1. Introduction
Potassium channels are tetrameric membrane proteins that selectively conduct K+ across cellular membranes. With 78 family members, K+ channels make up about half of the extended superfamily of 143 voltage-gated ion channels in the human genome, the third largest family of signaling molecules following G-protein coupled receptors and protein kinases.1 K+ channels have probably evolved from an ancestral gene encoding a simple 2 transmembrane segment (TM) protein like the bacterial KcsA channel. Subsequent gene duplication or addition of a 4 TM voltage sensor domain and/or intracellular domains for ligand binding have produced a large family of extraordinarily versatile signaling molecules: 15 inwardly rectifying 2 TM K+ channels (Kir),2 15 two-pore 4 TM K+ channels (K2P),3 8 calcium-activated 6 or 7 TM K+ channels (KCa),4 and 40 voltage-gated K+ channels (Kv).5 [For more information on the structure of K+ channels we refer the reader to the review by Declan Doyle in this issue of Chemical Reviews.] Each of these 78 K+ channels has a unique expression pattern allowing cells in a complex multicellular organism to “fine tune” their membrane potential and their excitability according to their respective physiological functions. Specific modulation of individual K+ channel types therefore offers an enormous potential for the development of physiological tool compounds and new drugs. To name just a few examples, K+ channel modulators are already clinically used as drugs for the treatment of type-2 diabetes and cardiac arrhythmia and are widely pursued in academia and the pharmaceutical industry as novel targets for epilepsy, memory disorders, chronic pain, cardiac and brain ischemia, hypertension, bladder over-reactivity, immunosuppression and cancer. In this review we will first give an overview of K+ channel pharmacology in general and then discuss the medicinal chemistry of the K+ channels which constitute targets for the treatment of neurological disorders (Kv7.2–7.5, KCa1.1, KCa2.1–2.3) and autoimmune diseases (Kv1.3, KCa3.1) in more detail.
1.1. K+ Channels as Drug Targets
The extracellular K+ concentration of 4 mM is about 40-times lower than the intracellular K+ concentration of 160 mM. The opening of K+ channels consequently generates an efflux of positive charge, which hyperpolarizes or repolarizes the cellular membrane. In excitable cells such as neurons or cardiac myocytes K+ channels are generally expressed together with voltage-gated Na+ (Nav) or Ca2+ (Cav) channels and are responsible for the repolarization after action potential firing. Pharmacological activation of K+ channels in excitable cells therefore reduces excitability whereas channel inhibition has the opposite effect and increases excitability. For example the Kv7.2/7.3 activator retigabine acts as an anticonvulsant while the unselective K+ blocker 4-aminopyridine (4-AP) induces seizures.6 In both excitable and none-excitable cells K+ channels further play an important role in Ca2+ signaling, volume regulation, secretion, proliferation and migration. In proliferating cells, such as lymphocytes or dedifferentiated smooth muscle cells, K+ channels are often found together with store-operated inward-rectifier Ca2+ channels like CRAC (calcium-release activated Ca2+ channel)7,8 or transient receptor potential channels like TRPC19 and provide the counterbalancing K+ efflux for the Ca2+ influx which is necessary for cellular activation. In this case K+ channel inhibitors like the KCa3.1 blocker TRAM-34 inhibit proliferation.10,11
However, despite the fact that all K+ channels in the human genome have been cloned and their biophysical properties characterized in great detail,1 it still often is a challenge to precisely determine which of the 78 K+ channels underlies a native K+ conductance in a specific cell type. In contrast to voltage-gated Na+ and Ca2+ channels, which contain 4 homologous domains in a single large polypeptide chain, K+ channels consist of 4 individual α-subunits arranged circumferentially around a central pore as a homo- or heterotetramer. Each subunit includes six transmembrane α-helical segments S1–S6 and a membrane-reentering P-loop (P) consisting of an extracellular S5-P linker (a turret), a pore helix (P-helix), an ascending limb containing the signature sequence TVGYG, and the extracellular linker P-S6.12 Four voltage-sensing domains S1–S4 are linked to the pore-forming domain composed of four S5-P-S6 sequences.
Within a K+ channel subfamily (see Table 1 – Table 4) like the Kv1-family or the Kv7-family (KCNQ) the α-subunits can heteromultimerize relatively freely leading to a wide variety of different channel tetramers with different biophysical and pharmacological properties.5 The properties of K+ channel α-subunit complexes can further be modified by association with intracellular or membrane-spanning β-subunits. For example, Kv1-family channels interact through their N-terminal T1 domain with Kvβ1-3 proteins, which form a second symmetric tetramer on the intracellular surface of the channel and modify the gating of the α-subunits. Kv4 channels interact with the so-called “K+ channel interacting proteins” KChIp1-4, which enhance surface expression and alter the functional properties of the Kv4 α-subunits.5 KCa1.1 (BK) channels associate in a tissue specific manner with four different 2 TM domain β-subunits β1-4 (KCNMB1-4),13 which influence Ca2+-sensitivity, inactivation and in the case of β4 render the channel complexes resistant to the KCa1.1 blockers charybdotoxin and iberiotoxin.14 The inward rectifiers Kir6.1 and Kir6.2 associate with 2 members of the ATP binding cassette proteins, the sulfonylurea receptors SUR1 or SUR2A/B to form ATP-sensitive K+ channels in the pancreas or cardiovascular system.15,16 In addition to “mixing” with α-subunits from the same channel subfamily and combining with different β-subunits, the α-subunits of many K+ channel gene families can be alternately spliced in order to generate additional K+ channel diversity. (The exception are the Kv1-family channels, which contain intronless coding regions.5) K+ channel properties can be further altered through post-translational modifications like phosphorylation,17,18 ubiquitinylation,19 and palmitoylation.20 In terms of drug discovery this molecular diversity constitutes a considerable challenge, but also offers opportunities for achieving tissue specificity by targeting tissue-specific β-subunits or for designing modulators that selectively target homotetramers over hetermultimers and vice versa.
Table 1.
Kv Channels
| Channel | Inhibitors | Activators |
|---|---|---|
| Kv1.1 (KCNA1) | DTX, DTX-κ, ShK, HgTX, KTX, 4- AP, TEA |
Unknown |
| Kv1.2 (KCNA2) | ChTx, DTX, MTX, NTX, HgTX, 4- AP, TEA |
Unknown |
| Kv1.3 (KCNA3) | ChTX, MgTX, ShK, OSK1, HgTX, KTX, NTX, correolide, PAP-1 |
Unknown |
| Kv1.4 (KCNA4) | UK78282, 4-AP | Unknown |
| Kv1.5 (KCNA5) | AVEO118, S9947, clofilium, 4-AP | Unknown |
| Kv1.6 (KCNA6) | HgTX, ShK, DTX, 4-AP | Unknown |
| Kv1.7 (KCNA7) | Flecainide, 4-AP | Unknown |
| Kv1.8 (KCNA10) | Ba2+, TEA, ketoconazole | cGMP |
| Kv2.1 (KCNB1) | Hanatoxin, TEA | Linoleic acid |
| Kv2.2 (KCNB2) | Quinine, TEA | Unknown |
| Kv3.1 (KCNC1) | 4-AP, TEA | Unknown |
| Kv3.2 (KCNC2) | ShK, verapamil, 4-AP, TEA | Unknown |
| Kv3.3 (KCNC3) | 4-AP, TEA | Unknown |
| Kv3.4 (KCNC4) | BDS-1, TEA | Unknown |
| Kv4.1 (KCND1) | 4-AP, TEA | Unknown |
| Kv4.2 (KCND2) | Heteropodatoxins, PATX1, PATX2 | Unknown |
| Kv4.3 (KCND3) | PATX1, PATX2, nicotine | Unknown |
|
Kv5.1 (KCNF1) |
Kv5 and Kv6 channels not functional alone, co-assemble with Kv2 subunits and act as modifiers or silencers | |
| Kv6.1 (KCNG1) | ||
| Kv6.2 (KCNG2) | ||
| Kv6.3 (KCNG3) | ||
| Kv6.4 (KCNG4) | ||
| Kv7.1 (KCNQ1, KVLQT) | L735821, chromanol 293B, mefloquine | L364373, mefenamic acid |
| Kv7.2 (KCNQ2) | XE991, linopirdine | Retigabine, BMS204352, S-1 |
| Kv7.3 (KCNQ3) | Linopirdine, XE991 | Retigabine, BMS204352, diclofenac |
| Kv7.4 (KCNQ4) | Linopirdine, XE991, bepridil | Retigabine, BMS204352, S-1 |
| Kv7.5 (KCNQ5) | Linopirdine, XE991 | Retigabine, BMS204352, S-1 |
| Kv8.1 (KCNV1) | Kv8 and Kv9 channels are not functional alone, co-assemble with Kv2 subunits and modify their properties | |
|
Kv8.2 (KCNV2) |
||
| Kv9.1 (KCNS1) | ||
| Kv9.2 (KCNS2) | ||
| Kv9.3 (KCNS3) | ||
| Kv10.1 (KCNH1, eag-1) | Quinidine | Unknown |
| Kv10.2 (KCNH5, eag-2) | Quinidine | Unknown |
| Kv11.1 (KCNH2, erg-1, HERG) | Astemizole, BeKM-1, ergtoxin, E4031, sertindole, dofetilide, terfenadine | Mallotoxin, RPR260243 |
| Kv11.2 (KCNH6, erg-2) | Sipatrigine | Unknown |
| Kv11.3 (KCNH7, erg-3) | Sertindole, pimozide | Unknown |
| Kv12.1 (KCNH8, elk-1) | Ba2+ | Unknown |
| Kv12.2 (KCNH3, elk-2) | None | Unknown |
| Kv12.3 (KCNH14, elk-3) | Ba2+ | Unknown |
Table 4.
K2P Channels
| Channel | Inhibitors | Activators |
|---|---|---|
| K2P1.1 (KCNK1, TWIK-1) | Unknown | Unknown |
| K2P2.1 (KCNK2, TREK-1) | Ba2+, quinidine | Arachidonic acid, volatile anesthetics riluzole |
| K2P3.1 (KCNK3, TASK-1) | Anandamide, Ba2+ | Halothane, isofluorane |
| K2P4.1 (KCNK4, TRAAK) | Gd+ | Arachidonic acid, lysopholipids, riluzole |
| K2P5.1 (KCNK5, TASK-2) | Clofilium, lidocaine, quinidine | Halothane |
| K2P6.1 (KCNK6, TWIK-2) | Ba2+, quinidine, volatile anesthetics | Arachidonic acid |
| K2P7.1 (KCNK7) | Does not form functional channels | |
| K2P9.1 (KCNK9, TASK-3) | Ruthenium red | Unknown |
| K2P10.1 (KCNK10, TREK-2) | Quinidine | Arachidonic acid, halothane, isoflurane, riluzole |
| K2P12.1 (KCNK12. THIK-2) | Does not form functional channels | |
| K2P13.1 (KCNK13. THIK-1) | Ba2+, halothane | Arachidonic acid |
| K2P15.1 (KCNK15. TASK-5) | Does not form functional channels | |
| K2P16.1 (KCNK16. TALK-1) | Ba2+, quinidine, chloroform | Isoflurane, nitric oxide |
| K2P17.1 (KCNK17. TASK-4) | Ba2+, chloroform | Nitric oxide |
| K2P18.1 (KCNK18) | Quinine, quinidine, free fatty acids, | Volatile anesthetics |
However, rational design of selective K+ channel modulators is difficult because there are currently no X-ray structures for medically important channels like Kv11.1 (hERG), Kv7.1–7.5, or KCa1.1. Investigators are therefore building homology models of these channels with inner-pore blockers based on the available X-ray structures of KcsA, MthK, KvAP, and Kv1.2 and data from mutational studies.21–27 Since usage of absolute numbers of residues, which are different in different channels, is inconvenient in a review we are using a universal labeling scheme.28 This scheme is shown in Figure 3A for Kv1.2, where residues facing either the inner pore or niches between the extracellular halves of S6s and P-helices are marked by asterisks. For well-known residues, we also show absolute numbers in brackets. The above-mentioned models visualize contacts between ligands and ligand-sensing residues, which were identified in mutational studies, and therefore are useful for designing new mutational experiments. However, experimental data on specific contacts between the functional groups of the ligands and the functional groups of the ligand-sensing residues is usually unavailable. Multiple drug-binding modes may coexist and their population is highly sensitive to ligand-receptor energy, and hence to the inner-pore geometry. The latter varies substantially in the available X-ray structures of open channels. For example, mutations of the Kv11.1-channel pore-facing Phei22(656) significantly decreased the affinity of the antiarrhytmic drug MK-499, cisapridine, and several other ligands.29 In matching positions of KvAP30 and Kv1.2,31 the distances between Cβ atoms in diagonally opposed α-subunits are 17.4, and 13.3 Å, respectively, and therefore vary by as much as 4 Å (the distances were measured in the biological-unit structures, PDB indexes 1ORQ and 2A79, respectively). Usually it is unclear which X-ray structure would be a better template for a homology model. Another problem is that C-type inactivation usually,32–35 but not universally32,36,37 enhances ligand binding and that the structural changes that ligand-binding sites undergo upon C-type inactivation are unknown.37 In these circumstances, the predicted free energy of the ligand-receptor complex cannot be the decisive criterion to choose the most promising binding mode for rational drug design. Additional (and very important) information that may help to choose the correct binding mode may be obtained from considering structure-activity relations in a series of ligands. For example, if a particular ligand-binding mode explains the structure-activity relationships, it may be favored over other binding modes that may be better in terms of ligand-receptor energy, but do not explain the structure-activity relations.
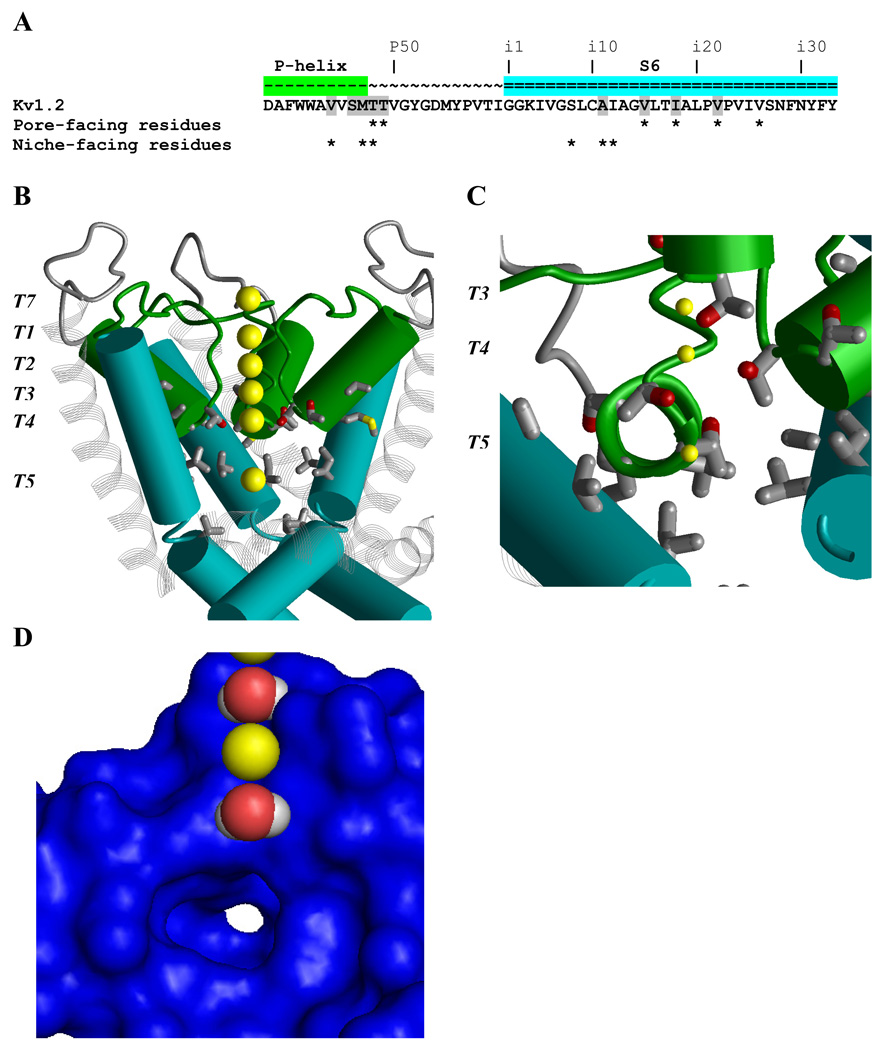
Figure 3. Ligand-binding regions in the inner pore and niches of K+ channels.
A, P-S6 sequence of Kv1.2 with residue labels.28 Positions where ligand-sensing residues have been identified in various Kv channels are highlighted. Residues whose Cα-Cβ bonds face the inner pore and/or the niches are marked by asterisks. B Side view of the Kv1.2 structure31 with P-helices (green), S6s (cyan) and S5s (gray strands). One subunit is removed for clarity. Yellow spheres show K+ ions in the center of the inner pore (K+T5), at the extracellular side of the channel (K+T7), and in the outer pore (K+T1, K+T2, K+T3, and K+T4). Sidechains of residues highlighted in A are shown. In the inner pore, the ligand-sensing positions are exemplified by Tp48 and Tp49 at the level of K+T4, Vi15 at the level of K+T5, and Ii18 and Vi22 at the helix kink. C, View from inside the pore along a P-helix shown by the ribbon. Side chains of Vp44, Sp46, Mp47, Tp48, and Tp49 in the helix represent ligand-sensing position in various Kv channels.28 K+T3, and K+T4, and K+T5 are shown by small yellow spheres. D, View at the fraction of the protein surface from inside the pore. K+T2 and K+T4 are replaced with water molecules. K+T5 is removed to show a niche with a white opening at the far end. In K+ channels, the niches contain ligand-sensing residues.
Another challenging aspect for developing K+ channel modulators is the fact that K+ channels like all ion channels are “moving targets” that undergo large conformational changes switching between open and closed states on a millisecond time scale. These changes in “gating state” are often accompanied by dramatic changes in the conformation of drug binding sites resulting in a phenomenon referred to as “state-dependent inhibition”. For ion channels in excitable cells firing action potentials like neurons, state-dependent inhibition translates into what is called “use-dependent inhibition” since channel block increases as channels are “used” during rapid cycling from closed into open states and back and the number of blocked channels increases with time. In order to determine the “true” effect of an ion channel modulator on a channel, it is therefore advisable to use functional assays rather than binding assays.
1.2. K+ Channel Pharmacology
Chemicals modulating K+ channel function fall into 3 general categories: metal ions, organic small molecules and venom-derived peptides. These substances can affect K+ channels by blocking the ion-conducting pore from the external or internal side or by modulating channel gating through binding to the voltage-sensor domain or auxiliary subunits. Table 1, Table 2, Table 3 and Table 4 contain a list of the most commonly used Kv, KCa, Kir and K2P channel inhibitors and activators. In this review we will not discuss the actions of metal ions such as Cs+, Ba2+, Cd2+, Pb2+, Co2+ and Ni2+ since most of these cations block K+ channels only in the millimolar range and are not very specific. Instead, we will first give a brief overview of venom-derived peptide toxins and small-molecule K+ channel modulators and then focus on the medicinal chemistry of K+ channel modulators, which hold promise for the treatment of diseases of the nervous or immune system.
Table 2.
KCa Channels
| Channel | Inhibitors | Activators |
|---|---|---|
| KCa1.1 (KCNMA1, BK, slo) | IbTX, ChTX, paxilline, slotoxine, TEA | NS1609, NS1619, BMS204352, DHS-1, estradiol |
| KCa2.1 (KCNN1, SK1) | UCL1684, apamin, tamapin, leiurotoxin, NS8593 | NS309, DC-EBIO, riluzole, EBIO |
| KCa2.2 (KCNN2, SK2) | Tamapin, apamin, leiurotoxin, UCL1684, Lei-Dab7, NS8593 | NS309, DC-EBIO, riluzole, EBIO, CyPPA |
| KCa2.3 (KCNN3, SK3) | Leiurotoxin, apamin, UCL1684, NS8593 | NS309, DC-EBIO, riluzole, EBIO, CyPPA |
| KCa3.1 (KCNN4, IK1, SK4) | MTX, ChTX, TRAM-34, ICA-17043 | NS309, DC-EBIO, riluzole, EBIO |
| KCa4.1 (KCNT1, slack) | TEA, quinidine, bepridil | Bithionol |
| KCa4.2 (KCNT2, slick) | TEA, quinidine | Unknown |
| KCa5.1 (KCNU1, slo3) | TEA | Unknown |
Table 3.
Kir Channels
| Channel | Inhibitors | Activators |
|---|---|---|
| Kir1.1 (KCNJ1, ROMK1) | Tertiapin, Ba2+, Cs+ | None |
| Kir2.1 (KCNJ2, IRK1) | Spermine, spermidine, Ba2+, Cs+ | Unknown |
| Kir2.2 (KCNJ12, IRK2) | Spermine, spermidine, Ba2+, Cs+ | Unknown |
| Kir2.3 (KCNJ4, IRK3) | Spermine, spermidine, Ba2+, Cs+ | Arachidonic acid, tenidap |
| Kir2.4 (KCNJ14, IRK4) | Ba2+, Cs+ | Unknown |
| Kir3.1 (KCNJ3, GIRK1) | Ba2+, Cs+ | Unknown |
| Kir3.2 (KCNJ6, GIRK2) | Tertiapin, halothane, Ba2+, Cs+ | Unknown |
| Kir3.3 (KCNJ9, GIRK3) | None | Unknown |
| Kir3.4 (KCNJ5, GIRK4) | Ba2+, Cs+, 4-AP, TEA | Unknown |
| Kir4.1 (KCNJ10, BIRK1) | Ba2+, Cs+ | Unknown |
| Kir4.2 (KCNJ15, Kir1.3) | Ba2+, Cs+ | Unknown |
| Kir5.1 (KCNJ16) | Ba2+, Cs+ | Unknown |
| Kir6.1 (KCNJ8) | Glibenclamide and other sulfonylureas (sites on associated SUR subunits and channel) | Diazoxide, pinacidil, nicorandil (for associated SUR subunits) |
| Kir6.2 (KCNJ11) | Glibenclamide and other sulfonylureas phentolamine (Kir6.2) | Diazoxide, pinacidil, cromokalim, nicorandil |
| Kir7.1 (KCNJ13, Kir1.4) | Ba2+, Cs+ | Unknown |
1.2.1. Peptides
Venomous animals such as snakes, spiders, scorpions, sea anemones, cone snails, and bees produce a large variety of peptide toxins, which target ion channels. The first peptide toxins that were found to inhibit K+ channels in the late 1970s and early 1980s were the bee venom apamin, which blocks KCa2 (SK) channels,38,39 and the scorpion toxins noxiustoxin (NTX)40 and charybdotoxin (ChTX),41 which were isolated from the venoms of the scorpions Centruroides noxius and Leiurus quinquestriatus. While NTX only blocks the Kv1-family channels Kv1.2 and Kv1.3, ChTX turned out to be relatively promiscuous: it inhibits both the KCa channels KCa1.1 and KCa3.1 as well as Kv1.2, Kv1.3 and Kv1.6.42 Apamin, ChTX and the Kv1-family channel blocking α-dendrotoxins43 from the venoms of the black mamba Dendroaspis polylepis and the green mamba Dendroaspis angusticeps quickly became popular neuroscience tools. They have been widely used to elucidate the physiological function of K+ channels even before the Drosophila Shaker channel in 198744 and the mammalian K+ channels starting with Kv1.1, Kv1.2 and Kv1.3 in 199045 were cloned. However, K+ channel blocking peptide toxins are not just pharmacological tools. They also proved tremendously useful for gaining structural information about the K+ channel proteins themselves and continue to be widely used to study K+ channel gating and subunit composition [see review on Chemical Approaches to Probing Voltage-gated Ion Channels in this issue of Chemical Reviews for some elegant examples]. Starting in the mid-1990s, the groups of Chris Miller, Roderick MacKinnon and George Chandy deduced the dimensions of the outer vestibule of Shaker and Kv1.3 using ChTX and the related scorpion toxins agiotoxin-2 and kaliotoxin, whose structures had been determined by NMR,46–48 as molecular calipers in mutant cycle analysis experiments.47,49 All three groups estimated that the outer entrance to the pore is 9–14 Å wide at the top and then taperes down to a width of 4–5 Å at a depth of 5–7 Å.47,49–52 These predicted dimensions proved to be remarkably accurate when compared with the crystal structure of the bacterial K+ channel KcsA published three years later in 1998.12
During the last 20 years many laboratories have identified about 200 K+ channel-targeting peptide toxins not only in snake and scorpion venoms, but also in sea anemones, marine cone snails and tarantulas (see53 for a systematic nomenclature). The toxins contain between 18 and 60 amino acid residues and are cross-linked by two to four disulfide bridges forming compact molecules, which are remarkably resistant to denaturation. Based on the arrangement and the number of β-strands and α-helices in their structures, the toxins can be categorized into eight different folds: βββ, hairpin-like and cross-like αα, 310αα (helical capping motif), 310ββ, and the three α/β scaffolds αββ, βαββ and 310ββα.54 Taken together, these peptide toxins and their synthetic derivatives constitute a large pharmacological armamentarium to target Kv and KCa channels with high potency and specificity. Interestingly, so far only one Kir blocking peptide, the bee venom toxin tertiapin,55 and no toxins targeting K2P channels56 have been identified.
