SUMMARY
The direct induction of apoptosis has emerged as a powerful anti-cancer strategy, and small molecules that either inhibit or activate certain proteins in the apoptotic pathway have great potential as novel chemotherapeutic agents. Central to apoptosis is the activation of the zymogen procaspase-3 to caspase-3. Caspase-3 is the key “executioner” caspase, catalyzing the hydrolysis of a multitude of protein substrates within the cell. Interestingly, procaspase-3 levels are often elevated in cancer cells, suggesting a compound that directly stimulates the activation of procaspase-3 to caspase-3 could selectively induce apoptosis in cancer cells. We recently reported the discovery of a compound, PAC-1, which enhances procaspase-3 activity in vitro and induces apoptotic death in cancer cells in culture and in mouse xenograft models. Described herein is the mechanism by which PAC-1 activates procaspase-3 in vitro. We show that zinc inhibits the enzymatic activity of procaspase-3, and that PAC-1 strongly activates procaspase-3 in buffers that contain zinc. PAC-1 and zinc form a tight complex with one another, with a dissociation constant of approximately 42 nM. The combined data indicate that PAC-1 activates procaspase-3 in vitro by sequestering inhibitory zinc ions, thus allowing procaspase-3 to autoactivate itself to caspase-3. The small molecule mediated activation of procaspases has great therapeutic potential and thus this discovery of the in vitro mechanism of action of PAC-1 is critical to the development and optimization of other procaspase-activating compounds.
Keywords: Procaspase-3, PAC-1, Apoptosis, Cancer, Zinc
INTRODUCTION
Although general anti-proliferative agents that induce death in all rapidly dividing cell types remain the backbone of many chemotherapeutic regimens, recent advances have suggested that “personalized” anti-cancer strategies hold considerable promise. In personalized medicine, a detailed understanding of the molecular aberrations in the cell is utilized to select a drug with an appropriate mechanism of action. The potential for such strategies has been powerfully demonstrated for cancers that have translocations, mutations, or aberrant expression levels of key proteins.1
One of the hallmarks of cancer is its resistance to apoptosis,2 a programmed form of cell death important in both the development and maintenance of higher organisms. This resistance enables cancerous cells to survive and divide even in the presence of endogenous pro-apoptotic stimuli. Morphologically, apoptosis is characterized by cell shrinkage and membrane blebbing, which facilitate the engulfment and recycling of cellular components without provoking the inflammatory response.3 In human cells, apoptosis occurs through a cascade of events involving two main pathways: the intrinsic pathway and the extrinsic pathway. Both pathways ultimately converge on the activation of the “executioner” procaspases (primarily procaspase-3, but also procaspase-7 and procaspase-6) to caspases, which are the cysteine proteases that cleave scores of protein substrates within the cell.4; 5; 6 Cancer cells are known to evade apoptosis through a variety of mechanisms involving mutation or altered expression levels of key proteins in the apoptotic cascade,7; 8 including mutation of p53,9 elevated expression of the anti-apoptotic proteins in the Bcl-2 family,10 and inactivation of Apaf-1.11 The net result of these alterations is a resistance to apoptosis, allowing for unchecked cellular proliferation.
In the hopes of reactivating damaged apoptotic cascades and inducing cell death, several proteins in the apoptotic pathway have been targeted by small molecules.12; 13; 14 Indeed, small molecules that disrupt the p53—MDM-2 interaction,15; 16 bind to Bcl-2,17 promote apoptosome formation,18 or inhibit XIAP19; 20 have shown potential in cell culture and/or pre-clinical models of cancer. We embarked on an effort to bypass the damaged apoptotic cascade in cancer cells and directly activate procaspase-3 to caspase-3 with a small molecule. A procaspase-3 activator would be predicted to induce cell death, and, due to the elevation of procaspase-3 in certain tumor types,21; 22; 23; 24; 25; 26 a procaspase-3 activating compound could have selectivity for killing cancer cells versus normal cells. We recently reported the discovery of a compound (called PAC-1, Figure 1(a)) that activates procaspase-3 to caspase-3 in vitro, induces death in cancer cell lines and primary tumor samples in cell culture, and retards tumor growth in vivo.27 PAC-1 appears to induce death in cancer cells via the direct activation of procaspase-3, as evidenced by both the order and timing of apoptotic events, and the correlation of PAC-1 potency with procaspase-3 levels in cancer cells isolated from primary colon tumors.27
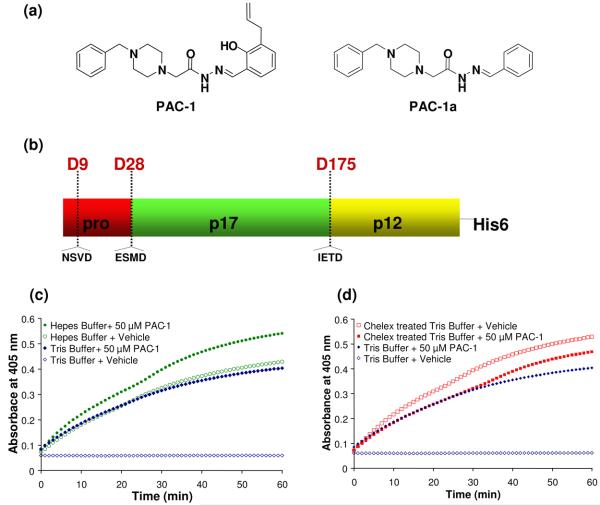
Figure 1.
(a) Chemical structures of PAC-1 and PAC-1a. (b) Procaspase-3 can be proteolyzed by caspases or granzymes at D9, D28, or D175. (c) Progress curves reporting on the procaspase-3 processing of the Ac-DEVD-pNA substrate as a function of time. In Tris buffer, procaspase-3 has very little activity on the Ac-DEVD-pNA substrate (open blue diamonds); this activity is dramatically enhanced in the presence of 50 μM PAC-1 (closed blue diamonds). The effect of PAC-1 on procaspase-3 activity is much less pronounced in a Hepes buffer (green circles). (d) Progress curves reporting on the procaspase-3 processing of the Ac-DEVD-pNA substrate as a function of time. Treatment of the Tris buffer with a Chelex® resin eliminates the activating effect of PAC-1. In a Chelex®-treated Tris buffer, the sample containing PAC-1 (closed red squares) has no greater activity over the vehicle-treated controls (open red squares). Shown for comparison is the data for procaspase-3 in Tris buffer that has not been Chelex® treated (blue diamonds), the same data as in Figure 1(c).
This preliminary study indicates that procaspase-3 activation, as well as proenzyme activation in general, could be an important anti-cancer strategy. However, the precise mechanism by which PAC-1 activates procaspase-3 is unknown. The further development of compounds that induce procaspase activation will be greatly aided by a detailed mechanistic understanding of the PAC-1-induced activation of procaspase-3, and the utility of PAC-1 as a probe in apoptotic research will be significantly enhanced by such information. Described herein are experiments demonstrating that PAC-1 activates procaspase-3 in vitro via sequestration of inhibitory zinc ions. Evidence is also presented suggesting that zinc binding is critical to the ability of PAC-1 to induce death in cancer cells in culture. These experiments represent the first detailed look at the in vitro mechanism of the PAC-1-mediated activation of procaspase-3 and have implications for both the discovery of other compounds that activate procaspases and for the role of zinc in regulating latent cellular procaspase activity.
RESULTS
To evaluate the effect of PAC-1 on procaspase-3 in vitro, two distinct biochemical assays were utilized. First, caspase-3 enzymatic activity was monitored at 405 nm through the cleavage of the peptidic Ac-DEVD-pNA substrate. Second, the maturation of the procaspase-3 zymogen was tracked by SDS-PAGE. This second method provides a biochemical readout orthogonal to the activity assay and allows direct visualization of procaspase-3 maturation.
Full length procaspase-3 is comprised of a ∼4 kDa (28 amino acid) prodomain, a 17 kDa large subunit, and a 12 kDa small subunit (Figure 1(b)). While caspase-8 and caspase-9 activate procaspase-3 in vivo by proteolysis between the p17 and p12 fragments (at D175), there are two additional sites where procaspase-3 is proteolyzed by caspase/granzyme-related enzymes: between the pro and p17 domains (at D28), and inside the prodomain (at D9) (see Figure 1(b)).28; 29 In vitro, it is known that caspase-3 will cleave procaspase-3 at D9, D28 and D175.30; 31 Interestingly, the procaspase-3 zymogen is itself also catalytically active.21; 30 In vitro procaspase-3 will cleave itself to the active caspase-3,21 and procaspase-3 (either wild-type or the caspase-resistant D9A/D28A/D175A triple mutant) will also process synthetic chromogenic/fluorogenic peptidic caspase-3 substrates.30 As inferred by studies on the triple mutant, the kcat/KM of procaspase-3 is approximately 200-fold lower than caspase-3 on peptidic substrates, with the major effect being a reduction in kcat.30 Thus, in vitro procaspase-3 is both an enzyme and a substrate; the cellular relevance of the procaspase-3 enzymatic activity is unknown.
