pathogen-infected cells and tumor cells. Although many genes in this pathway have been extensively characterized (e.g., perforin, granzymes A and B), the role of granzyme C is less clear. We therefore developed a granzyme C-specific mAb and used flow cytometry to examine the expression of granzyme B and C in the lymphocyte compartments of wild-type and mutant GzmB−/− cre mice, which have a small deletion in the granzyme B gene. We detected granzyme B and C expression in CD4+ and CD8+ T cells activated with CD3/CD28 beads or MLRs. Stimulation of NK cells in vitro with IL-15 also induced expression of both granzymes. Granzyme C up-regulation was delayed relative to granzyme B in wild-type lymphocytes, whereas GzmB−/− cre cells expressed granzyme C earlier and more abundantly on a per-cell basis, suggesting that the deleted 350-bp region in the granzyme B gene is important for the regulation of both granzymes B and C. Quantitative RT-PCR revealed that granzyme C protein levels were regulated by mRNA abundance. In vivo, a population of wild-type CD8αα+ intraepithelial lymphocytes constitutively expressed granzyme B and GzmB−/− cre intraepithelial lymphocytes likewise expressed granzyme C. Using a model of a persistent murine CMV infection, we detected delayed expression of granzyme C in NK cells from infected hosts. Taken together, these findings suggest that granzyme C is activated with persistent antigenic stimulation, providing nonredundant backup protection for the host when granzyme B fails.
Cytotoxic T lymphocytes, such as activated CD8+ T cells and NK cells, use a variety of mechanisms to induce the death of virus-infected and tumor cells. These include secretion of proapoptotic cytokines (e.g., TNF-α and IFN-γ), engagement of cell death receptors (e.g., Fas), and granule exocytosis of perforin and granzymes (1–4). When a fully primed CTL recognizes a target cell that is to be killed, an immunological synapse is formed between the two cells. Cytotoxic granules contained within the lymphocytes polarize toward the synapse, fuse with the CTL plasma membrane, and then release their contents into the synaptic cleft (4). Perforin then facilitates the delivery of granzymes into the cytosol of the target cell, where they cleave a variety of substrates to initiate cell death (3).
The granzyme genes are organized into clusters located on three chromosomes (5). Granzymes A and K are clustered together on human chromosome 5 and mouse chromosome 13, respectively. Granzyme M is tightly linked to a cluster of highly related myeloid serine protease genes on human chromosome 19 and mouse chromosome 10. The human granzyme B gene cluster, found on chromosome 14, contains granzyme B and is followed downstream by granzyme H and cathepsin G (which is exclusively expressed in early myeloid cells). In the murine granzyme B gene cluster on chromosome 14, granzyme C, the closest murine homolog of human granzyme H, is also found directly downstream from granzyme B. Six additional granzymes (5′ F, L, N, G, D, and E 3′) are present between granzyme B and cathepsin G in the murine gene cluster, but the precise specificities and functions of these “orphan” granzymes are not yet known.
Granzyme B, the most thoroughly characterized of the granzymes, cleaves a variety of procaspases, BID, inhibitor of caspase-activated DNase, and other important intracellular substrates to initiate classical apoptotic pathways (3, 6–10). Granzyme C-induced cell death is accompanied by phosphatidylserine externalization, nuclear condensation, ssDNA nicking, and mitochondrial depolarization, but virtually nothing is known about the cellular substrates of this enzyme (11). Interestingly, it was recently shown that cell death induced by human granzyme H, an enzyme with chymotrypsin-like activity, similarly resulted in nuclear condensation and loss of mitochondrial membrane potential (12, 13). Given their high amino acid sequence similarity (61%), locations directly downstream from granzyme B, and similar functional characteristics, these data suggest that granzyme C is the functional murine counterpart to granzyme H. Finally, we have previously shown that the human granzyme H 5′ flanking region is regulated similarly to that of granzyme C in transgenic animals, again suggesting that these genes share regulatory homology (14).
Analysis of granzyme-deficient mouse strains has shown distinct roles for granzymes in antiviral and antitumor immune responses. Granzyme A-deficient mice have an increased susceptibility to ectromelia virus infections (15). Our group recently reported that granzyme B-deficient mice are particularly sensitive to challenge with murine CMV (MCMV)3 (16). We have also recently demonstrated that granzyme B-deficient mice are resistant to challenge with syngeneic and allogeneic tumors (17) due to granzyme B-dependent suppression of antitumor responses mediated by CD4+Foxp3+ regulatory T cells. Alternative, noncytotoxic activities have been proposed for granzyme A in mediating immune responses, but have not been described for granzymes B or C (18).
Although a granzyme C knockout mouse has not yet been generated, there is evidence that suggests that granzyme C plays a role in CTL-mediated cytotoxicity. We previously compared the cytotoxic function of two granzyme B-deficient strains, one in which expression of granzyme C and F were decreased due to a retained PGK-neo cassette and one in which expression of these down-stream genes was restored after removal of this cassette (19). CTLs from mice with diminished granzyme C and F expression had a corresponding reduction in cytotoxicity. More recently, Getachew et al. (20) used small interfering RNA-mediated knockdown of granzyme C in CTLs derived from MLRs to demonstrate a role for this gene during prolonged T cell responses.
The kinetics and patterns of granzyme expression in cytotoxic lymphocytes are dependent on the conditions under which these cells are activated. Kelso et al.(21) previously reported the differential expression of granzyme transcripts in polyclonally activated T cells. By performing nested RT-PCR on single CD8+ T cells that were sorted from in vitro-activated cultures at various time points, they found that up-regulation of granzyme B transcripts preceded the induction of granzyme A and C mRNAs. Furthermore, individual T cells had different patterns of expression of these genes. However, until now, there has been no thorough characterization of granzyme C protein expression at the single-cell level.
In this study, we report flow cytometric analyses of granzyme B and C protein expression in activated T and NK cells using a novel granzyme C-specific mAb. Our findings demonstrate the differential expression of granzymes in cytotoxic lymphocytes and shed insight into regulatory mechanisms that control the expression of these genes.
Materials and Methods
Mice
Wild-type (WT) 129/SvJ, C57BL/6, BALB/c, and Rag1−/− (B6) mice were obtained from The Jackson Laboratory. GzmB−/− cluster and GzmB−/− cre mice have been previously described and were derived in the 129/SvJ background (19, 22). All mice were maintained in specific pathogen-free housing and all experiments were conducted in accordance with institutional animal care and use guidelines.
Hamster mAb production
Armenian hamsters were immunized with purified recombinant granzyme C in CFA and boosted in IFA. Hamsters showing ELISA seropositivity for granzyme C were boosted and hybridomas were generated as previously described (23). Hybridoma supernatants were screened by flow cytometric analysis of fixed and permeabilized lymphokine-activated killer (LAK) cells. A PE-conjugated goat anti-Armenian hamster IgG secondary Ab (Jackson ImmunoResearch Laboratories) was used for detection. Multiple positive cell lines were identified. One of these, SFC1D8, was selected for further characterization and repeatedly cloned by limiting dilution. Hamster IgG was purified from SFC1D8 hybridoma supernatants by protein A affinity chromatography and subsequently labeled with Alexa Fluor 488 (Invitrogen Protein Labeling Kit). With the exception of the original screening of hybridoma clones, all subsequent staining was performed using this directly conjugated form of SFC1D8.
Abs and reagents
Abs used include anti-mouse NK1.1 (PK136), CD3 (145-2C11), CD4 (RM4-5), CD8 (53-6.7), CD16/32 (2.4G2; BD Biosciences), NKp46 (29A1.4; eBioscience), and granzyme B (GB12; Caltag Laboratories). Mouse CD3/CD28 Dynabeads were obtained from Invitrogen. Cytokines were obtained from R&D Systems (recombinant murine IL-15) or Chiron (recombinant human IL-2). All cytokines were endotoxin free and stored at −80°C after reconstitution in PBS plus 0.1% BSA.
Cell isolation and stimulation
LAK cell and MLR preparations were performed as previously described (22, 24). All cells were cultured in K10 medium (RPMI 1640, 10% FCS, 10 mM HEPES, 1% nonessential amino acids, 1% sodium pyruvate, 1% l-glutamine, 1 × penicillin/streptomycin, and 0.57 µM 2-ME). For CD3/CD28 activations, bulk splenocytes (2 × 106 cells/well) from WT or GzmB−/− cre mice were processed into single-cell suspensions and cultured in 6-well plates with Dynabeads at a 1:1 ratio. At indicated time points, cells were harvested for analysis. IL-15 stimulation of splenocytes was performed as previously described (16). CD8+ T cells were purified from the resting spleens of WT and GzmB−/− cre mice with a CD8+ T cell isolation kit, followed by cell separation on the AutoMACS according to the manufacturer’s instructions (Miltenyi Biotec). For purification of NK cells, splenocytes were surface stained with anti-NKp46 and anti-CD3, and NKp46+CD3− cells were isolated on a Reflection (iCyt) cell sorter (routinely ≥95% pure).
Preparation of intraepithelial cells (IELs)
For isolation of intestinal IELs, small and large intestines were harvested, Peyer’s patches were removed, and tissue was cut open longitudinally. The washed tissue was cut into 1-cm segments. Intraepithelial lymphocytes were released from associated epithelium with two consecutive washes of HBSS with shaking at 220 rpm for 20 min at 37°C. Lymphocytes were then passed through sterile 70-µm nylon mesh and analyzed.