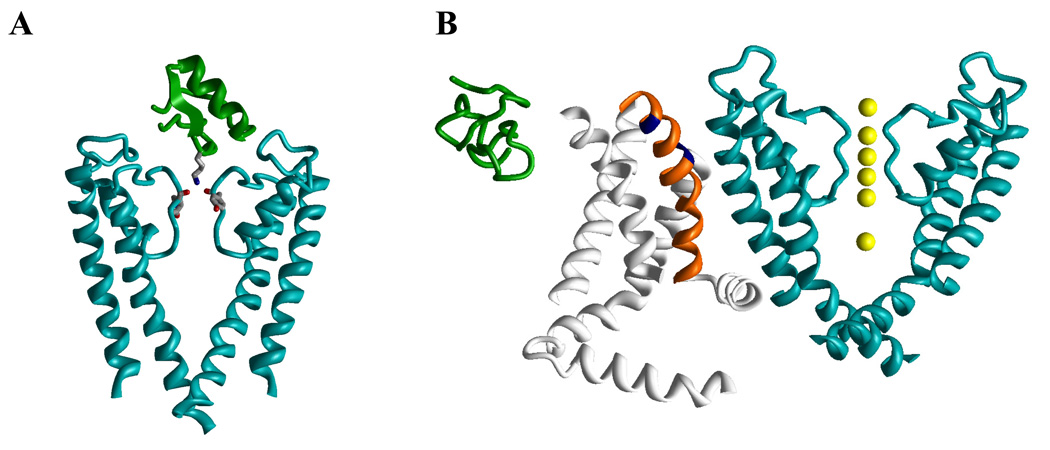
Venom-derived peptide toxins produce effects on K+ channels by two different mechanisms (Fig. 1). The toxins from scorpions, sea anemones, snakes and cone snails bind to the outer vestibule of K+ channels and in most cases insert a lysine side chain (position 27 in charybdotoxin) into the channel pore to occlude it.54,57–59 This mechanism has often been compared with a cork being inserted into a wine bottle. The toxin molecule forms several secondary contacts with channel residues in the outer vestibule to further stabilize the binding. Figure 1A illustrates this binding mode by showing the complex between ChTX and KcsA determined by NMR.60 Only two of the four KcsA subunits are shown to allow a view into the pore revealing that the Lys residue of the toxin binds to the backbone carbonyls groups of four tyrosines from the signature-sequence GYG motifs in the ion channel pore. Spider toxins like hanatoxin (HaTX), which was isolated from the venom of the Chilean rose tarantula Grammostola spatulata,61 in contrast, interact with the voltage sensor domain of Kv channels and increase the stability of the closed state.62,63 The resulting rightward shift in activation voltage and acceleration of deactivation means that the channel is “harder” to open (i.e. membrane requires more depolarization) and closes faster. However, in contrast to channels inhibited by pore-blocking toxins, channels inhibited by so-called “gating modifier” toxins can still be opened by strong depolarizations. Gating-modifier toxins differ from pore blocking toxins in two more aspects: the stoichiometry of the toxin-channel interaction and the location of the binding site. While pore blocking toxins bind with a 1:1 stoichiometry to the receptor site involving multiple amino acid residues in the outer vestibule of the channel, experiments with saturating concentrations of gating-modifier toxins suggest that 3 to 4 toxin molecules bind to the four voltage-sensor domains of a single channel molecule.62,64 The combined evidence from studies on the kinetics of inhibition, mutagenesis and membrane partition experiments further suggests that the amphiphilic gating-modifier toxins, which contain a cluster of hydrophobic residues on one face of the molecule, partition into the membrane when they bind to the voltage sensor.64,65 This nonspecific membrane partitioning makes it difficult to design specific gating modifier molecules for use as drugs as pointed out by Maria Garcia in a commentary on the work by Lee and MacKinnon characterizing the binding of the tarantula toxin VSTX1 to the bacterial KvAP channel.65,66 Figure 1B shows the NMR structures of hanatoxin167 on the same scale as the X-ray structure of Kv1.2.31 For clarity, only one of the four voltage-sensing domains (gray with orange S4) and only two of the four pore-domain subunits (cyan) are shown. Two positions in S4, whose mutations affect the toxin binding in Kv2.163 are marked in blue. However, it should be remembered that this figure only is a cartoon illustrating the binding mode since the structure of the toxin-channel complex has not been experimentally determined.
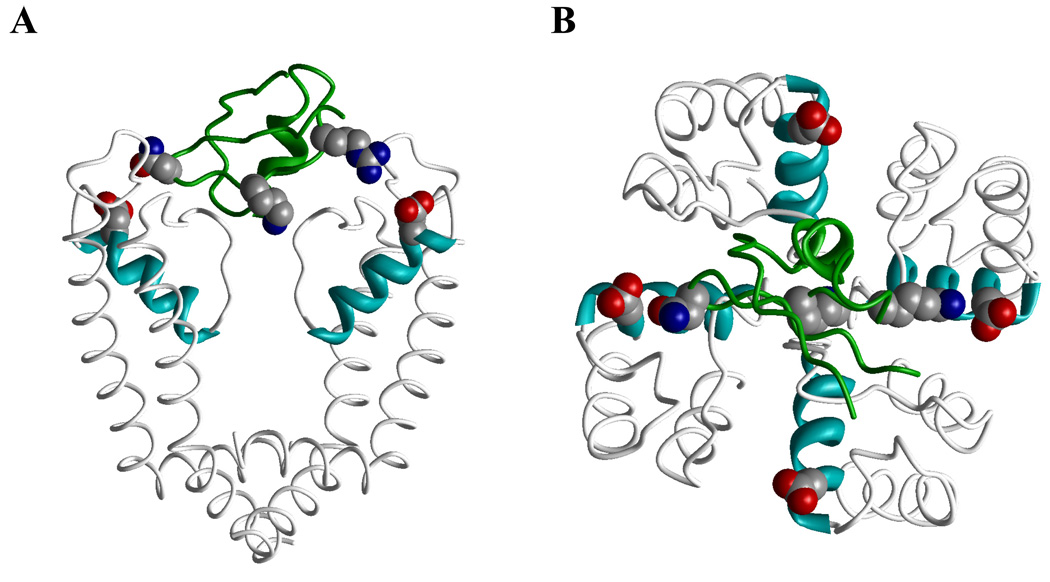
Figure 1. Action of peptide toxins on potassium channels.
A, Complex of KcsA with charybdotoxin (PDB index 2A9H). B, NMR structure of hanatoxin1 (PDB 1D1H) on the same scale as the X-ray structure of Kv1.2 (PDB 2A79).
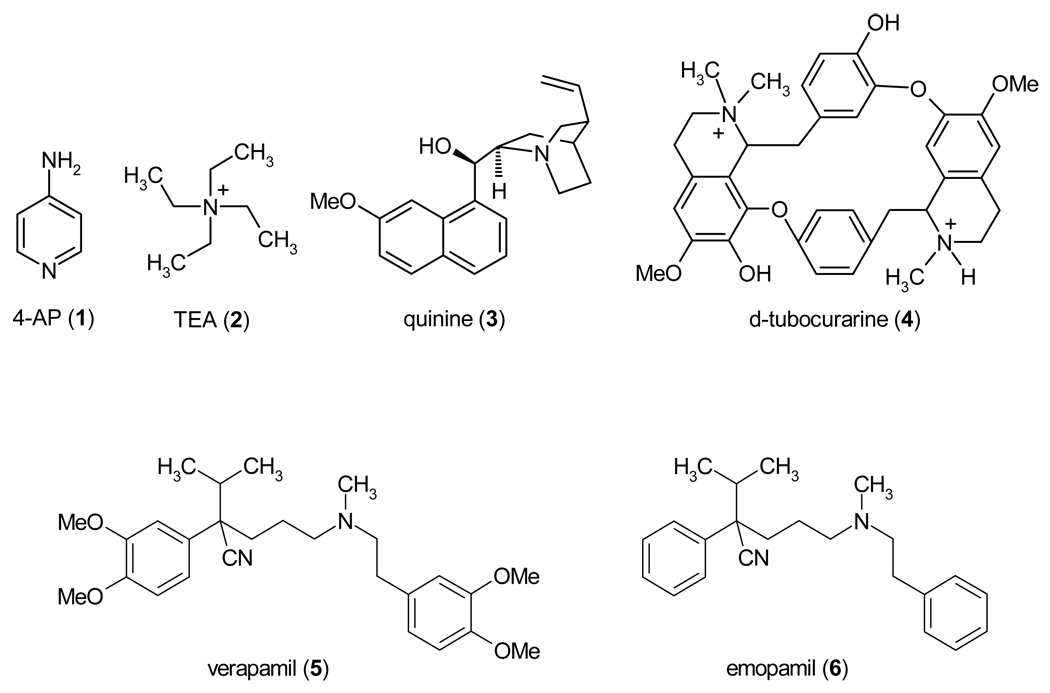
1.2.2. Small Molecules
In the early days of studying ion channels, the two agents that were used to pharmacologically identify K+ channels were 4-aminopyridine (4-AP, 1) and tetraethylammonium (TEA, 2), which inhibit many K+ channels in the high-micromolar or millimolar range, but have little or no effect on Na+ and Ca2+ channels. For some K+ channels like Kv3.1, which have no other known peptide or small molecule inhibitors, 4-AP (1) and TEA (2) still remain the only available blockers (see Table 1–Table 4). Other broadly active K+ channel blockers include quinine (3), d-tubocurarine (4) and verapamil (5). These drugs are organic cations and block open K+ channels by binding in the inner pore. However, before discussing their binding sites, it is necessary to mention binding sites for inorganic cations in K+ channels. Seven K+ binding sites are seen in the crystal of the KcsA-FAB complex in high K+ concentration.68 Sometimes these sites are designated S1 – S7, but since S1 – S6 are generally used for transmembrane helical segments in P-loop channels, we use designations T1 to T7 (Fig. 3B). Occupancy of sites T1 – T4 in the selectivity filter by ions and water molecules alternates during the permeation process69 and may depend on the channel state and physiological conditions. A metal ion in the center of the cavity (site T5) is stabilized by electrostatic interactions with P-helixes.70 Another site at the cytoplasmic entrance of the selectivity filter is seen in the complex of KcsA with Cs+.71 We designate this site T4’. Ions at sites T4’ and T5 do not form multiple coordinating bonds with the channel and may oscillate in broader regions than octa-coordinated ions in sites T1 – T4. Therefore, T4’ and T5 designate regions rather than points.
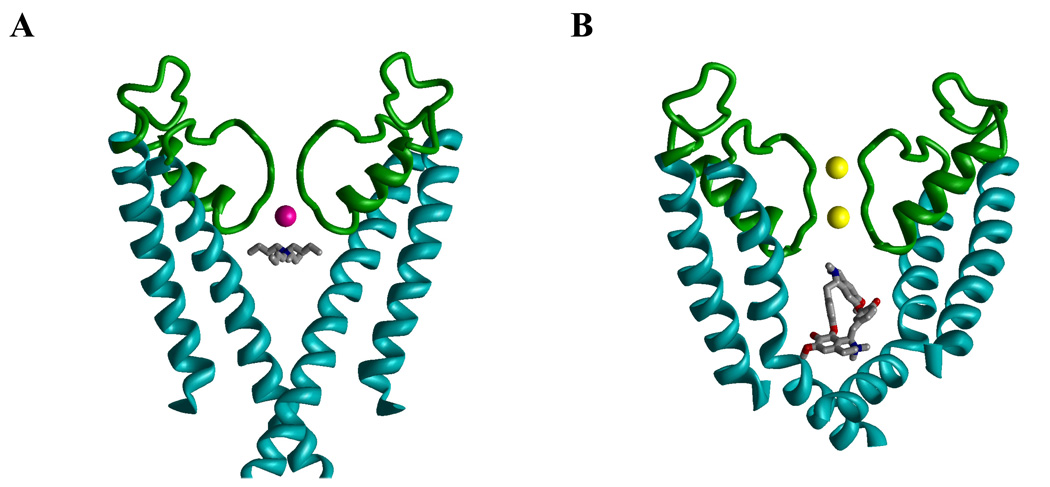
Tetraalkylammonium compounds like TEA (2) have long been known to block K+ channels from the cytoplasmic side.72,73 The X-ray structures of co-crystals of KcsA and tetrabutylammonium (TBA) show the TBA molecule trapped between sites T4’ and T5 below the selectivity filter of the closed channel (Fig. 4A).68,74–76 Reconstruction of the experimental electron-density maps from molecular-dynamics data indicates that the exact position of TBA along the pore axis depends on how the selectivity filter is occupied by permeating ions.75 Understandably, electrostatic repulsion between an ion in site T4, which is most proximal to the inner pore, shifts TBA towards the cytoplasmic direction.75 In KcsA, the shift of the trapped TBA in the cytoplasmic direction is limited by the closed activation gate (Fig. 4A), but in the open channel the effect of the selectivity-filter occupancy on the binding mode of a cationic ligand may be more pronounced as predicted by a model of a local anesthetic in an open Na+ channel.77 In the absence of X-ray structures of open K+ channels with an open-channel blocker bound, structures of such complexes can only be suggested from studies, which combine experimental and molecular-modeling approaches. Docking of the dicationic d-tubocurarine (4) in the Kv1.2-based homology model of Kv1.3 with K+ ions in sites T1 and T3 (Fig. 4B)25 predicted that an ammonium nitrogen occupies approximately the same position between sites T4’ and T5 as the nitrogen atom of TBA in KcsA.68,74–76 The second ammonium group and ether oxygens of d-tubocurarine (4) are not involved in strong electrostatic or H-bonding interaction,25 which can explain the rather low affinity of the drug to Kv1.3.
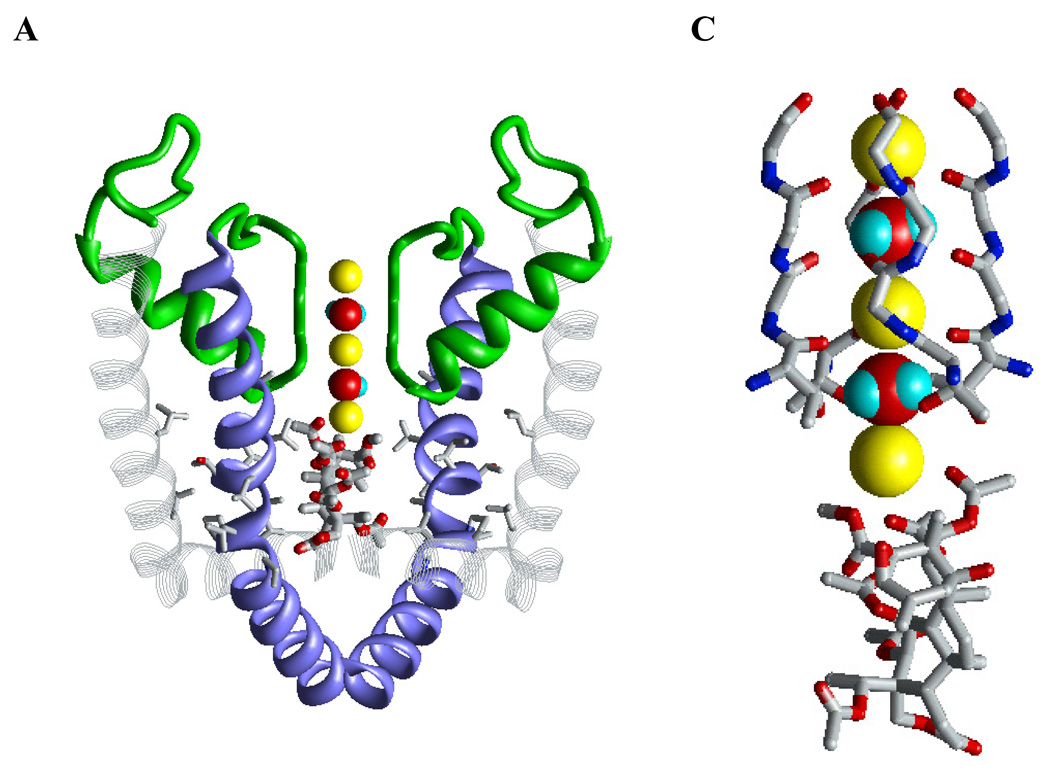
Figure 4. Cationic ligands in the inner pore of K+ channels.
A, X-ray structure (PDB Index 2BOB) of tetrabutylammonium trapped in the closed conformation of KcsA. The thallium ion in position 4 of the selectivity filter is colored magenta. B, Kv1.2-based model of the open Kv1.3 with d-tubocurarine. K+ ions in positions 1 and 3 are yellow. In both complexes, the ammonium group of the ligand is at the focus of the macrodipoles of the P-loop helices (green).
N-terminal parts of S6s and P-loops line four “niches” in the inner pore (Fig. 3C) that can harbor terminal moieties of some ligands, e.g. four methyl groups of TBA. Verapamil (5), a cardiovascular drug targeting L-type Ca2+ channels, also blocks the inner pore of the open Kv1.3 and cysteine substitution of Alai11(413), which lines the niche, dramatically reduced verapamil potency.33 Intriguingly, verapamil (5) blocks the mutant with 1:1 stoichiometry, but its derivative emopamil (6), which lacks all four methoxy groups, blocks the mutant with 2:1 stoichiometry,33 To explain this observation, Dreker and Grissmer proposed that the unsubstituted phenyl rings of emopamil (6) penetrate the niches deeper than the dimethoxy substituted phenyl rings of verapamil (5) and that the inner pore therefore can accommodate two molecules of emopamil (6), but only one molecule of verapamil (5). Despite the fact that the protonation state of the tertiary amino groups of the two emopamil (6) molecules and their position in the inner pore remain unknown, this study suggests that residues lining the niches interact with drugs. In some K+ channels, the niches have polar residues, which may be targets for subtype-specific ligands. For example, Kv1.2 channels are insensitive to the antiarrhythmic drug propafenone, but replacement of the Vp44VSp46 motif with the TIT motif from the propafenone-sensitive channel Kv2.1 confers propafenone sensitivity to Kv1.2.78
Hydrophobic cations like TBA, d-tubocurarine (4), and verapamil (5) block K+ channels by physically occluding the inner pore and inserting their ammonium group into the ion permeation pathway. Another mechanism is described for 4-AP (1), which blocks the Shaker channel by promoting activation-gate closure.79–81 Interestingly, 4-AP (1) demonstrates some features characteristic for hydrophobic cations: it enters the open Shaker, competes with TEA (2) for binding in the open channel, and remains trapped in the closed channels.80 However, the small size of 4-AP (2) and lack of groups ionizable at physiological pH does not allow the drug to physically occlude the permeation pathway.
In addition to the above described relatively low-affinity and unselective small molecule K+ channel blockers, medicinal chemistry efforts in both the pharmaceutical industry and in academia have identified a number of small molecule K+ channel modulators, which are potent and selective enough to be developed into drugs. Some of these compounds like the Kv7 channel activator flupirtine are already in clinical use and several other compounds are currently in various stages of clinical trials. However, it is not our intent to review all ongoing K+ channel drug discovery efforts in all therapeutic areas in this article. In the following two sections we will focus on K+ channel modulators for the treatment of neurological disorders and autoimmune diseases and use these compounds as examples of the evolving medicinal chemistry of K+ channels.
2. K+ Channel Modulators for the Treatment of Neurological Diseases
K+ channels have equilibrium potential close to −80 mV. They are therefore ideally suited to set the resting membrane potential, a task they perform in most cells. In neurons and other excitable cells, K+ channels are also crucial for determining the shape, the duration, and the frequency of action potential firing. In order to adjust these functions to the specific requirements of a particular neuron, the different K+ channels often show subfamily-specific patterns of cellular and subcellular localization. For example, Kv1 channels are predominantly found on axons and nerve terminals, Kv2 channels in the soma and in dendrites, Kv3 channels in dendritic or axonal domains, and Kv4 channel in somatodendritic membranes.82
The neuronal K+ channels, we are focusing on in this review, the voltage-gated Kv7.2–7.5 channels and the calcium-activated KCa1.1 and KCa2.1–2.3 channels also serve very specific physiological functions in certain types of neurons. Kv7.2–7.5 channels underly the so-called M-current in hippocampal and cortical neurons, which plays a critical role in determining the sub-threshold excitability and responsiveness to synaptic inputs. The Ca2+-activated KCa1.1 and KCa2 channels are involved in the neuronal afterhyperpolarization (AHP), which prevents the immediate initiation of a second action potential and KCa channels thus act as modulators of action potential frequency. Though not necessarily existent together in all neuronal subtypes, three components of the AHP have been described: a fast component lasting 50 msec (fAHP) carried by KCa1.1, a medium component decaying in 200 ms (mAHP) partly carried by KCa2 channels, and a slow component of unknown molecular identity, that decays in roughly 1–2 sec (sAHP). As discussed in the following, activation or inhibition of neuronal Kv7 and KCa channels offers exciting therapeutic possibilities for intervening in epilepsy, ataxia, dementia, stroke, and neurophatic pain.