Buffer dependence of PAC-1-mediated activation of procaspase-3
A starting point for our mechanistic studies was the observation that the in vitro activation of procaspase-3 by PAC-1 varied considerably depending on the buffer composition. Caspases are typically assessed in complex buffers containing multiple components, including EDTA and DTT. In such buffers the in vitro activating effect of PAC-1 on procaspase-3 is low on an absolute scale, 3-4 fold over background procaspase-3 levels.27 However, when procaspase-3 activation is assessed in simplified buffers (50 mM Tris, 300 mM NaCl, pH = 7.2) large activation of procaspase-3 by PAC-1 is observed as demonstrated by the enzyme's ability to cleave the Ac-DEVD-pNA substrate. The progress curves for these experiments are displayed in Figure 1(c); in the Tris/NaCl buffer, procaspase-3 has very little activity, and the activity is greatly enhanced by the addition of PAC-1. Considerably less PAC-1—mediated activation is observed in a Hepes buffer, mostly because procaspase-3 is already quite active in this buffer (Figure 1(c)). PAC-1a (Figure 1(a)) is a derivative of PAC-1 that had previously been shown to have no effect on procaspase-3 activation in vitro, and no death-inducing effect on cultured cancer cells.27 In accord with this previous data, PAC-1a had no activating effect in any of the buffer conditions in Figure 1(c) (see Supporting Information). It had been previously shown that caspase-3, and certain other caspases, are inhibited by the divalent metal ion zinc.32; 33; 34; 35; 36 Given the known ability of ortho-hydroxy N-acyl hydrazones (such as contained within PAC-1) to chelate metals,37 we began to suspect that an interaction of PAC-1 with a divalent metal in the assay buffer could be part of its mechanism of action. We thus set out to test the hypothesis that PAC-1 activates procaspase-3 via sequestration of inhibitory metals.
Metal removal abolishes the activating effect of PAC-1
Aqueous buffers can be treated with commercial resins that will bind to and remove divalent metal ions. One such resin is Chelex®, polystyrene beads that display imidoacetate functional groups. Chelex® will bind tightly to divalent transition metals, and poorly to smaller cations; treatment of a buffer at 5 grams Chelex® per 100 mL for 1 hour quantitatively removes transition metals. The ability of PAC-1 to activate procaspase-3 was thus assessed before and after treatment of the Tris/NaCl buffer with the Chelex® resin. As shown by the progress curves in Figure 1(d), in the Chelex®-treated buffer procaspase-3 has considerable activity, and the addition of PAC-1 has no activating effect, and is in fact slightly inhibitory. PAC-1a also had no activating effect in any of the buffer conditions in Figure 1(d) (see Supporting Information).
Caspase-3 and procaspase-3 are inhibited by divalent metals
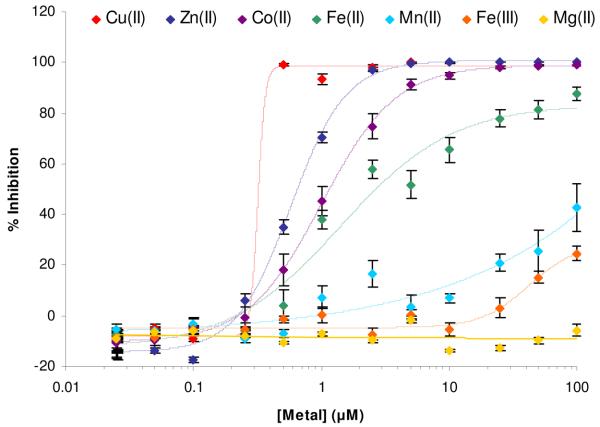
To further probe the metal-dependence of PAC-1 activity, we examined the effect of various metal ions on caspase-3 activity. As shown by the graph in Figure 2, Zn2+ and Co2+ are both potent inhibitors of caspase-3 activity. Sub-stoichiometric levels of Cu2+ appear to inhibit caspase-3, perhaps not surprising given the known redox chemistry of this metal and the importance of the active site sulfhydryl in caspase-3. Other metal ions (manganese, iron, magnesium) had a lesser effect on caspase-3 inhibition. The data for caspase-3 and Zn2+ is consistent with data in the literature,32; 33 while the effect of other metal ions on caspase-3 activity has not been previously reported, to our knowledge. Thus the variable activity of PAC-1 in different buffers (Figures 1(c), 1(d)) can be explained by the presence of metal ions in those buffers. For example, our ICP-MS analysis of the Trizma base (minimum 99% titration purity, from Sigma, analyzed as a solid) showed that a 50 mM Tris buffer made from this solid contain 100 nM zinc, 0.3 nM copper, 780 nM iron, and 250 nM cobalt.
Figure 2.
The effect of various metal ions on the catalytic activity of caspase-3. CuSO4, ZnSO4, CoCl2, FeCl2, MnCl2, FeCl3, MgCl2 were added to caspase-3 (500 nM) over the given concentration range. The activity of caspase-3 was then assessed by monitoring the cleavage of the Ac-DEVD-pNA substrate. Error bars represent standard error.
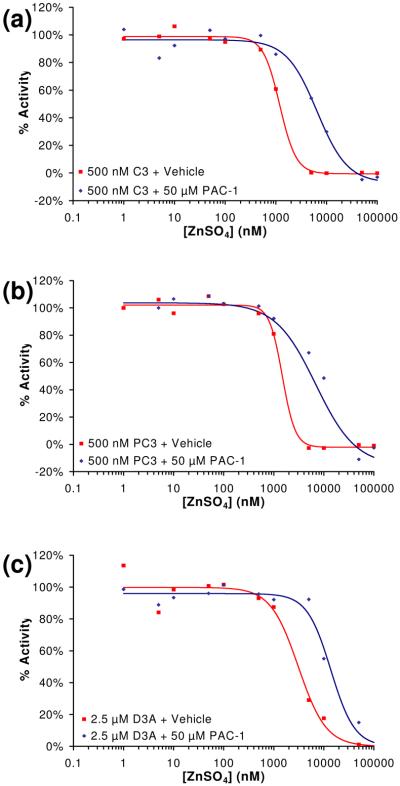
Although 90% of cellular zinc is believed to be sequestered in tightly-bound complexes within proteins (metalloenzymes, zinc fingers, etc), it is believed that approximately 10% of zinc exists in labile, loosely-bound pools.38 Given that zinc appears to be the most physiologically relevant metal of those inhibitors identified in Figure 2, we choose to focus on zinc. The effect of a range of concentrations of zinc on caspase-3 and procaspase-3 enzymatic activity was examined. All buffers were first treated with Chelex® resin to remove any trace metal contaminants. As procaspase-3 will slowly autoproteolyze itself to active caspase-3, we also created the triple mutant of procaspase-3 in which all three caspase cleavage sites have been removed (D9A/D28A/D175A). Consistent with data in the literature,30 this procaspase-3 triple mutant is proteolytically stable and has activity ∼200-fold less than caspase-3 (data not shown). Enzyme assays in the presence of zinc indicate that this metal powerfully inhibits caspase-3 (Figure 3(a)), procaspase-3 (Figure 3(b)), and the procaspase-3(D9A/D28A/D175A) mutant (Figure 3(c)). Because procaspase-3 is considerably less active than caspase-3, a higher concentration of the procaspase-3(D9A/D28A/D175A) mutant was used in these experiments. To the best of our knowledge this is the first demonstration that zinc inhibits procaspase-3 activity in vitro.
Figure 3.
(a) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit caspase-3 (C3, 500 nM) activity in vitro is reduced in the presence of PAC-1 (50 μM). (b) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit procaspase-3 (PC-3, 500 nM) activity in vitro is reduced in the presence of PAC-1 (50 μM). (c) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit the procaspase-3(D9A/D28A/D175A) mutant (D3A, 2.5 μM) activity in vitro is reduced in the presence of PAC-1 (50 μM). Data shown is representative of three trials.
PAC-1 addition reactivates zinc-inhibited caspase-3 and procaspase-3
Experiments were also conducted to assess the ability of PAC-1 to relieve the zinc-mediated inhibition of caspase-3 and procaspase-3 activity. A concentration of 50 μM PAC-1 was used for these experiments. The results from these experiments are displayed in Figures 3(a), 3(b), and 3(c) for caspase-3, procaspase-3, and the procaspase-3(D9A/D28A/D175A) mutant, respectively. PAC-1 relieves the zinc-mediated inhibition of caspase-3, procaspase-3 and the procaspase-3(D9A/D28A/D175A) mutant, as indicated by the shift in the ZnSO4 inhibition curves in the presence of PAC-1 (Fig. 3(a), 3(b), 3(c)).