Intracellular staining and flow cytometry
One × 106 cells were washed and resuspended in staining buffer (PBS, 0.5% BSA, and 0.5 mM EDTA). Samples were labeled with primary-conjugated Abs against cell surface markers, fixed, permeabilized (Foxp3 staining kit; eBioscience), and stained with primary-conjugated antigranzyme B Ab and anti-granzyme C Ab. Sample data were acquired on a Cytek-modified FACScan (BD Biosciences) flow cytometer and analyzed with FlowJo (Tree Star) software.
Quantitative real-time RT-PCR
Total RNA was isolated from resting and activated CD8+ T cells and NK cell samples (2.5 × 105) with the RNeasy Micro Kit (Qiagen). Quantitative RT-PCR (qRT-PCR) was performed as described for granzyme B and C (19). Comparisons were made by the comparative threshold cycle (Ct) method, with β-actin serving as the comparator. Data are presented as fold change relative to resting NK or T cell samples, which are set to a value of 1.
MCMV infection
A salivary gland stock of Smith strain MCMV was prepared from BALB/c mice that had been i.p. injected with tissue culture-propagated MCMV, and the titer was determined via standard plaque assay using permissive NIH3T12 fibroblasts (American Type Culture Collection) (25). Rag1−/− mice were injected i.p. with 2 × 104 PFU/mouse of a salivary gland MCMV stock. Twenty-one days after infection, splenocytes were harvested and analyzed.
Results
Characterization of granzyme expression in CD3/CD28 bead-activated CD4+ and CD8+ T cells
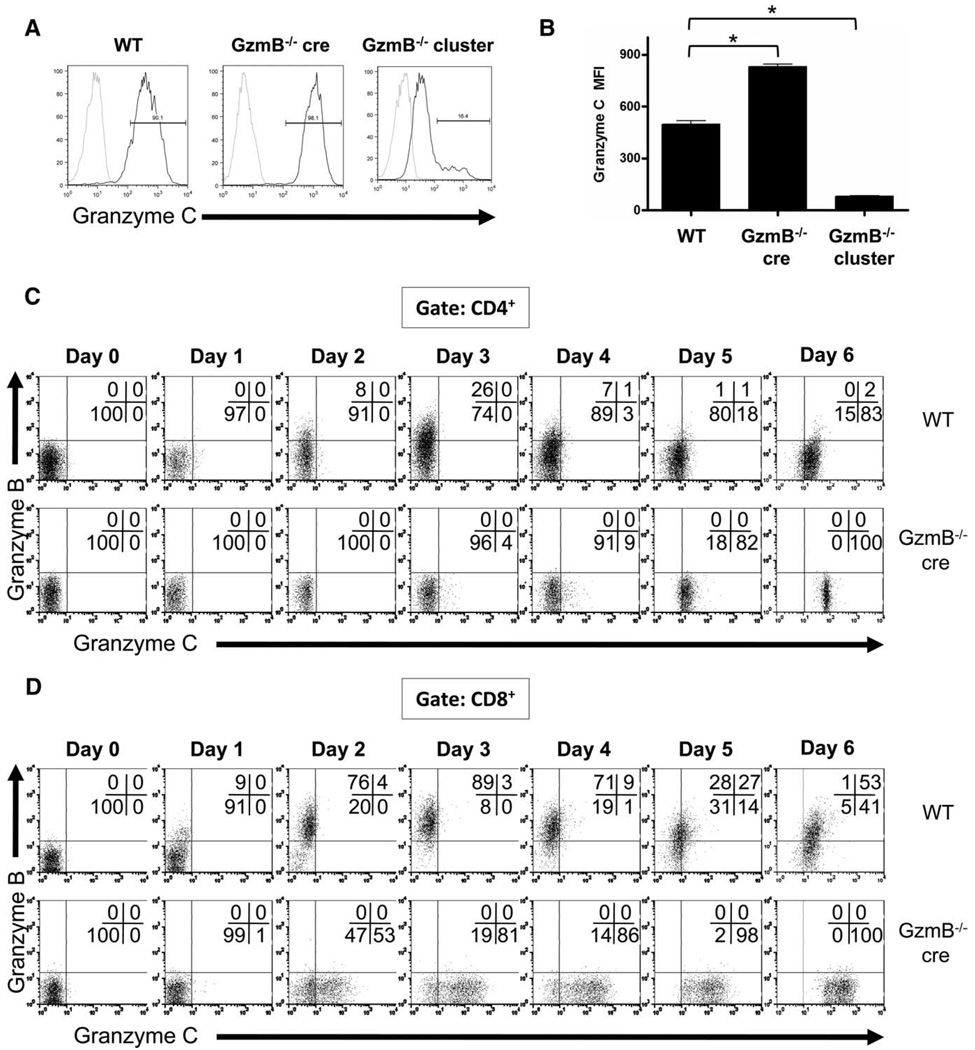
To define the expression of granzymes at the single-cell level in activated lymphocytes, we generated a granzyme C-specific mAb. Hybridoma clones were obtained after immunization of Armenian hamsters with purified recombinant granzyme C and CFA. Supernatants from these hybridoma clones were screened using flow cytometric analysis of LAK cells derived from 129/SvJ WT mice and two types of strain-matched granzyme B-deficient mice (granzyme B−/− cluster and granzyme B−/− cre mice). GzmB−/− cluster mice have a PGK-neo cassette retained within the granzyme B gene, which produces a neighborhood effect that reduces the expression of downstream granzymes, including granzyme C (19, 26). This PGK-neo cassette was removed by cre-mediated recombination in targeted embryonic stem cells, which were then used to create GzmB−/− cre mice. As a result, no neighborhood effect is observed in these mice. In fact, greater amounts of granzyme C mRNA and protein were detected in activated CTLs from GzmB−/− cre mice compared with their WT counterparts (19). Since no knockout mice for granzyme C currently exist, we used the differential expression of granzyme C in these mice to define the specificity of candidate hybridoma clones in a flow-based assay. Several reactive clones were identified. One of the clones stained GzmB−/− cre LAK cells with a higher mean fluorescence intensity (MFI) than WT cells and also stained GzmB−/− cluster LAK cells with a significantly lower MFI than WT cells (Fig. 1, A and B). This pattern of staining is consistent with that of a granzyme C-specific Ab. However, this Ab did not recognize either native or recombinant murine granzyme C with Western blotting of proteins separated using SDS-PAGE, suggesting that it recognizes a folded epitope (data not shown). This was confirmed by direct spotting of serially diluted, nondenatured recombinant murine granzymes A, B, and C followed by detection with the antigranzyme C mAb (19, 27, 28). Under conditions where the Ab could detect 12.5 ng of granzyme C, 500 ng of recombinant granzyme B and 1000 ng of granzyme A yielded no signal with this Ab. These data show that this Ab has minimal cross-reactivity with granzymes A and B, the most closely related granzyme family member by amino acid sequence similarity (66% identical) (supplemental Fig. 14). Granzyme F also has a similar expression pattern to granzyme C in LAK cells, which raises the possibility that this Ab may potentially cross-react with granzyme F in the flow-based assay (19). Because granzymes C and F share less sequence similarity (59.9% amino acid identity) than granzymes C and B (66.1% amino acid identity) and because there was minimal cross-reactivity between granzymes C and B in both the flow-based and dot blot assays, it is not likely that this Ab cross-reacts with granzyme F. In the absence of a granzyme C-deficient mouse, the flow cytometric analysis of LAK cells derived from mice with genetically defined amounts of granzyme C expression, coupled with dot blot analysis of available granzymes in recombinant form, strongly suggests that this reagent is granzyme C specific. Using this Ab, along with a commercially available granzymeB-specific Ab (GB12) (16), we characterized the expression of granzymes B and C in polyclonally activated T cells over a 6-day time course.
FIGURE 1.
Characterization of granzyme protein expression in activated lymphocytes using a novel granzyme C-specific mAb. LAK cells were generated by culturing splenocytes from WT, granzyme B−/− cre, or granzyme B−/− cluster-deficient mice in K10 medium supplemented with high-dose IL-2 (1000 U/ml). After 10 days of culture, LAK cells were harvested, fixed, permeabilized, and stained for intracellular granzyme C followed by staining with a PE-conjugated anti-hamster IgG secondary Ab. A secondary Ab alone condition (gray) was included as a negative control. Representative histograms are shown in A. A summary graph plotting MFI of granzyme C expression (mean ± SD) from three independent experiments is shown in B. *, p < 0.0001. WT and granzyme B−/− cre splenocytes were cultured in K10 medium with CD3/CD28 beads and harvested for flow cytometric analysis at various times during activation. T cell expression of granzymes B and C are shown. Results from an individual experiment are shown, gating on CD4+ T cells (C) and CD8+ T cells (D).
Unfractionated splenocytes from WT and GzmB−/− cre mice were cultured with CD3/CD28 beads and harvested at various times for flow cytometric analysis. Representative flow plots of granzyme B and C expression in CD4+ T cells are shown in Fig. 1C and a summary graph is shown in Fig. 2A. Although the percentage of granzyme B-expressing WT CD4+ T cells peaked on day 3 of activation at ~25%, the proportion of granzyme C-expressing CD4+ T cells continued to increase throughout the 6-day time course. Approximately 75% of all WT CD4+ T cells in culture had detectable amounts of granzyme C protein on day 6 of activation. Notably, the rise in granzyme C expression lagged behind the induction of granzyme B protein by ~2 days. As expected, no granzyme B was detectable at any time using GzmB−/− cre-derived splenocytes. However, a greater proportion of CD4+ T cells were granzyme C+ on days 5 and 6. By day 6, 100% of GzmB−/− cre CD4+ T cells expressed granzyme C. Analysis of Foxp3+ regulatory and Foxp3− effector CD4+ T cells showed that this activation protocol preferentially expanded effector T cells; almost all of the CD4+ T cells were Foxp3− at the end of the time course (data not shown). In addition, based on the fluorescence intensity of granzyme C staining, the amount of granzyme C protein in GzmB−/− cre CD4+ T cells on a per-cell basis was ~5-fold greater than in WT cells (Fig. 1C).