2.1. Kv7.2–7.5 (KCNQ) Channels
Kv7 (KCNQ) channels are “classical” Kv channels with six transmembrane segments, including a voltage-sensor segment S4 and a long intracellular C-terminus, which contains binding sites for regulatory molecules such as calmodulin and phosphatidylinositol-4,5-biphosphate (PIP2).83 The family consists of five members, Kv7.1–7.5, which share between 30 and 65% amino acid identity.5 While Kv7.1 (KCNQ1, KvLQT) channels are found in the heart, peripheral epithelia and smooth muscle, Kv7.2–7.5 (KCNQ2-5) channels are expressed in hippocampal and cortical neurons and in regions of the nervous system involved in neuropathic pain such as the dorsal and ventral horn of the spinal cord and dorsal root and trigeminal ganglion neurons. In these neurons Kv7.2–7.5 channels underlie the so-called M current, a noninactivating current, which exhibits significant conductance in the voltage-range of action potential generation. Consequently, the M current tends to allow the firing of single action potentials, but opposes sustained membrane depolarization and repetitive firing. The M current has therefore often been likened to a “brake” on neuronal firing. The current was first described by Brown and Adams in 1980 in frog sympathetic neurons and named M current (IM) because of its inhibition by muscarinic agonists.84 Today it is relatively well established that several common neurotransmitters such as acetylcholine, substance P, or bradykinin close M channels by stimulating G-protein coupled receptors like M1 muscarinic acetylcholine receptors or bradykinin B2R receptors.83 Although down-stream signaling from these receptors is complex, the primary mechanism for receptor-induced M channel closure seems to be the fall in membrane PIP2 levels following the receptor mediated activation of phospholipase-Cβ, which hydrolyzes PIP2 in the inner leaflet of the plasma membrane. When PIP2 levels fall below the levels, which are required for Kv7 channel activity,85,86 the channels close resulting in membrane depolarization and leading to increased neuronal excitability.83
As demonstrated by the group of David McKinnon, the “classical” M channel in sympathetic neurons consists of Kv7.2/Kv7.3 heteromultimers.87 However, Kv7.4 and Kv7.5 can also form functional M channels and have been proposed to be involved in certain neuronal pathways like the auditory system (Kv7.4) or the cerebral cortex (Kv7.5).83 In keeping with the physiological importance of the M current, homozygous Kv7.2−/− mice die within a few hours after birth, while heterozygous Kv7.2+/− mice with decreased expression of Kv7.2 showed hypersensitivity to pentylenetetrazole-induced seizures.88 In contrast, mice where Kv7.2 expression was drastically reduced but not completely abolished through a dominant-negative suppression approach, exhibited spontaneous seizures, behavioral hyperactivity and morphological changes in the hippocampus.89 Reduction of Kv7.2 expression after the completion of hippocampal development in adult mice produced deficits in hippocampus-dependent spatial memory suggesting a critical role for M channels in cognitive performance.89 In humans missense or deletion mutations in Kv7.2 or Kv7.3 cause benign familial neonatal convulsions (BFNC), an autosomal dominant epilepsy of infancy.90–92 Although the seizures in BFNC typically resolve by 3 months of age, BFNC is associated with an increased incidence of various forms of epilepsy later in life suggesting a strong link between Kv7.2 and Kv7.3 mutations and epilepsy.6 Loss of function mutations in Kv7.4 in contrast result in hearing loss presumable due to an important function of Kv7.4 in sensory outer hair cells.93
2.1.1. Kv7 Channel Blockers
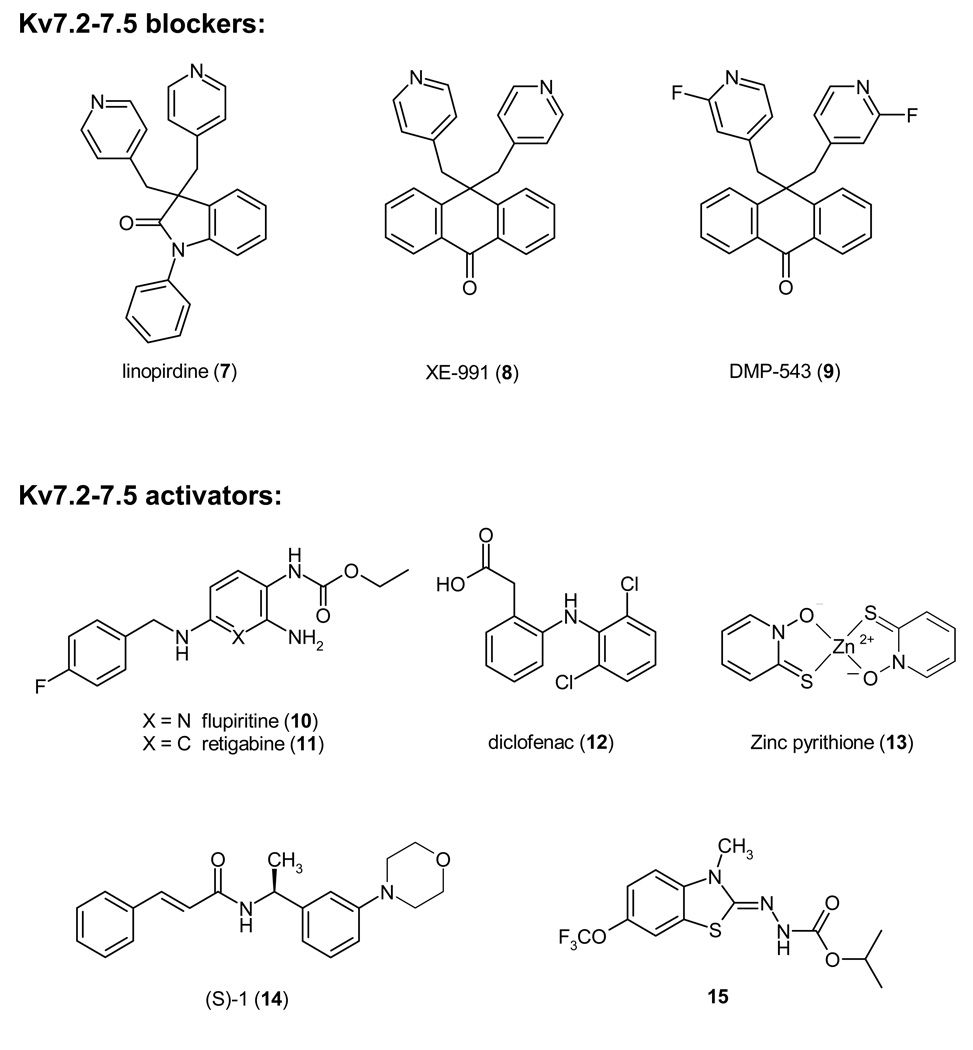
In the late 1980s DuPont initiated clinical trials with the phenylindolinone DuP 996, which is now generally called linopirdine (7), for Alzheimer’s disease.94 Linopirdine was regarded as a promising drug because it enhanced the release of acetylcholine in cholinergic nerve terminals in the brain only when its release was triggered and improved learning and memory in rodents and primates. Although clinical trials with linopirdine remained largely inconclusive95 or showed that it did not improve memory performance in elderly subjects,96 linopirdine became a valuable pharmacological tool. In 1995 Aiken et al. reported that linopirdine blocks the M current in rat CA1 pyramidal neurons and suggested that IM blockade might be responsible for the enhancement of neurotransmitter release by linopirdine because the two effects had similar IC50/EC50 values.97 This observation was later confirmed when Kv7.2 and Kv7.3 were cloned in 1998 and linopirdine was found to inhibit Kv7.2 channels or heteromultimers consisting of Kv7.2 and Kv7.3 with the same IC50 of 4 µM87 as the M current in sympathetic neurons.98 Two more potent anthracenone analogs of linopirdine are XE991 (8) and DPM-543 (9), which enhance [3H]-acetylcholine release from rat brain slices about 5–10 fold more potently than linopirdine99 and in the case of XE991 (8) inhibit Kv7.2/Kv7.3 heteromultimers with an IC50 of 600 nM.87 However, after the failure of linopirdine the pharmaceutical industry ceased to show interest in Kv7 channel inhibitors and no further compounds were developed. The impairment in hippocampus-dependent spatial memory observed in the above-mentioned mice, where Kv7.2 was conditionally suppressed,89 also raises doubts about whether Kv7 channel inhibition indeed constitutes a valid target for cognition enhancement. Interestingly, so far no venom-derived peptides have been identified that block Kv7 channels.
2.1.2. Kv7 Channel Activators
In contrast to Kv7 channel blockers, Kv7 activators are currently generating a lot of interest as potential drugs for the treatment of epilepsy and neuropathic pain after several clinically used drugs were retrospectively shown to activate Kv7 channels.6,100,101 In the early 1990s Asta Medica submitted a collection of compounds including flupirtine (10), a non-opioid centrally acting analgesic, which was marketed in Europe since 1984 for the treatment for postoperative and cancer pain,102 to the NIH Anticonvulsant Screening Program, which randomly tests compounds submitted by academic laboratories or pharmaceutical companies in mouse models of epilepsy (http://www.ninds.nih.gov/funding/research/asp/). Since flupirtine was found to have potent anticonvulsant effects, chemists at Asta Medica in collaboration with scientists from the NIH Antiepileptic Drug Development Program optimized the molecule for anticonvulsant activity by removing the basic nitrogen atom in the pyridine ring and reported desazaflupirtine (D-23129) in 1996, now called retigabine (11), as an orally active broad spectrum anticonvulsant.103 Retigabine subsequently entered clinical trials at Valeant Pharmaceuticals Inc. It recently was reported to reduce seizure frequency in a dose-dependent manner in Phase-2 clinical trials104 and is currently undergoing two large Phase-3 clinical trials,105 which are expected to be completed in summer 2008. After retigabine (11) was initially believed to enhance GABAergic transmission, it was independently shown by three groups in 2000 to activate Kv7.2/Kv7.3 channels with an EC50 of about 1 µM by shifting the voltage-dependence of activation by 20 to 30 mV to more negative potentials and by markedly slowing channel deactivation kinetics.106–108 Subsequent studies demonstrated that retigabine (11) also activates Kv7.2 and Kv7.3 homotetramers as well as Kv7.4 and Kv7.3/Kv7.5 channels with roughly the same potency, but only affects the cardiac Kv7.1 channel at 100-fold higher concentrations.109 Through site-directed mutagenesis Wuttke et al. located retigabine’s binding site in a putative hydrophobic pocket formed upon channel opening between the cytoplasmic parts of S5 and S6 involving Trp236 in S5 and Glyi14(301) in S6, which is proposed to be the “gating-hinge”.110 Two other “old drugs”, which were recently reported by a group from Tel Aviv University to activate Kv7.2/Kv7.3 channels are the cyclooxygenase (COX) inhibitors meclofenac (EC50 25 µM) and diclofenac (12, EC50 2.6 µM).111 Both compounds enhanced the native M-current in cultured cortical neurons and diclofenac (12) effectively suppressed electroshock-induced seizures in mice with an ED50 of 43 mg/kg.111 These findings suggest that activation of neuronal Kv7 channels could contribute to the analgesics effects of diclofenac.
Reflecting the great interest of the pharmaceutical industry Kv7 activators belonging to many different chemotypes have been reported in the last 5 years. These include: benzanilides, benzisoxazoles, indazoles, oxindoles including the KCa1.1 activators BMS-204352 (24), quinolinones, 2,4-disubstituted pyrimidine-5-carboxamides, phenylacrylamides, 5-carboxamide-thiazoles, benzothiazoles, quinazolinones and salicylic acid derivatives. The interested reader is referred to several recent review articles and book chapters on neuronal Kv7 activators.100,101,112 Out of these compounds we here only show the morpholino-substituted phenylacrylamide (S)-1 (14, EC50 3 µM for Kv7.2), which is effective in a rat model of migraine,113 the recently reported zinc pyrithione (13, EC50 1.5 µM for Kv7.2), which was identified at the High Throughput Biology Center at Johns Hopkins University,114 and the benzamide ICA-27243 (15), which was recently described by Wickenden et al. at Icagen. 115 ICA-27243 (15) activates Kv7.2/K7.3 channels with an EC50 of 200–400 nM and suppresses electroshock-induced seizures in rodents following oral administration. 115 Another Kv7 channel activator of undisclosed structure from Icagen (ICA-105665) has recently entered Phase-1 clinical trials for the treatment of epilepsy.
Taken together, the neuronal Kv7.2–7.5 channels constitute promising new targets for the treatment of epilepsy, pain of various etiologies, and possibly migraine and anxiety.116 It is currently not clear whether highly subtype specific activators will exhibit any advantages over the currently available more broadly active compounds, but it is feasible that subtype selective activators might cause less dizziness and somnolence than retigabine.
2.2. KCa1.1 (BK, Maxi-K) Channels
KCa1.1 channels are activated by intracellular Ca2+, depolarization, or a combination of both Ca2+ and voltage and are characterized by an unusually high single channel conductance of 250 pS, which is responsible for the different names the channel has received: BK for “big”, large-conductance, or Maxi-K. The channel was first cloned in 1991 from a Drosophila mutant117 (named slowpoke because of its lethargic phenotype) and two years later by the group of Lawrence Salkoff from mouse brain.118 In contrast to other Kv and KCa channels, KCa1.1 has seven transmembrane segments (S0–S6) instead of six, an extremely long intracellular C-terminus containing four additional hydrophobic segments (S7–S10) and a so-called “calcium-bowl”, which together with the RCK (regulators of conductance of K+) domain in S7 and S8 is responsible for the complex calcium-dependent gating of the channel (see Salkoff et al.13 for a recent review). Another salient feature that distinguishes KCa1.1 from other K+ channels are two rings of negative charges in the inner and outer vestibules, which have been proposed to increase K+ concentration in the vicinity of the selectivity filter and could explain KCa1.1’s high single channel conductance.119,120 KCa1.1 channels in different tissues can vary significantly in their sensitivity to Ca2+, inactivation properties, and even pharmacology due to alternative splicing of the α-subunit and co-assembly with four different auxiliary β-subunits.13 While β1 is primarily expressed in smooth muscle, hair cells and some neurons, β2 is found in ovary and endocrine tissue, β3 in testis, and β4 is the most abundant β-subunit in the brain.121
KCa1.1 channels are widely expressed throughout the body, however their physiological role is currently best understood in neurons, smooth muscle, secretory endocrine cells and sensory receptors. In the brain KCa1.1 is found in the soma, dendrites, and presynaptic terminals of neurons122,123 and is thought to underlie the fast afterhyperpolarization current and to regulate synaptic transmission by limiting the Ca2+ influx through Cav channels. In peripheral tissue, KCa1.1 is involved in regulating the tone of vascular, uterine, gastrointestinal, airway and bladder smooth muscle. However, contrary to initial reports by Ahluwalia et al.,124 KCa1.1 is not essential for innate immunity. Two subsequently published studies demonstrate that neither human nor mouse neutrophils express KCa1.1 and that pharmacological inhibition or genetic deletion of KCa1.1 has no effect on neutrophile function.125–127 KCa1.1 knockout mice are viable and exhibit a number of surprisingly mild phenotypes including ataxia, high-frequency hearing loss, hypertension, bladder over-reactivity, and erectile dysfunction.128–132 In humans a mutation resulting in increased KCa1.1 channel function has been found to be responsible for a rare form of generalized epilepsy.133 This seemingly counterintuitive phenotype is most likely due to faster repolarization caused by increased KCa1.1 function, which reduces the refractory period following action potentials. However, despite this human gain-of-function mutation, which sounds a note of caution against indiscriminately activating KCa1.1, KCa1.1 activators have often been proposed to be ideally suited to potentiate existing negative feed-back mechanisms in both neurons and vascular smooth muscle.
2.2.1. KCa1.1 Channel Blockers
KCa1.1 is most commonly pharmacologically characterized by its sensitivity to the scorpion toxins ChTX and iberiotoxin. While ChTX also blocks Kv1.2, Kv1.3, Kv1.6, and KCa3.1,42 iberiotoxin (IbTX, IC50 ~10 nM) is selective for KCa1.1.134 Other more recently identified and less commonly used peptide blockers of KCa1.1 are slotoxin (IC50 1.5 nM)135 and BmP09 from the Chinese scorpion Buthus martensi Karsch (IC50 27 nM).136 An interesting feature of peptide inhibition of KCa1.1 is that the β-subunit participates in the formation of the high-affinity toxin binding site either through a direct interaction between four residues in its large extracellular loop and the toxin bound in the channel pore or through an allosteric effect on the α-subunit.137,138 The presence of different β-subunits can thus drastically change the sensitivity of KCa1.1 to ChTX and IbTX. While β1 enhances toxin binding, complexes of 1 β4 with KCa1.1 are resistant to both toxins.14
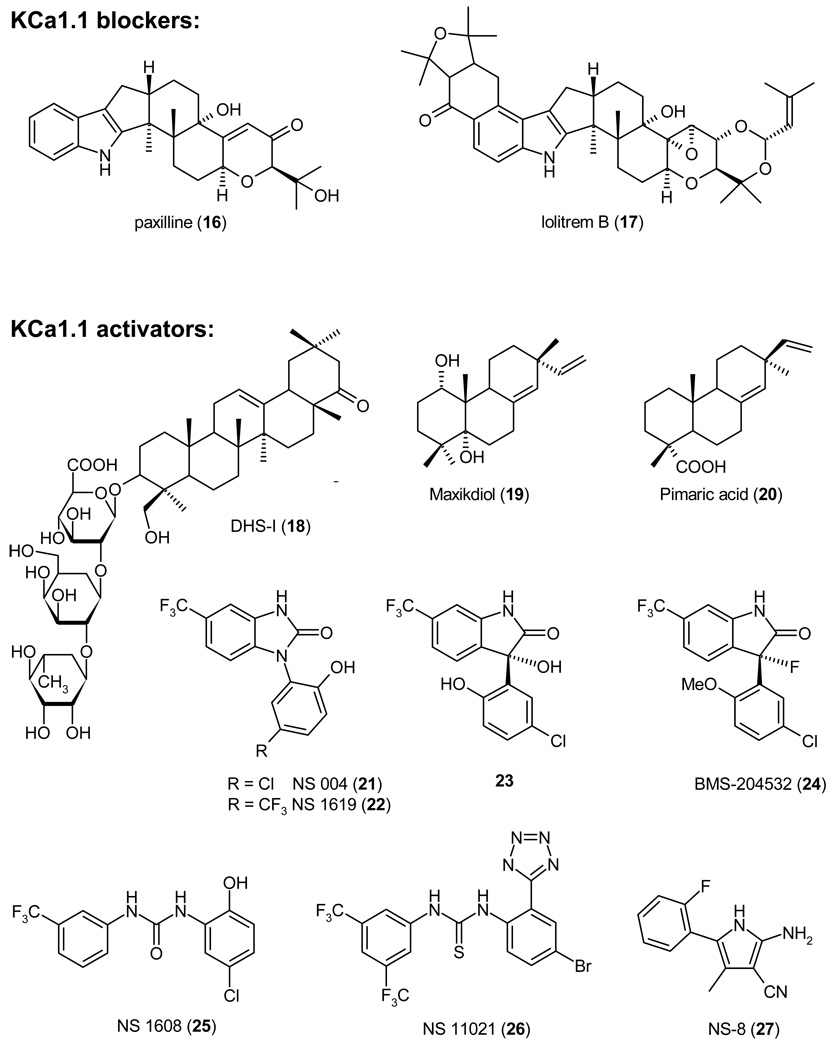
KCa1.1 blockers have been suggested for the possible treatment of depression and memory impairment. However, no major efforts have been seemingly done yet to identify non-peptide KCa1.1 blockers. The exception is some work performed by the Membrane Biochemistry and Biophysics group at Merck in the early 1990, where Maria Garcia and coworkers investigated whether the tremorgenic indole alkaloids, which are produced by fungi of the Penicillium, Aspergillus and Claviceps genera and which are known to cause staggers syndromes in animals that feed on contaminated grain, might block KCa1.1.139 The group found that several indole diterpenes including the now commonly used paxilline (16, IC50 2 – 50 nM depending on the intracellular Ca2+ concentration),140,141 indeed potently inhibit KCa1.1, but concluded that their tremorgenicity might be unrelated to KCa1.1 block.139 Paxilline binds to KCa1.1 at an intracellular site that is involved in channel gating and seems to be coupled to the calcium binding sites because of the antagonism between paxilline and Ca2+.140 However, the exact location of this site has not been identified. A more recently reported indole diterpene, which potently inhibits KCa1.1, is the mycotoxin lolitrem B (17, IC50 3.7 nM) from the endophyte Neotyphodium lolii, which commonly infects rye grass seeds.142
2.2.2. KCa1.1 Channel Activators
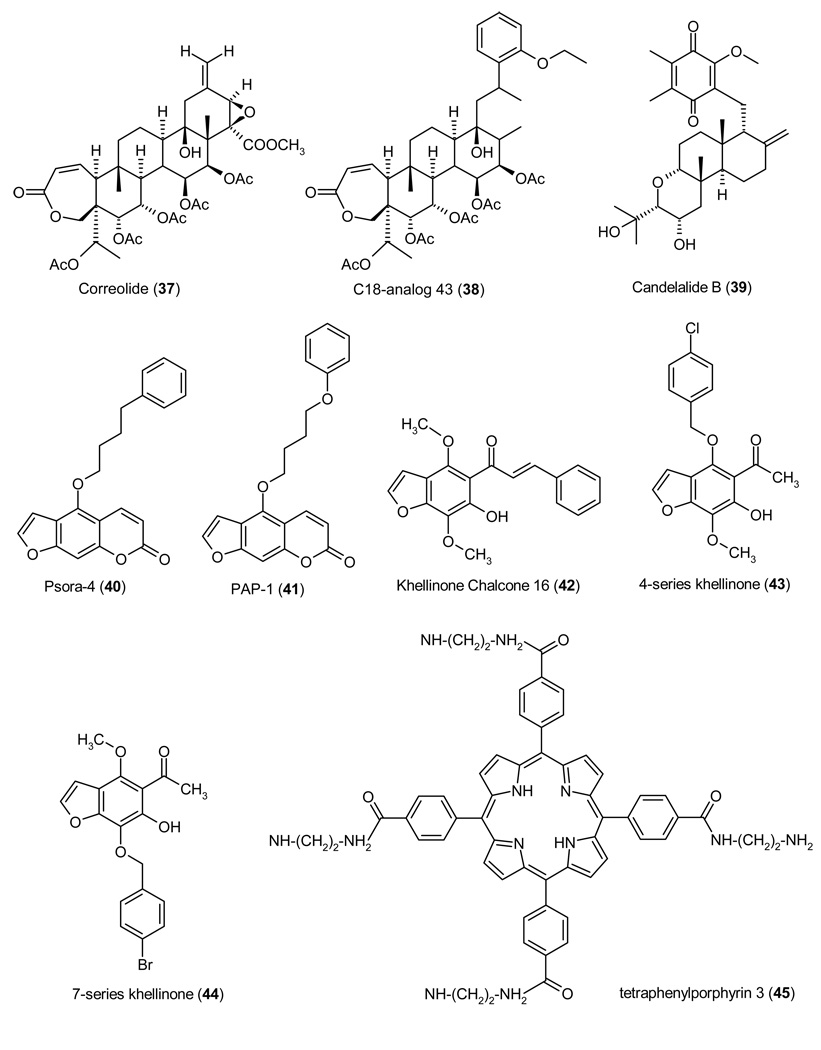
In contrast to the relatively small number of KCa1.1 inhibitors, a large number of both natural and synthetic KCa1.1 activators have been reported. Two KCa1.1 activators, BMS-204352 (24) and NS-8 (27), even advanced into clinical trials for stroke and overactive bladder. In 1993 the same group at Merck that had identified the KCa1.1 blockers IbTX and paxilline reported the isolation of three KCa1.1 activators from Desmodium adscendens, a medicinal herb used in Ghana as a treatment for asthma.143 NMR and mass spectroscopic analysis revealed the three active components: the known triterpenoid glycosides dehydrosoyasaponin I (DHS-I, 18), and its derivatives soyasaponin I, and soyasaponin III. DHS-I was reported to be a non-competitive inhibitor of 125I-ChTX binding to smooth muscle membranes containing BK channels, suggesting a binding site located outside of the pore but allosterically coupled to it. Subsequent studies performed by McManus et al. on oocytes injected with either KCa1.1 alone or KCa1.1 together with β1 revealed that DHS-I stimulates KCa1.1 activity only in the presence of the β-subunit.144 However, DHS-I (18) is difficult to study in physiological preparations despite its low nanomolar potency because it is poorly membrane-permeable and needs to be applied intracellularly in electrophysiological experiments to reach its intracellular binding site.143 A diterpenoid that activates KCa1.1 less potently than DHS-I is the 1,5-dihydroxyisoprimane maxikdiol (19), which was identified in 1994 at Merck in the fermentation broth of an unidentified coelomycite.145 A SAR study of diterpenoids structurally related to maxikdiol more recently showed that relatively small structural changes have a significant effect on the activity of terpenoid KCa1.1 activators.146 While maxikdiol (19) is only active if applied intracellularly and requires the presence of the β1 subunit, the pine resin acid pimaric acid (20) activates KCa1.1 channels consisting of only α-subunits (EC50 ~3 µM) and is freely membrane-permeable. Interestingly, sandaracopimaric acid, the diastereomer of pimaric acid was equipotent at increasing KCa1.1 activity, while abietic acid, which lacks the extracyclic double-bond, was ineffective.146 Another natural product that activates KCa1.1 is the benzopyran mallotoxin (EC50 0.5 µM) also known as rottlerin.147 However, mallotoxin is not specific for KCa1.1 and also activates Kv11.1 (hERG) channels.148 An endogenous compound that activates KCa1.1 channels by binding to an extracellular site on the β1-subunit is the female hormone 17β-estradiol (EC50 2.4 µM).149 This KCa1.1 activation has been suggested to be at least partially responsible for the direct vasorelexant effect of estrogens.
The first synthetic KCa1.1 activators were the benzimidazolones NS 004 (21) and NS 1619 (22), which were identified by the Danish company NeuroSearch A/S in 1992 and which have become widely used pharmacological tool compounds. In 1994 Olesen et al. reported that NS 004 (21) and NS 1619 (22) increased KCa1.1 currents in cerebellar granule and aortic smooth muscle cells at concentrations between 3 and 30 µM by shifting their current-voltage relationship towards more negative membrane potentials.150,151 In a subsequent study the same group showed that opening of the benzimidazolone ring into the diphenylurea NS 1608 (25) resulted in a 10-fold more potent KCa1.1 activator (EC50 2.1 µM).141 NS 1608 (25) caused KCa1.1 channels expressed in HEK-293 cells to open at less depolarized potentials (maximal shift of the I/V curve by 70 mV) and to deactivate more slowly. The effect of NS 1619 (22) and NS 1608 (25) is independent of the presence of internal Ca2+ demonstrating that the compound does not act by increasing the Ca2+ sensitivity of the channel. In contrast to DHS-I (18), NS 1619 (22) directly activates the α-subunit and does not require the presence of a β-subunit. This difference in the action of NS 1619 (22) and DHS-1 (18) has subsequently often been used to determine whether a β-subunit is present in native tissue. For example, Papassotiriou et al. concluded that BK channels in endothelium are composed of α-subunits without associated β-subunits because the current could be activated by NS 1619 but not by DHS-I,152 while Tanaka et al. argued that the BK channels in human coronary smooth muscle cells consist of both α- and β-subunits due to their sensitivity to DHS-I.153 A recently described more potent and selective diphenylurea KCa1.1 activator from NeuroSearch is the tetrazole substitute thiourea NS 11021 (26).154 NS 11021 activates KCa1.1 with an EC50 of 400 nM and in contrast to NS 1609 exerts no effect on L- or T-type Ca2+ channels at 30 µm. However, NS 11021 (26) is not perfectly selective and activates Kv7.4 and α7 nicotinic acetylcholine receptors at concentrations of 10–30 µM154 It remains to be seen if NS 11021 will replace the earlier NeuroSearch KCa1.1 activators as the most frequently used tool compound.