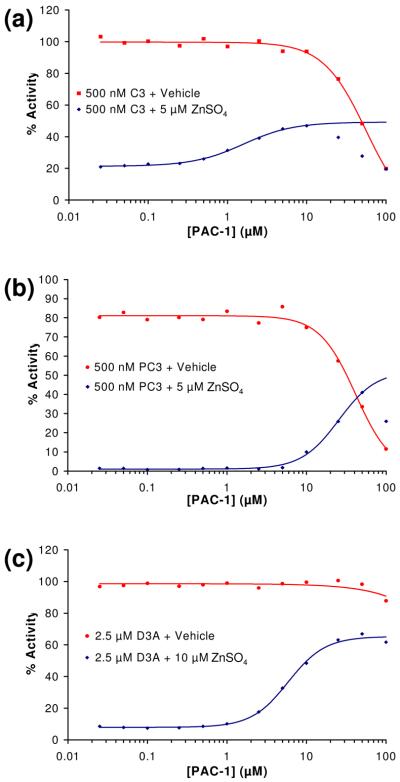
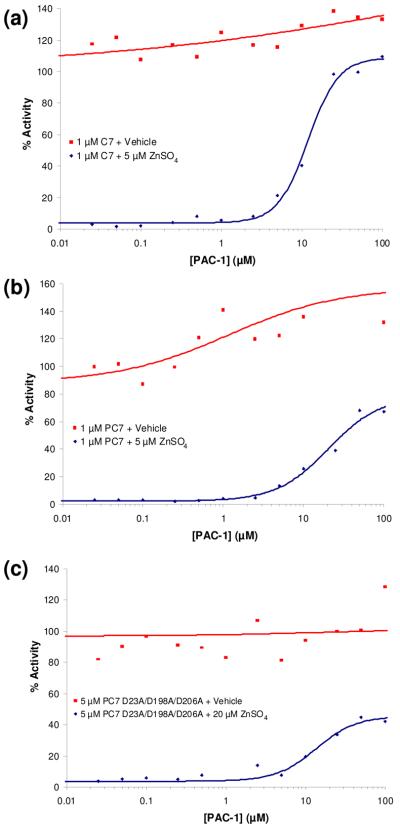
PAC-1 activates procaspase-3 and caspase-3 in a dose-dependant manner
Next, the ability of PAC-1 to activate procaspase-3 and caspase-3 in a dose-dependant manner was assessed in the presence and absence of zinc. For these experiments, concentrations of PAC-1 from 0.025 μM to 100 μM were evaluated, and all buffers were treated with Chelex® resin prior to addition of PAC-1 or zinc. The results of these experiments are displayed in Figure 4(a), 4(b), and 4(c) for caspase-3, procaspase-3, and the procaspase-3(D9A/D28A/D175A) mutant, respectively. As expected, in the presence of zinc and very low concentrations of PAC-1, the procaspase-3/caspase-3 enzymes are powerfully inhibited. However, as PAC-1 concentration is increased, the activity of the enzymes in the buffer containing zinc is increased to 40-60% of the maximal rate (that is, the rate of the enzyme in the absence of zinc). Interestingly, very high concentrations of PAC-1 actually inhibit the enzymes. Analogous experiments to those presented in Figure 4(a), 4(b) and 4(c) show that PAC-1a had no activating effect on caspase-3, procaspase-3, and the procaspase-3(D9A/D28A/D175A) triple mutant (see Supporting Information).
Figure 4.
(a) PAC-1 enhances caspase-3 (C3) activity when assayed in a buffer containing 5 μM ZnSO4. (b) PAC-1 enhances procaspase-3 (PC-3) activity when assayed in a buffer containing 5 μM ZnSO4. (c) PAC-1 enhances the procaspase-3(D9A/D28A/D175A) (D3A) mutant activity when assayed in a buffer containing 5 μM ZnSO4. As shown in panels (a), (b) and (c), PAC-1 actually inhibits these three enzymes at high compound concentrations. Data shown is representative of three trials.
PAC-1 activates caspase-3 to cleave procaspase-3
As mentioned, caspase-3 will cleave procaspase-3 in vitro, a process that is easily observed by SDS-PAGE.30; 31 In addition, the automaturation of procaspase-3 has also been observed in vitro by SDS-PAGE.21 Based on our above results in the activity assay, we predicted that zinc would inhibit the procaspase-3 cleavage by caspase-3, and the procaspase-3 automaturation, and that this inhibition would be relieved by PAC-1. A potential complication in the analysis is that wild-type procaspase-3 is both the substrate and the enzyme in these automaturation experiments. In order to separate these two roles, the procaspase-3(D9A/D28A/D175A) triple mutant was utilized as the version that functions solely as the enzyme (that is, it cannot be cleaved by caspases), and procaspase-3(C163A) was utilized as the version that functions solely as the substrate. The mutation of the active site cysteine 163 of caspase-3/procaspase-3 abolishes catalytic activity,39; 40; 41; 42 thus this protein can still be cleaved by caspases, but itself has no enzymatic activity.
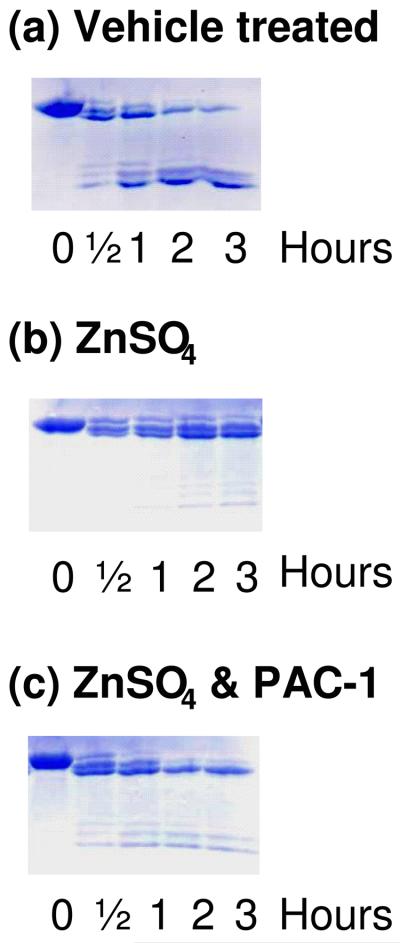
To assess the effect of zinc and PAC-1 on the ability of caspase-3 to cleave procaspase-3, procaspase-3(C163A) was incubated with caspase-3 for defined periods of time. As shown in Figure 5(a), in the absence of zinc caspase-3 will process procaspase-3(C163A) in this in vitro experiment, consistent with the data in the literature.30; 31 As shown in Figure 5(b), this processing is greatly inhibited by the addition of zinc. Finally, as shown in Figure 5(c), addition of PAC-1 restores the ability of caspase-3 to cleave procaspase-3. This is shown both by the appearance of the p12 and p17 active fragments, and by the processing of procaspase-3 to successively shorter forms. During in vitro activation of procaspase-3 by caspase-3, it is known that the cleavage is such that bands are observed corresponding to the full length procaspase-3 (∼34 kDa), the Δ1-9 procaspase-3 (∼32 kDa), and the Δ1-28 procaspase-3 (∼30 kDa), in addition to the active caspase-3 fragments;30; 31 this can be observed in the gels in Figure 5.
Figure 5.
Zinc inhibits the ability of caspase-3 to cleave procaspase-3, an effect that is relieved by addition of PAC-1. (a) Procaspase-3 (C163A) (50 μM) was incubated with caspase-3 (10 μM); (b) Procaspase-3 (C163A) (50 μM) was incubated with caspase-3 (10 μM) and ZnSO4 (50 μM); (c) Procaspase-3 (C163A) (50 μM) was incubated with caspase-3 (10 μM), ZnSO4 (50 μM), and 50 μM PAC-1 (50 μM). Samples were analyzed by SDS-PAGE and coomassie staining.
PAC-1 induces procaspase-3 automaturation
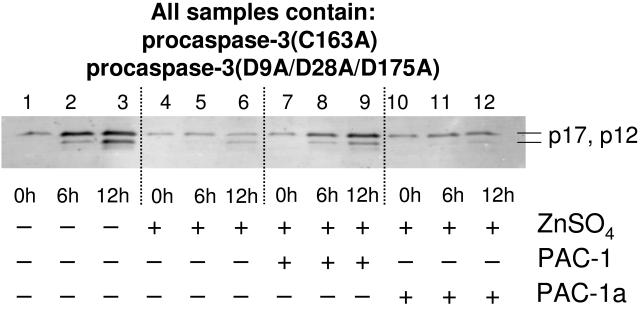
To investigate the effect of zinc and PAC-1 on the ability of procaspase-3 to autoactivate itself to caspase-3, an experiment was devised whereby procaspase-3(C163A) was used as the protein substrate, and procaspase-3(D9A/D28A/D175A) was used as the enzyme. In the event, procaspase-3(C163A) (50 μM) was incubated with procaspase-3(D9A/D28A/D175A) (10 μM) in the presence/absence of zinc and PAC-1, for 0, 6, and 12 hours. The lowered catalytic activity of procaspase-3 produces only low levels of product in these experiments, thus detection was performed by Western blotting. When procaspase-3(C163A) is incubated with procaspase-3(D9A/D28A/D175A), caspase-3 p17 and p12 fragments are observed within 12 hours (Figure 6, lanes 1-3). Running this same experiment in the presence of zinc significantly inhibits zymogen processing (Figure 6, lanes 4-6). However, upon addition of PAC-1, an increase in the levels of the processed fragments is once again observed (Figure 6, lanes 7-9); PAC-1a has no such effect (Figure 6, lanes 10-12). This experiment demonstrates that PAC-1 enhances the activity of the procaspase-3 zymogen, allowing it to cleave another molecule of procaspase-3. In the cell, of course, active caspase-3 would be generated through this process, and PAC-1 could then also enhance the activity of caspase-3 as part of a feed-forward cycle.
Figure 6.
Zinc inhibits the ability of procaspase-3 to cleave another molecule of procaspase-3, an effect that is relieved by addition of PAC-1. Procaspase-3(D9A/D28A/D175A) (10 μM) was incubated with procaspase-3 (C163A)(50 μM) (Lanes 1-3) and 100 μM ZnSO4 (Lanes 4-6) or 100 μM ZnSO4 and 50 μM PAC-1 (lanes 7-9) for up to 12 hours at 37 °C. Samples were analyzed by SDS-PAGE and Western blotting using antibodies against the p12 and p17 caspase-3 fragments. In the absence of zinc, procaspase-3 (D9A/D28A/D175A) processes the procaspase-3 (C163A) substrate within 12 hours. In the presence of zinc, this automaturation is inhibited (lanes 4-6), an effect that is relieved by the addition of PAC-1 (lanes 7-9), but not PAC-1a (lanes 10-12).