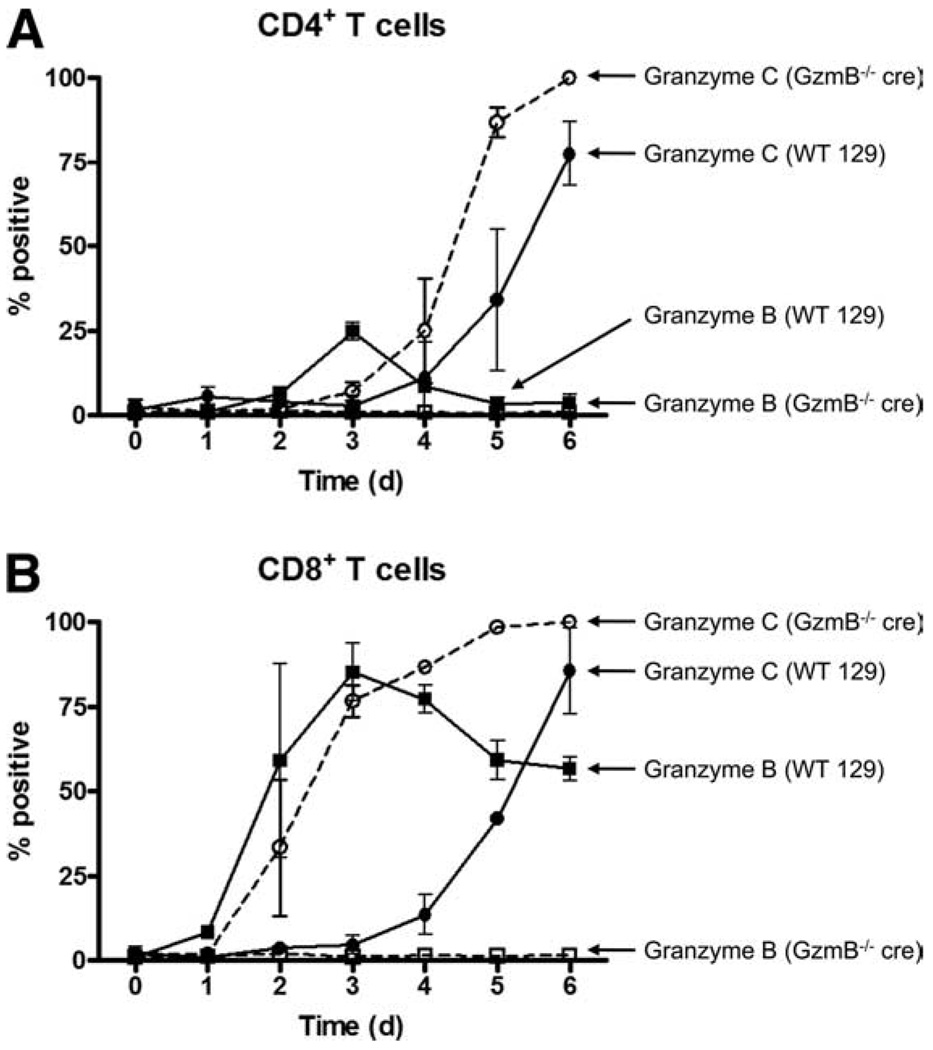
FIGURE 2.
Kinetics of granzyme B and granzyme C protein expression in CD3/CD28-activated T cells. WT and granzyme B−/− cre splenocytes were cultured in K10 medium with CD3/CD28 beads and harvested for flow cytometric analysis at various times during activation. The percentage of CD4+ T cells positive for granzymes B and C is summarized in A and the percentage of CD8+ T cells positive for granzymes B and C is summarized in B. Summary results (mean ± SD) represent pooled data from three independent experiments.
Representative flow plots of granzyme B and C expression in CD3/CD28 bead-activated CD8+ T cells is shown in Fig. 1D and a summary graph is shown in Fig. 2B. Like WT CD4+ T cells, the percentage of granzyme B-expressing WT CD8+ T cells peaked on day 3 of activation at ~80%, whereas the proportion of granzyme C-expressing CD8+ T cells began rising above baseline on day 4, reaching ~80% on day 6. For CD8+ T cells from GzmB−/− cre cultures, the proportion of granzyme C-expressing CD8+ T cells began rising on day 2 and continued to increase until all CD8+ T cells in culture were granzyme C+. Again, as was seen for CD4+ T cells, a higher granzyme C MFI (~5-fold on day 6) was observed in GzmB−/− cre CD8+ T cells.
Characterization of granzyme expression in CD4+ and CD8+ T cells activated during MLRs
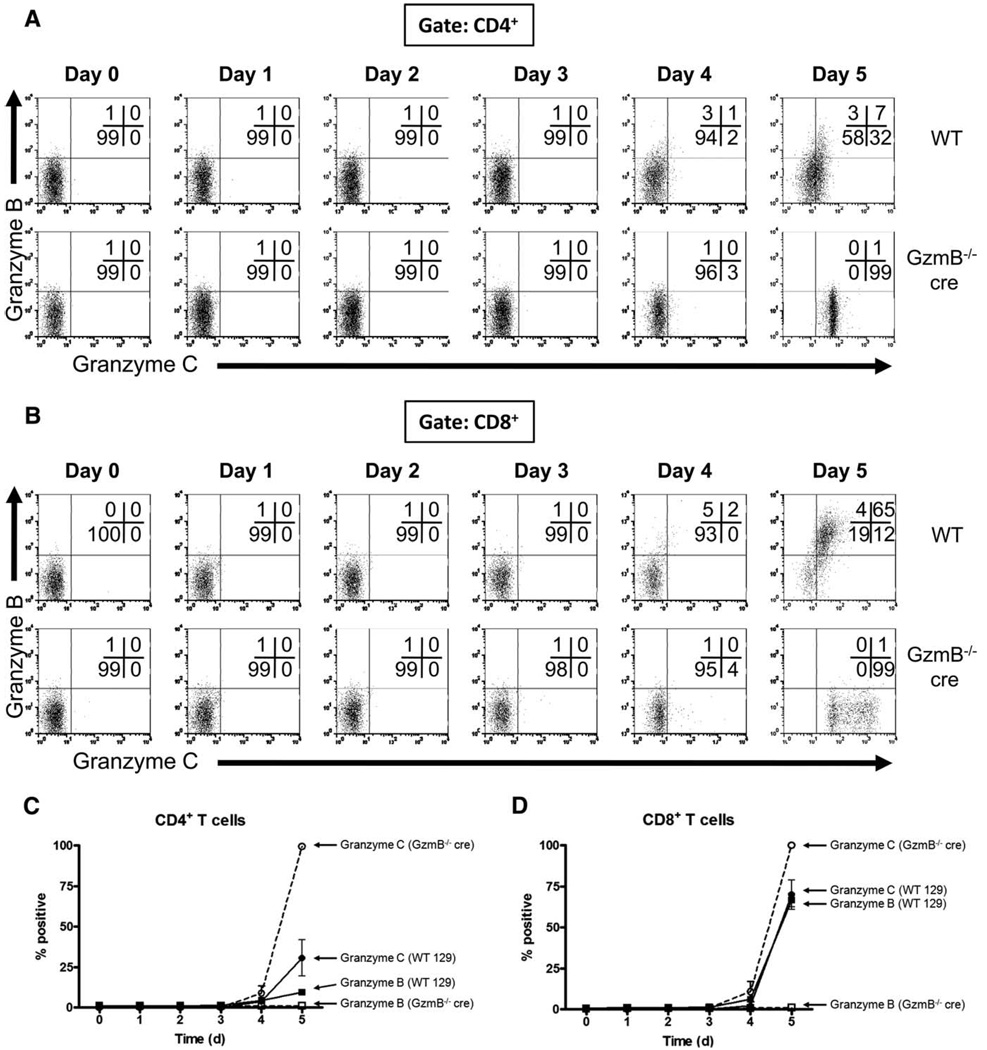
In these experiments, we used allogeneic mismatch as another stimulus for T cell activation. Our group had previously reported the expression of granzyme C by Western blot analysis of bulk populations harvested from MLRs (19). A flow-based assay using our mAb allows for a more detailed analysis of discrete T cell populations as a function of time. Splenocytes from 129/SvJ WT and GzmB−/− cre mice (H-2b) were cultured with irradiated splenocytes from BALB/c mice (H-2d) and harvested at various times for flow cytometric analysis. Representative flow plots gated on CD4+ T cells are shown in Fig. 3A and a summary graph is shown in Fig. 3C. No granzyme B or C was detectable during the first 3 days of culture in either WT or GzmB−/− cre MLRs. The proportion of granzyme B- and C-expressing CD4+ T cells began rising on day 4 and peaked on day 5 with ~10% granzyme B+ and ~30% granzyme C+ in WT cultures. All CD4+ T cells from GzmB−/− cre MLRs were granzyme C+ by day 5. Further analysis of CD4+ effector and regulatory subsets showed that granzyme C expression was restricted to the Foxp3− compartment (data not shown).