Scientists at Briston-Myers Squibb demonstrated that one of the nitrogen atoms of the benzimidazolones could be replaced by a carbon and generated a number of aryloxindole KCa1.1 activators exemplified by 23.155 However, 23 and its des-hydroxy analog were not further pursued as neuroprotective drug candidates in favor of the more metabolically stable BMS-204352 (24, Flindokalner or MaxiPost), which entered clinical trials for stroke. BMS-204352 (24) activates KCa1.1 channels expressed in HEK cells with an EC50 of 350 nM in a strongly Ca2+ dependent manner.156 With a calculated logP of 5.1 BMS- 204352 enters the brain quickly and reaches roughly 10-fold higher concentrations in the brain than in plasma. In permanent middle cerebral artery occlusion, an animal model of stroke, BMS-204352 reduced infarct areas measured at 24 hours in both normotensive and hypertensive rats by 20–30% when administered 2 hours after the beginning of brain ischemia at doses between 1 µg/kg and 1 mg/kg.156 However, BMS 204352 (24) displayed an interesting inverted-U shaped dose-response relationship and at doses of 3 mg/kg no longer reduced infarct sizes. It is currently not clear if this effect might be related to the activation of Kv7.2–7.5 channels seen at higher concentrations of BMS-204352.157,158 Based on the promising results in rodent models of stroke, BMS-204352 (24) entered clinical trials and was found to be well tolerated and safe in both Phase-1 and Phase-2 studies. However, BMS-204352 failed to show efficacy compared to placebo in a Phase-3 study involving about 2000 patients and was therefore discontinued.159 The idea behind using KCa1.1 activators as neuroprotectants was to hyperpolarize neurons and protect them from “excitotoxic” cell death. Following an ischemic stroke, neurons surrounding the core of the infarct tend to die through a combination of low oxygen tension, excessive excitatory amino acid release, and elevation in intracellular Ca2+ (“Ca2+-overload”). KCa1.1 activators are thought to be able to block Ca2+ entry through Cav channels and NMDA receptors by hyperpolarizing neurons and to thus prevent neurotransmitter release and Ca2+-overload.156 The reasons for why BMS-204352 (24) has failed to show benefit in humans might be numerous, but the major reason was probably the timing of the administration after the infarct. In rats BMS-204352 was always administered 2 hours after the occlusion. However, for logistic reasons this was not possible in stroke patients and the drug was given at various times within the first 48 hours after stroke onset. Other CNS indications for which KCa1.1 activators have been proposed include epilepsy and pain. However, very little has been published in this area and it is currently not clear how useful KCa1.1 activators will be for these indications and whether they exhibit any advantages over the Kv7 channel activators.
In addition to CNS indications, KCa1.1 activators could also be useful for a number of peripheral indications such as urinary incontinence, erectile dysfunction, asthma and hypertension because of their ability to relax smooth muscle. One KCa1.1 activator, the arylpyrrole NS-8 (27) identified by Nippon Shinyaku, even advanced into Phase-2 clinical trials for overactive bladder160 but its development was stopped in January 2007 due to lack of sufficient efficacy at the expected therapeutic dosage.
2.3. KCa2 (SK) Channels
Small-conductance Ca2+-activated K+ channels were first described in 1982161 and later found to be encoded by three closely related genes, when they were cloned by Kohler et al. in 1996.162 Similar to the Kv channels, KCa2.1 (SK1), KCa2.2 (SK2) and KCa2.3 (SK3) channels have six transmembrane segments (S1–S6) and intracellular N- and C-termini.4 However, KCa2 channels only contain two positively charged amino acids in the S4 segment and are therefore insensitive to changes in membrane voltage. Instead, KCa2 channels are highly sensitive to increases in intracellular Ca2+. The latter activates the channels with EC50s of 300–700 nM163 by binding to calmodulin, which is constitutively associated with the intracellular C-terminus of the channel in a 1:1 stoichiometry.164 In expression systems both homomeric and heteromeric KCa2 channels are formed,165,166 but the existence of heteromeric KCa2 channels has so far not been confirmed in native tissues. Similar to KCa1.1 channels, KCa2 channels can be alternatively spliced to generate channels with modified calmodulin binding as in the case of KCa2.1,167 with an extended N-terminus dubbed SK2-L as in the case of KCa2.2,168 or without the N-terminus and first transmembrane segment as in the case of KCa2.3.169,170 Alternative splicing of KCa2.3 resulting in the insertion of fifteen additional amino acids in the outer pore region can also produce KCa2.3 channels, which are insensitive to the peptide toxins apamin and leiurotoxin-I.171
Despite their small conductance of only a few pS, KCa2 channels are powerful modulators of electrical excitability and exert profound physiological effects both within and outside the nervous system. In the mature rat brain, KCa2.1, KCa2.2 and KCa2.3 have a partially overlapping but clearly distinct distribution patterns. While KCa2.1 and KCa2.2 are co-expressed in the neocortex, hippocampus, and thalamus,163 KCa2.3 is primarily expressed in subcortical regions and in many monoaminergic neurons including the dopaminergic neurons of the substantia nigra.172,173 In all these neurons KCa2 channels underlie the apamin-sensitive medium afterhyperpolarization (mAHP).174,175 However, depending on the type of neuron and the other ion channels it expresses, the function of KCa2 channels varies from contributing to the instantaneous firing rate, over setting the tonic firing frequency, to regulating burst firing and potentially catecholamine release.163 Mice lacking KCa2.1, KCa2.2, or KCa2.3 are viable and do not exhibit any overt neurological phenotypes. 175,176 KCa2.2−/− mice lack the apamin-sensitive mAHP current in CA1 hippocampal neurons,175 while mice conditionally over-expressing KCa2.2 exhibit reduced synaptic plasticity and show impaired hippocampal learning and memory.176 Conditional KCa2.3 over-expression, in contrast, resulted in a greater bladder capacity, abnormal breathing patterns in response to hypoxia, and compromised parturition with protracted labor suggesting that KCa2.3 channels play a role in neurons regulating breathing patterns and uterine and bladder smooth muscle tone.177,178 Furthermore, KCa2.3 polymorphisms such as polyglutamine repeats and a truncation mutation,179 which can act as a dominant-negative suppressor,180 have been associated with schizophrenia181,182 and cerebellar ataxia.183
Outside of the nervous system KCa2 channels are involved in blood pressure regulation184 and metabolism. The role of KCa2 channels in blood pressure regulation seems to involve both catecholamine release from chromaffin cells in the adrenal gland185,186 as well as participation of KCa2.3 channels in the endothelium-derived hyperpolarization factor (EDHF) response.187 For more details on other peripheral indications the interested reader is referred to a recent extensive review on the physiology and pharmacology of KCa2 channels.188
2.3.1. KCa2 Channel Blockers
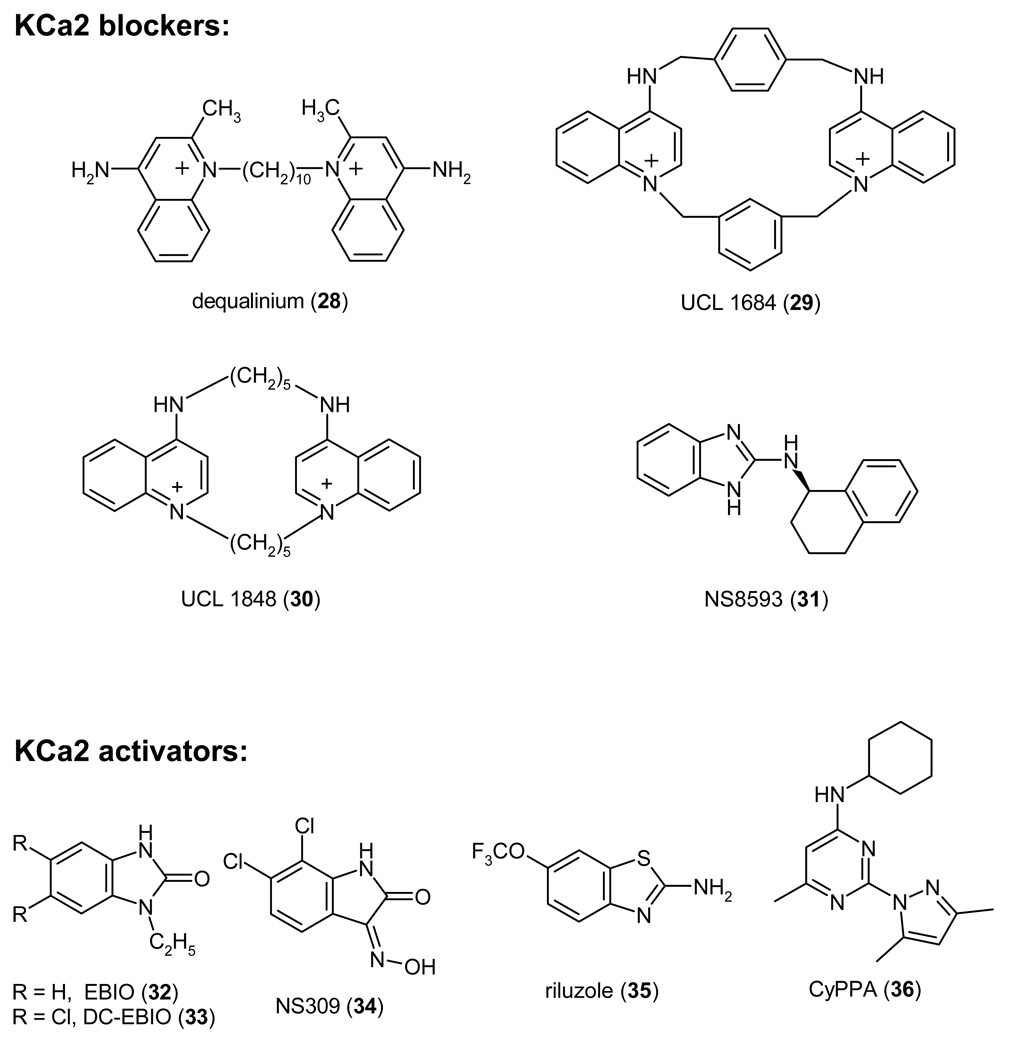
Since the early 1980s, the 18-amino-acid bee-venom toxin apamin has been the main pharmacological tool to distinguish KCa2 channels from the “apamin-insensitive” KCa channels KCa1.1 and KCa3.1.39 Following the cloning of the three KCa2 channels, apamin was found to be most potent on KCa2.2 channels (IC50 30–200 pM) and to block KCa2.1 (IC50 1–12 nM) and KCa2.3 (IC50 1–20 nM) channels with 10–50-fold lower affinity.162,163 Other pore blocking peptide toxins, which inhibit KCa2 channels, are the 31-amino acid scorpion toxins leiurotoxin-I (scyllatoxin)189,190 and tamapin.191 Both toxins have roughly the same potency as apamin and show comparable (leiurotoxin-I) or somewhat greater (tamapin) preference for KCa2.2. A common structural feature of apamin and leiurotoxin-I is the presence of a RXCQ motif,192,193 which is changed to RXCE in tamapin. This motif seems to be crucial for high-affinity blockade of KCa2 channels because other scorpion toxins, which lack this motif, show little or no effect on KCa2.2 and KCa2.3 currents.194 By replacing Met7 in the RMCQ sequence of leiurotoxin-I with the unnatural amino acid diaminobutanoic acid (Dab), Shakkottai et al. generated Lei-Dab7, which blocks KCa2.2 with 650-fold higher potency than KCa2.3 and KCa2.1.194 However, currently there are no natural toxins or analogs that selectively inhibit KCa2.1 or KCa2.3.
Apamin has been an invaluable tool for determining both the physiological role and the therapeutic potential of KCa2 channels. Intrathecal or systemic administration of apamin has been shown to enhance learning and memory responses in both mice and rats.195–198 Taken together with the observation that mice over-expressing KCa2.2 exhibit impaired hippocampal learning and memory,176 these findings suggest that selective KCa2.2 blockers might be able to function as “memory enhancers” and improve cognitive performance in dementia. However, higher doses of systemically administered apamin induce seizures cautioning that the therapeutic window for this application might be narrow. Based on the fact that apamin reduces immobility time as effectively as amitriptyline in the mouse forced swimming tests,199 KCa2 blockers have also been suggested for the treatment of depression and Parkinson’s disease.173 Selective KCa2.3 blockers might be particularly interesting in this respect because KCa2.3 underlies the mAHP in dopaminergic neurons of the substantia nigra.172,173
KCa2 channels are also potently blocked by a number of compounds containing two permanently charged or protonatable nitrogens, which are incorporated into aromatic rings connected through relatively variable linkers and separated by a distance of a little less than 6Å.200,201 Work on these compounds started in the early 80s when it was noticed that d-tubocurarine (4) and the antiseptic dequalinium (28) inhibited small-conductance KCa channels in the micromolar range.202–204 Using dequalinium (28) as a template Ganellin, Jenkinson and Galanakis at the Departments of Pharmacology and Chemistry at the University College of London (UCL) carefully explored the SAR around dequalinium in a large series of papers205–211 and eventually designed the bis-quinolinium cyclophane UCL 1684 (29), which blocks KCa2 channels as potently and as selectively as apamin. Like apamin, UCL 1684 inhibits KCa2.2 (IC50 200 pM) 3-fold and 50-fold more potently than KCa2.1 and KCa2.3.212,213 The subsequently published UCL 1848 (30), in which the aromatic xylyl linkers of UCL 1684 (29) were replaced by aliphatic pentylene groups,214,215 is even more potent and blocks KCa2.2 channel with an IC50 of 110 pM.166 Because of their relatively large molecular weight and permanent charge, these bis-quinolinium cyclophanes are not viable drug candidates. However they have become popular tool compounds because they wash out better than apamin from physiological preparations.
A recently reported KCa2 channel blocker of a completely different chemical structure is the benzimidazole derivative NS8593 (31).216 In contrast to apamin and the UCL compounds, which can all displace apamin and are therefore presumably also pore blockers, NS8593 (31) is an inhibitory gating modulator that decreases the Ca2+ sensitivity of KCa2 channels by shifting their Ca2+ activation curve roughly 10-fold to the right. At an intracellular Ca2+ concentration of 500 nM NS8593 inhibits KCa2.1, KCa2.2, and KCa2.3 with IC50 values of 420, 600 and 730 nM, respectively.216 It is currently not known what the in vivo effects of NS8593 (31) are and whether it exhibits less toxicity than the pore blocker apamin. Since NS8593 reduces the apparent Ca2+ sensitivity of KCa2 channels, it could potentially be more active in relatively slow-firing neurons with low Ca2+ concentrations and less effective in fast-firing neurons with presumably higher Ca2+ levels.216 KCa2 channel blockade has further been proposed to contribute to the therapeutic effects and possibly the side effects of tricyclic and phenothiazine antidepressants like amitryptiline, imipramine and trifluoperazine, which block KCa2.2 channels at concentrations of 8 to 60 µM.217,218 However, it is currently not clear if this really is the case.
Taken together, selective KCa2.2 blockers could potentially be useful as cognition enhancers in dementias like Alzheimer’s disease, while KCa2.3 blockers might be helpful for the treatment of depression and Parkinson’s disease. However, in the absence of any drug-like and brain penetrable small-molecule KCa2 channel blockers, these exciting possibilities remain untested. Another challenge for medicinal chemists will be the design of subtype-specific KCa2 blockers and only the future will show if compounds suitable for clinical development will emerge out of the ongoing efforts.
2.3.2. KCa2 Channel Activators
KCa2 channels are activated by several relatively simple heterocyclic molecules like ethylbenzimidazolone (EBIO, 32, EC50 87–450 µM) and its more potent derivative dichloro-EBIO (33, EC50 12–27 µM).219,220 In contrast to the KCa1.1 activating benzimidazolones NS 004 (21) and NS 1619 (22), which seem to activate KCa1.1 channels in a Ca2+ independent fashion, KCa2 channel activators increase KCa2 channel function by increasing Ca2+ sensitivity and therefore absolutely require the presence of at least 30–50 nM of Ca2+ in order to exert their effects.220 The structurally related oxime NS309 (34) is significantly more potent and activates all three KCa2 channels at submicromolar concentrations.221,222 In contrast to EBIO (32), NS309 (34) selectively activates the mAHP in hippocampal pyramidal neurons and has no effect on the slow afterhyperpolarization,222 which is carried by a currently not identified ion channel. Unfortunately, NS309 inhibits the cardiac K+ channel Kv11.1 (hERG) with an IC50 of 1 µM,221 which precludes its possible clinical use. Other compounds that show some structural similarity to EBIO (32) are the centrally acting muscle relaxants chlorzoxazone and zoxazolamine, which both activate KCa2.2 at high micromolar concentrations,219,223 and the more potent neuroprotectant riluzole (35), which activates KCa2.1, KCa2.2, and KCa2.3 with EC50s of 2–10 µM.188,224 Similar to EBIO and NS309, the benzothiazole riluzole (35) shifts the Ca2+ sensitivity of KCa2 channels causing them to open at lower intracellular Ca2+ concentrations.225 A recently reported KCa2 channel activator of a chemically different structure is the aminopyrimidine CyPPA (36) from Neurosearch, which activates KCa2.3 and KCa2.2 currents with EC50s of 6 and 14 µM, but has no effect on KCa2.1 or KCa3.1 at concentrations up to 100 µM.226 Although the potency of this compound is not high, its design demonstrates that it is in principle possible to obtain subtype-specific KCa2 channel activators.
In contrast to KCa2 blockers, which in general increase neuronal excitability, KCa2 channel activators reduce excitability and have therefore been proposed for the treatment of CNS disorders that are characterized by hyperexcitability. Based on the observation that over-expression of the KCa2.3 isoform SK3-1B, which suppressed KCa2 channels in deep cerebellar neurons of transgenic mice and thus increased hyperexcitability of these neurons, lead to severe ataxia with in-coordination, tremor, and altered gait,227 KCa2 activators have been suggested for the symptomatic treatment of cerebellar ataxia.227,228 Since there are currently no approved treatments for cerebellar ataxia, a Phase-2 clinical trial with the unselective KCa2 channel activator riluzole (35) is currently ongoing to test this exciting hypothesis (ClinicalTrials.gov identifier NCT00202397). KCa2 channel activators could potentially also be used as antiepileptic drugs. EBIO (32) inhibits epileptiform activity in an in vitro hippocampal slice model of epilepsy229 and reduces seizures induced by either electroshock or pentylenetetrazole when injected subcutaneously at concentrations of 10–80 mg/kg into mice.230 However, at 80 mg/kg EBIO induced profound sedation causing the authors of this study to conclude that KCa2 channel activation probably has a smaller therapeutic window than most currently used antiepileptic drugs. Unfortunately, the fact that EBIO also increases the slow APH and not only the KCa2 channel mediated medium APH,220,222 makes it hard to interpret both the in vitro and the in vivo results with EBIO. In future it will be interesting to see whether more potent and selective KCa2 activators than EBIO are effective in epilepsy models. It further remains to be seen whether KCa2 activators impair learning and memory as suggested by the phenotype of mice over-expressing KCa2.2.176 Based on findings made in mice over-expressing KCa2.3, selective KCa2.3 activation further constitutes a potential new therapeutic target for the treatment of hypertension and urinary incontinence.177,184 Taken together, KCa2 channel activation offers many therapeutic possibilities that await the discovery of more potent and ideally KCa2 channel subtype specific activators in order to be fully explored.
2.4. Other K+ Channel Modulators with CNS Activity
Neuronal K+ channels also seem to be the targets of several drugs, which have been clinically used for a long time. The two-pore K+ channels K2P2.1 (TREK-1) and several other K2P channels are activated by the volatile anesthetics halothane and isoflurane, which seem to bind to a region in the C-terminus of these channels.231 Based on the fact that K2P2.1−/− mice are resistant to anesthesia by volatile anesthetics, it seems likely that K2P2.1 channel activation indeed contributes considerably to the physiological effect of volatile anesthetics.232 K2P2.1−/− mice further show increased sensitivity to ischemia and epilepsy induction in comparison to K2P2.1+/+ mice suggesting that K2P2.1 activators might be useful as anticonvulsants and neuroprotective agents. Interestingly, the neuroprotectant riluzole (35) activates K2P2.1 and K2P4.1 channels at concentrations of 10–100 µM233 and it has been suggested that this effect contributes to riluzole’s ability to reduce infarct areas in mice subjected to focal cerebral ischemia.234 See Mathie et al. for a recent comprehensive review about the therapeutic potential of K2P channels.56
Two other K+ channel blockers, which have long been used for the treatment of multiple sclerosis and Lambert-Eaton syndrome are 4-AP (1) and its derivative 3,4-diaminopyridine.42 The use of 4-AP was originally based on the observation that 4-AP could improve conductivity in experimentally demyelinated nerve fibers.235 However, more recent work by Smith et al. demonstrated that at clinically used doses 4-AP fails to restore conduction in demyelinated rat axons, but potentiates synaptic transmission by increasing transmitter release at synapses.236 The potential targets for 4-AP include all 4-AP sensitive K+ channels in the Kv1, Kv3 and Kv4 families and it is currently not clear which of these channels is the primary target. It is further possible that inhibition of Kv1.3 in T cells contributes to the clinically observed effect.42,237 Unfortunately, 4-AP has a very narrow therapeutic window and its usefulness as a drug for multiple sclerosis is limited because of its propensity to induce seizures at higher doses.42
3. K+ Channel Blockers for the Treatment of Autoimmune Diseases and Inflammation
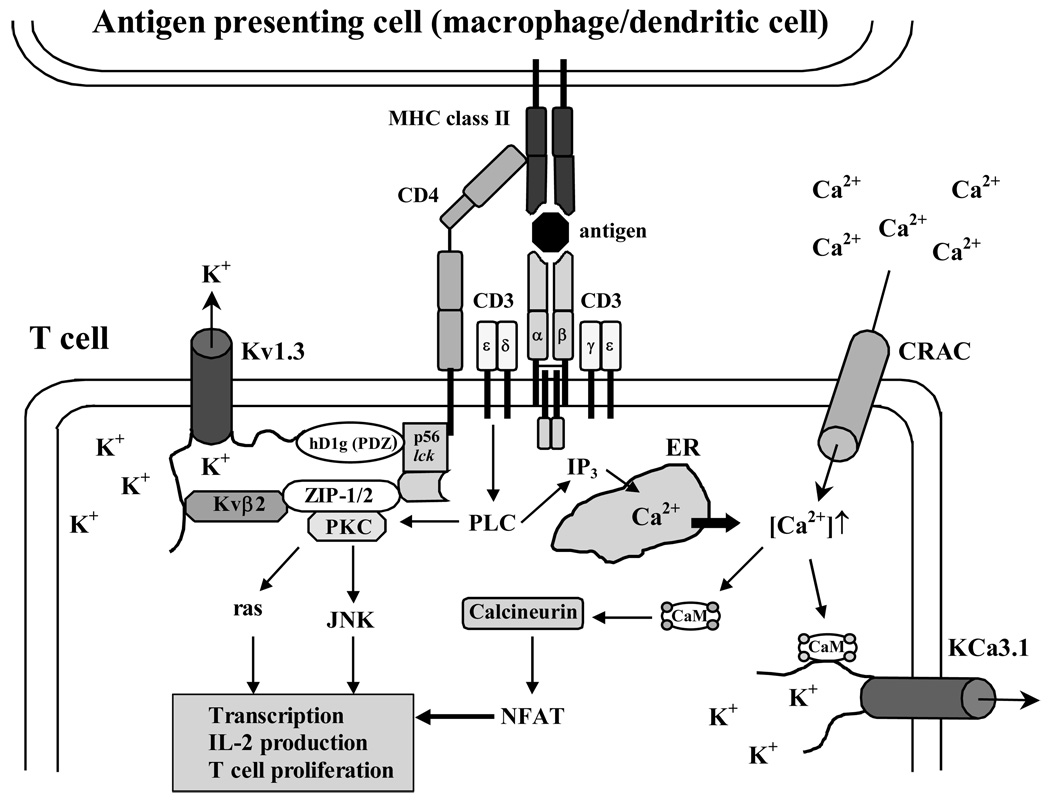
K+ channels do not only play an important role in the central nervous and cardiovascular system, they are also critically involved in regulating calcium signaling, proliferation, secretion and migration in non-excitable cells. In the immune system K+ channels are expressed in cells of both the innate and the adaptive immune system. The evolutionary older innate immune system, involves a number of mechanisms and cell types that defend us from infections by recognizing a variety of microbe-derived molecules such as LPS (lipopolysaccaride), peptidoglycans, bacterial lipoproteins, unmethylated CpG DNA, or double-stranded RNA through so-called pattern recognition receptors. Cells belonging to the innate immue system include natural killer cells, mast cells, eosinophils, basophils, and the phagocytic cells: monocytes/macrophages, neutrophils and dendritic cells. These cells can generate and release toxic molecules such as reactive oxygen species, engulf or kill microbes, and in the case of dendritic cells and macrophages present antigen to T cells. They are therefore also called antigen-presenting cells (APCs). Most innate immune cells seem to express K+ channels, although their function in the different cells types is currently not well understood and their expression insufficiently characterized. Human natural killer cells were reported to express Kv1.3-like Kv channels more than 20 years ago,238,239 but have not been studied after the cloning of the various K+ channels and it is currently not clear which other K+ channels they express. Human mast cells express KCa3.1 and probably also Kir channels.240,241 Human eosinophils require proton channels and do not seem to exhibit any significant K+ conductance.125 Human basophils have not been studied. Monocytes/macrophages are particularly confusing and depending on the species, the source of cells, the culture conditions, and their activation and differentiation status have been reported to express Kv1.3, Kv1.5, KCa1.1, KCa3.1, and Kir2.1.242–254 The Kv current in macrophages is most likely a heteromultimer consisting of Kv1.3 and Kv1.5.255,256 Microglia, which are brain-resident macrophages, resembles macrophages in many ways and have been shown to express Kir2.1, Kv1.3, Kv1.5, and the KCa channels KCa2.3 and KCa3.1.257–262 Neutrophils require proton channels for their oxidative burst125 and have recently been described to also express KCa2.3.263 The expression of K+ channels in dendtritic cells has not been studied.