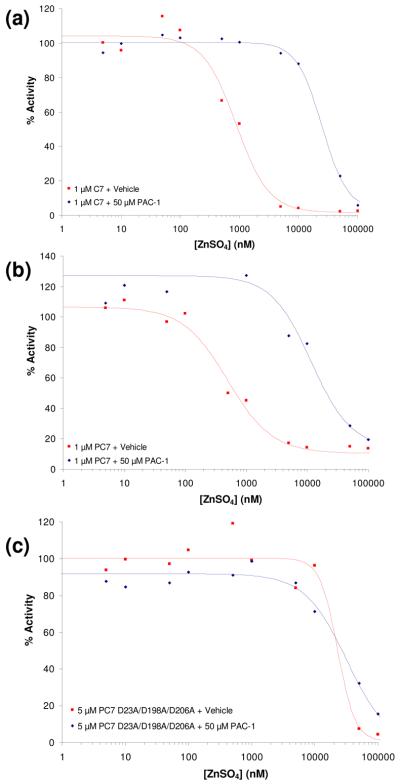
PAC-1 addition reactivates zinc-inhibited caspase-7 and procaspase-7
Caspase-7 is another major cellular executioner caspase and it shares a high degree of structural homology with caspase-3.43; 44 Similar to procaspase-3, the procaspase-7 zymogen is expressed with a short pro-domain (23 amino acids) followed by large and small subunits.45 Similar to procaspase-3, procaspase-7 is activated by upstream initiator caspases,46; 47 and caspase-7 is inhibited by zinc.33 Experiments were thus conducted to assess the ability of PAC-1 to relieve the zinc-mediated inhibition of caspase-7 and procaspase-7 activity. A concentration of 50 μM PAC-1 was used for these experiments. The results from these experiments are displayed in Figures 7(a), 7(b), and 7(c) for caspase-7, procaspase-7, and the procaspase-7(D23A/D198A/D206A) mutant, respectively. The triple mutant of procaspase-7 was created to render this zymogen resistant to caspase-mediated proteolysis; this mutant protein has the aspartic acids of two canonical caspase cleavage sites altered (D23 and D198), in addition to a third site (D206) that is known to be a substrate for further processing.44; 48; 49 Analysis of this triple mutant by SDS-PAGE shows it to be fully stable over the course of 12 hours. This procaspase-7(D23A/D198A/D206A) mutant is thus analogous to the procaspase-3(D9A/D28A/D175A) triple mutant, and provides a version of procaspase-7 that is stable during the time course of the biochemical experiments described herein. PAC-1 relieves the zinc-mediated inhibition of caspase-7, procaspase-7 and the procaspase-7(D23A/D198A/D206A) mutant, as indicated by the shift in the ZnSO4 inhibition curves in the presence of PAC-1 (Fig. 7(a), 7(b), 7(c)). To the best of our knowledge this is the first demonstration that zinc inhibits procaspase-7 activity in vitro.
Figure 7.
(a) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit caspase-7 (C7, 1 μM) activity in vitro is reduced in the presence of PAC-1 (50 μM). (b) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit procaspase-7 (PC-7, 1 μM) activity in vitro is reduced in the presence of PAC-1 (50 μM). (c) As assessed by the cleavage of the Ac-DEVD-pNA substrate, the ability of zinc to inhibit the procaspase-7(D23A/D198A/D206A) mutant (5 μM) activity in vitro is reduced in the presence of PAC-1 (50 μM). Data shown is representative of at least three trials.
PAC-1 activates procaspase-7 and caspase-7 in a dose-dependant manner
In analogy to our experiments with procaspase-3/caspase-3 (Figure 4), the ability of PAC-1 to activate procaspase-7 and caspase-7 in a dose-dependant manner was assessed in the presence and absence of zinc. For these experiments, concentrations of PAC-1 from 0.025 μM to 100 μM were evaluated, and all buffers were treated with Chelex® resin prior to addition of PAC-1 or zinc. The results of these experiments are displayed in Figure 8(a), 8(b), and 8(c) for caspase-7, procaspase-7, and the procaspase-7(D23A/D198A/D206A) mutant, respectively. As expected, in the presence of zinc and very low concentrations of PAC-1, the procaspase-7/caspase-7 enzymes are powerfully inhibited. However, as PAC-1 concentration is increased, the activity of the enzymes in the buffer containing zinc is increased to 40-100% of the maximal rate (that is, the rate of the enzyme in the absence of zinc). Interestingly, in contrast to the data with procaspase-3/caspase-3, very high concentrations of PAC-1 do not inhibit the procaspase-7/caspase-7 enzymes.
Figure 8.
(a) PAC-1 enhances caspase-7 (C7) activity when assayed in a buffer containing 5 μM ZnSO4. (b) PAC-1 enhances procaspase-7 (PC-7) activity when assayed in a buffer containing 5 μM ZnSO4. (c) PAC-1 enhances the procaspase-7(D23A/D198A/D206A) mutant activity when assayed in a buffer containing 5 μM ZnSO4. Data shown is representative of at least three trials.
PAC-1 binds tightly to Zn2+
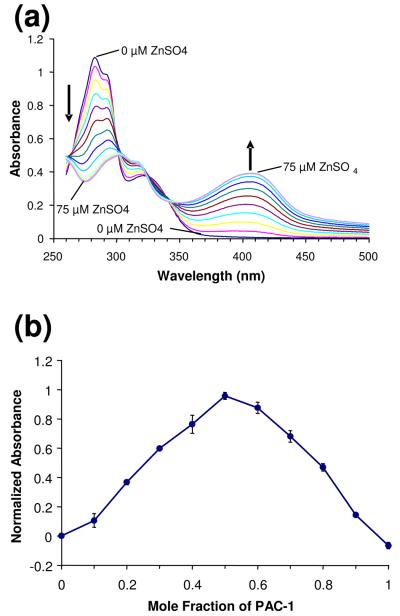
Although compounds containing the ortho-hydroxy N-acyl hydrozone motif have been previously shown to bind to metals,50 some of these compounds do not bind Zn2+ tightly and instead have Kd values in the micromolar range.51 We thus set out to determine the affinity of PAC-1 for zinc. As shown in Figure 9(a), titration of PAC-1 with increasing amounts of ZnSO4 results in a change in the UV-visible spectrum of this compound. This shift in molar absorptivity upon Zn2+ binding provides a convenient method to determine both the stoichiometry of PAC-1—Zn2+ binding and the dissociation constant of this interaction.
Figure 9.
(a) Titration of ZnSO4 into a solution of PAC-1 (50 μM in 50 mM Hepes, 100 mM KNO3, pH 7.2 buffer) causes a change in the UV-visible spectra of PAC-1. Shown are ZnSO4 concentrations of 0 to 75 μM in 5 μM increments. (b) Analysis of the PAC-1—Zn2+ binding interaction and stoichiometry by the continuous variations method. Based on the location of the peak value, the stoichiometry was determined to be 1:1, and the Kd was determined to be 42 nM. Error bars represent standard deviation from the mean.
A modified version of the method of continuous variations was used for this determination.52 The absorbance at 410 nm was acquired for various mole fractions of ZnSO4 and PAC-1 with a total concentration of 50 μM. These values were then normalized versus the maximal absorbance possible at the stoichiometric point (i.e. when all of the Zn2+ is bound by excess PAC-1), which was set as 1. Application of the continuous variations method revealed that PAC-1 binds Zn2+ in a 1:1 stoichiometry, as evidenced by the peak at 0.5 mole fraction in the graph in Figure 9(b). Also apparent from Figure 9(b) is the very strong nature of this small molecule—metal interaction. At the 1:1 stoichiometry PAC-1 has almost entirely shifted its absorption to the zinc-bound state (96% of PAC-1 is Zn2+-bound at a 1:1 ratio of PAC-1— Zn2+). Using this 96% value and the equation: log Ka = 0.3010 − log M + log ymax − 2 log (1 − ymax), we obtain a Kd = 42 nM for the PAC-1— Zn2+ interaction. While the Kd for the PAC-1—Zn2+ interaction is 42 nM, as shown in Figure 9(a) an absorbance change is still observed with micromolar concentrations of zinc. This is due to the high concentration of total ligand (50 μM) in this experiment relative to the Kd. Based on the concentrations of zinc and PAC-1 utilized in this experiment, the Adair equation53 predicts a half maximal population average site occupancy for the PAC-1—Zn2+ complex at ∼30 μM zinc, consistent with the data obtained (Figure 9(a)). Titration of ZnSO4 into PAC-1a gave no spectral change as monitored by UV/vis spectroscopy, consistent with the notion that PAC-1a does not bind Zn2+ (see Supporting Information).
To confirm this result, a second method was used to determine the PAC-1—Zn2+ dissociation constant. In this experiment, a preformed complex of PAC-1 and Zn2+ was titrated with EGTA (a metal binder with a known affinity for Zn2+), and the absorbance at 410 nm was monitored (see Supporting Information). Using the literature value for the EGTA—Zn2+ association constant,54 the PAC-1—Zn2+ dissociation constant was calculated as 55 nM, in general agreement with Kd of 42 nM calculated through the method of continuous variations.