FIGURE 3.
Characterization of granzyme protein expression in T cells activated during MLRs. Splenocytes from WT or granzyme B−/− cre-deficient mice were cultured with irradiated BALB/c splenocytes (2000 cGy) in K10 medium supplemented with IL-2 (50 U/ml). MLR cultures were harvested for flow cytometric analysis at various times during activation. T cell expression of granzymes B and C are shown. Results from an individual experiment are shown, gating on CD4+ T cells (A) and CD8+ T cells (B). The percentage of CD4+ T cells positive for granzymes B and C is summarized in C and the percentage of CD8+ T cells positive for granzymes B and C is summarized in D. Summary results (mean ± SD) represent data pooled from three independent experiments.
Flow plots and summary graphs for MLR-activated CD8+ T cells are shown in Fig. 3, B and D, respectively. Similar to MLR-activated CD4+ T cells, neither granzyme B nor C was detected in CD8+ T cells during the first 3 days of culture, and there was only a slight increase in the percentage of WT CD8+ T cells that were a substantial increase in granzyme B+ on day 4. After 5 days of culture, however, there was a substantial increase in granzyme B and C double-positive cells, and there were few granzyme single-positive T cells. Analysis of MLR cultures in an extended time course showed that there was little change in the proportion of granzyme B- and granzyme C-expressing CD4+ and CD8+ T cells from day 5 through day 8 (supplemental Fig. 2). Interestingly, GzmB−/− cre CD4+ and CD8+ T cells had distinct patterns of granzyme C positivity. Although CD4+ T cells are homogeneously granzyme C positive, the population of granzyme C-positive CD8+ T cells is distributed over a 1-log range in fluorescence intensity, suggesting that there may be greater heterogeneity in CD8+ T cell subsets activated in culture relative to CD4+ T cells. Unlike CD3/CD28-activated T cells, no lag in granzyme C expression was observed relative to granzyme B, although expression of granzyme B and C protein during MLR was delayed and less robust. We attribute these differences to the strength of the activation signal, since allogeneic mismatch stimulates only a small proportion of the total T cells in culture, whereas CD3/CD28 activation stimulates virtually the entire T cell population (29). Granzyme C was maximally expressed in GzmB−/− cre CD8+ T cells on day 5 and little to no granzyme C was observed at earlier time points.
Differential granzyme B and C protein expression in WT and GzmB−/− cre CD8+ T cells is regulated by mRNA abundance
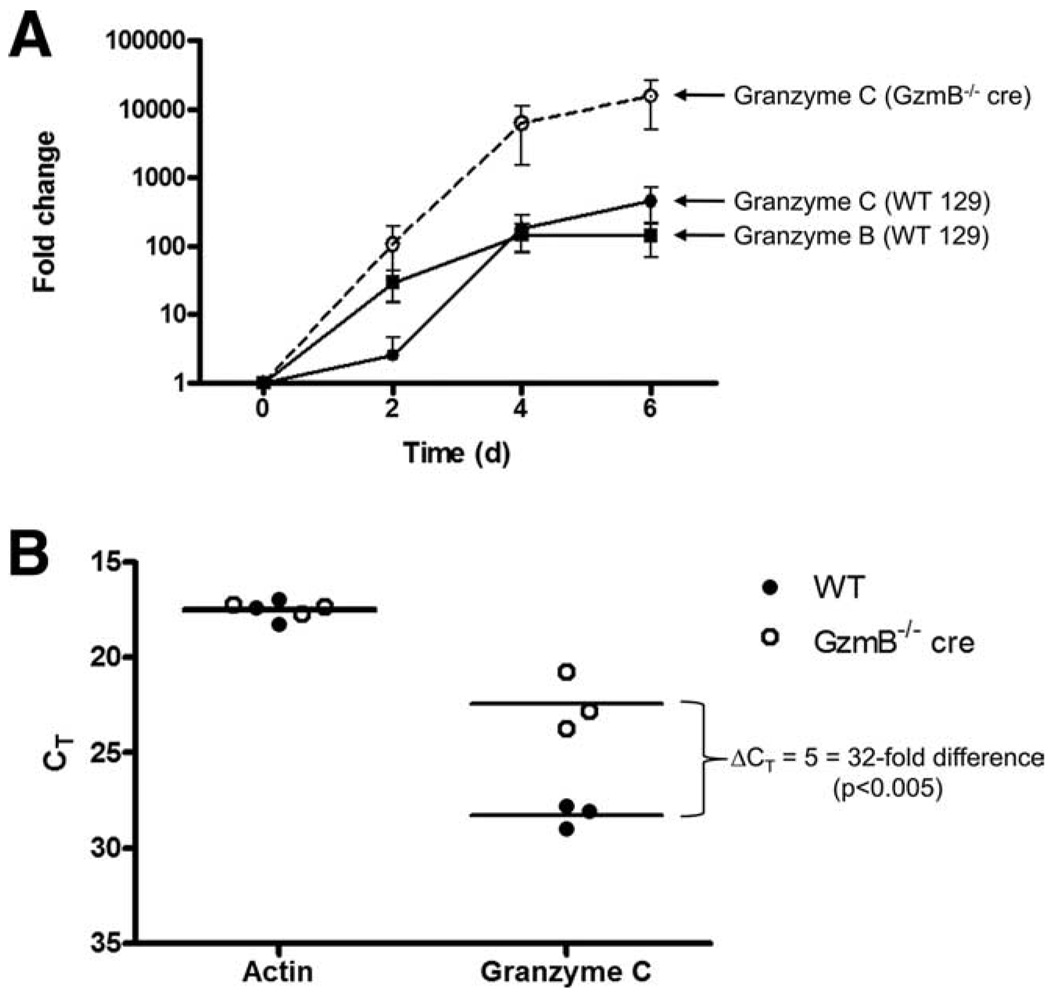
Our flow cytometric analysis of polyclonally activated T cells revealed that granzyme C lags behind the induction of granzyme B in WT T cells and that the induction of granzyme C in GzmB−/− cre T cells occurs earlier and is more abundant on a per-cell basis when compared with WT T cells. These data suggest that granzymes B and C may be regulated by distinct mechanisms during T cell activation and subsequent differentiation into a cytotoxic lymphocyte. To determine whether this process is regulated at the level of mRNA abundance, we performed quantitative real-time RT-PCR on CD3/CD28-activated CD8+ T cells obtained from WT or GzmB−/− cre mice (Fig. 4A). The abundance of granzyme B mRNA between days 0 and 2 in WT CD8+ T cells increased by at least a factor of 10, whereas the fold change for granzyme C was much less on day 2. By day 4, the fold changes for granzyme B and granzyme C mRNAs were comparable. In contrast, mRNA levels of granzyme C in GzmB−/− cre CD8+ T cells increased on day 2 and the fold changes from baseline on days 4 and 6 were higher than that of WT cells. Consistent with the flow data, there is a reproducible lag in the induction of granzyme C mRNA abundance compared with granzyme B in WT CD8+ T cells, and the fold increase in granzyme C mRNA abundance for GzmB−/− cre CD8+ T cells was significantly higher than that observed in WT T cells at all time points after activation. These data suggest that the differential expression of granzymes in CD3/CD28-activated T cells is regulated by mRNA abundance.
FIGURE 4.
Quantitative real-time RT-PCR comparing WT and granzyme B−/− cre T cell expression of granzyme C mRNA. CD8+ T cells were cultured in K10 medium with CD3/CD28 beads and RNA was harvested for qRT-PCR at various times during activation. The mean ± SEM expression from three independent experiments is shown as fold change of activated compared with naive T cells (A). The comparative Ct method was used and granzyme C expression was normalized to β-actin. Raw threshold cycles for actin and granzyme C mRNA on day 4 are plotted on an inverted scale in B.
To demonstrate that the difference in fold change observed on day 4 in granzyme C mRNA between WT and GzmB−/− cre CD8+ T cells is due to higher granzyme C mRNA abundance in GzmB−/− cre T cells (and not due to altered actin mRNA abundance, which was used to normalize expression levels), we plotted the raw threshold cycles detected in each biological replicate for actin and granzyme C in Fig. 4B. Although the threshold cycles for actin were indistinguishable for both genotypes, granzyme C mRNA detection in GzmB−/− cre CD8+ T cells preceded that of WT cells by five cycles (i.e., a 32-fold difference in the absolute abundance in granzyme C transcripts on day 4). Taken together, these data suggest that the granzyme B null mutation in GzmB−/− cre mice (caused by the removal of a 350-bp AvrII fragment that extends from base +70 from the transcription initiation site, and including the remainder of exon 1, and 283 bases of intron 1) perturbs the regulation of granzyme C expression.