In contrast to the innate immune system, K+ channel expression has been studied much more systematically in the adaptive immune system, which provides vertebrates with the ability to remember specific antigens (= immunological memory). Following repeated encounters with the same antigen, both T and B cells can differentiate into more reactive and long-lived memory cells. In the B cell lineage, memory cells produce high affinity antibodies, while memory T cells produce special cytokines and are more effective at providing help to B cells and macropages (CD4+ = helper T cells) or at killing target cells (CD8+ = cytotoxic T cells.) In parallel with this differentiation from naïve into memory cells, expression of the voltage-gated Kv1.3 and the Ca2+-activated KCa3.1 channel changes in both human T and B cells during activation and differentiation. In both lineages naïve and early memory cells (IgD+ B cells and CCR7+ T cells) express about 200–300 Kv1.3 channels per cell and very few KCa3.1 channels in the resting state. Following activation, naïve and early memory cells up-regulate KCa3.1 to 500 channels per cell with no change in Kv1.3 expression.264–266 “Late” memory cells (CCR7− effector memory T cells and IgD−CD27+ memory B cells), in contrast, increase Kv1.3 to 1500 or 2000 channels per cell with no increase in KCa3.1 expression.265,266 In consequence of this differential expression pattern, KCa3.1 is the functionally dominating K+ channel in naïve and early memory T and B cells, while Kv1.3 is the dominating K+ channel in effector memory T cells and class-switched memory B cells.7,266
The exact reason for why T and B cells change K+ channel expression during differentiation is currently not known, but it has been hypothesized that different Kv1.3 to KCa3.1 ratios allow cells to generate a differently shaped calcium signal, which in turn allows cells to express different genes.7 Figure 8 summarizes our current understanding of the roles of Kv1.3 and KCa3.1 during the activation of naïve human T cells. Antigen presented on the surface of an antigen-presenting cell activates the T cell receptor/CD3 complex. Through a number of tyrosine kinases, which are not shown in the cartoon, this leads to the down-stream activation of phospholipase-C, which catalyses the hydrolysis of membrane phosphatidylinosito-4,5-biphosphate into the two second messengers: inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 binds to and opens IP3 receptors on the membrane of the endoplasmic reticulum (ER), resulting in an initial small rise in cytoplasmic Ca2+. However, T cells are small and do not have sufficiently large Ca2+ stores in the ER to sustain T cell activation. T cells therefore require additional Ca2+ influx from the extracellular space through a voltage-independent Ca2+ channels called CRAC for Ca2+-release activated Ca2+ channel.267,268 The CRAC channel, which is encoded by the ORAI1 or CRACM1 gene,8,269 is activated by the stromal interacting molecule 1 (STIM1), which “senses” the decrease in the Ca2+ content of the ER through a Ca2+-binding EF-hand motif.270,271 However, the exact mechanism by which STIM1 activates the CRAC channel is currently not completely understood. Calcium influx through CRAC and the resulting increase in cytosolic Ca2+ lead to the translocation of NFAT (nuclear factor of activated T cells) and other transcription factors to the nucleus. The initiation of new transcription then ultimately results in cytokine production and T cell proliferation.268,272,273 How are Kv1.3 and KCa3.1 involved in this process? CRAC is an inward-rectifier Ca2+ channel that can only bring Ca2+ into cells at negative membrane potentials and closes at more positive potentials. The opening of Kv1.3 and KCa3.1 and the resulting K+ efflux generates a negative membrane potential of approximately −60 to −70 mV and thus facilitates and maintains Ca2+ entry for the entire duration of T cell activation.7,268,273 In other words, Kv1.3 and KCa3.1 provide the counterbalancing K+ efflux for the Ca2+ influx during T cell activation, and therefore constitute pharmacological targets for inhibiting T cell activation.7,274 However, based on the differential expression of KCa3.1 and Kv1.3 in naïve versus memory T and B cells, KCa3.1 blockers are probably more suitable for treating acute immune responses, while Kv1.3 blockers constitute promising new immunosuppressants for autoimmune diseases such as multiple sclerosis, rheumatoid arthritis, and psoriasis, where effector memory T cells are involved in the pathogenesis.237,275
Figure 8. Cartoon showing involvement of Kv1.3, KCa3.1, and CRAC channels in the activation of a CD4+ T cell by an antigen-presenting cell (APC).
Engagement of the T-cell receptor/CD3 complex through at an antigenic peptide presented in the context of major histocompatibility complex (MHC) class II activates protein kinase C (PKC) and generates IP3, which liberates Ca2+ from intracellular stores. The rise in [Ca2+]i activates the phosphatase calcineurin, which then dephosphorylates the transcription factor nuclear factor of activated T cells (NFAT) enabling it to accumulate in the nucleus and bind to the promoter of the interleukin-2 (IL-2) gene. Parallel activation of the c-Jun N-terminal kinase (JNK) and ras by PKC results in the activation of other transcription factors and initiates transcription of various genes and finally T cell proliferation. CaM: calmodulin; hD1g: human homologue of the Drosophila disc-large tumor suppressor protein.
3.1. Kv1.3 Channels
Kv1.3 is a classical Shaker-type K+ channel with 6 transmembrane segments including a voltage sensor in S4. The channel was cloned in 1990276,277 and shown to encode the Kv channel that had been discovered in human T cells in 1984.278,279 Apart from T cells, Kv1.3 channels are expressed in B cells,266 macrophages,254,280 microglia,258,261 fat cells,281 oligodendrocytes,282 and the olfactory bulb.283,284 In the brain Kv1.3 is generally only found as part of a heteromultimer with other Kv1-family subunits.285
Based on the observation that unspecific K+ channel blockers like 4-AP (1) and TEA (2) inhibited T-cell proliferation,278 George Chandy at the University of California, Irvine proposed Kv1.3 as a new potential target for immunosuppression in 1984. Investigators at Merck corroborated this hypothesis in 1997 by demonstrating that continuous infusion of the Kv1.3 blocking peptide margatoxin prevented delayed type hypersensitivity in mini-pigs.286 Initially, Kv1.3 blockers were viewed as general immunosuppressants and often proposed for the prevention of transplant rejection because it was assumed that they inhibited calcium influx in all T cell subsets equally well. They therefore seemed to simply constitute potential alternatives to the general immunosuppressants cyclosporine and FK506.287 However, the above described more recent discovery that only CCR7−effector memory T cells rely on Kv1.3, while CCR7+ naïve T and central memory T cells rely on KCa3.1, has changed this concept. Kv1.3 blockers are now regarded as immunomodulators that can selectively inhibit human effector memory T cells and spare naïve and central memory T cells.7,275 One fact that complicates Kv1.3 drug development is a species difference in lymphocyte K+ channel expression between mice and other animals. While Kv1.3 is up-regulated in activated human, rat, pig, and primate effector memory T cells,265,288,289 mouse effector memory T cells do not up-regulate Kv1.3 expression.290 In addition to Kv1.3 and KCa3.1, mouse T cells express Kv1.1, Kv1.2, and Kv1.6 in CD4+ T cells291 and Kv3.1 in CD8+ T cells.290,292 As expected from this species difference, Kv1.3−/−mice exhibit no defects in the immune system.293 The other phenotype observed in these mice, a decrease in body weight and improved insulin sensitivity,294,295 might also be a species difference. While mouse adipocytes certainly express Kv1.3 protein, electrophysiological studies performed with neonatal brown fat cells281,296 and cultured white adipocytes from rats and adult humans297,298 show Kv currents with properties that do not fit the pharmacological and biophysical characteristics of a current carried by Kv1.3 homotetramers (e.g. no use-dependence or ChTX sensitivity).
Similar to all other Kv channels, Kv1.3 activates in response to membrane depolarization. However, Kv1.3 shows two unique biophysical properties: C-type inactivation and a pronounced use-dependence during repetitive depolarizing pulses. With an inactivation time constant of 250 ms Kv1.3 inactivates slower than classical A-type currents but faster than classical delayed-rectifiers, which inactivate with time constants of more than 1 second. This type of inactivation, called C-type inactivation, is distinct from the N-terminal ball-and-chain mechanism of Shaker and is caused through a rearrangement of the external vestibule of the channel.299 The structural changes that occur during C-type inactivation are probably profound since Kv1.3 requires a considerable time, 30–60 sec depending on the length of the depolarizing pulses, in order to be ready to open again. Consequently, if a channel ensemble is subjected to fast depolarizing pulses, e.g. a 200 ms pulse every second, fewer and fewer channels are ready to open again with every pulse, a phenomenon that is called use-dependence. C-type inactivation is slowed down considerably by increasing the extracellular K+ concentration, presumably because K+ interacts with residues in the external vestibule that are involved in C-type inactivation.300 When testing Kv1.3 blockers, it is important to take these two properties of the channel into consideration. When using manual or automated patch-clamp, the pulse protocol has to be chosen appropriately in order to obtain a stable control current. It is further important to be aware of the fact that all assays, which use high extracellular K+ in order to depolarize the membrane and thus activate Kv1.3 channels, slow down C-type inactivation and can make Kv1.3 blockers that bind to the C-type inactivated state appear less potent than they would be under physiological conditions.
3.1.1. Peptidic Kv1.3 Blockers
Kv1.3 seems to be quite a “popular” target for venom-derived peptide toxins and is blocked by a large number of scorpion, snake, sea anemone, and cone snail toxins at picomolar to nanomolar concentrations. The scorpion toxins are particularly numerous and include charybdotoxin (IC50 3 nM),301,302 kaliotoxin (IC50 650 pM),303 margatoxin (IC50 110 pM),304,305 agitoxin-2 (IC50 200 pM),48 noxiustoxin (IC50 1 nM),306 Heterometrus spinnifer toxin 1 (IC50 12 pM).307 hongotoxin (IC50 86 pM),308 Pandinus imperator toxin 1 and 2 (IC50 11 nM and 50 pM),309 anuroctoxin (IC50 730 pM),310 and Orthochirus scrobiculosus toxin 1 (OSK1, IC50 14 pM).311 The most potent and selective toxin inhibitor of Kv1.3 is currently the OSK1 derivative OSK1-Lys16 Asp20, which blocks Kv1.3 with an IC50 value of 3 pM and shows >300-fold selectivity over closely related channels.311
As mentioned in section 1.2.1, the group of George Chandy used kaliotoxin (KTX) as a molecular probe to deduce the dimensions of the outer vestibule and the depths of the pore of Kv1.3.47,49 Using an early molecular model of Shaker by Durrell and Guy312 and the results from their mutant cycle analysis this group then generated a molecular model of the S5-P-S6 region of the Kv1.3 channel.47 In collaboration with Raymond Norton’s group in Melbourne, KTX was manually docked in this model by guiding Lys27 into the pore such that it would lie near the selectivity filter, and the toxin was then rotated about the central pore axis until Arg24 in the toxin aligned with Aspp38(386) in Kv1.3 in a salt bridge suggested by the mutagenesis. 47 Using high-resolution solid-state NMR spectroscopy, Lange et al. demonstrated that high-affinity binding of KTX to a chimeric KcsA-Kv1.3 channel involves structural rearrangements in both molecules.313 In particular, significant chemical-shift changes upon complex formation were observed for the channel residues Aspp38 and GYGDp54, which are directly involved in the KTX binding. Figure 9 shows a schematic presentation of the Kv1.3-KTX complex reconstructed from the Lange et al. model.313 The X-ray structure of Kv1.231 is superimposed on the solid-state NMR structure of KTX314 (PDB index 1SWX). The KTX backbone is green with space-filled residues R24, K27 and N30. The pore domain of Kv1.3 is shown as white backbones, cyan P-helices and space-filled Aspp38(386). It should be noted that side-chain conformations taken from non-bound structures of KTX and Kv1.3 and shown in Figure 9 are likely to differ from those in the KTX-Kv1.3 complex. Furthermore, since the reconstruction was not energy-minimized, details of the mutual disposition of KTX and Kv1.3 may differ from those obtained by Lange et al.315 with the help of NMR-derived constraints and molecular dynamics. Despite these limitations, the reconstruction shows that KTX blocks the ion permeation by inserting Lys27 in the outer pore, while R24 and N30 stabilize the complex by interacting with two of the four Aspp38 residues in the channel P-helices.
Figure 9.
Schematic presentation of the Kv1.3-KTX complex reconstructed from the model of Lange et al.312 A, Side view with two channel subunits removed for clarity. B, Top view with cytoplasmic half of the channel removed for clarity.
In the mid-1990s Kv1.3 blocking peptides were also discovered in the two Caribbean sea anemones Stichodactyla heliantus and Bunadosoma granulifera, which contain ShK and BgK, respectively.316–319 Of these two peptides the 35 residue ShK with reported IC50s ranging from 0.9 to 11 pM320–322 is the more potent and more widely studied molecule. However, in addition to potently blocking Kv1.3, ShK also displays picomolar affinity for Kv1.1 (IC50 25 pM) and Kv1.6 (200 pM) and blocks Kv3.2323,324 and KCa3.1 (IC50 28 nM)320 in the nanomolar range. The NMR solution structure of ShK325 shows a 310αα fold54 and a salt bridge between Asp5 and Lys30, which is conserved in all sea anemone K+ channel toxins.318,325 By performing extensive SAR around ShK in combination with mutant cycle analysis, George Chandy and co-workers refined their previous Kv1.3 vestibule model47 using restrained molecular dynamics simulations and the KcsA structure as a template.320 Interestingly, docking models of ShK and its more Kv1.3-selective derivative ShK-Dap22, which contains diaminopropionic acid (DAP) in place of the critical Lys22, show that the two peptides bind with a different orientation.326 While ShK occupies the pore with its positively charged Lys22 similar to the above described scorpion toxin KTX, the equivalent Dap22 in ShK-Dap22 interacts with Hisp56(404) and Aspp38 (386). Based on these models, attempts were made by Baell, Harvey and Norton at the Biomedical Research Institute in Australia to design type-III ShK peptidomimetics. However, their most potent compounds, which mimic ShK’s Lys22-Tyr23 diad plus Arg11 or Arg24, block Kv1.3 only with IC50s of 75 or 95 µM.327,328
ShK and its recently described more Kv1.3-selective analog ShK(L5) in which L-phosphotyrosine is attached to the N-terminus via an aminoethyloxyethyloxy-acetyl linker324 served as important tools to further validate Kv1.3 as a target for the treatment of effector memory T cell-mediated autoimmune disease. Both compounds suppress delayed type hypersensitivity and effectively prevent or treat adoptive-transfer experimental autoimmune encephalomyelitis in Lewis rats,288,324 an animal model of multiple sclerosis induced by the transfer of CD4+ myelin specific memory T cells. ShK-L5 has further been shown to significantly reduce the number of affected joints and to improve radiological and histopathological findings in pristane induced arthritis in Dark Agoti rats.237 Based on these experiments an analogue of ShK-L5 is currently in pre-clinical development for the treatment of multiple sclerosis and possibly other autoimmune diseases. The short plasma half-life of the peptides does not seem to constitute a major obstacle since ShK-L5 was extremely effective in rats with once daily subcutaneous administration.237 This could be due to the fact that the peptide partitions into a deep compartment that prevents its rapid elimination through filtration in the kidney.324 Alternatively, it may be possible that once daily suppression of activated effector memory T cells is enough to inhibit an on-going immune reaction. Interestingly, the recently published D-diastereomer of ShK,329 which is resistant to proteolysis, only has a slightly longer plasma half-life demonstrating that plasma clearance of peptide toxins (MW ~4000 d), is predominantly renal since these toxins are freely filtered in the kidney (size exclusion of the filter MW ~60 kd). D-allo-ShK (IC50 36 nM) retains biological activity but is more than 1000-fold less effective than naitive ShK (IC50 11 pM). Models of D-allo-ShK in the Kv1.3 pore suggest that it makes different contacts with the vestibule, some of which are less favorable than for native ShK.329
The high affinity of peptide toxins to Kv1.3 also makes these toxins attractive as fluorophore-tagged tools for channel visualization in living cells. This is especially useful for flow cytometry since there currently is no good monoclonal antibody that recognizes an extracellular epitope of Kv1.3 and that could be used to identify Kv1.3high effector memory T cells. By attaching fluorescein-6-carboxyl (F6CA) through a hydrophilic aminoethyloxyethyloxy-acetyl linker to the N-terminal Arg-1 of ShK, Beeton et al. generated such a probe and demonstrated that it specifically stained Kv1.3 expressing cells and was useful in flow cytometry.330 Using a similar approach, Pragle et al. visualized Kv1-family channels in unpermeabilized rat brain sections with hongotoxin conjugated to an Alexa dye via Cys19 on the “backside” of the toxin.331
3.1.2. Small Molecule Kv1.3 Blockers
Similarly to the other Kv1-family channels, Kv1.3 is blocked by 4-AP (1) and TEA (2) at micromolar to millimolar concentrations and by a number of promiscuous small molecule ion channel inhibitors like quinine (3), d-tubocuraine (4), verapamil (5), diltiazem, and nifedipine.7,332 The more potent and selective Kv1.3 blockers, which were described during the last 10 years, fall into two general groups. The first group includes typical combinatorial library compounds like the dihydroquinolone CP-339818, the piperidine UK-78282, and phenyl-stilbene A, which are of a relatively simple structure and are rich in nitrogen and halogen atoms. The second group consists of natural products or natural-product-derivatives like the terpenoids correolide and candelalide B, the psoralens and the khellinones, which are rich in oxygen atoms and have a more complex stereochemistry. Since we and others have previously extensively reviewed Kv1.3 blockers,7,42,274,275,332,333 we concentrate here on the more recently described compounds, their mode of action and potential to be developed into drugs.
Using a high-throughput 86Rb-flux assay scientists at Merck identified the nor-triterpene correolide (37), which contains an unusual α,β-unsaturated-7-membered lactone ring, in extracts from the bark and roots of the Costa Rican tree Spachea correa.334 Correolide blocks Kv1.3 with an IC50 of 90 nM264,335 and two correolide derivatives, which cause less hyperactivity than correolide in vivo, have been shown to suppress delayed type hypersensitivity in minipigs.336 However, because of their lack of selectivity over Kv1.2, Kv1.5, Kv1.6 and especially Kv1.1,337 correolide (37) and its derivatives increase the peristaltic activity of the gastrointestinal tract by increasing acetylcholine and tachykinin release.338,339 Since correolide (37) contains a total of 15 chiral carbon atoms, chemists at Merck have attempted to simplify it by synthesizing a number of derivatives in which the E-ring was removed.340 The most potent of these compounds, the C18-analog 43 (38), inhibited Kv1.3 with an EC50 of 37 nM in 86Rb-flux assays and suppressed T cell proliferation 15-times more potently than correolide.340 However, the Kv1.3-specificity of this compound has not been reported and it still requires correolide (37) as a starting material. The Merck group also isolated three novel diterpenoid pyrones called candelalides A–C from the fermentation broth of Sesquicillium candelabrum and reported that the most potent of these compounds, candelalide B (39), blocks 86Rb efflux through Kv1.3 channels with an IC50 of 1.2 µM.341
Through site-directed mutagenesis Hanner et al. revealed several correolide-sensing residues in segments S5 and S6 of Kv1.3, including the pore-facing residues Vali15, Alai19, and Vali22 and concluded that correolide (37) binds in the inner pore between the selectivity filter and the activation gate.34 As described in section 1.2.2, residues in positions i15, i19 and i22 are also involved in the binding of permanently charged hydrophobic cations such tetraalkylammonium to KcsA68,74–76 and Shaker.73,342 The center of the drug-binding region coincides with site T5 for permeating metal ions, which in the absence of drugs are stabilized there by electrostatic interactions with P-helices.70 How can a nucleophilic molecule like correolide (37) lacking a positive charge have the same binding site as cationic ligands in the region of the channel, which is close to the selectivity filter and hence evolved to provide stabilization for permeating metal ions? Furthermore, correolide (37) has an elongated shape and should bind inside the pore with a vertex approaching the selectivity filter and the long axis collinear to the pore axis.343 In this orientation, multiple oxygen atoms at the sides of correolide would lack H-bonding partners because the pore-facing positions in the inner helices have only hydrophobic residues. To address this problem, Bruhova and Zhorov used Monte Carlo-minimization to search for the correolide binding site from 20,000 starting positions/orientations of the drug in models of Kv1.3 based on the X-ray structure of Kv1.2.343 When a drug blocks the ion permeation pathway, ions obviously do not permeate, but can remain in the channel and occupy some of the sites T1–T5 in an unknown pattern. Cationic blockers targeting the inner pore would compete with metal ions and displace them from sites T4’ and T5, where metal ions make few or no direct contacts with the protein atoms, but nucleophilic drugs like correolide (37) could bind a metal ion in the pore. To explore this possibility, two models of Kv1.3 were generated with K+ binding sites T2/T4 or T1/T3/T5 loaded with K+ ions. During energy optimization of the T2/T4 model, correolide coordinated the K+ ion in site T4. In the T1/T3/T5 model, the third K+ ion was initially placed in site T5. During energy optimization, this K+ ion was readily chelated by three acetoxy groups of correolide (Fig. 11A and B), shifted upwards into site T4’ and occurred approximately in the same position as the ammonium groups of TBA and d-tubocurarine (Fig. 4). In both models, correolide (37) further directly interacted with the pore-facing residues, whose mutation decreased correolide binding in experiments by Hanner et al.34 A model in which correolide coordinates the K+ ion at site T4’ may also explain why correolide derivatives without the epoxy group and the seven-membered ring retain the channel-blocking activity.340
Figure 11. Kv1.2-based model of the open Kv1.3 with correolide (37)342.