Induction of apoptotic cell death by PAC-1 is inhibited by Zn
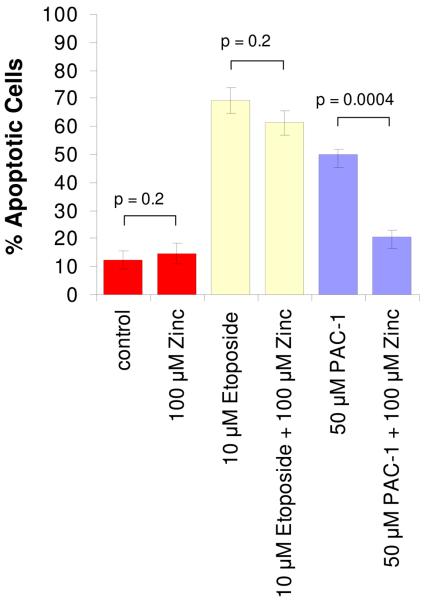
Although the goal of this study was to determine the mechanism by which PAC-1 activates procaspase-3 in vitro, we have also conducted preliminary experiments investigating the relevance of this mechanism to the death-inducing effect of PAC-1 in cell culture. Thus, the effect of exogenous zinc on the ability of PAC-1 to induce apoptotic death in cancer cells in culture was assessed. For this experiment, U-937 cells (human lymphoma cell line) were incubated with vehicle, ZnSO4 (100 μM), etoposide (10 μM), etoposide plus ZnSO4, PAC-1(50 μM), and PAC-1 plus ZnSO4; after 24 hours apoptotic cell death was assessed by FITC-Annexin V and propidium iodide staining and flow cytometry. Apoptotic cells are those that stain positive for FITC-Annexin V, but negative for propidium iodide. The results of this experiment are displayed in Figure 10. Treatment of cells with vehicle or with ZnSO4 induces little apoptosis, whereas treatment with etoposide induces apoptosis in almost 70% of the cells, a value that is not significantly reduced by the presence of ZnSO4. In contrast, the apoptotic death induced by PAC-1 is significantly reduced by the addition of ZnSO4. Comparison of the etoposide and PAC-1 data suggests that the ZnSO4-mediated inhibition of the PAC-1 cytotoxicity is not merely a consequence of a general anti-apoptotic effect of ZnSO4.
Figure 10.
PAC-1 induces apoptosis in U-937 cells, an effect that is reduced in the presence of ZnSO4. U-937 cells were treated with the indicated concentrations of PAC-1 and/or ZnSO4 for 24 hours, at which point cell death was assessed by Annexin V/propidium iodide staining and flow cytometry. As also shown, ZnSO4 has little effect on the toxicity of etoposide in the same assay. Error bars represent standard error, n = 3.
DISCUSSION
The data presented in this manuscript demonstrate that PAC-1 activates procaspase-3 in vitro through the sequestration of inhibitory metals. While it had been previously shown that modest levels of activation of procaspase-3 (and no activation of caspase-3) by PAC-1 are observed in complex buffer systems,27 we now show that in buffers consisting simply of Tris and NaCl, powerful PAC-1-mediated activation of procaspase-3 and caspase-3 is observed. Treatment of these buffers with Chelex® removes inhibitory divalent metals, allowing the enzymes to be fully active and thus abolishing the PAC-1-mediated activation. Consistent with this hypothesis is the fact that PAC-1 binds quite tightly to zinc, with a Kd = 42 nM. A structurally related compound that does not bind zinc, PAC-1a, does not activate procaspase-3 in vitro, and does not induce death of cancer cells in culture.
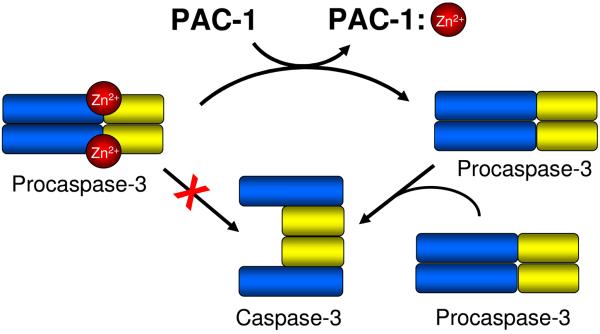
Also reported herein is the first in vitro demonstration that the catalytic activity of procaspase-3 is inhibited by zinc. We show that zinc will inhibit the procaspase-3-catalyzed processing of peptidic substrates (Figures 3(b), 4(b)), and the autoactivation of procaspase-3, where the enzymatic and the substrate properties of procaspase-3 were bifurcated (Figure 6). As wild-type procaspase-3 can be contaminated with active caspase-3, the D9A/D28A/D175A triple mutant of procaspase-3 was created and evaluated; this mutant is known to be fully resistant to caspase cleavage.30 Importantly, experiments with this triple mutant of procaspase-3 show that zinc is also an inhibitor of this enzyme (Figures 3(c), 4(c)), and in the presence of zinc PAC-1 is able to enhance the activity of this mutant against both peptidic (Figures 3(c), 4(c)) and protein (Figure 6) substrates. The mechanism of PAC-1-mediated activation of procaspase-3 in vitro that thus emerges from this work is shown in Figure 11: PAC-1 binds to zinc, allowing procaspase-3 to be enzymatically active and to subsequently activate another molecule of procaspase-3 to form caspase-3.
Figure 11.
The proposed mechanism for the PAC-1-induced activation of procaspase-3 in vitro. Consistent with data showing that zinc inhibits the catalytic activity of procaspase-3, PAC-1 binds tightly to zinc, and PAC-1 relieves the zinc-mediated inhibition of procaspase-3, it is proposed that PAC-1 binds to inhibitory zinc. This zinc sequestration enables procaspase-3 to function as an enzyme and proteolytically cleave another molecule of procaspase-3, converting it to caspase-3.
We also present data consistent with the notion that PAC-1 induces death in cultured cancer cells through the activation of procaspase-3 by metal sequestration, as pretreatment of PAC-1 with zinc reduces the ability of PAC-1 to induce apoptotic death in U-937 cells (Figure 10). It is of course possible that exogenous zinc is simply binding to PAC-1 and preventing it from entering the cell. Minimally, the data in Figure 10 suggests that PAC-1 is capable of binding zinc in cell culture media. More importantly, this experiment together with in vitro data for PAC-1a demonstrates a direct correlation between the cytotoxic potential of PAC-1 and its ability to bind zinc. Additionally, zinc has been previously shown to co-localize with procaspase-3 inside cells.38; 55 Based on the zinc—procaspase-3 co-localization data and other data, it has been previously suggested that zinc binds to and inhibits procaspase-3 inside the cell.38; 56 Although more experiments are required to determine the mechanism by which PAC-1 induces death in cancer cells in culture, these data, combined with our in vitro data showing PAC-1-mediated activation of procaspase-3 by metal sequestration, leads us to the following hypothesis: PAC-1 enters cells where, with a Kd of 42 nM, it competes favorably for the labile zinc pool to reduce the levels of zinc available to inhibit procaspase-3, thus resulting in procaspase-3 autoactivation to caspase-3. Once caspase-3 is generated it can activate more procaspase-3, caspase-3 substrates are cleaved, and apoptosis occurs.
Examination of the literature lends ample support to this hypothesis. There are multiple reports that describe the ability of zinc chelators to induce apoptosis.57 Perhaps the most detailed and convincing studies have been performed with the zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN). In several different cell lines and systems, TPEN has been found to strongly induce apoptosis.58; 59; 60; 61; 62 Other studies have shown that small changes to the labile zinc content of the cell make cells more susceptible to apoptosis.63 In light of the in vitro data presented herein with PAC-1, and the evidence showing that procaspase-3 is inhibited by zinc, TPEN may be exerting its pro-apoptotic effect in a similar manner to PAC-1, through the sequestration of inhibitory zinc. It should be noted that it is also possible that zinc is inhibiting caspase-3 in the cancer cell, and that PAC-1, TPEN, and other zinc chelators induce apoptosis by relieving this inhibition of caspase-3. The in vitro data presented in Figures 3, 4, and 5 indicate that PAC-1 does indeed activate zinc-inhibited caspase-3. Although in a number of cancer types the cellular procaspase-3/caspase-3 ratio is skewed heavily toward procaspase-3,21; 24; 64 there is solid evidence in some cancer cell lines and primary isolates for the presence significant levels of caspase-3.22; 23; 65; 66
The results described herein have ramifications for the development of zinc chelators as anti-cancer agents and for the regulatory role played by zinc in apoptosis. As shown in Figure 4(a) and 4(b), PAC-1 actually inhibits caspase-3/procaspase-3 activity at high concentrations, thus PAC-1 analogues that lack this effect (but still strongly chelate zinc) might be more potent proapoptotic agents. Furthermore, the addition of PAC-1 to caspase-3/procaspase-3 in Zn-containing buffer restores enzymatic activity to 40-60% of the levels in the absence of Zn (Figures 4(a), 4(b), 4(c)), thus it is possible that PAC-1 analogues that more fully activate caspase-3/procaspase-3 may be more potent anti-cancer agents. Finally, the data presented herein support the notion that cellular zinc plays an antiapoptotic role by inhibiting latent procaspase-3 activity. The identification of more potent versions of PAC-1, the evaluation of PAC-1 and derivatives in multiple animal models of cancer, and the role of zinc in procaspase-3 inhibition in vivo are all being actively investigated in our laboratory and the results will be presented in due course.