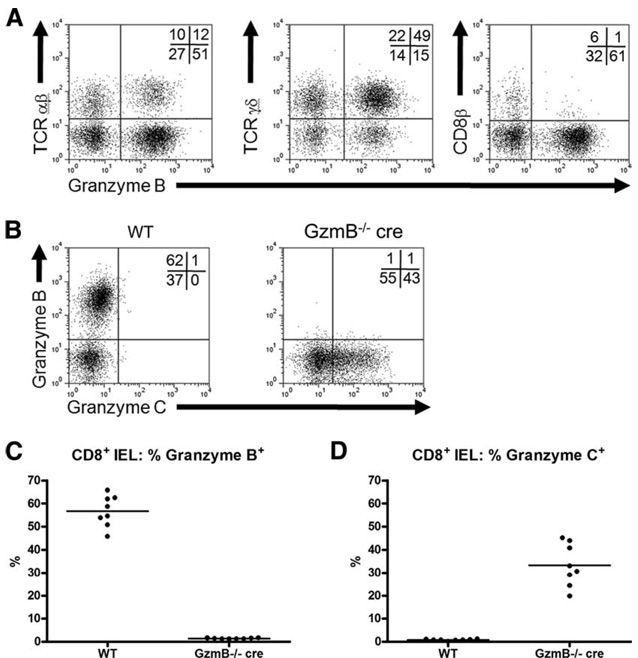
Granzyme C is constitutively expressed in CD8αα+ IELs of GzmB−/− cre mice
To further define the conditions under which granzymes B and C are expressed in vivo, we isolated IELs from WT and GzmB−/− cre mice. Within this lymphoid compartment, there are well-defined populations of thymus-dependent and thymus-independent T cells expressing the α/α homodimeric form of CD8 along with either αβ or γδ TCRs. Other groups have reported that these cells constitutively express perforin, granzyme A, and granzyme B mRNA (30, 31). These cells are able to kill target cells in redirected cytotoxicity assays as well as lymph node-derived CD4+ T cell blasts (32). We therefore analyzed these T cell subsets by flow cytometry to determine whether they express granzyme B and C protein. Immunophenotyping analysis gated on CD8α+ IELs revealed that the majority of the cells express γδ TCRs, while the rest express αβ TCRs. Almost all of the gated cells are CD8β−, thereby confirming that these cells express the CD8α/α homodimer (Fig. 5A). Within each of these two subsets, there are discrete populations of granzyme B+ and granzyme B− T cells, with the majority of the gated population expressing granzyme B (Fig. 5, B and C). Costaining experiments further showed that no granzyme C was detected in WT CD8αα+ T cells. However, ~30% of CD8αα+ IELs harvested from GzmB−/− cre mice were granzyme C+.
FIGURE 5.
Characterization of constitutive granzyme protein expression in CD8αα+ IELs. IELs were harvested from the intestines of WT and granzyme B−/− cre-deficient mice. Cells were stained and analyzed by flow cytometry for the expression of TCRαβ, TCRγδ, CD8β, and granzyme B. Representative flow plots, gated on CD8α+ IELs, are shown in A. Coexpression of granzymes B and C in WT and granzyme B−/− cre-deficient CD8α+ IELs is shown in B. The percentage of CD8αα− IELs positive for granzymes B and C is summarized in C and D, respectively. Summary results represent data from eight mice per genotype.
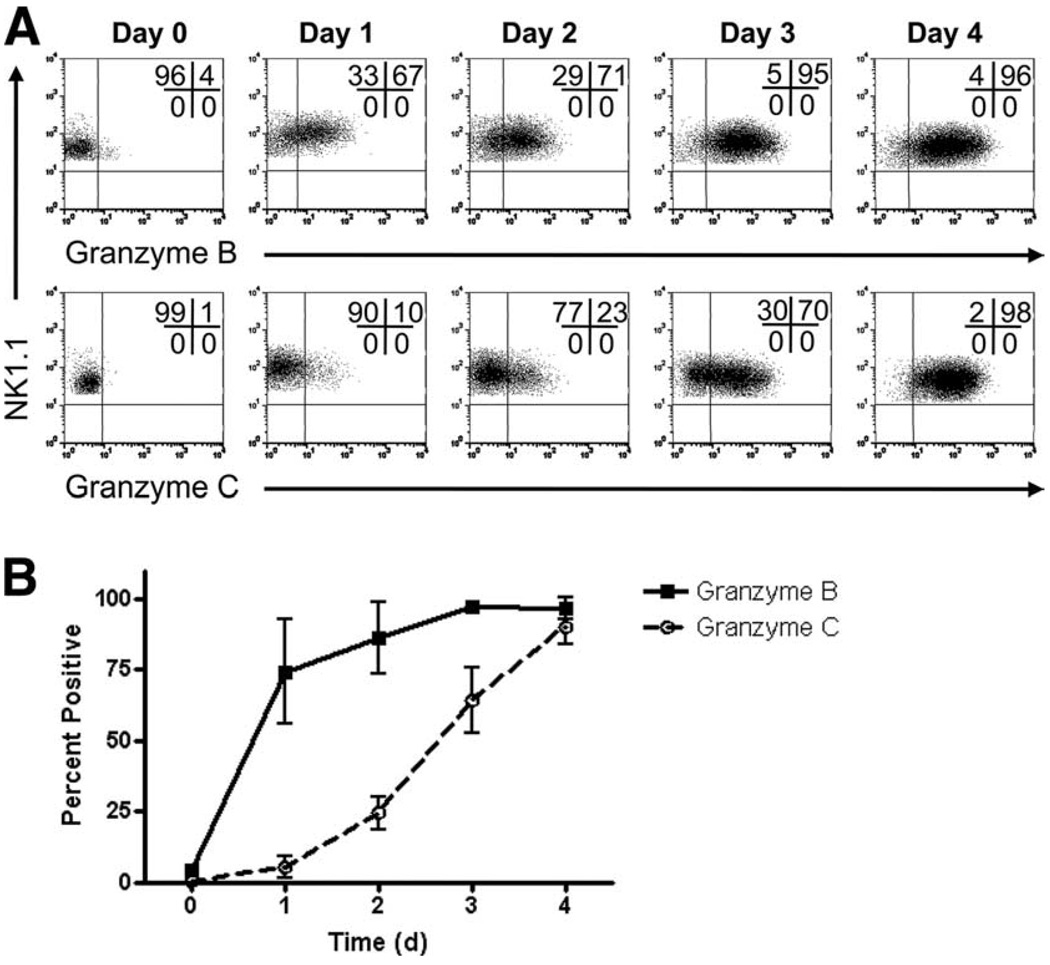
Characterization of granzyme C expression in IL-15-stimulated NK cells
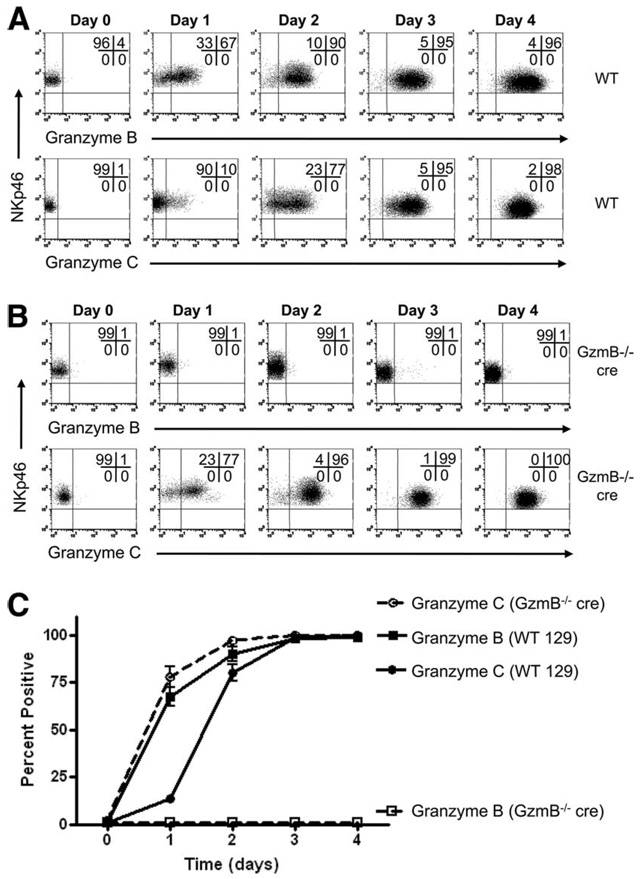
Our group previously demonstrated that resting murine NK cells express abundant amounts of granzyme A protein but not granzyme B or perforin, although transcripts for granzyme A, granzyme B, and perforin were all detected in resting NK cells (16). Granzyme B and perforin protein were induced upon cytokine stimulation, with IL-15 having the most potent effect. To define the kinetics of granzyme C expression in NK cells under these conditions, bulk spleen cells were stimulated with IL-15 over a 4-day time course. Splenocytes from WT B6 mice were cultured and harvested daily for flow cytometric analysis of granzyme B and C expression gated on NK1.1+CD3− NK cells (Fig. 6A). A summary graph is shown in Fig. 6B. Consistent with our previously published observations, there was little to no granzyme B protein expression in resting NK cells, but after 1 day of culture in the presence of IL-15, the majority of NK cells were granzyme B+. The proportion of granzyme B+ NK cells continued to rise, peaking on days 3 and 4 at ~95%. Similar to T cells, NK cell expression of granzyme C lagged behind granzyme B. Whereas most NK cells were granzyme B+ after 1 day of culture, the majority did not become granzyme C+ until day 3. By day 4, almost all NK cells expressed both granzymes B and C. Similar kinetics were also observed in NKp46+CD3− NK cells from WT 129/SvJ mice (Fig. 7A). However, granzyme C up-regulation was not delayed in GzmB−/− cre NK cells (Fig. 7B). Notably, the kinetics of granzyme C expression in GzmB−/− cre NK cells was nearly identical to that of granzyme B expression in WT NK cells (Fig. 7C).
FIGURE 6.