A, Side view of the model with only two of the four S5-P-S6 chains shown for clarity. The selectivity-filter area is loaded with two K+ ions (positions 1 and 3) and two water molecules (positions 2 and 4). The third K+ ion was initially placed in position 5. During energy optimization, it was chelated by three acetoxy groups of correolide, shifted upwards and occurred approximately in the same position as the ammonium groups of TBA and d-tubocurarine (Figure 4). Sticks show side chains of correolide-sensing residues34 in the inner helixes (blue) and outer helices (gray strands). B, Close-up view of the complex, in which the signature-sequence TVGYGp53 backbones and Thrp49 side chains are shown as sticks. Ions K+T1 and K+T3 are octa-coordinated by the backbone carbonyls. K+T4’, is coordinated by three acetoxy groups of correolide and a water molecule in site T4. In addition, four Thrp49 sidechain oxygens are within 4 Å from K+T4’, and P-helices’ macrodipoles can additionally stabilize this ion.
The direct experimental validation of this ternary-complex model would require a high resolution X-ray structure. However, there is a simple analogy. When a proton binds to a nucleophilic amino group of a ligand, the proton-ligand complex is considered as a protonated ligand, which can bind to a nucleophilic group of the protein. Similarly, when a ligand binds K+, the complex may be considered as a K+-bound ligand that can bind to a nucleophilic site of the ion channel. Many X-ray structures in which a metal ion is coordinated between a ligand and a protein can be found in the Protein databank. In most cases, the ternary complexes involve a transition metal with well-defined coordination geometry and strong coordinating bonds. In contrast, ternary complexes involving an alkali or alkaline earth metal ions are weak. However, the high local concentration of permeating ions in ion channels should promote the formation of ternary complexes. Such complexes have long been predicted to explain paradoxes in SAR344–346 and the ability of some ion-channel drugs to bind metal ions has been experimentally demonstrated.347–349 A number of ion channel models with pore-bound ligands chelating metal ions further support the possible involvement of metal ions in ligand-receptor complexes.22,343,350–353 For Na+ channels a recent model explained the intriguing observation that two molecules of the nucleophilic local anesthetic benzocaine bind to a site, where a single molecule of a cationic local anesthetic ligand like lidocaine binds.354 According to a classical concept, batrachotoxin, veratridine, and other Na+ channel agonists activate Na+ channels allosterically.355 Based on the ternary-complex idea, a new mechanism of action of these important pharmacological tools was proposed, according to which a Na+ channel agonist molecule binds in the central pore, but leaves a path for ion permeation between its nucleophilic face and a nucleophilic residue in the inner pore.353 Subsequent mutational, electrophysiological, and ligand-binding experiments confirmed important predictions of the new model of action of sodium agonists.356–358
Two other classes of oxygen-rich natural products that potently block Kv1.3 channels are psoralens and khelliones bearing lipophilic phenylalkyl or phenoxyalkyl side-chains.275,332 Following up on anecdotal reports that tea prepared from leaves of Ruta graveolens, the common rue, had beneficial effects in multiple sclerosis, the groups of Wolfram Hänsel and Eilhard Koppenhöfer at the University of Kiel in Germany extracted the low-affinity Kv1.3 blocker 5-methoxypsoralen from Ruta and demonstrated that it reduced visual field defects in single-case studies in multiple sclerosis patients.359 However, 5-methoxypsoralen, which is a drug used for the treatment of psoriasis, was not ideal for the treatment of multiple sclerosis because of its phototoxicity. Our own group therefore performed detailed SAR investigations around 5-methoxypsoralen with the aim of eliminating the phototoxicity and increasing potency and selectivity for Kv1.3.360 After identifying Psora-4 (40), the first nanomolar Kv1.3 blocker (IC50 3 nM) in 2003,361 we designed PAP-1 (41) through a classical medicinal chemistry approach in 2005.35 Although PAP-1 (IC50 2 nM) is only slightly more potent than Psora-4, it is less lipophilic and more selective than Psora-4 and therefore more suitable for in vivo use. PAP-1 is 25–125 fold selective over Kv1.1, Kv1.2, Kv1.4, Kv1.5, Kv1.6 and Kv1.7 and more than 1000-fold selective over more distantly related K+ channels like Kv2.1, Kv3.1, Kv4.2, Kv11.1 (hERG), and the KCa channels.35 PAP-1 further displays no in vitro and in vivo toxicity, is orally available, and has a half-life of 3 hours in rats35,237,362 and 6 hours in rhesus macaques.289 As a first proof of efficacy, Schmitz et al. demonstrated that PAP-1 (41) suppresses delayed type hypersensitivity, a reaction mediated by skin homing CD4+ effector memory T cells, when administered at 3 mg/kg intraperitoneally.35 Azam at al.362 next tested whether PAP-1 (41) could also suppress allergic contact dermatitis (ACD), an inflammatory skin reaction that is mediated by CD8+ effector memory T cells and resembles psoriasis in some aspects. In keeping with the selective effect of Kv1.3 blocker on effector memory T cells, PAP-1 (41) did not prevent antigen presentation during the sensitization phase of ACD, but potently suppressed oxazolone-induced inflammation by inhibiting the infiltration of CD8+ T cells and reducing the production of the inflammatory cytokines INF-γ, IL-2 and IL-17, when administered intraperitoneally or orally during the elicitation phase.362 PAP-1 was equally effective when applied topically in a cream demonstrating that it penetrates skin well and could potentially be developed into a topical for the treatment of psoriasis.362 Oral PAP-1 treatment at 50 mg/kg further significantly reduces diabetes incidence and delays diabetes onset in a rat model of type-1 diabetes.237
Starting with the 4,7-dimethoxy substituted benzofuran khellinone isolated from Ammi visnaga, which can be regarded as a lactone ring-opened version of the psoralen ring system, Baell et al.363,364 synthesized three other classes of Kv1.3 blockers through multiple parallel synthesis: 3-substituted khellinone chalcones (42), khellinone dimers and khellinone derivatives alkylated at either the 4- or 7-position (43 and 44). All three compound classes inhibit Kv1.3 with IC50s of 100–400 nM and display moderate 3–10 fold selectivities over other Kv1-family channels, are not cytotoxic, and suppress human T cell proliferation at low micromolar concentrations.363,364 A more potent khellinone derivative of undisclosed structure is currently in preclinical development at the Australian company Bionomics. Interestingly, the different khellinone derivatives and the psoralens exhibit different mechanisms of block despite their structural similarity. While the psoralens Psora-4 (40) and PAP-1 (41) block Kv1.3 by binding to the C-type inactivated state and therefore only reach their full blocking potency after repeated channel openings,35 the 3-substituted khellinone chalcones like chalcone-16 (42) and the 4-substituted khellinones (43) seem to be open channel blockers and reach full blocking potency on the first depolarizing pulse.363,364 Intriguingly, the 7-series khellinones (44) display blocking kinetics that are a mixture between open-channel and use-dependent block.364 The compound classes further differ in the stoichiometry of their interaction with Kv1.3. Psora-4, PAP-1, the khellinone chalcones and the 7-substituted khelliones exhibit Hill coefficient of 2, suggesting that 2 blocker molecules bind to one channel molecule. Although cooperativity has not yet been rigorously proven for all these compounds, it is highly likely based on detailed binding studies performed by Schmalhofer et al. with di-substituted cyclohexyl type Kv1.3 blockers, which demonstrate the presence of two receptor sites on the Kv1.3 channel protein that display positive allosteric cooperativity.365 In contrast, the khellinone dimers and the 4-substituted khellinones have Hill coefficients close to unity indicating a 1:1 interaction.
Dirk Trauner’s group at the University of California, Berkeley took a peptidomimetic approach for designing Kv1.3 blockers. Using the tetraphenylporphyrin system (45) as a scaffold Gradl et al.366 put four positively charged groups at the optimal distance to form salt bridges with four Aspp38 residues in the outer vestibule of Kv1.3. However, no mutational or computational studies have yet been performed to confirm the binding mode of the tetraphenylporphyrins. The tetraphenylporphyrins displace radiolabeled peptides from Kv1.3 with Kd values of 20–150 nM and inhibit Kv1.3 currents in patch-clamp experiments at low micromolar concentrations. The tetraphenylporphyrins may not be ideal drug candidates because of their permanent charge and their relatively high molecular weight. However, these cross-like compounds, which have been suggested to simultaneously form four salt bridges with four Aspp38 residues, may serve as the basis for the attachments of fluorophores as alternatives to fluorophore-tagged peptides or antibodies. The tetraphenylporphyrins could also be used for the synthesis of metalloporphyrins for imaging and crystallographic studies.366
3.2. KCa3.1 Channels
A calcium-dependent K+ efflux, which is now known to be carried by KCa3.1, was first described in 1958 by the Hungarian scientist Gardos in human erythrocytes.367 KCa3.1 is therefore also often referred to as the “Gardos channel”. Other names for the channel include SK4, KCNN4 and “intermediate-conductance” KCa because its single-channel conductance is larger than the conductance of the KCa2 channels but smaller than the conductance of KCa1.1 channels. KCa3.1 was cloned nearly simultaneously by three groups in 1997368–370 and found to show about 42–44% sequence identity to the KCa2 channels (see 2.3). The channel is voltage-independent and like the KCa2 channels binds calmodulin in its C-terminus, which renders the channel sensitive to submicromolar Ca2+ concentrations.371 KCa3.1 is primarily expressed in placenta, lung, salivary gland, distal colon and lymphoid organs, but is mostly absent from cardiac and neuronal tissue.264,369,370,372 In naïve T and B cells,264,266 fibroblasts,373 dedifferentiated vascular smooth muscle cells,11,374 and vascular endothelial cells375 KCa3.1 is an important regulator of proliferation that exerts its effect by hyperpolarizing the cell membrane and thus facilitating Ca2+-entry, a prerequisite for cell proliferation. In all these cell types stimulation with either growth factors (in the case of fibroblast or vascular smooth muscle cells) or antigen or mitogen (in the case of T and B cells) leads to transcriptional up-regulation of KCa3.1 expression11,264,376 and KCa3.1 blockade suppresses proliferation. In tissues that are involved in salt and fluid transport like the colon, the lung, and salivary glands, KCa3.1 activity is found on the basolateral side of the respective epithelium, where it recycles K+ together with Kv7.1 and thus helps to facilitate chloride secretion.377,378 In vascular endothelia KCa3.1 mediates the so-called endothelium-derived hyperpolarization (EDHF) response together with KCa2.3.379,380 KCa3.1 channels have further been shown to be involved in the migration of macrophages,253 microglia,262 vascular smooth muscle cells381,382 and mast cells.241 In microglia KCa3.1 also seems to play a role in the oxidative burst, nitric oxide production and microglia-mediated neuronal killing.261,383 Two independently generated KCa3.1−/− mice384,385 were both viable, of normal appearance, produced normal litter sizes, did not show any gross abnormalities in any of their major organs, and exhibited rather mild phenotypes: impaired volume regulation in erythrocytes and lymphocytes384 and a reduced EDHF response together with a mild ~7 mmHg increase in blood pressure.385 So far no human disease involving KCa3.1 mutations have been described. Readers interested in more details on the expression and physiological function of KCa3.1 are referred to a recent review.188
3.2.1. Peptidic KCa3.1 blockers
The most potent peptidic blocker of KCa3.1 is maurotoxin (MTX, IC50 1 nM), a 34-residue scorpion toxin with an αββ fold.386,387 However, MTX has even higher affinity (IC50 100 pM) for Kv1.2.386,388 Another scorpion toxin that inhibits KCa3.1 is ChTX (IC50 5 nM), which has been traditionally used to distinguish “charybdotoxin-sensitive” from “apamin-sensitive” KCa channels despite the fact that ChTX also inhibits Kv1.3 and KCa1.1 (see 2.2.1). Mutational studies by Rauer et al.,321,389 which used ChTX and the sea anemone toxins ShK and BgK as molecular calipers, demonstrated that the outer vestibules of KCa3.1 and Kv1.3 have very similar dimensions, which partly explains this cross-reactivity. Based on this information, the authors designed the ChTX analog ChTX-Glu32, whose negatively charged Glu32 is repelled by negatively charged residues in the outer vestibule of Kv1.3, resulting in 30-fold selectivity for KCa3.1 over Kv1.3.389 However, this modification did not increase selectivity over KCa1.1.
3.2.2. Small Molecule KCa3.1 Blockers
Since we recently reviewed the pharmacology of KCa3.1 in detail188 we here only briefly describe the most commonly used KCa3.1 blockers and comment on their therapeutic applications. Compounds that inhibit KCa3.1 in the micromolar range include the chinchona alkaloid quinine (IC50 100 µM), the vasodilator cetiedil (IC50 25 µM) and the L-type Ca2+ channel blockers nifidipine (IC50 4 µM) and nitrendipine (IC50 1 µM).188 The first nanomolar KCa3.1 blocker that was identified is the azole antimycotic clotrimazole (46), which inhibits both the Gardos channel in human erythrocytes and the cloned KCa3.1 channel with IC50s of 70 to 250 nM.188,390,391 Using clotrimazole (46) as a tool compound, Carlo Brugnara and coworkers at Harvard University demonstrated that pharmacological KCa3.1 blockade has beneficial effects in a transgenic mouse model of sickle-cell anemia.392 The group further provided evidence that clotrimazole could reduce erythrocyte dehydration in a small number of patients with sickle cell anemia.393 However, clotrimazole (46) itself was not an ideal drug for long-term use because of its acute inhibition and chronic induction of human cytochrome P450-dependent enzymes leading to liver damage. Three groups, including us therefore used clotrimazole (46) as a template for the design of triarylmethane based KCa3.1 blockers, which are free of cytochrome P450 inhibition.188 By replacing clotrimazole’s azole ring, which is responsible for the compounds strong P450 inhibition, with a pyrazole ring our own group identified TRAM-34 (47), a compound that inhibits KCa3.1 with an IC50 of 20 nM and displays 200–1000 fold selectivity over Kv and other KCa channels.10 Through site-directed mutagenesis we later demonstrated that TRAM-34 (47) and clotrimazole (46) block KCa3.1 by interacting with Thrp49(250) in the pore loop and Vali15(275) in S6.394 Scientists at Icagen developed the fluorinated triphenyl acetamide ICA-17043 (48), which inhibits KCa3.1 with an IC50 of 11 nM.395 ICA-17043 had been found to be both effective and safe in a Phase-2 trial for sickle-cell anemia,396 but recently failed in Phase-3 clinical trials apparently due to lack of efficacy. A chemically different class of KCa3.1 blockers are the 4-phenyl-4H-pyrans397 and the related cyclohexadiens.398 Using the dihydropyridine nifedipine as a template Urbahns et al. at Bayer replaced the NH in the dihydropyridine ring system, which is required for Cav channel blockade, first with an O atom in 49 (IC50 8 nM)397 and then with CH2 in the cyclohexadiene 50 (IC501.5 nM).398 However, the cyclohexadienes showed a tendency to 3,6/3,5 double bond isomerization and the authors therefore also synthesized a series of corresponding cyclohexadiene lactones exemplified by 51 (IC50 8 nM), in which the second ring prevents isomerization.398 Compound 51 was subsequently shown to exhibit good selectivity over ion channels and to reduce infarct volumes, brain edema and intracranial pressure following traumatic brain injury in rats.399 The authors of this study demonstrated that KCa3.1 messenger RNA increases in the brain following injury, but did not determine whether this increase was on microglia, brain infiltrating macrophages and T cells, or possibly on cells of the blood brain barrier endothelium. However, taken together with reports by the group of Lyanne Schlichter showing that TRAM-34 (47) reduces microglia-mediated neuronal killing both in vitro and in an optic nerve transection model in vivo,383 this study suggests the possibility of using KCa3.1 blockers for the treatment of traumatic brain injury and possibly ischemic stroke and other neurodegenerative disorders with an inflammatory component.
Based on the expression of KCa3.1 in T cells, B cells, macrophages, and mast cells, KCa3.1 blockers have also been suggested as novel immunosuppressants.7,188 Our own group demonstrated that TRAM-34 (47) suppresses the proliferation of human CCR7+ T cells and IgD+ B cells in vitro,10,266 while scientists at Schering-Plough reported that TRAM-34 effectively prevents experimental autoimmune encephalomyelitis (EAE) induced by immunization with MOG peptide in mice.400 While KCa3.1 blockers might not be optimal for the treatment of diseases such as multiple sclerosis where the brain-infiltrating T cells are of a Kv1.3high effector memory phenotype,401,402 they could potentially be useful for the treatment of rheumatoid arthritis, transplant rejection, asthma and primary biliary cirrhosis (PBC). A small clinical trial, conducted by Wotjtulewski at al. in 1980 reported that clotrimazole (46) was superior to ketoprofen in improving rheumatoid arthritis.403 In T cell proliferation assays, TRAM-34 (47) further synergizes with cyclosporine suggesting that it could be used in combination therapy in transplant rejection in order to reduce cyclosporine toxicity.10 KCa3.1 blockade with TRAM-34 (47) also inhibits human lung mast cell migration241 and anti-mitochondrial antibody secretion by B cells from PBC patients404 suggesting that KCa3.1 blockade might be useful for the therapy of asthma and PBC. KCa3.1 blockade also constitutes a potential new therapeutic approach to cardiovascular diseases such as restenosis and possibly atherosclerosis based on KCa3.1’s role in driving vascular smooth muscle cell and fibroblast proliferation.11,376,382,405 In proof of this concept, TRAM-34 (47) and clotrimazole (46) significantly reduce intimal hyperplasia following balloon catheter injury in rats11 demonstrating that KCa3.1 blockers can prevent restenosis, a common complication of angioplasty. Finally, KCa3.1 blockers might also be useful to inhibit tumor angiogenesis because of their proven ability to inhibit angiogenesis in the mouse madrigel plug assay.375
From a drug-design standpoint, both the triarylmethane- and the phenyl-pyran/cyclohexadiene-type KCa3.1 blockers are perfect examples of the power of what Camille Wermuth has termed the “SOSA” approach, the selective optimization of the side activity of an “old” drug for new target.406 In both cases careful consideration of the previously known SAR for cytochrome P450 inhibition or Cav1.2 blockade made it possible to design potent and selective KCa3.1 blockers.
3.2.3. KCa3.1 Activators
Presumably because of their similar mode of activation through a C-terminally bound calmodulin, KCa3.1 channels are activated by the same compounds that activate KCa2 channels (see 2.2.2). The only reported exception so far is the aminopyrimidine CyPPA (36), which activates KCa2.3 and KCa2.2 currents but not KCa3.1 and KCa1.1.226 All other KCa2 activators, namely EBIO (32), DC-EBIO (33), NS309 (34), and riluzole (35) activate KCa3.1 about 5-fold more potently than KCa2 channels.188 The most potent KCa3.1 activator is currently NS309 (34) with an EC50 of 27 nM.188,221 Therapeutically KCa3.1 activators might be useful for the treatment of cystic fibrosis because they could potentially increase the diminished chloride secretion onto the lung surface by activating the basolateral KCa3.1 channel on the lung epithelium.407 Based on the expression of KCa3.1 in vascular endothelium and the role of KCa3.1 in the EDHF response, KCa3.1 activators might also be able to lower blood pressure. However, as discussed above (sections 2.3 and 3.2) the EDHF response is carried by both KCa2.3 and KCa3.1 channels and it would be necessary to have subtype specific activators before attempting to make any predictions about which of the two channels is physiologically more important. It is currently not clear what the effects, if any, of KCa3.1 activation will be on the immune system since none of the existing activators have been tested on immune cell function in vitro or in vivo.
4. Conclusion and Perspectives
During recent years remarkable progress has been made in our understanding of the physiological and pathophysiological role of K+ channels. All 78 K+ channels in the human genome have been cloned and a large and steadily increasing body of literature is available describing their tissue distribution, subcellular localization, involvement in signaling, and role in disease processes. However, due to the difficulties of targeting ion channels in general, chemistry efforts in this area have considerably lacked behind the large-scale medicinal chemistry programs targeting G-protein coupled receptors and protein kinases, and there are currently no clinically used drugs, that have been rationally developed to target a particular K+ channel. With the recent advent of high- or at least medium-throughput electrophysiology this situation is currently changing rapidly and academic screening centers and pharmaceutical companies are increasingly screening for K+ channel modulators as physiological tool compounds and as potential drug candidates. K+ modulators are particularly attractive for the treatment of neurological disorders, autoimmune diseases and inflammation. It is therefore to be expected, that drugs modulating the channels discussed here (Kv7.2–7.5, KCa1.1, and KCa2.1–2.3 for neurological disorders and Kv1.3 and KCa3.1 for autoimmune diseases and inflammation) will be developed within the next 5–10 years. The success of these efforts is often assumed to critically depend on the ability of medicinal chemists to identify subtype selective modulators. However, the example of retigabine shows that a relatively low potency and not subtype selective compound can be developed as a drug, and only the future will show if more potent and selective Kv7.2–7.5 channel activators will offer any advantages.
The discovery of K+ channel modulating drugs is also increasingly assisted by structural information. The X-ray structures of K+ channels in the open and closed states have revolutionized our knowledge about how drugs target K+ channels during the last 10 years. And although a co-crystals of KcsA with TBA currently is the the only visualized example of a drug bound in the inner pore of a K+ channel, results of numerous mutational, electrophysiological, and ligand-binding experiments are increasingly interpreted in structural terms using homology modeling and ligand docking. However, it should be noted that the inner-pore dimensions between the X-ray structures of MthK, KvAP, and Kv1.2, which were all crystallized in the open state, vary widely. It is therefore advisable to interpret homology models of medicinally important K+ channels like Kv11.1 (hERG), which are based on these templates, with caution. This restricts reliability of predicted ligand-receptor complexes, which are based only on energy criteria. Another complication is the huge diversity in the chemical structure of drugs modulating the same channel protein. In particular, the paradoxical observations that both cationic and nucleophilic ligands target the same region in the inner pore, which evolved to enable fast flow of K+ ions via the low-dielectric energy barrier in the membrane environment, suggest that metal ions may be invisible, but important receptor components for nucleophilic ligands. Future high-resolution structures of medically important ion channels in the closed, open, and slow-inactivated states should make it possible to perform more precise energy calculations and more reliable structure-based drug design. Meanwhile, homology modeling and ligand docking is likely to become an increasingly popular approach to rationalize huge and often controversial data on structure-activity relations of available ligands. In some cases, this approach may provide important information for structure-assisted design of new potent and specific drugs.
Figure 2. Structures of unselective K+ channel blockers.
Figure 5. Kv7 channel modulators.
Figure 6. KCa1.1 modulators.
Figure 7. KCa2 channel modulators.
Figure 10. Kv1.3 blockers.
Figure 12. KCa3.1 blockers.
Acknowledgments
This work was supported by grants from NIH to H.W. (NS053426-01 and GM076063-01) and by grants from NSERC and CIHR to B.Z.