MATERIALS AND METHODS
Materials
All reagents were obtained from Fisher unless otherwise indicated. All buffer solutions were made with Milli-Q purified water. PAC-1 and PAC-1a (Figure 1(a)) were synthesized as described.27 Ac-DEVD-pNA was synthesized as described.67 Luria Broth (LB) was obtained from EMD. Etoposide was obtained from Sigma. Caspase Assay Buffer contains 50 mM HEPES pH 7.4, 100 mM NaCl, 10 mM DTT, 0.1 mM EDTA Disodium Salt, 0.10% CHAPS, 10% glycerol. Ni NTA Binding Buffer contains 50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole. Ni NTA Wash Buffer contains 50 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole. Ni NTA Elution Buffer contains 50 mM Tris pH 8.0, 300 mM NaCl, 500 mM imidazole. The C-terminal 6xHis-tagged procaspase-3 and procaspase-7 proteins were expressed and purified as described below. The 2xSDS sample buffer contains 100 mM Tris-Cl (pH 6.8), 200 mM DTT, 4% (w/v) SDS, 0.2% bromophenol blue, 20% (v/v) glycerol.
Strains and Plasmids
Procaspase-3 and caspase-3 were expressed using the pHC332 expression plasmid. The uncleavable mutant (D9A/D28A/D175A) of procaspase-3 was created by successive quick change mutations on pHC332 as previously described.30 Procaspase-7 and caspase-7 were expressed from a pET23b plasmid. The uncleavable mutant (D23A/D198A/D206A) of procaspase-7 was created by successive quick change mutations using the following primers: gcaaatgaagattcagtggctgctaagccagaccg for D23A, ggcatccaggccgctagcgggcccatcaatg for D198A, and ccatcaatgacacggccgctaatcctc for D206A.48
Recombinant expression and purification of procaspase-3/-7
Expression of procaspase-3/-7 was preformed in the BL21(DE3) strain of Escherichia coli (Novagen) similar to that previously described.27 Briefly, plasmids encoding for the C-terminally 6xHis-tagged procaspase-3/-7 protein were transformed into electrocompetent BL21. The transformants were selected for by growth in ampicillin. Individual colonies were picked and used to inoculate 10 mL seed cultures of LB with ampicillin (100 μg/mL) for expression. Seed cultures were used to inoculate 2 L cultures of LB which were grown at 37°C to an OD600 = 0.6. Protein expression was induced via the addition of IPTG to a final concentration of 0.5 mM; after addition of IPTG the culture was incubated at 37°C with shaking for 20 additional minutes. Bacterial cultures were then pelleted by centrifugation (at 5000 × g, 5 min, 4°C) and lysed by pulse sonication on ice for five minutes in the presence of 30 mL Ni NTA binding Buffer. Bacterial debris was removed by centrifugation (at 35,000 × g, 30 min, 4°C) and the 6xHis-tagged protein was purified from the supernatant with 1 mL 50% slurry Ni NTA resin (Qiagen). Protein was batch loaded onto the resin for 1 hr at 4°C. The bound resin was applied to a disposable column and the resin was washed with 15 mL of Ni NTA Wash Buffer. The protein was then eluted from the column with 10 mL of Ni NTA Elution Buffer. Fractions (∼1 mL) were collected and the presence of protein was determined by reaction with the Bradford reagent (BioRad). Fractions containing protein were pooled and flash frozen in liquid nitrogen. Protein was stored at −80 °C.
Zinc-Free Preparation
Protein was expressed and purified as above until fractions were pooled. Protein was not flash frozen at this point, but fractions containing protein were pooled and applied to a PD-10 column (GE Healthcare) charged with HEPES buffer that had been treated with Chelex® resin. The HEPES buffer consisted of 50 mM HEPES, 300 mM NaCl, pH = 7.4. The pH was monitored after treatment with the chelex resin and was found to have not changed. The protein was eluted using an additional 3.5 mL of Chelex®-treated HEPES buffer and the concentration was determined using the Edelhoch method for protein quantitation by absorbance at 280 nm. Protein isolated directly from the column typically had a concentration of ∼50 μM; for experiments described herein requiring zinc-free conditions, the protein was typically diluted to the desired concentration in HEPES Buffer that had been treated with Chelex® resin. Protein was flash frozen in 500 μL aliquots in liquid nitrogen and stored at −80°C.
Expression of various caspase forms
Expression of procaspase-3, procaspase-3 (D9A/D28A/D175A), procaspase-7, and procaspase-7 (D23A/D198A/D206A) were performed as described above. Expression of caspase-3/-7 was performed as above except that the induction time with IPTG was for 4 hours rather than 20 minutes. Purification and zinc-free preparations were performed the same for all caspase forms.
Buffer Analysis
Aliquots of standard preparation of procaspase-3 were thawed and diluted to 500 nM in either HEPES, TRIS buffer or chelex treated TRIS buffer. A 45 μL volume of protein was added to each well of a 384 well plate. Vehicle or compound was added to each well to a final concentration of 50 μM. The plate was incubated at 37°C for 2 hours. A 5 μL volume of a 2 mM stock of Ac-DEVD-pNA was added to the plates to a final concentration of 200 μM. The absorbance of each well at 405 nm was monitored in a plate reader every minute for 60 minutes and the progress curve was plotted.
Metal inhibition
Aliquots of zinc-free procaspase-3 were thawed and diluted to 500 nM in Chelex®-treated HEPES buffer. A 45 μL volume of protein was added to each well of a 384 well plate. Various concentrations of different metals were added to each well and the plate was incubated for 2 hours at 37°C. A 5 μL volume of a 2 mM stock of Ac-DEVD-pNA was added to the plates to a final concentration of 200 μM. The absorbance of each well at 405 nm was monitored in a plate reader every minute for 60 minutes. The initial slope of each well was determined from the linear portion of the progress curve. This typically included all data points collected. The slopes were converted to a percent of the inhibition in the absence of metal and were plotted vs. concentration. Data was generated in triplicate.
Assay of Procaspase-3/-7 and Caspase-3/-7 Activity
Stocks of zinc-free enzyme were prepared with or without the appropriate amount of zinc and were incubated at room temperature for at least 30 minutes prior to addition to compound. Compound dilutions were made from the 10 mM DMSO stock to the appropriate concentration in the specified assay buffer and DMSO to maintain a constant amount of DMSO in every well (never above 5%). These were placed into wells of a 384 well plate (20 μL), and then 25 μL of an appropriately diluted enzyme stock in assay buffer was added to the compound wells (1 μM procaspase-3, 1 μM caspase-3, or 5 μM D9A/D28A/D175A procaspase-3, 2 μM procaspase-7, 2 μM caspase-7, or 10 μM D23A/D198A/D206A procaspase-7). Controls containing DMSO and assay buffer without compound or without both compound and enzyme were included for each experiment. For experiments in the presence of zinc, a control containing no zinc was also included. The plates were then incubated for 2 hours at 37 °C. Then, to each well of the plate was added 5 μL of a 2 mM stock of Ac-DEVD-pNA in assay buffer and the absorbance at 405 nm was read every minute for one hour using a Spectra Max Plus 384 plate reader (Molecular Devices, Sunny Vale, CA). The slope of each well was used to determine the activity as compared to the controls. The data was plotted as percent activity versus compound concentration, and fitted to a logistic dose response curve using OriginPro (OriginLab Software, Northampton, MA) or Table Curve (SYSTAT Software, Richmond, CA) if appropriate. Data was generated in triplicate and repeated at least three times. The data presented is the average of the triplicate data points and is representative of the replicate experiments.
Assay of zinc inhibition of procaspase-3/-7 and caspase-3/-7 activity
Aliquots of frozen zinc-free protein were thawed and diluted in HEPES buffer (that had been treated with Chelex® resin) to the appropriate concentration. A 40 μL solution of protein was added to each well of 384 well plate and 5 μL of various concentrations of zinc were added. Appropriate wells also received PAC-1 to a final concentration of 50 μM. The plate was incubated at 37 °C for 2 hours. A 5 μL solution of a 2 mM stock of Ac-DEVD-pNA was added to each well to a final concentration of 200 μM. The absorbance at 405 nm was monitored every minute for 60 minutes and the initial slope determined from the linear portion of the progress curve. The slopes were converted to percent of maximal activity (the slope in the absence of zinc) and the data was fitted with a logistical dose response curve. Data was generated in triplicate and repeated at least three times. The data presented is the average of the triplicate data points and is representative of the replicate experiments.
SDS-PAGE analysis of caspase-3 activation
Aliquots of zinc-free caspase-3 and zinc-free procaspase-3 (C163A) were thawed and diluted to the appropriate concentration. A 22.5 μL solution of each protein was added to each of fifteen 0.5 mL tubes. A 5 μL solution of 500 μM ZnSO4 in HEPES Buffer was added to ten tubes to obtain a final ZnSO4 concentration of 50 μM. The remaining five tubes received 5 μL of buffer. The appropriate compound was added (0.5 μL of a 5 mM DMSO stock solution) to each tube and the final concentrations were: 10 μM caspase-3, 50 μM procaspase-3 (C163A), 50 μM ZnSO4 and 50 μM compound. The tubes were incubated at 37 °C for up to 3 hours. The reaction was stopped by the addition of 2x SDS loading dye and boiling for 5 minutes. A 15 μL volume of each sample was loaded on a 4-20% 15-well pre-cast polyacrylamide gel (BioRad). The gel was run at 150 V for 45 minutes and stained with coomassie brilliant blue. The experiment was run three times each on two different batches of protein.