Granzyme C protein is expressed later than granzyme B protein in IL-15-activated NK cells from WT C57BL/6 mice. Splenocytes were isolated from WT C57BL/6 mice, stained, and analyzed by flow cytometry for the expression of intracellular granzyme B and C at rest (day 0) or after 1, 2, 3, or 4 days of activation with recombinant murine IL-15 (100 ng/ml). Representative flow plots (gated on NK1.1+CD3− NK cells) demonstrating the expression of granzyme B (upper panels) and granzyme C (lower panels) in NK cells is shown in A. A summary of the percentage of granzyme Band C-positive NK cells from three independent experiments (mean ± SD) is shown in B, demonstrating that granzyme C protein is expressed after granzyme B following IL-15 activation.
FIGURE 7.
NK cell granzyme C protein activation in GzmB−/− cre mice is accelerated. Splenocytes were isolated from WT 129/SvJ or GzmB−/− cre 129/SvJ mice, stained, and analyzed by flow cytometry for the expression of intracellular granzymes B and C at rest (day 0) or after 1, 2, 3, or 4 days of activation with recombinant murine IL-15 (100 ng/ml). Representative flow plots (gated on NKp46+CD3− NK cells) demonstrate the expression of granzyme B (upper panels) and granzyme C (lower panels) in WT (A) or GzmB−/− cre (B) 129/SvJ mice. Notably, in GzmB−/− cre mice, granzyme C protein is activated in a similar fashion as granzyme B in WT mice. The percentage (mean ± SD) of granzyme B- and C-positive NK cells from WT and GzmB−/− cre mice is summarized in C. These results are from three independent experiments.
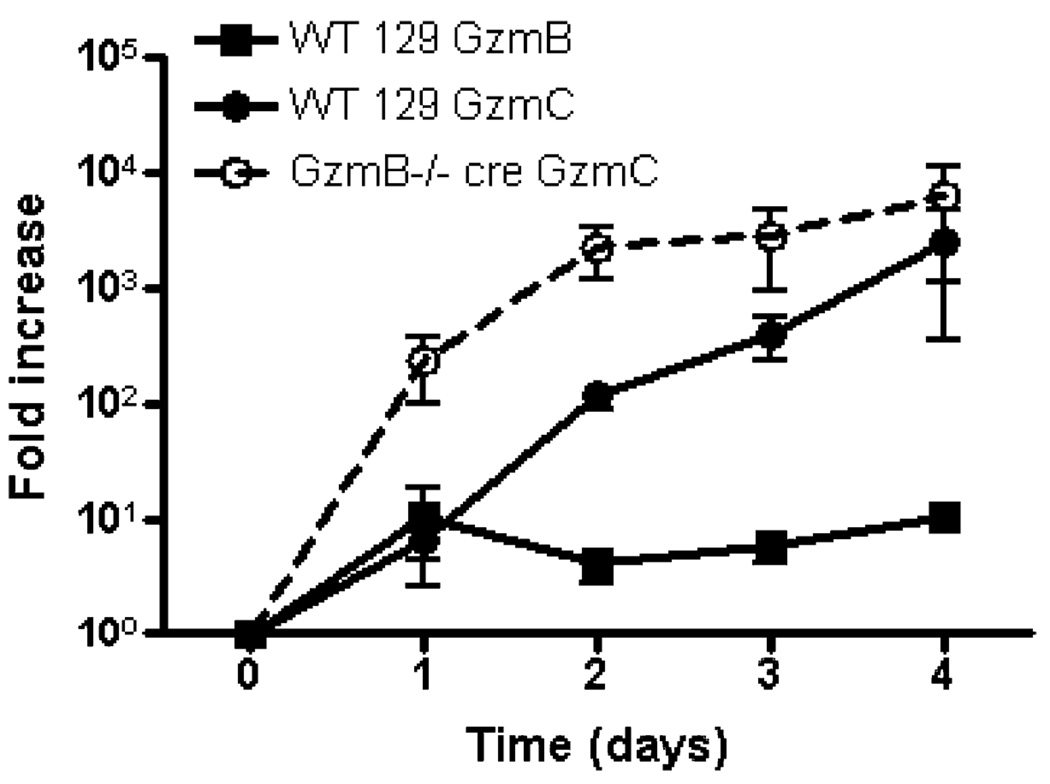
Granzyme C protein expression in WT and GzmB−/− cre NK cells is regulated by mRNA abundance
Next, we used quantitative real-time RT-PCR to examine the expression of granzyme B and C mRNAs in flow-sorted NK cells during a time course of IL-15 activation (Fig. 8). Consistent with our published findings, there is a modest increase in granzyme B mRNA after 1 day of IL-15 stimulation (16). WT NK cells had a similar increase in granzyme C mRNA abundance on day 1. Although granzyme C mRNA levels continued to rise throughout the 3-day time course, granzyme B mRNA abundance stabilized after 1 day. For GzmB−/− cre NK cells, there was a greater fold increase in granzyme C mRNA at each time point after IL-15 activation. Microarray analyses revealed that no granzyme C transcripts were detectable in resting NK cells, which stands in distinct contrast to that of granzyme B and perforin, which have abundant mRNA in resting NK cells, but no protein expression (16). After NK cell activation with IL-15, granzyme B and perforin mRNAs are rapidly translated, arming the cells to kill their targets. Taken together, these data suggest that the regulation of granzyme C in activated NK cells is fundamentally different from that of granzyme B or perforin.
FIGURE 8.
Comparison of IL-15-induced WT vs GzmB−/− cre NK cell granzyme B and C mRNA levels. Flow-sorted NK cells (≥95% NKp46+CD3−) were cultured in K10 medium with recombinant murine IL-15 (100 ng/ml), and total RNA was harvested for qRT-PCR at various times during activation. The data shown are fold change (mean ± SEM) of granzyme B and granzyme C mRNA in IL-15-activated NK cells, compared with fresh (day 0) NK cells, in three independent experiments. The comparative Ct method was used and granzyme B and C mRNA abundance was normalized to β-actin.
Persistent viral infection induces NK cell expression of granzyme C in vivo
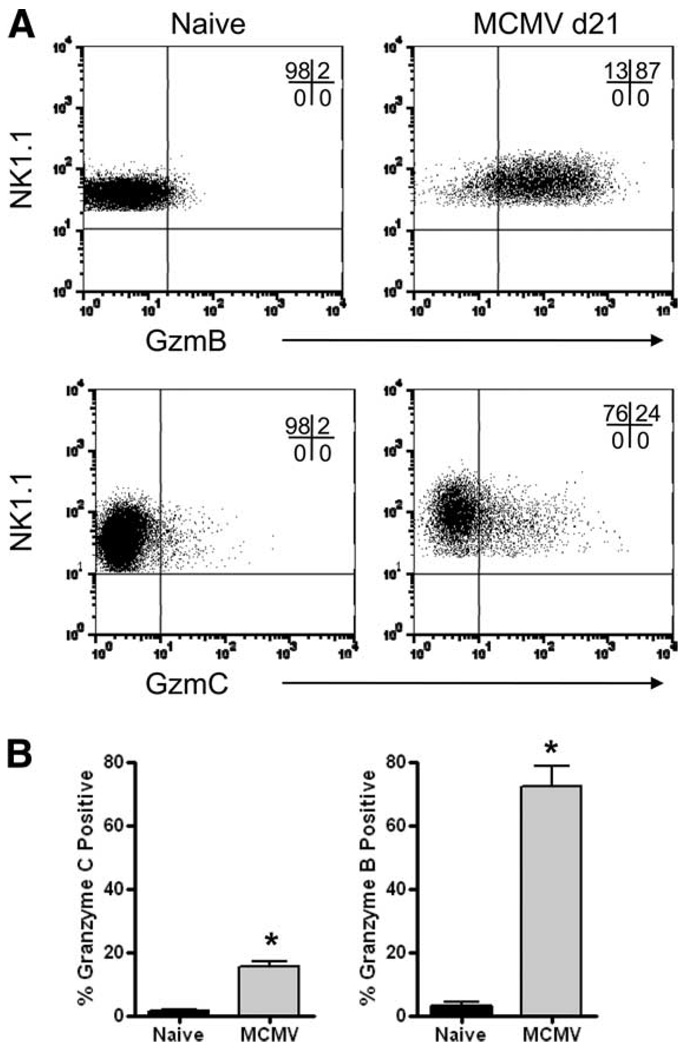
To determine whether NK cells express granzyme C in vivo during a viral infection, we challenged WT B6 mice with a sublethal dose (5 × 104 PFU) of Smith strain MCMV and analyzed the spleens of infected mice by flow cytometry. In this well-characterized mouse model of an acute infection, there is an early NK cell-dependent phase of viral clearance. We previously reported that granzyme B and perforin are rapidly up-regulated in NK cells during the first 8 days following infection (16). However, we were unable to detect granzyme C during this time frame (data not shown). Because granzyme C expression is relatively delayed and has been postulated to serve as a fail-safe mechanism in killing target cells, we hypothesized that a sublethal dose of MCMV that is rapidly cleared may not provide sufficient time and/or activation signals for the induction of granzyme C in NK cells. To address this hypothesis, we infected immunodeficient B6 Rag1−/− mice with a lower dose (2 × 104 PFU) of MCMV. In this model, NK cells were able to mount a response to the viral challenge initially, but weeks later, the infected mice died due to consequences of uncontrolled MCMV replication (33, 34). The selection pressure exerted by Ly49H+ NK cells in the absence of adaptive immunity generated escape mutants that ultimately resulted in the death of these immunodeficient hosts. We hypothesized that a viral infection that persists beyond the initial NK cell-dependent phase of clearance may induce the expression of granzyme C in NK cells. Splenocytes from B6 Rag1−/− mice were harvested on day 21 after infection and analyzed for granzyme B and C expression in NK1.1+CD3− NK cells. Representative flow plots are shown in Fig. 9A and summary graphs are depicted in Fig. 9B. As expected, NK cells from uninfected mice had little or no detectable granzyme B or C. Approximately 70% of NK cells from infected hosts were granzyme B+ and a smaller proportion (~15%) expressed granzyme C. Thus, although immunocompetent hosts challenged with a sublethal dose of MCMV failed to induce NK cell expression of granzyme C, we did detect granzyme C expression in NK cells from immunodeficient Rag1−/− mice that rely on innate NK cells for transient control of a persistent and ultimately lethal MCMV infection.