Abbreviations
- AHP
afterhyperpolarization
- 4-AP
4-aminopyridine
- BFNC
benign familial neonatal convulsions
- BgK
Bunadosoma granulifera K+ channel toxin
- CaV
volgate-gated Ca2+ channel
- ChTX
charybdotoxin
- CNS
central nervous system
- COX
cyclooxygenase
- CRAC
calcium release activated Ca2+ channel
- DAP
diaminopropionic acid
- DC-EBIO
dichloroethylenbenzimidazolone
- DHS
dehydrosoyasaponin
- EAE
experimental autoimmune encephamolyelitis
- EBIO
ethylenbenzimidazolone
- EDHF
endothelium-derived hyperpolarization factor
- F6CA
fluorescein-6-carboxyl
- GABA
γ-aminobutyric acid
- HaTX
hanatoxin
- hERG
human ether-à-go-go related gene (Kv11.1)
- IbTX
iberiotoxin
- Im
M current (neuronal K+ current inhibited by muscarinic agonists)
- K2P
two-pore K+ channel
- KTX
kaliotoxin
- KCa
calcium-activated K+ channel
- Kir
inward-rectifier K+ channel
- KV
voltgage-gated K+ channel
- mAHP
medium afterhyperpolarization
- MgTX
margatoxin
- MTX
maurotoxin
- PBC
primary biliary cirrhosis
- PIP2
phosphatidylinositol-4,5-biphosphate
- NaV
voltage-gated Na+ channel
- NMDA
N-methyl-D-aspartic acid
- RCK
regulator of conductance of K+
- ShK
Stichodactyla helianthus K+ channel toxin
- SK
small-conductance KCa
- SOSA
selective optimization of a side activity
- SUR
sulfonylurea receptor
- TBA
tetrabutylammonium
- TEA
tetraethylammonium
- TM
transmembrane segment
- VSTX1
voltage sensor toxin 1 from Grammostola spatulata
References
- 1.Catterall W, Gutman GA. Pharmcol. Rev. 2005;57:385. [Google Scholar]
- 2.Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA. Pharmacol. Rev. 2005;57:509. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein SA, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S. Pharmacol. Rev. 2005;57:527. doi: 10.1124/pr.57.4.12. [DOI] [PubMed] [Google Scholar]
- 4.Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H. Pharmacol. Rev. 2005;57:463. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]
- 5.Gutman GA, Chandy KG, Grissmer S, Lazdunski M, McKinnon D, Pardo LA, Robertson GA, Rudy B, Sanguinetti MC, Stuhmer W, Wang X. Pharmacol. Rev. 2005;57:473. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- 6.Meldrum BS, Rogawski MA. Neurotherapeutics. 2007;4:18. doi: 10.1016/j.nurt.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandy KG, Wulff H, Beeton C, Pennington M, Gutman GA, Cahalan MD. Trends Pharmacol. Sci. 2004;25:280. doi: 10.1016/j.tips.2004.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. Nature. 2006;441:179. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 9.Kumar B, Dreja K, Shah SS, Cheong A, Xu SZ, Sukumar P, Naylor J, Forte A, Cipollaro M, McHugh D, Kingston PA, Heagerty AM, Munsch CM, Bergdahl A, Hultgardh-Nilsson A, Gomez MF, Porter KE, Hellstrand P, Beech DJ. Circ. Res. 2006;98:557. doi: 10.1161/01.RES.0000204724.29685.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wulff H, Miller MJ, Haensel W, Grissmer S, Cahalan MD, Chandy KG. Proc. Natl. Acad. Sci. USA. 2000;97:8151. doi: 10.1073/pnas.97.14.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler R, Wulff H, Eichler I, Kneifel M, Neumann D, Knorr A, Grgic I, Kampfe D, Si H, Wibawa J, Real R, Borner K, Brakemeier S, Orzechowski HD, Reusch HP, Paul M, Chandy KG, Hoyer J. Circulation. 2003;108:1119. doi: 10.1161/01.CIR.0000086464.04719.DD. [DOI] [PubMed] [Google Scholar]
- 12.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 13.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. Nat. Rev. Neurosci. 2006;7:921. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 14.Meera P, Wallner M, Toro L. Proc. Natl. Acad. Sci. USA. 2000;97:5562. doi: 10.1073/pnas.100118597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haider S, Antcliff JF, Proks P, Sansom MS, Ashcroft FM. J. Mol. Cell. Cardiol. 2005;38:927. doi: 10.1016/j.yjmcc.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Ashcroft FM. J. Clin. Invest. 2005;115:2047. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jerng HH, Pfaffinger PJ, Covarrubias M. Mol. Cell. Neurosci. 2004;27:343. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Park KS, Mohapatra DP, Misonou H, Trimmer JS. Science. 2006;313:976. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 19.Henke G, Maier G, Wallisch S, Boehmer C, Lang F. J. Cell. Physiol. 2004;199:194. doi: 10.1002/jcp.10430. [DOI] [PubMed] [Google Scholar]
- 20.Gubitosi-Klug RA, Mancuso DJ, Gross RW. Proc. Natl. Acad. Sci. USA. 2005;102:5964. doi: 10.1073/pnas.0501999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitcheson JS, Chen J, Lin M, Culberson C, Sanguinetti MC. Proc. Natl. Acad. Sci. USA. 2000;97:12329. doi: 10.1073/pnas.210244497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerche C, Bruhova I, Lerche H, Steinmeyer K, Wei AD, Strutz-Seebohm N, Lang F, Busch AE, Zhorov BS, Seebohm G. Mol. Pharmacol. 2007;71:1503. doi: 10.1124/mol.106.031682. [DOI] [PubMed] [Google Scholar]
- 23.Osterberg F, Aqvist J. FEBS Lett. 2005;579:2939. doi: 10.1016/j.febslet.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 24.Sanguinetti MC, Mitcheson JS. Trends Pharmacol. Sci. 2005;26:119. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Rossokhin A, Teodorescu G, Grissmer S, Zhorov BS. Mol. Pharmacol. 2006;69:1356. doi: 10.1124/mol.105.017970. [DOI] [PubMed] [Google Scholar]
- 26.Stansfeld PJ, Gedeck P, Gosling M, Cox B, Mitcheson JS, Sutcliffe MJ. Proteins. 2007;68:568. doi: 10.1002/prot.21400. [DOI] [PubMed] [Google Scholar]
- 27.Tseng GN, Sonawane KD, Korolkova YV, Zhang M, Liu J, Grishin EV, Guy HR. Biophys. J. 2007;92:3524. doi: 10.1529/biophysj.106.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhorov BS, Tikhonov DB. J. Neurochem. 2004;88:782. doi: 10.1111/j.1471-4159.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 29.Sanguinetti MC, Tristani-Firouzi M. Nature. 2006;440:463. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Lee A, Ruta V, Cadene M, Chait BT, MacKinnon R. Nature. 2003;423:33. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 31.Long SB, Campbell EB, Mackinnon R. Science. 2005;309:897. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 32.Brock MW, Mathes C, Gilly WF. J. Gen. Physiol. 2001;118:113. doi: 10.1085/jgp.118.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreker T, Grissmer S. Mol. Pharmacol. 2005;68:966. doi: 10.1124/mol.105.012401. [DOI] [PubMed] [Google Scholar]
- 34.Hanner M, Green B, Gao YD, Schmalhofer WA, Matyskiela M, Durand DJ, Felix JP, Linde AR, Bordallo C, Kaczorowski GJ, Kohler M, Garcia ML. Biochemistry. 2001;40:11687. doi: 10.1021/bi0111698. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz A, Sankaranarayanan A, Azam P, Schmidt-Lassen K, Homerick D, Hansel W, Wulff H. Mol. Pharmacol. 2005;68:1254. doi: 10.1124/mol.105.015669. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Seebohm G, Sanguinetti MC. Proc. Natl. Acad. Sci. USA. 2002;99:12461. doi: 10.1073/pnas.192367299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panyi G, Deutsch C. J Gen Physiol. 2007;129:403. doi: 10.1085/jgp.200709758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banks BE, Brown C, Burgess GM, Burnstock G, Claret M, Cocks TM, Jenkinson DH. Nature. 1979;282:415. doi: 10.1038/282415a0. [DOI] [PubMed] [Google Scholar]
- 39.Hugues M, Romey G, Duval D, Vincent JP, Lazdunski M. Proc. Natl. Acad. Sci. USA. 1982;79:1308. doi: 10.1073/pnas.79.4.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbone E, Wanke E, Prestipino G, Possani LD, Maelicke A. Nature. 1982;296:90. doi: 10.1038/296090a0. [DOI] [PubMed] [Google Scholar]
- 41.Miller C, Moczydlowski E, Latorre R, Phillips M. Nature. 1985;313:316. doi: 10.1038/313316a0. [DOI] [PubMed] [Google Scholar]
- 42.Judge SI, Bever CT., Jr Pharmaco.l Ther. 2006;111:224. doi: 10.1016/j.pharmthera.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Harvey AL, Robertson B. Curr. Med. Chem. 2004;11:3065. doi: 10.2174/0929867043363820. [DOI] [PubMed] [Google Scholar]
- 44.Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Science. 1987;237:749. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 45.Chandy KG, Williams CB, Spencer RH, Aguilar BA, Ghanshani S, Tempel BL, Gutman GA. Science. 1990;247:973. doi: 10.1126/science.2305265. [DOI] [PubMed] [Google Scholar]
- 46.Bontems F, Roumestand C, Boyot P, Gilquin B, Doljansky Y, Menez A, Toma F. Eur. J. Biochem. 1991;196:19. doi: 10.1111/j.1432-1033.1991.tb15780.x. [DOI] [PubMed] [Google Scholar]
- 47.Aiyar J, Withka JM, Rizzi JP, Singleton DH, Andrews GC, Lin W, Boyd J, Hanson DC, Simon M, Dethlefs B, Lee C-L, Hall JE, Gutman GA, Chandy KG. Neuron. 1995;15:1169. doi: 10.1016/0896-6273(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 48.Garcia ML, Garcia-Calvo M, Hidalgo P, Lee A, MacKinnon R. Biochemistry. 1994;33:6834. doi: 10.1021/bi00188a012. [DOI] [PubMed] [Google Scholar]
- 49.Aiyar J, Rizzi JP, Gutman GA, Chandy KG. J. Biol. Chem. 1996;271:31013. doi: 10.1074/jbc.271.49.31013. [DOI] [PubMed] [Google Scholar]
- 50.Hidalgo P, MacKinnon R. Science. 1995;268:307. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 51.Ranganathan R, Lewis JH, MacKinnon R. Neuron. 1996;16:131. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 52.Gross A, MacKinnon R. Neuron. 1996;16:399. doi: 10.1016/s0896-6273(00)80057-4. [DOI] [PubMed] [Google Scholar]
- 53.Tytgat J, Chandy KG, Garcia ML, Gutman GA, Martin-Eauclaire MF, van der Walt JJ, Possani LD. Trends Pharmacol. Sci. 1999;20:444. doi: 10.1016/s0165-6147(99)01398-x. [DOI] [PubMed] [Google Scholar]
- 54.Mouhat S, Jouirou B, Mosbah A, De Waard M, Sabatier JM. Biochem. J. 2004;378:717. doi: 10.1042/BJ20031860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin W, Lu Z. Biochemistry. 1998;37:13291. doi: 10.1021/bi981178p. [DOI] [PubMed] [Google Scholar]
- 56.Mathie A, Veale EL. Curr. Opin. Investig. Drugs. 2007;8:555. [PubMed] [Google Scholar]
- 57.MacKinnon R, Miller C. Science. 1989;245:1382. doi: 10.1126/science.2476850. [DOI] [PubMed] [Google Scholar]
- 58.MacKinnon R, Heginbotham L, Abramson T. Neuron. 1990;5:767. doi: 10.1016/0896-6273(90)90335-d. [DOI] [PubMed] [Google Scholar]
- 59.Chandy KG, Cahalan MD, Pennington M, Norton RS, Wulff H, Gutman GA. Toxicon. 2001;39:1269. doi: 10.1016/s0041-0101(01)00120-9. [DOI] [PubMed] [Google Scholar]
- 60.Yu L, Sun C, Song D, Shen J, Xu N, Gunasekera A, Hajduk PJ, Olejniczak ET. Biochemistry. 2005;44:15834. doi: 10.1021/bi051656d. [DOI] [PubMed] [Google Scholar]
- 61.Swartz KJ, MacKinnon R. Neuron. 1995;15:941. doi: 10.1016/0896-6273(95)90184-1. [DOI] [PubMed] [Google Scholar]
- 62.Swartz KJ, MacKinnon R. Neuron. 1997;18:665. doi: 10.1016/s0896-6273(00)80306-2. [DOI] [PubMed] [Google Scholar]
- 63.Swartz KJ. Toxicon. 2007;49:213. doi: 10.1016/j.toxicon.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips LR, Milescu M, Li-Smerin Y, Mindell JA, Kim JI, Swartz KJ. Nature. 2005;436:857. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 65.Lee SY, MacKinnon R. Nature. 2004;430:232. doi: 10.1038/nature02632. [DOI] [PubMed] [Google Scholar]
- 66.Garcia ML. Nature. 2004;430:153. doi: 10.1038/430153a. [DOI] [PubMed] [Google Scholar]
- 67.Takahashi H, Kim JI, Min HJ, Sato K, Swartz KJ, Shimada I. J. Mol. Biol. 2000;297:771. doi: 10.1006/jmbi.2000.3609. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001;414:43. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 69.Zhou M, MacKinnon R. J. Mol. Biol. 2004;338:839. doi: 10.1016/j.jmb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 70.Roux B, MacKinnon R. Science. 1999;285:100. doi: 10.1126/science.285.5424.100. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y, MacKinnon R. Biochemistry. 2004;43:4978. doi: 10.1021/bi049876z. [DOI] [PubMed] [Google Scholar]
- 72.French RJ, Shoukimas JJ. Biophys. J. 1981;34:271. doi: 10.1016/S0006-3495(81)84849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi KL, Mossman C, Aube J, Yellen G. Neuron. 1993;10:533. doi: 10.1016/0896-6273(93)90340-w. [DOI] [PubMed] [Google Scholar]
- 74.Lenaeus MJ, Vamvouka M, Focia PJ, Gross A. Nat. Struct. Mol. Biol. 2005;12:454. doi: 10.1038/nsmb929. [DOI] [PubMed] [Google Scholar]
- 75.Faraldo-Gomez JD, Kutluay E, Jogini V, Zhao Y, Heginbotham L, Roux B. J. Mol. Biol. 2007;365:649. doi: 10.1016/j.jmb.2006.09.069. [DOI] [PubMed] [Google Scholar]
- 76.Yohannan S, Hu Y, Zhou Y. J. Mol. Biol. 2007;366:806. doi: 10.1016/j.jmb.2006.11.081. [DOI] [PubMed] [Google Scholar]
- 77.Tikhonov DB, Zhorov BS. Biophys. J. 2007;93:1557. doi: 10.1529/biophysj.106.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Madeja M, Leicher T, Friederich P, Punke MA, Haverkamp W, Musshoff U, Breithardt G, Speckmann EJ. Mol. Pharmacol. 2003;63:547. doi: 10.1124/mol.63.3.547. [DOI] [PubMed] [Google Scholar]
- 79.Armstrong CM, Loboda A. Biophys. J. 2001;81:895. doi: 10.1016/S0006-3495(01)75749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.del Camino D, Kanevsky M, Yellen G. J. Gen. Physiol. 2005;126:419. doi: 10.1085/jgp.200509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Judge SI, Yeh JZ, Goolsby JE, Monteiro MJ, Bever CT., Jr Mol. Pharmacol. 2002;61:913. doi: 10.1124/mol.61.4.913. [DOI] [PubMed] [Google Scholar]
- 82.Trimmer JS, Rhodes KJ. Annu. Rev. Physiol. 2004;66:477. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- 83.Delmas P, Brown DA. Nat. Rev. Neurosci. 2005;6:850. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 84.Brown DA, Adams PR. Nature. 1980;283:673. doi: 10.1038/283673a0. [DOI] [PubMed] [Google Scholar]
- 85.Suh BC, Hille B. Neuron. 2002;35:507. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Craciun LC, Mirshahi T, Rohacs T, Lopes CM, Jin T, Logothetis DE. Neuron. 2003;37:963. doi: 10.1016/s0896-6273(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 87.Wang HS, Pan Z, Shi W, Brown BS, Wymore RS, Cohen IS, Dixon JE, McKinnon D. Science. 1998;282:1890. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe H, Nagata E, Kosakai A, Nakamura M, Yokoyama M, Tanaka K, Sasai H. J. Neurochem. 2000;75:28. doi: 10.1046/j.1471-4159.2000.0750028.x. [DOI] [PubMed] [Google Scholar]
- 89.Peters HC, Hu H, Pongs O, Storm JF, Isbrandt D. Nat Neurosci. 2005;8:51. doi: 10.1038/nn1375. [DOI] [PubMed] [Google Scholar]
- 90.Biervert C, Schroeder BC, Kubisch C, Berkovic SF, Propping P, Jentsch TJ, Steinlein OK. Science. 1998;279:403. doi: 10.1126/science.279.5349.403. [DOI] [PubMed] [Google Scholar]
- 91.Charlier C, Singh NA, Ryan SG, Lewis TB, Reus BE, Leach RJ, Leppert M. Nature Genet. 1998;18:53. doi: 10.1038/ng0198-53. [DOI] [PubMed] [Google Scholar]
- 92.Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, Bjerre I, Quattlebaum T, Murphy JV, McHarg ML, Gagnon D, Rosales TO, Peiffer A, Anderson VE, Leppert M. Nat. Genet. 1998;18:25. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- 93.Kubisch C, Schroeder BC, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch TJ. Cell. 1999;96:437. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 94.Saletu B, Darragh A, Salmon P, Coen R. Br. J. Clin. Pharmacol. 1989;28:1. doi: 10.1111/j.1365-2125.1989.tb03500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rockwood K, Beattie BL, Eastwood MR, Feldman H, Mohr E, Pryse-Phillips W, Gauthier S. Can. J. Neurol. Sci. 1997;24:140. doi: 10.1017/s031716710002148x. [DOI] [PubMed] [Google Scholar]
- 96.Borjesson A, Karlsson T, Adolfsson R, Ronnlund M, Nilsson L. Neuropsychobiol. 1999;40:78. doi: 10.1159/000026602. [DOI] [PubMed] [Google Scholar]
- 97.Aiken SP, Lampe BJ, Murphy PA, Brown BS. Br. J. Pharmacol. 1995;115:1163. doi: 10.1111/j.1476-5381.1995.tb15019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamas JA, Selyanko AA, Brown DA. Eur. J. Neurosci. 1997;9:605. doi: 10.1111/j.1460-9568.1997.tb01637.x. [DOI] [PubMed] [Google Scholar]
- 99.Zaczek R, Chorvat RJ, Saye JA, Pierdomenico ME, Maciag CM, Logue AR, Fisher BN, Rominger DH, Earl RA. J. Pharmacol. Exp. Ther. 1998;285:724. [PubMed] [Google Scholar]
- 100.Dalby-Brown W, Hansen HH, Korsgaard MP, Mirza N, Olesen SP. Curr. Top. Med. Chem. 2006;6:999. doi: 10.2174/156802606777323728. [DOI] [PubMed] [Google Scholar]
- 101.Munro G, Dalby-Brown W. J. Med. Chem. 2007;50:2576. doi: 10.1021/jm060989l. [DOI] [PubMed] [Google Scholar]
- 102.Friedel HA, Fitton A. Drugs. 1993;45:548. doi: 10.2165/00003495-199345040-00007. [DOI] [PubMed] [Google Scholar]
- 103.Rostock A, Tober C, Rundfeldt C, Bartsch R, Engel J, Polymeropoulos EE, Kutscher B, Loscher W, Honack D, White HS, Wolf HH. Epilepsy Res. 1996;23:211. doi: 10.1016/0920-1211(95)00101-8. [DOI] [PubMed] [Google Scholar]
- 104.Porter RJ, Partiot A, Sachdeo R, Nohria V, Alves WM. Neurology. 2007;68:1197. doi: 10.1212/01.wnl.0000259034.45049.00. [DOI] [PubMed] [Google Scholar]
- 105.Porter RJ, Nohria V, Rundfeldt C. Neurotherapeutics. 2007;4:149. doi: 10.1016/j.nurt.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Main MJ, Cryan JE, Dupere JR, Cox B, Clare JJ, Burbidge SA. Mol. Pharmacol. 2000;58:253. doi: 10.1124/mol.58.2.253. [DOI] [PubMed] [Google Scholar]
- 107.Rundfeldt C, Netzer R. Neurosci. Lett. 2000;282:73. doi: 10.1016/s0304-3940(00)00866-1. [DOI] [PubMed] [Google Scholar]
- 108.Wickenden AD, Yu W, Zou A, Jegla T, Wagoner PK. Mol. Pharmacol. 2000;58:591. doi: 10.1124/mol.58.3.591. [DOI] [PubMed] [Google Scholar]
- 109.Tatulian L, Delmas P, Abogadie FC, Brown DA. J. Neurosci. 2001;21:5535. doi: 10.1523/JNEUROSCI.21-15-05535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wuttke TV, Seebohm G, Bail S, Maljevic S, Lerche H. Mol. Pharmacol. 2005;67:1009. doi: 10.1124/mol.104.010793. [DOI] [PubMed] [Google Scholar]
- 111.Peretz A, Degani N, Nachman R, Uziyel Y, Gibor G, Shabat D, Attali B. Mol. Pharmacol. 2005;67:1053. doi: 10.1124/mol.104.007112. [DOI] [PubMed] [Google Scholar]
- 112.Mc-Naughthon-Smith GA, Wickenden AD. In: Voltage-Gated Ion Channels as Drug Targets. Triggle DJ, Gopalakrishnan M, Rampe D, Zheng W, editors. Weinheim: Wiley-VCH Verlag GmbH; 2006. p. 355. [Google Scholar]
- 113.Wu YJ, Boissard CG, Greco C, Gribkoff VK, Harden DG, He H, L'Heureux A, Kang SH, Kinney GG, Knox RJ, Natale J, Newton AE, Lehtinen-Oboma S, Sinz MW, Sivarao DV, Starrett JE, Jr, Sun LQ, Tertyshnikova S, Thompson MW, Weaver D, Wong HS, Zhang L, Dworetzky SI. J. Med. Chem. 2003;46:3197. doi: 10.1021/jm034073f. [DOI] [PubMed] [Google Scholar]
- 114.Xiong Q, Sun H, Li M. Nat. Chem. Biol. 2007;3:287. doi: 10.1038/nchembio874. [DOI] [PubMed] [Google Scholar]
- 115.Wickenden AD, Krajewski JL, London B, Wagoner PK, Wilson WA, Clark S, Roeloffs R, McNaughton-Smith G, Rigdon GC. Mol. Pharmacol. 2007 Dec 18; doi: 10.1124/mol.107.043216. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 116.Korsgaard MP, Hartz BP, Brown WD, Ahring PK, Strobaek D, Mirza NR. J. Pharmacol. Exp. Ther. 2005;314:282. doi: 10.1124/jpet.105.083923. [DOI] [PubMed] [Google Scholar]
- 117.Atkinson NS, Robertson GA, Ganetzky B. Science. 1991;253:551. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- 118.Butler A, Tsunoda S, McCobb DP, Wei A, Salkoff L. Science. 1993;261:221. doi: 10.1126/science.7687074. [DOI] [PubMed] [Google Scholar]
- 119.Brelidze TI, Niu X, Magleby KL. Proc. Natl. Acad. Sci. USA. 2003;100:9017. doi: 10.1073/pnas.1532257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nimigean CM, Chappie JS, Miller C. Biochemistry. 2003;42:9263. doi: 10.1021/bi0348720. [DOI] [PubMed] [Google Scholar]
- 121.Orio P, Rojas P, Ferreira G, Latorre R. News Physiol. Sci. 2002;17:156. doi: 10.1152/nips.01387.2002. [DOI] [PubMed] [Google Scholar]