Western blot analysis of procaspase-3 activation
Aliquots of zinc-free procaspase-3 (D9A/D28A/D175A) and zinc-free procaspase-3 (C163A) were thawed and diluted to the appropriate concentration. A 22.5 μL solution of each protein was added to each of twelve 0.5 mL tubes. A 5 μL solution of 1 mM ZnSO4 in HEPES Buffer was added to nine tubes to obtain a final ZnSO4 concentration of 100 μM. The remaining three tubes received 5 μL of buffer. The appropriate compound was added (0.5 μL of a 5 mM DMSO stock solution) to each tube and the final concentrations were: 10 μM procaspase-3(D9A/D28A/D175A) and 50 μM procaspase-3 (C163A); 100 μM ZnSO4 and 50 μM compound were added to certain experiments as indicated in Figure 6. The tubes were incubated at 37 °C for up to 12 hours. The reaction was stopped by the addition of 2x SDS loading dye and boiling for 5 minutes. A 15 μL volume of each sample was loaded on a 4-20% 15-well pre-cast polyacrylamide gel (BioRad). The gel was run at 150 V for 45 minutes and transferred to a PVDF membrane (Millipore) using a wet transfer method (40V for 1 hour). The blot was blocked in 10% milk in PBST for 4 hours at 4 °C. The blot was then washed three times for 3 minutes with PBST to remove excess milk. The primary antibody (rabbit polyclonal) against the active fragments of caspase-3 (calbiochem) was added at a dilution of 1:2000 and incubated at 4°C for 4 hours. The blot was then washed 2 times with PBST and a 1:2000 dilution of a goat-antirabbit alexa fluor 647 conjugated antibody was applied to the blot and allowed to incubate at 4 °C overnight. The blot was then washed 3 times with PBST and imaged on a Typhoon fluorescence scanner (Molecular Devices, Sunnyvale, CA). The experiment was repeated four times on two different batches of protein and the blot shown is representative of the results.
Zinc Titration
Compounds were diluted from their 10 mM DMSO stocks into 3 mL of buffer A (50 mM Hepes, 100 mM KNO3, pH 7.4) to give a final concentration of 50 μM in a quartz cuvette. The absorbance spectrum between 260 and 500 nm was then acquired using a Varian Carey 4000 Spectrophotometer (Palo Alto, CA). Then 5 μL of a 3 mM ZnSO4 solution in buffer A was added. The solution was mixed and allowed to equilibrate for 30 minutes. The absorbance spectrum was acquired again. This process was continued until the ZnSO4 concentration had reached 75 μM (addition of 75 μL).
Continuous Variations Method
A series of buffer A solutions of PAC-1 and ZnSO4 were prepared such that the sum of metal and ligand concentration was constant (50 μM). The mole fraction of PAC-1 was varied between 0.1 and 1.0. A series of buffer A solutions of PAC-1 alone at the same concentrations was also prepared. A final set of solutions was prepared for maximal absorbance readings using 250 μM PAC-1 with or without 25 μM ZnSO4. In a 96-well plate, 200 μL of each solution was added to different wells in triplicate. The absorbance at 410 nm was then acquired using a SpectraMax 384 plate reader (Molecular Devices, Sunnyvale, CA). The maximal absorbance was calculated by subtraction of the absorbance of 250 μM PAC-1 alone from the absorbance of 250 μM PAC-1 with 25 μM ZnSO4. The normalized absorbance for each mole fraction was then calculated by subtracting the absorbance of PAC-1 at the appropriate concentration from the corresponding PAC-1/Zn mixed sample and dividing by the corrected maximal absorbance value. The normalized absorbance was then plotted versus mole fraction. The peak of the plot gives the stoichiometry of the interaction (based on the mole fraction) and was used to calculate the binding constant from the following equation (as ymax):52
EGTA Competition Titration
In a quartz cuvette, a solution containing of 32 μM PAC-1 and 9.5 μM ZnSO4 in buffer A was titrated with 5 μL aliquots of a 2.9 mM solution of EGTA in buffer A. Upon each addition of EGTA aliquot, the solution was allowed to equilibrate for 30 minutes prior to reading the absorbance spectrum between 260 and 500 nm on a Varian Carey 4000 Spectrophotometer. The absorbance at 410 nm was used to calculate the [EGTA·Zn]. The absorbance data was first converted to the concentration of [PAC-1·Zn] using a calibration curve, then this value was subtracted by the total [Zn]. The plot of [EGTA·Zn] versus total [EGTA] was fit to the Hill equation using OriginPro (OriginLab Software, Northampton, MA). The KD for PAC-1 was found by dividing the K from the fit by the KD of EGTA (3.78×108 M−1).
Cell Death Assays
U-937 cells were cultured in RPMI 1640 growth media with 10% FBS and 1% Penn-Strep. One million cells were added to each well of a 12 well plate. Compound stocks in DMSO were added directly to the growth medium maintaining a constant amount of DMSO (< 1%) in each well. Cells were incubated with compound at 37°C and 5% CO2 for 24 hours. After incubation with compound, cells were pelleted and the media was removed by aspiration. The cells were washed once in PBS. The cells were again pelleted and resuspended in 100 μL of Annexin V binding buffer (10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2, 0.1% BSA). A 10 μL solution of FITC conjugated Annexin V was added to the cells and incubated on ice for 15 minutes. An additional 380 μL of Annexin V binding buffer and a 10 μL solution of a 1mg/mL stock of propidium iodide was added to each sample. The cells were analyzed by flow cytometry and percent apoptosis was determined. The experiment was performed three times and the data presented is the average of each experiment.
Acknowledgement
We are grateful to the National Institutes of Health (R01-CA120439) for the support of this work. Q.P.P. was partially supported by a Chemistry-Biology Interface Training Grant from the National Institutes of Health (Ruth L. Kirschstein National Research Service Award 1 T32 GM070421 from the National Institute of General Medical Sciences). D.C.W. was supported by Ruth L. Kirschstein National Research Service Award 3F31CA130138-01S1.
References
- 1.Papadopoulos N, Kinzler KW, Vogelstein B. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat Biotechnol. 2006;24:985–95. doi: 10.1038/nbt1234. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Blatt NB, Glick GD. Signaling pathways and effector mechanisms pre-programmed cell death. Bioorg. Med. Chem. 2001;9:1371–1384. doi: 10.1016/s0968-0896(01)00041-4. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Diff. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–50. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 6.Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 7.Lowe SW, Cepero E, Evan G. Intrinsic tumor suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 8.Igney FH, Krammer PH. Death and anti-death: tumor resistance to apoptosis. Nature Rev. Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. Achilles' heel of cancer. Nature. 2001;412:865–866. doi: 10.1038/35091170. [DOI] [PubMed] [Google Scholar]
- 10.Kirkin V, Joos S, Zornig M. The role of Bcl-2 family members in tumorigenesis. Biochim Biophys Acta. 2004;1644:229–49. doi: 10.1016/j.bbamcr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordon-Cardo C, Lowe SW. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 12.Bremer E, van Dam G, Kroesen BJ, de Leij L, Helfrich W. Targeted induction of apoptosis for cancer therapy: current progress and prospects. Trends Mol Med. 2006;12:382–93. doi: 10.1016/j.molmed.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Green DR, Kroemer G. Pharmacological manipulation of cell death: clinical applications in sight? J Clin Invest. 2005;115:2610–7. doi: 10.1172/JCI26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–85. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 15.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 16.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–93. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen JT, Wells JA. Direct activation of the apoptosis machinery as a mechanism to target cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7533–7538. doi: 10.1073/pnas.1031631100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Nikolovska-Coleska Z, Yang CY, Xu L, Liu M, Tomita Y, Pan H, Yoshioka Y, Krajewski K, Roller PP, Wang S. Structure-based design of potent, conformationally constrained Smac mimetics. J Am Chem Soc. 2004;126:16686–7. doi: 10.1021/ja047438+. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, Bayly CI, Gareau Y, Houtzager VM, Kargman S, Keen SLC, Rowland K, Seiden IM, Thornberry NA, Nicholoson DW. Maintenance of caspase-3 proenzyme dormancy by an intrinsic “safety catch” regulatory tripeptide. Proc. Natl. Acad. Sci. 2001;98:6132–6137. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoriev MY, Pozharissky KM, Hanson KP, Imyanitov EN, Zhivotovsky B. Expression of caspase-3 and -7 does not correlate with the extent of apoptosis in primary breast carcinomas. Cell Cycle. 2002;1:337–42. [PubMed] [Google Scholar]
- 23.O'Donovan N, Crown J, Stunell H, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Caspase 3 in breast cancer. Clin Cancer Res. 2003;9:738–42. [PubMed] [Google Scholar]
- 24.Krepela E, Prochazka J, Liul X, Fiala P, Kinkor Z. Increased expression of Apaf-1 and procaspase-3 and the functionality of intrinsic apoptosis apparatus in non-small cell lung carcinoma. Biol Chem. 2004;385:153–68. doi: 10.1515/BC.2004.034. [DOI] [PubMed] [Google Scholar]