FIGURE 9.
NK cells express granzyme C protein in vivo following persistent MCMV infection. Rag1−/− C57BL/6 mice were infected with MCMV (2 × 104 PFU i.p.) and splenic NK cells were evaluated for granzyme B and C protein expression at day 21 after infection. Representative flow plots from control littermates and day 21 postinfection Rag1−/− mice, gated on NK1.1+ cells, are shown in A. In the top panel, a PerCP-Cy5.5-NK1.1- conjugated Ab was used, and in the lower panel, an allophycocyanin-NK1.1 conjugate was used. The percentage of NK cells positive for granzyme B and C (mean ± SD) is summarized from three independent experiments in B. In total, six uninfected mice and nine MCMV-infected mice were analyzed. *, p < 0.05.
Discussion
In this report, we developed a novel mAb to characterize the expression of granzyme C in cytotoxic lymphocytes at singlecell resolution. Allogeneic mismatch and coculture with CD3/CD28 beads induces granzyme C expression in both CD4+ and CD8+ T cells. Activation of NK cells with IL-15, a cytokine previously shown to induce potent perforin-dependent cytotoxicity in NK cells, induces granzyme C mRNA and protein expression. Granzyme C activation was delayed relative to granzyme B in WT T and NK cells, while granzyme C was expressed earlier and was more abundant on a per-cell basis in GzmB−/− cre CTLs. The expression of granzyme C protein was regulated at the level of mRNA abundance in CD3/CD28-activated T cells and in IL-15-activated NK cells. In addition to in vitro-activated lymphocytes, we also detected granzyme C expression in vivo in CD8αα+ intestinal IELs harvested from GzmB−/− cre mice as well as in NK cells from immunodeficient mice that had been challenged with a persistent and lethal viral infection.
The delayed expression of granzyme C protein relative to granzyme B is in concordance with previous studies that measured mRNA abundance of various components of the perforin/granzyme pathway during T cell activation. Kelso et al. (21) activated purified naive CD8+ T cells with immobilized Abs to CD3, CD8, and CD11a and used single-cell PCR to demonstrate that up-regulation of granzyme B mRNA preceded granzyme C by 1 day. The fact that this delay in granzyme C expression was observed across multiple lymphocyte subsets and activation methods suggests that the granzyme genes are subject to distinct regulatory mechanisms. Furthermore, these data also support the notion that orphan granzymes serve as fail-safe mechanisms for the induction of target cell death.
Recently, Thiele and colleagues (20) provided functional data in support of this hypothesis. By comparing CTLs from WT mice and mice deficient for dipeptidyl peptidase I (DPPI, a protease that is required for the functional activation of granzymes A and B, but not granzyme C), they showed that DPPI−/− CTLs that had been primed over a period of 5 days exhibited reduced cytotoxicity. However, after restimulating the cells for an additional 3 days, cytotoxic function in DPPI−/− CTLs had returned to WT levels. The restoration of cytotoxicity in DPPI−/− CTLs correlated with late granzyme C up-regulation. Furthermore, knockdown of granzyme C during restimulation decreased the cytotoxic activity of DPPI−/− (but not WT) CTLs. Together, these data suggest that granzyme C can maintain cytotoxic activity during late T cell responses in the absence of granzymes A and B.
These observations complement our expression studies of NK cells activated during MCMV infection. Under conditions where granzyme B and perforin are rapidly induced following challenge of WT mice with a sublethal dose of MCMV, no granzyme C was detected. However, when mice that lack an adaptive immune system were challenged with a dose that allowed the virus to persist and eventually kill the host, a population of granzyme C-expressing NK cells arose relatively late during the course of the infection. These findings suggest that the persistence of the virus in the host may provide sufficient time for the up-regulation of granzyme C in NK cells. Alternatively, additional signals generated during the prolonged course of the infection may be required for granzyme C to be expressed.
The GzmB−/− cre mice were made after our group discovered that activated CTLs from GzmB−/− cluster mice also had diminished expression of granzymes C and F. The null mutation in GzmB−/− cluster mice was originally generated by replacing a 350-bp AvrII fragment (containing most of exon 1, including the start codon, and 283 bp of intron 1) with a PGK promoter-driven neomycin phosphotransferase cassette. Pham et al. (26) reported that activation of GzmB−/− cluster CTLs resulted in the robust induction of PGK-neo mRNA at the expense of the expression of downstream granzymes, suggesting that PGK-neo had been captured by regulatory elements in the granzyme B gene cluster (26). We postulated that the retained PGK-neo cassette exerted a “neighborhood effect” on the downstream granzyme genes in the cluster.
Consistent with that hypothesis, we found that deletion of a loxp-flanked PGK-neo cassette targeted to the same location resulted in the earlier, more abundant expression of granzyme C mRNA and protein relative to WT CTLs. Additionally, although CD8αα+ IELs from WT mice expressed granzyme B but not granzyme C, GzmB−/− cre IELs had constitutive expression of granzyme C in vivo. In the case of CD3/CD28 activation, we found that up-regulation of granzyme C in GzmB−/− cre CD8+ T cells occurred 2 days earlier than WT CD8+ T cells. Interestingly, the kinetics of granzyme C expression in GzmB−/− cre CTLs was identical to that of granzyme B expression in WT CTLs (true for both CD3/CD28-mediated T cell activation and IL-15-stimulated NK cell activation). Since the only difference between WT and GzmB−/− cre mice is the presence or absence of the 350-bp AvrII fragment (and the retained loxp site), we hypothesize that deletion of this fragment causes a recently defined locus control region (LCR) to “skip” granzyme B and proceed to activate the next gene downstream, granzyme C; this gene must contain regulatory elements that permit the LCR to activate it (35). Although we have demonstrated that levels of granzyme C protein expression are regulated by mRNA abundance, it remains unclear whether regulation of transcriptional activity or mRNA half-life account for the differential expression of granzyme C mRNA in WT and GzmB−/− cre lymphocytes. Alternatively, GzmB−/− cre mice may generate signals that activate additional granzymes as a fail-safe mechanism when these animals fail to clear viruses or tumors, or granzyme B mRNA or protein may repress granzyme C expression, a process that could be overcome during persistent stimulation.
Bleackley and colleagues (35) have recently developed strong evidence for a LCR that regulates gene expression in the granzyme B gene cluster. These investigators identified a DNA element located upstream from granzyme B that is involved in the control of its transcription (35). In their studies, a DNase I-hypersensitive site (HS2) was detected 3.9 kb upstream from the transcriptional start site of granzyme B in activated T cells. When this DNA element was included in transgenes that were transferred to a CTL line, HS2 conferred position-independent expression of granzyme B. Transgenic mice expressing a tagged version of granzyme B (to distinguish transgenic and endogenous granzyme B) with or without HS2 were also made. Splenocytes from transgenic mice containing HS2 resulted in a 10-fold greater amount of transgene mRNA when compared with transgenic mice lacking this element. Since this element confers position independence of granzyme B expression in vivo, these features are most consistent with that of a LCR.
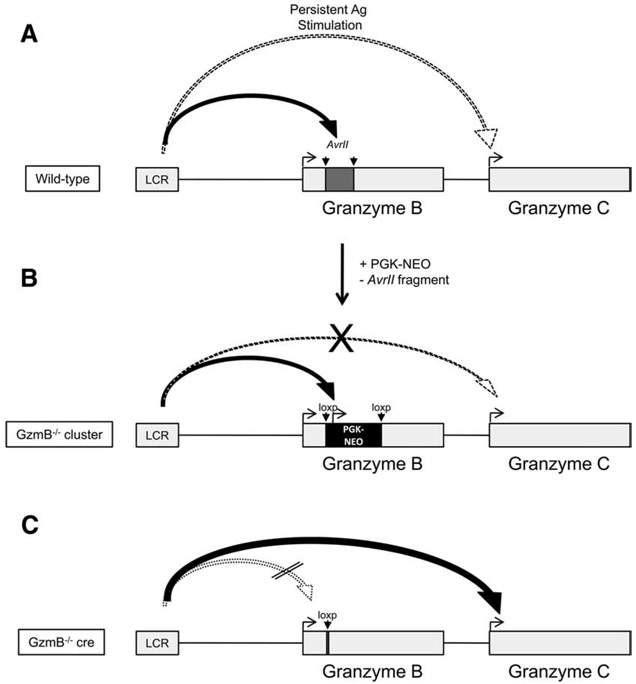
The interpretation of the findings described here is informed by many studies of the β-globin cluster LCR elements. Similar to the β-globin genes, the granzyme genes are structurally organized in clusters and expression of genes within these clusters are temporally regulated (36). In WT CTLs, we have observed the delayed expression of granzyme C relative to granzyme B in both T and NK cells (Fig. 10). The two granzyme B-deficient mouse strains that were developed in our laboratory have provided some clues into how this differential expression occurs. The retained PGK-neo cassette in GzmB−/− cluster mice is thought to function as a transcriptional “sink” within that locus, thereby diminishing the transcriptional activity of downstream granzymes C and F. When the PGK-neo cassette is deleted in GzmB−/− cre mice, the interaction between the 5′ LCR and the PGK promoter is presumably lost, causing the LCR to scan downstream to the next gene, granzyme C. Knockout mouse models and analysis of regulatory components upstream of these genes will be required to confirm these hypotheses.