- 25.Fink D, Schlagbauer-Wadl H, Selzer E, Lucas T, Wolff K, Pehamberger H, Eichler HG, Jansen B. Elevated procaspase levels in human melanoma. Melanoma Res. 2001;11:385–93. doi: 10.1097/00008390-200108000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Estrov Z, Thall PF, Talpaz M, Estey EH, Kantarjian HM, Andreeff M, Harris D, Van Q, Walterscheid M, Kornblau SM. Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood. 1998;92:3090–7. [PubMed] [Google Scholar]
- 27.Putt KS, Chen GW, Pearson JM, Sandhorst JS, Hoagland MS, Kwon J-T, Hwang S-K, Jin H, Churchwell MI, Cho M-H, Doerge DR, Helferich WG, Hergenrother PJ. Small molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nature Chem Biol. 2006;2:543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 28.Han Z, Hendrickson EA, Bremner TA, Wyche JH. A sequential two-step mechanism for the production of the mature p17:p12 form of caspase-3 in vitro. J Biol Chem. 1997;272:13432–6. doi: 10.1074/jbc.272.20.13432. [DOI] [PubMed] [Google Scholar]
- 29.Stennicke HR, Jurgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X, Zhou Q, Ellerby HM, Ellerby LM, Bredesen D, Green DR, Reed JC, Froelich CJ, Salvesen GS. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–90. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 30.Bose K, Pop C, Feeney B, Clark AC. An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3. Biochemistry. 2003;42:12298–310. doi: 10.1021/bi034998x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feeney B, Pop C, Swartz P, Mattos C, Clark AC. Role of loop bundle hydrogen bonds in the maturation and activity of (Pro)caspase-3. Biochemistry. 2006;45:13249–63. doi: 10.1021/bi0611964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry DK, Smyth MJ, Stennicke HR, Salvesen GS, Duriez P, Poirier GG, Hannun YA. Zinc is a potent inhibitor of the apoptotic protease, caspase-3. A novel target for zinc in the inhibition of apoptosis. J Biol Chem. 1997;272:18530–3. doi: 10.1074/jbc.272.30.18530. [DOI] [PubMed] [Google Scholar]
- 33.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–23. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 34.Aiuchi T, Mihara S, Nakaya M, Masuda Y, Nakajo S, Nakaya K. Zinc ions prevent processing of caspase-3 during apoptosis induced by geranylgeraniol in HL-60 cells. J Biochem (Tokyo) 1998;124:300–3. doi: 10.1093/oxfordjournals.jbchem.a022111. [DOI] [PubMed] [Google Scholar]
- 35.Chai F, Truong-Tran AQ, Ho LH, Zalewski PD. Regulation of caspase activation and apoptosis by cellular zinc fluxes and zinc deprivation: A review. Immunol Cell Biol. 1999;77:272–8. doi: 10.1046/j.1440-1711.1999.00825.x. [DOI] [PubMed] [Google Scholar]
- 36.Maret W, Jacob C, Vallee BL, Fischer EH. Inhibitory sites in enzymes: zinc removal and reactivation by thionein. Proc Natl Acad Sci U S A. 1999;96:1936–40. doi: 10.1073/pnas.96.5.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charkoudian LK, Franz KJ. Fe(III)-coordination properties of neuromelanin components: 5,6-dihydroxyindole and 5,6-dihydroxyindole-2-carboxylic acid. Inorg Chem. 2006;45:3657–64. doi: 10.1021/ic060014r. [DOI] [PubMed] [Google Scholar]
- 38.Truong-Tran AQ, Grosser D, Ruffin RE, Murgia C, Zalewski PD. Apoptosis in the normal and inflamed airway epithelium: role of zinc in epithelial protection and procaspase-3 regulation. Biochem Pharmacol. 2003;66:1459–68. doi: 10.1016/s0006-2952(03)00498-2. [DOI] [PubMed] [Google Scholar]
- 39.Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci U S A. 2002;99:4197–202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou H, Yang R, Hao J, Wang J, Sun C, Fesik SW, Wu JC, Tomaselli KJ, Armstrong RC. Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J Biol Chem. 2003;278:8091–8. doi: 10.1074/jbc.M204783200. [DOI] [PubMed] [Google Scholar]
- 41.Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3:e183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–27. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 43.Juan TS, McNiece IK, Argento JM, Jenkins NA, Gilbert DJ, Copeland NG, Fletcher FA. Identification and mapping of Casp7, a cysteine protease resembling CPP32 beta, interleukin-1 beta converting enzyme, and CED-3. Genomics. 1997;40:86–93. doi: 10.1006/geno.1996.4548. [DOI] [PubMed] [Google Scholar]
- 44.Lippke JA, Gu Y, Sarnecki C, Caron PR, Su MS. Identification and characterization of CPP32/Mch2 homolog 1, a novel cysteine protease similar to CPP32. J Biol Chem. 1996;271:1825–8. doi: 10.1074/jbc.271.4.1825. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F, Fritz LC, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci U S A. 1996;93:7464–9. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stennicke HR, Salvesen GS. Catalytic properties of the caspases. Cell Death Differ. 1999;6:1054–9. doi: 10.1038/sj.cdd.4400599. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci. 2004;13:1979–87. doi: 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denault J-B, Salvesen GS. Human caspase-7 activity and regulation by its N-terminal peptide. J. Biol. Chem. 2003;278:34042–24050. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- 49.Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. Embo J. 2005;24:645–55. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charkoudian LK, Pham DM, Franz KJ. A pro-chelator triggered by hydrogen peroxide inhibits iron-promoted hydroxyl radical formation. J Am Chem Soc. 2006;128:12424–5. doi: 10.1021/ja064806w. [DOI] [PubMed] [Google Scholar]
- 51.Sivaramaiah S, Reddy PR. Direct and Derivative Spectrophotometric Determination of Zinc with 2,4-Dihydroxybenzaldehyde Isonicotinoyl Hydrazone in Potable Water and Pharmaceutical Samples. J. Anal. Chem. 2005;60:828–832. [Google Scholar]
- 52.Likussar W, Boltz DF. Theory of continuous variations plots and a new method for spectrophotometric determination of extraction and formation constants. Anal. Chem. 1971;43:1265–1272. [Google Scholar]
- 53.Adair GS, Bock AB, Field H. The hemoglobin system. VI. The oxygen dissociation curve of hemoglobin. J. Biol. Chem. 1925;63:259. [Google Scholar]
- 54.Fahrni CJ, O'Halloran TV. Aqueous coordination chemistry of quinoline-based fluorescence probes for the biological chemistry of zinc. J. Am. Chem. Soc. 1999;121:11448–11458. [Google Scholar]
- 55.Carter JE, Truong-Tran AQ, Grosser D, Ho L, Ruffin RE, Zalewski PD. Involvement of redox events in caspase activation in zinc-depleted airway epithelial cells. Biochem Biophys Res Commun. 2002;297:1062–70. doi: 10.1016/s0006-291x(02)02292-1. [DOI] [PubMed] [Google Scholar]
- 56.Truong-Tran AQ, Carter J, Ruffin RE, Zalewski PD. The role of zinc in caspase activation and apoptotic cell death. Biometals. 2001;14:315–30. doi: 10.1023/a:1012993017026. [DOI] [PubMed] [Google Scholar]
- 57.Zhao R, Planalp RP, Ma R, Greene BT, Jones BT, Brechbiel MW, Torti FM, Torti SV. Role of zinc and iron chelation in apoptosis mediated by tachpyridine, an anti-cancer iron chelator. Biochem Pharmacol. 2004;67:1677–88. doi: 10.1016/j.bcp.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 58.Donadelli M, Dalla Pozza E, Costanzo C, Scupoli MT, Scarpa A, Palmieri M. Zinc depletion efficiently inhibits pancreatic cancer cell growth by increasing the ratio of antiproliferative/proliferative genes. J Cell Biochem. 2007 doi: 10.1002/jcb.21613. [DOI] [PubMed] [Google Scholar]
- 59.Hyun HJ, Sohn JH, Ha DW, Ahn YH, Koh JY, Yoon YH. Depletion of intracellular zinc and copper with TPEN results in apoptosis of cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2001;42:460–5. [PubMed] [Google Scholar]
- 60.Hyun HJ, Sohn J, Ahn YH, Shin HC, Koh JY, Yoon YH. Depletion of intracellular zinc induces macromolecule synthesis- and caspase-dependent apoptosis of cultured retinal cells. Brain Res. 2000;869:39–48. doi: 10.1016/s0006-8993(00)02340-4. [DOI] [PubMed] [Google Scholar]
- 61.Truong-Tran AQ, Ruffin RE, Zalewski PD. Visualization of labile zinc and its role in apoptosis of primary airway epithelial cells and cell lines. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1172–83. doi: 10.1152/ajplung.2000.279.6.L1172. [DOI] [PubMed] [Google Scholar]
- 62.Hashemi M, Ghavami S, Eshraghi M, Booy EP, Los M. Cytotoxic effects of intra and extracellular zinc chelation on human breast cancer cells. Eur J Pharmacol. 2007;557:9–19. doi: 10.1016/j.ejphar.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Zalewski PD, Forbes IJ, Betts WH. Correlation of apoptosis with change in intracellular labile Zn(II) using zinquin [(2-methyl-8-p-toluenesulphonamido-6-quinolyloxy)acetic acid], a new specific fluorescent probe for Zn(II) Biochem J. 1993;296(Pt 2):403–8. doi: 10.1042/bj2960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kania J, Konturek SJ, Marlicz K, Hahn EG, Konturek PC. Expression of survivin and caspase-3 in gastric cancer. Dig Dis Sci. 2003;48:266–71. doi: 10.1023/a:1021915124064. [DOI] [PubMed] [Google Scholar]
- 65.Yang S, Liu J, Thor AD, Yang X. Caspase expression profile and functional activity in a panel of breast cancer cell lines. Oncol Rep. 2007;17:1229–35. [PubMed] [Google Scholar]
- 66.Persad R, Liu C, Wu T-T, Houlihan PS, Hamilton SR, Diehl AM, Rashid A. Overexpression of caspase-3 in hepatocellular carcinomas. Mod Pathol. 2004;17:861–867. doi: 10.1038/modpathol.3800146. [DOI] [PubMed] [Google Scholar]
- 67.Goode DR, Sharma AK, Hergenrother PJ. Using peptidic inhibitors to systematically probe the S1' site of caspase-3 and caspase-7. Org. Lett. 2005;7:3529–3532. doi: 10.1021/ol051287d. [DOI] [PubMed] [Google Scholar]