FIGURE 10.
Model of granzyme B and C regulation in WT, GzmB−/− cre, and GzmB−/− cluster-derived cytotoxic lymphocytes. In WT cytotoxic lymphocytes (A), the LCR first interacts with regulatory elements within or near the AvrII fragment of granzyme B to induce transcription. Granzyme C is induced only after prolonged activation. GzmB−/− cluster-derived cytotoxic lymphocytes (B) have the normal AvrII fragment replaced with a PGK-neo cassette and fail to express granzyme B due to disruption of the gene. In addition, granzyme C expression is diminished due to the neighborhood effect created by the retained PGK-neo cassette. In GzmB−/− cre CTLs (C), granzyme C is expressed earlier and more abundantly (similarly to granzyme B in WT cells), because missing regulatory elements in granzyme B cause the LCR to “scan” downstream and activate the next gene in the cluster. Removal of the PGK-neo cassette eliminates the neighborhood effect.
Granzyme C and its human ortholog, granzyme H, can potently induce cell death via mechanisms that are distinct from that of granzyme B (11, 12). The diversity of death pathways targeted by granzymes allows the host to respond to a variety of immunological challenges. In the setting of granzyme B deficiency or inhibition, as has been demonstrated during certain viral infections, persistent antigenic stimulation may cause the delayed activation of downstream granzymes that act via novel mechanisms to remove the threat (37). The gene regulatory mechanisms controlling this late switch in granzyme gene expression were serendipitously uncovered by deleting the AvrII fragment in the granzyme B gene. These findings provide an important clue for the location of the critical elements and will be the subject of future studies.
Supplementary Material
Acknowledgments
We thank the Siteman Cancer Center High Speed Cell Sorter Core and the WUSM Hybridoma Center for invaluable support. Mieke Hoock provided expert animal husbandry. We also thank Dan George, Maggie Young, Nicole Grieselhuber, and Andy Bredemeyer for helpful advice and technical assistance.
Footnotes
This work was supported by grants from the National Institutes of Health (DK49786 to T.J.L. and AI059083 to A.R.F.). The granzyme C mAb was developed in part with the Washington University School of Medicine Hybridoma Center (supported by the Department of Pathology and Immunology and National Institutes of Health Grant P30 AR048335).
Abbreviations used in this paper: MCMV, murine CMV; WT, wild type; LAK, lymphokine-activated killer; IEL, intraepithelial lymphocyte; qRT-PCR, quantitative RT-PCR; Ct, threshold cycle; MFI, mean fluorescence intensity; DPPI, dipeptidyl peptidase I; LCR, locus control region.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat. Rev. Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 3.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 4.Stinchcombe JC, Griffiths GM. Secretory mechanisms in cell-mediated cytotoxicity. Annu. Rev. Cell Dev. Biol. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- 5.Grossman WJ, Revell PA, Lu ZH, Johnson H, Bredemeyer AJ, Ley TJ. The orphan granzymes of humans and mice. Curr. Opin. Immunol. 2003;15:544–552. doi: 10.1016/s0952-7915(03)00099-2. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DA, Du C, Xu M, Wang X, Ley TJ. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 7.Barry M, Heibein JA, Pinkoski MJ, Lee SF, Moyer RW, Green DR, Bleackley RC. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heibein JA, Goping IS, Barry M, Pinkoski MJ, Shore GC, Green DR, Bleackley RC. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members bid and Bax. J. Exp. Med. 2000;192:1391–1402. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton VR, Davis JE, Cancilla M, Johnstone RW, Ruefli AA, Sedelies K, Browne KA, Trapani JA. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 2000;192:1403–1414. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas DA, Scorrano L, Putcha GV, Korsmeyer SJ, Ley TJ. Granzyme B can cause mitochondrial depolarization and cell death in the absence of BID, BAX, and BAK. Proc. Natl. Acad. Sci. USA. 2001;98:14985–14990. doi: 10.1073/pnas.261581498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson H, Scorrano L, Korsmeyer SJ, Ley TJ. Cell death induced by granzyme. C. Blood. 2003;101:3093–3101. doi: 10.1182/blood-2002-08-2485. [DOI] [PubMed] [Google Scholar]
- 12.Fellows E, Gil-Parrado S, Jenne DE, Kurschus FC. Natural killer cell-derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood. 2007;110:544–552. doi: 10.1182/blood-2006-10-051649. [DOI] [PubMed] [Google Scholar]
- 13.Edwards KM, Kam CM, Powers JC, Trapani JA. The human cytotoxic T cell granule serine protease granzyme H has chymotrypsin-like (chymase) activity and is taken up into cytoplasmic vesicles reminiscent of granzyme B-containing endosomes. J. Biol. Chem. 1999;274:30468–30473. doi: 10.1074/jbc.274.43.30468. [DOI] [PubMed] [Google Scholar]
- 14.MacIvor DM, Pham CT, Ley TJ. The 5′ flanking region of the human granzyme H gene directs expression to T/natural killer cell progenitors and lymphokine-activated killer cells in transgenic mice. Blood. 1999;93:963–973. [PubMed] [Google Scholar]
- 15.Mullbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, Museteanu C, Simon MM. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. Proc. Natl. Acad. Sci. USA. 1996;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, Ley TJ. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, Kim S, Raja SM, Shi L, Simon MM, Froelich CJ. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Revell PA, Grossman WJ, Thomas DA, Cao X, Behl R, Ratner JA, Lu ZH, Ley TJ. Granzyme B and the downstream granzymes C and/or F are important for cytotoxic lymphocyte functions. J. Immunol. 2005;174:2124–2131. doi: 10.4049/jimmunol.174.4.2124. [DOI] [PubMed] [Google Scholar]
- 20.Getachew Y, Stout-Delgado H, Miller BC, Thiele DL. Granzyme C supports efficient CTL-mediated killing late in primary alloimmune responses. J. Immunol. 2008;181:7810–7817. doi: 10.4049/jimmunol.181.11.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelso A, Costelloe EO, Johnson BJ, Groves P, Buttigieg K, Fitzpatrick DR. The genes for perforin, granzymes A-C and IFN-γ are differentially expressed in single CD8+ T cells during primary activation. Int. Immunol. 2002;14:605–613. doi: 10.1093/intimm/dxf028. [DOI] [PubMed] [Google Scholar]
- 22.Heusel JW, Wesselschmidt RL, Shresta S, Russell JH, Ley TJ. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 23.Sheehan KC, Calderon J, Schreiber RD. Generation and characterization of monoclonal antibodies specific for the human IFN-γ receptor. J. Immunol. 1988;140:4231–4237. [PubMed] [Google Scholar]
- 24.Shresta S, MacIvor DM, Heusel JW, Russell JH, Ley TJ. Natural killer and lymphokine-activated killer cells require granzyme B for the rapid induction of apoptosis in susceptible target cells. Proc. Natl. Acad. Sci. USA. 1995;92:5679–5683. doi: 10.1073/pnas.92.12.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 26.Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. USA. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham CT, Thomas DA, Mercer JD, Ley TJ. Production of fully active recombinant murine granzyme B in yeast. J. Biol. Chem. 1998;273:1629–1633. doi: 10.1074/jbc.273.3.1629. [DOI] [PubMed] [Google Scholar]
- 28.Shresta S, Graubert TA, Thomas DA, Raptis SZ, Ley TJ. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 29.Sherman LA, Chattopadhyay S. The molecular basis of allorecognition. Annu. Rev. Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 30.Guy-Grand D, Malassis-Seris M, Briottet C, Vassalli P. Cytotoxic differentiation of mouse gut thymodependent and independent intraepithelial T lymphocytes is induced locally: correlation between functional assays, presence of perforin and granzyme transcripts, and cytoplasmic granules. J. Exp. Med. 1991;173:1549–1552. doi: 10.1084/jem.173.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shires J, Theodoridis E, Hayday AC. Biological insights into TCRγδ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–434. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- 32.Kawamoto Y, Sasaki K, Kato Y, Kojima K, Tsuji T, Miyama A. Rapid killing of murine lymph node T blasts by intestinal intraepithelial lymphocytes in vitro. Eur. J. Immunol. 1996;26:653–658. doi: 10.1002/eji.1830260322. [DOI] [PubMed] [Google Scholar]
- 33.French AR, Pingel JT, Kim S, Yang L, Yokoyama WM. Rapid emergence of escape mutants following infection with murine cytomegalovirus in immunodeficient mice. Clin. Immunol. 2005;115:61–69. doi: 10.1016/j.clim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 34.French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, Koszinowski U, Jonjic S, Yokoyama WM. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 2004;20:747–756. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Duggan BL, Cabilio NR, Dickie P, Witmer J, Goping IS, Underhill A, Bleackley RC. A novel lineage-specific hypersensitive site is essential for position independent granzyme B expression in transgenic mice. Biochem. Biophys. Res. Commun. 2008;368:357–363. doi: 10.1016/j.bbrc.2008.01.065. [DOI] [PubMed] [Google Scholar]
- 36.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 37.Quan LT, Caputo A, Bleackley RC, Pickup DJ, Salvesen GS. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.