- 122.Knaus HG, Schwarzer C, Koch RO, Eberhart A, Kaczorowski GJ, Glossmann H, Wunder F, Pongs O, Garcia ML, Sperk G. J. Neurosci. 1996;16:955. doi: 10.1523/JNEUROSCI.16-03-00955.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marrion NV, Tavalin SJ. Nature. 1998;395:900. doi: 10.1038/27674. [DOI] [PubMed] [Google Scholar]
- 124.Ahluwalia J, Tinker A, Clapp LH, Duchen MR, Abramov AY, Pope S, Nobles M, Segal AW. Nature. 2004;427:853. doi: 10.1038/nature02356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Femling JK, Cherny VV, Morgan D, Rada B, Davis AP, Czirjak G, Enyedi P, England SK, Moreland JG, Ligeti E, Nauseef WM, DeCoursey TE. J. Gen. Physiol. 2006;127:659. doi: 10.1085/jgp.200609504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Essin K, Salanova B, Kettritz R, Sausbier M, Luft FC, Kraus D, Bohn E, Autenrieth IB, Peschel A, Ruth P, Gollasch M. Am. J. Physiol. Cell. Physiol. 2007;293:C45. doi: 10.1152/ajpcell.00450.2006. [DOI] [PubMed] [Google Scholar]
- 127.DeCoursey TE. Am. J. Physiol. Cell. Physiol. 2007;293:C30. doi: 10.1152/ajpcell.00093.2007. [DOI] [PubMed] [Google Scholar]
- 128.Sausbier M, Hu H, Arntz C, Feil S, Kamm S, Adelsberger H, Sausbier U, Sailer CA, Feil R, Hofmann F, Korth M, Shipston MJ, Knaus HG, Wolfer DP, Pedroarena CM, Storm JF, Ruth P. Proc. Natl. Acad. Sci. USA. 2004;101:9474. doi: 10.1073/pnas.0401702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meredith AL, Thorneloe KS, Werner ME, Nelson MT, Aldrich RW. J. Biol. Chem. 2004;279:36746. doi: 10.1074/jbc.M405621200. [DOI] [PubMed] [Google Scholar]
- 130.Sausbier M, Arntz C, Bucurenciu I, Zhao H, Zhou XB, Sausbier U, Feil S, Kamm S, Essin K, Sailer CA, Abdullah U, Krippeit-Drews P, Feil R, Hofmann F, Knaus HG, Kenyon C, Shipston MJ, Storm JF, Neuhuber W, Korth M, Schubert R, Gollasch M, Ruth P. Circulation. 2005;112:60. doi: 10.1161/01.CIR.0000156448.74296.FE. [DOI] [PubMed] [Google Scholar]
- 131.Ruttiger L, Sausbier M, Zimmermann U, Winter H, Braig C, Engel J, Knirsch M, Arntz C, Langer P, Hirt B, Muller M, Kopschall I, Pfister M, Munkner S, Rohbock K, Pfaff I, Rusch A, Ruth P, Knipper M. Proc. Natl. Acad. Sci. USA. 2004;101:12922. doi: 10.1073/pnas.0402660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Werner ME, Zvara P, Meredith AL, Aldrich RW, Nelson MT. J. Physiol. 2005;567:545. doi: 10.1113/jphysiol.2005.093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Du W, Bautista JF, Yang H, Diez-Sampedro A, You SA, Wang L, Kotagal P, Luders HO, Shi J, Cui J, Richerson GB, Wang QK. Nat. Genet. 2005;37:733. doi: 10.1038/ng1585. [DOI] [PubMed] [Google Scholar]
- 134.Galvez A, Gimenez-Gallego G, Reuben JP, Roy-Contancin L, Feigenbaum P, Kaczorowski GJ, Garcia ML. J. Biol. Chem. 1990;265:11083. [PubMed] [Google Scholar]
- 135.Garcia-Valdes J, Zamudio FZ, Toro L, Possani LD. FEBS Lett. 2001;505:369. doi: 10.1016/s0014-5793(01)02791-0. [DOI] [PubMed] [Google Scholar]
- 136.Yao J, Chen X, Li H, Zhou Y, Yao L, Wu G, Zhang N, Zhou Z, Xu T, Wu H, Ding J. J. Biol. Chem. 2005;280:14819. doi: 10.1074/jbc.M412735200. [DOI] [PubMed] [Google Scholar]
- 137.Hanner M, Schmalhofer WA, Munujos P, Knaus HG, Kaczorowski GJ, Garcia ML. Proc. Natl. Acad. Sci. USA. 1997;94:2853. doi: 10.1073/pnas.94.7.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hanner M, Vianna-Jorge R, Kamassah A, Schmalhofer WA, Knaus HG, Kaczorowski GJ, Garcia ML. J. Biol. Chem. 1998;273:16289. doi: 10.1074/jbc.273.26.16289. [DOI] [PubMed] [Google Scholar]
- 139.Knaus HG, McManus OB, Lee SH, Schmalhofer WA, Garcia-Calvo M, Helms LM, Sanchez M, Giangiacomo K, Reuben JP, Smith AB, 3rd, et al. Biochemistry. 1994;33:5819. doi: 10.1021/bi00185a021. [DOI] [PubMed] [Google Scholar]
- 140.Sanchez M, McManus OB. Neuropharmacology. 1996;35:963. doi: 10.1016/0028-3908(96)00137-2. [DOI] [PubMed] [Google Scholar]
- 141.Strobaek D, Christophersen P, Holm NR, Moldt P, Ahring PK, Johansen TE, Olesen SP. Neuropharmacology. 1996;35:903. doi: 10.1016/0028-3908(96)00096-2. [DOI] [PubMed] [Google Scholar]
- 142.Dalziel JE, Finch SC, Dunlop J. Toxicol. Lett. 2005;155:421. doi: 10.1016/j.toxlet.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 143.McManus OB, Harris GH, Giangiacomo KM, Feigenbaum P, Reuben JP, Addy ME, Burka JF, Kaczorowski GJ, Garcia ML. Biochemistry. 1993;32:6128. doi: 10.1021/bi00075a002. [DOI] [PubMed] [Google Scholar]
- 144.McManus OB, Helms LM, Pallanck L, Ganetzky B, Swanson R, Leonard RJ. Neuron. 1995;14:645. doi: 10.1016/0896-6273(95)90321-6. [DOI] [PubMed] [Google Scholar]
- 145.Singh SB, Goetz MA, Zink DL, Dombrowski AW, Polishook JD, Garcia ML, Schamlhofer W, McManus OB, Kaczorowski GJ. J. Chem. Soc. (Perkin 1) 1994;22:3349. [Google Scholar]
- 146.Imaizumi Y, Sakamoto K, Yamada A, Hotta A, Ohya S, Muraki K, Uchiyama M, Ohwada T. Mol. Pharmacol. 2002;62:836. doi: 10.1124/mol.62.4.836. [DOI] [PubMed] [Google Scholar]
- 147.Zakharov SI, Morrow JP, Liu G, Yang L, Marx SO. J. Biol. Chem. 2005;280:30882. doi: 10.1074/jbc.M505302200. [DOI] [PubMed] [Google Scholar]
- 148.Zeng H, Lozinskaya IM, Lin Z, Willette RN, Brooks DP, Xu X. J. Pharmacol. Exp. Ther. 2006;319:957. doi: 10.1124/jpet.106.110593. [DOI] [PubMed] [Google Scholar]
- 149.Valverde MA, Rojas P, Amigo J, Cosmelli D, Orio P, Bahamonde MI, Mann GE, Vergara C, Latorre R. Science. 1999;285:1929. doi: 10.1126/science.285.5435.1929. [DOI] [PubMed] [Google Scholar]
- 150.Olesen SP, Munch E, Moldt P, Drejer J. Eur. J. Pharmacol. 1994;251:53. doi: 10.1016/0014-2999(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 151.Olesen SP, Munch E, Watjen F, Drejer J. Neuroreport. 1994;5:1001. doi: 10.1097/00001756-199404000-00037. [DOI] [PubMed] [Google Scholar]
- 152.Papassotiriou J, Kohler R, Prenen J, Krause H, Akbar M, Eggermont J, Paul M, Distler A, Nilius B, Hoyer J. FASEB J. 2000;14:885. [PubMed] [Google Scholar]
- 153.Tanaka Y, Meera P, Song M, Knaus HG, Toro L. J. Physiol. (Lond) 1997;502:545. doi: 10.1111/j.1469-7793.1997.545bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bentzen BH, Nardi A, Calloe K, Madsen LS, Olesen SP, Grunnet M. Mol. Pharmacol. 2007;72:1033. doi: 10.1124/mol.107.038331. [DOI] [PubMed] [Google Scholar]
- 155.Hewawasam P, Erway M, Moon SL, Knipe J, Weiner H, Boissard CG, Post-Munson DJ, Gao Q, Huang S, Gribkoff VK, Meanwell NA. J. Med. Chem. 2002;45:1487. doi: 10.1021/jm0101850. [DOI] [PubMed] [Google Scholar]
- 156.Gribkoff VK, Starrett JE, Dworetzky SI, Hewawasam P, Boissard CG, Cook DA, Frantz SW, Heman K, Hibbard JR, Huston K, Johnson G, Krishnan BS, Kinney GG, Lombardo LA, Meanwell NA, Molinoff PB, Myers RA, Moon SL, Ortiz A, Pajor L, Pieschl RL, Post-Munson DJ, Signor LJ, Srinivas N, Taber MT, Thalody G, Trojnacki JT, Wiener H, Yeleswaram K, Yeola SW. Nature Med. 2001;7:471. doi: 10.1038/86546. [DOI] [PubMed] [Google Scholar]
- 157.Schroder RL, Jespersen T, Christophersen P, Strobaek D, Jensen BS, Olesen SP. Neuropharmacology. 2001;40:888. doi: 10.1016/s0028-3908(01)00029-6. [DOI] [PubMed] [Google Scholar]
- 158.Dupuis DS, Schroder RL, Jespersen T, Christensen JK, Christophersen P, Jensen BS, Olesen SP. Eur. J. Pharmacol. 2002;437:129. doi: 10.1016/s0014-2999(02)01287-6. [DOI] [PubMed] [Google Scholar]
- 159.Jensen BS. CNS Drug. Rev. 2002;8:353. doi: 10.1111/j.1527-3458.2002.tb00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tanaka M, Sasaki Y, Kimura Y, Fukui T, Hamada K, Ukai Y. B.J.U. Int. 2003;92:1031. doi: 10.1111/j.1464-410x.2003.04512.x. [DOI] [PubMed] [Google Scholar]
- 161.Hugues M, Schmid H, Romey G, Duval D, Frelin C, Lazdunski M. EMBO J. 1982;1:1039. doi: 10.1002/j.1460-2075.1982.tb01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Science. 1996;273:1709. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 163.Stocker M, Hirzel K, D'Hoedt D, Pedarzani P. Toxicon. 2004;43:933. doi: 10.1016/j.toxicon.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 164.Xia XM, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen JE, Ishii T, Hirschberg B, Bond CT, Lutsenko S, Maylie J, Adelman JP. Nature. 1998;395:503. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- 165.Ishii TM, Maylie J, Adelman JP. J. Biol. Chem. 1997;272:23195. doi: 10.1074/jbc.272.37.23195. [DOI] [PubMed] [Google Scholar]
- 166.Monaghan AS, Benton DC, Bahia PK, Hosseini R, Shah YA, Haylett DG, Moss GW. J. Biol. Chem. 2004;279:1003. doi: 10.1074/jbc.M308070200. [DOI] [PubMed] [Google Scholar]
- 167.Zhang BM, Kohli V, Adachi R, Lopez JA, Udden MM, Sullivan R. Biochemistry. 2001;40:3189. doi: 10.1021/bi001675h. [DOI] [PubMed] [Google Scholar]
- 168.Strassmaier T, Bond CT, Sailer CA, Knaus HG, Maylie J, Adelman JP. J. Biol. Chem. 2005;280:21231. doi: 10.1074/jbc.M413125200. [DOI] [PubMed] [Google Scholar]
- 169.Tomita H, Shakkottai VG, Gutman GA, Sun G, Bunney WE, Cahalan MD, Chandy KG, Gargus JJ. Mol. Psychiatry. 2003;8:524. doi: 10.1038/sj.mp.4001271. [DOI] [PubMed] [Google Scholar]
- 170.Kolski-Andreaco A, Tomita H, Shakkottai VG, Gutman GA, Cahalan MD, Gargus JJ, Chandy KG. J. Biol. Chem. 2004;279:6893. doi: 10.1074/jbc.M311725200. [DOI] [PubMed] [Google Scholar]
- 171.Wittekindt OH, Visan V, Tomita H, Imtiaz F, Gargus JJ, Lehmann-Horn F, Grissmer S, Morris-Rosendahl DJ. Mol. Pharmacol. 2004;65:788. doi: 10.1124/mol.65.3.788. [DOI] [PubMed] [Google Scholar]
- 172.Wolfart J, Neuhoff H, Franz O, Roeper J. J. Neurosci. 2001;21:3443. doi: 10.1523/JNEUROSCI.21-10-03443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Waroux O, Massotte L, Alleva L, Graulich A, Thomas E, Liegeois JF, Scuvee-Moreau J, Seutin V. Eur. J. Neurosci. 2005;22:3111. doi: 10.1111/j.1460-9568.2005.04484.x. [DOI] [PubMed] [Google Scholar]
- 174.Villalobos C, Shakkottai VG, Chandy KG, Michelhaugh SK, Andrade R. J. Neurosci. 2004;24:3537. doi: 10.1523/JNEUROSCI.0380-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bond CT, Herson PS, Strassmaier T, Hammond R, Stackman R, Maylie J, Adelman JP. J. Neurosci. 2004;24:5301. doi: 10.1523/JNEUROSCI.0182-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hammond RS, Bond CT, Strassmaier T, Ngo-Anh TJ, Adelman JP, Maylie J, Stackman RW. J. Neurosci. 2006;26:1844. doi: 10.1523/JNEUROSCI.4106-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Herrera GM, Pozo MJ, Zvara P, Petkov GV, Bond CT, Adelman JP, Nelson MT. J. Physiol. 2003;551:893. doi: 10.1113/jphysiol.2003.045914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bond CT, Sprengel R, Bissonnette JM, Kaufman WA, Pribnow D, Neelands T, Storck T, Baetscher M, Jerecic J, Maylie J, Knaus HG, Seeburg P, Adelman JP. Science. 2000;289:1942. doi: 10.1126/science.289.5486.1942. [DOI] [PubMed] [Google Scholar]
- 179.Bowen T, Williams N, Norton N, Spurlock G, Wittekindt OH, Morris-Rosendahl DJ, Williams H, Brzustowicz L, Hoogendoorn B, Zammit S, Jones G, Sanders RD, Jones LA, McCarthy G, Jones S, Bassett A, Cardno AG, Owen MJ, O'Donovan MC. Mol. Psychiatry. 2001;6:259. doi: 10.1038/sj.mp.4000128. [DOI] [PubMed] [Google Scholar]
- 180.Miller MJ, Rauer H, Tomita H, Rauer H, Gargus JJ, Gutman GA, Cahalan MD, Chandy KG. J. Biol. Chem. 2001;276:27753. doi: 10.1074/jbc.C100221200. [DOI] [PubMed] [Google Scholar]
- 181.Chandy KG, Fantino E, Wittekindt O, Kalman K, Tong LL, Ho TH, Gutman GA, Crocq MA, Ganguli R, Nimgaonkar V, Morris-Rosendahl DJ, Gargus JJ. Mol. Psychiatry. 1998;3:32. doi: 10.1038/sj.mp.4000353. [DOI] [PubMed] [Google Scholar]
- 182.Bowen T, Guy CA, Craddock N, Cardno AG, Williams NM, Spurlock G, Murphy KC, Jones LA, Gray M, Sanders RD, McCarthy G, Chandy KG, Fantino E, Kalman K, Gutman GA, Gargus JJ, Williams J, McGuffin P, Owen MJ, O'Donovan MC. Mol. Psychiatry. 1998;3:266. doi: 10.1038/sj.mp.4000400. [DOI] [PubMed] [Google Scholar]
- 183.Figueroa KP, Chan P, Schols L, Tanner C, Riess O, Perlman SL, Geschwind DH, Pulst SM. Arch. Neurol. 2001;58:1649. doi: 10.1001/archneur.58.10.1649. [DOI] [PubMed] [Google Scholar]
- 184.Taylor MS, Bonev AD, Gross TP, Eckman DM, Brayden JE, Bond CT, Adelman JP, Nelson MT. Circ. Res. 2003;93:124. doi: 10.1161/01.RES.0000081980.63146.69. [DOI] [PubMed] [Google Scholar]
- 185.Nagayama T, Masada K, Yoshida M, Suzuki-Kusaba M, Hisa H, Kimura T, Satoh S. Am. J. Physiol. 1998;274:R1125. doi: 10.1152/ajpregu.1998.274.4.R1125. [DOI] [PubMed] [Google Scholar]
- 186.Nagayama T, Fukushima Y, Hikichi H, Yoshida M, Suzuki-Kusaba M, Hisa H, Kimura T, Satoh S. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000;279:R1731. doi: 10.1152/ajpregu.2000.279.5.R1731. [DOI] [PubMed] [Google Scholar]
- 187.Kohler R, Hoyer J. Kidney Int. 2007;72:145. doi: 10.1038/sj.ki.5002303. [DOI] [PubMed] [Google Scholar]
- 188.Wulff H, Kolski-Andreaco A, Sankaranarayanan A, Sabatier JM, Shakkottai V. Curr. Med. Chem. 2007;14:1437. doi: 10.2174/092986707780831186. [DOI] [PubMed] [Google Scholar]
- 189.Castle NA, Strong PN. FEBS Lett. 1986;209:117. doi: 10.1016/0014-5793(86)81095-x. [DOI] [PubMed] [Google Scholar]
- 190.Auguste P, Hugues M, Grave B, Gesquiere JC, Maes P, Tartar A, Romey G, Schweitz H, Lazdunski M. J. Biol. Chem. 1990;265:4753. [PubMed] [Google Scholar]
- 191.Pedarzani P, D'Hoedt D, Doorty KB, Wadsworth JD, Joseph JS, Jeyaseelan K, Kini RM, Gadre SV, Sapatnekar SM, Stocker M, Strong PN. J. Biol. Chem. 2002;277:46101. doi: 10.1074/jbc.M206465200. [DOI] [PubMed] [Google Scholar]
- 192.Sabatier JM, Zerrouk H, Darbon H, Mabrouk K, Benslimane A, Rochat H, Martin-Eauclaire MF, Van Rietschoten J. Biochemistry. 1993;32:2763. doi: 10.1021/bi00062a005. [DOI] [PubMed] [Google Scholar]
- 193.Sabatier JM, Fremont V, Mabrouk K, Crest M, Darbon H, Rochat H, Van Rietschoten J, Martin-Eauclaire MF. Intern. J. Peptide and Protein Res. 1994;43:486. doi: 10.1111/j.1399-3011.1994.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 194.Shakkottai VG, Regaya I, Wulff H, Fajloun Z, Tomita H, Fathallah M, Cahalan MD, Gargus JJ, Sabatier JM, Chandy KG. J. Biol. Chem. 2001;276:43145. doi: 10.1074/jbc.M106981200. [DOI] [PubMed] [Google Scholar]
- 195.Messier C, Mourre C, Bontempi B, Sif J, Lazdunski M, Destrade C. Brain Res. 1991;551:322. doi: 10.1016/0006-8993(91)90950-z. [DOI] [PubMed] [Google Scholar]
- 196.Ikonen S, Riekkinen P., Jr Eur. J. Pharmacol. 1999;382:151. doi: 10.1016/s0014-2999(99)00616-0. [DOI] [PubMed] [Google Scholar]
- 197.Deschaux O, Bizot JC, Goyffon M. Neurosci. Lett. 1997;222:159. doi: 10.1016/s0304-3940(97)13367-5. [DOI] [PubMed] [Google Scholar]
- 198.Stackman RW, Hammond RS, Linardatos E, Gerlach A, Maylie J, Adelman JP, Tzounopoulos T. J. Neurosci. 2002;22:10163. doi: 10.1523/JNEUROSCI.22-23-10163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Galeotti N, Ghelardini C, Caldari B, Bartolini A. Br. J. Pharmacol. 1999;126:1653. doi: 10.1038/sj.bjp.0702467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Galanakis D, Ganellin CR, Chen JQ, Gunasekera D, Dunn PM. Bioorg. Med. Chem. Lett. 2004;14:4231. doi: 10.1016/j.bmcl.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 201.Dilly S, Graulich A, Farce A, Seutin V, Liegeois JF, Chavatte P. J. Enzyme. Inhib. Med. Chem. 2005;20:517. doi: 10.1080/14756360500210989. [DOI] [PubMed] [Google Scholar]
- 202.Jenkinson DH, Haylett DG, Cook NS. Cell Calcium. 1983;4:429. doi: 10.1016/0143-4160(83)90019-2. [DOI] [PubMed] [Google Scholar]
- 203.Cook NS, Haylett DG. J. Physiol. 1985;358:373. doi: 10.1113/jphysiol.1985.sp015556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Castle NA, Haylett DG, Morgan JM, Jenkinson DH. Eur. J. Pharmacol. 1993;236:201. doi: 10.1016/0014-2999(93)90590-e. [DOI] [PubMed] [Google Scholar]
- 205.Galanakis D, Davis CA, Del Rey Herrero B, Ganellin CR, Dunn PM, Jenkinson DH. J. Med. Chem. 1995;38:595. doi: 10.1021/jm00004a005. [DOI] [PubMed] [Google Scholar]
- 206.Galanakis D, Calder JA, Ganellin CR, Owen CS, Dunn PM. J. Med. Chem. 1995;38:3536. doi: 10.1021/jm00018a013. [DOI] [PubMed] [Google Scholar]
- 207.Galanakis D, Ganellin CR, Dunn P, Jenkinson DH. Arch. Pharm. Pharm. Med. Chem. 1996;329:524. doi: 10.1002/ardp.19963291203. [DOI] [PubMed] [Google Scholar]
- 208.Galanakis D, Davis CA, Ganellin CR, Dunn PM. J. Med. Chem. 1996;39:359. doi: 10.1021/jm950520i. [DOI] [PubMed] [Google Scholar]
- 209.Galanakis D, Ganellin CR, Malik S, Dunn PM. J. Med. Chem. 1996;39:3592. doi: 10.1021/jm950838a. [DOI] [PubMed] [Google Scholar]
- 210.Rosa JC, Galanakis D, Ganellin CR, Dunn PM, Jenkinson DH. J. Med. Chem. 1998;41:2. doi: 10.1021/jm970571a. [DOI] [PubMed] [Google Scholar]
- 211.Campos Rosa J, Galanakis D, Piergentili A, Bhandari K, Ganellin CR, Dunn PM, Jenkinson DH. J. Med. Chem. 2000;43:420. doi: 10.1021/jm9902537. [DOI] [PubMed] [Google Scholar]
- 212.Strobaek D, Jorgensen TD, Christophersen P, Ahring PK, Olesen SP. Br. J. Pharmacol. 2000;129:991. doi: 10.1038/sj.bjp.0703120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fanger CM, Rauer H, Neben AL, Miller MJ, Rauer H, Wulff H, Rosa JC, Ganellin CR, Chandy KG, Cahalan MD. J. Biol. Chem. 2001;276:12249. doi: 10.1074/jbc.M011342200. [DOI] [PubMed] [Google Scholar]
- 214.Chen JQ, Galanakis D, Ganellin CR, Dunn PM, Jenkinson DH. J. Med. Chem. 2000;43:3478. doi: 10.1021/jm000904v. [DOI] [PubMed] [Google Scholar]
- 215.Hosseini R, Benton DC, Dunn PM, Jenkinson DH, Moss GW. J. Physiol. 2001;535:323. doi: 10.1111/j.1469-7793.2001.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Strobaek D, Hougaard C, Johansen TH, Sorensen US, Nielsen EO, Nielsen KS, Taylor RD, Pedarzani P, Christophersen P. Mol. Pharmacol. 2006;70:1771. doi: 10.1124/mol.106.027110. [DOI] [PubMed] [Google Scholar]
- 217.Dreixler JC, Bian J, Cao Y, Roberts MT, Roizen JD, Houamed KM. Eur. J. Pharmacol. 2000;401:1. doi: 10.1016/s0014-2999(00)00401-5. [DOI] [PubMed] [Google Scholar]
- 218.Terstappen GC, Pellacani A, Aldegheri L, Graziani F, Carignani C, Pula G, Virginio C. Neurosci. Lett. 2003;346:85. doi: 10.1016/s0304-3940(03)00574-3. [DOI] [PubMed] [Google Scholar]
- 219.Syme CA, Gerlach AC, Singh AK, Devor DC. Am. J. Physiol. Cell. Physiol. 2000;278:C570. doi: 10.1152/ajpcell.2000.278.3.C570. [DOI] [PubMed] [Google Scholar]
- 220.Pedarzani P, Mosbacher J, Rivard A, Cingolani LA, Oliver D, Stocker M, Adelman JP, Fakler B. J. Biol. Chem. 2001;276:9762. doi: 10.1074/jbc.M010001200. [DOI] [PubMed] [Google Scholar]
- 221.Strobaek D, Teuber L, Jorgensen TD, Ahring PK, Kjaer K, Hansen RS, Olesen SP, Christophersen P, Skaaning-Jensen B. Biochim. Biophys. Acta. 2004;1665:1. doi: 10.1016/j.bbamem.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 222.Pedarzani P, McCutcheon JE, Rogge G, Jensen BS, Christophersen P, Hougaard C, Strobaek D, Stocker M. J. Biol. Chem. 2005;280:41404. doi: 10.1074/jbc.M509610200. [DOI] [PubMed] [Google Scholar]
- 223.Cao Y, Dreixler JC, Roizen JD, Roberts MT, Houamed KM. J. Pharmacol. Exp. Ther. 2001;296:683. [PubMed] [Google Scholar]
- 224.Grunnet M, Jespersen T, Angelo K, Frokjaer-Jensen C, Klaerke DA, Olesen SP, Jensen BS. Neuropharmacology. 2001;40:879. doi: 10.1016/s0028-3908(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 225.Cao YJ, Dreixler JC, Couey JJ, Houamed KM. Eur. J. Pharmacol. 2002;449:47. doi: 10.1016/s0014-2999(02)01987-8. [DOI] [PubMed] [Google Scholar]
- 226.Hougaard C, Eriksen BL, Jorgensen S, Johansen TH, Dyhring T, Madsen LS, Strobaek D, Christophersen P. Br. J. Pharmacol. 2007;151:655. doi: 10.1038/sj.bjp.0707281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shakkottai VG, Chou CH, Oddo S, Sailer CA, Knaus HG, Gutman GA, Barish ME, LaFerla FM, Chandy KG. J. Clin. Invest. 2004;113:582. doi: 10.1172/JCI20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Orr HT. J. Clin. Invest. 2004;113:505. doi: 10.1172/JCI21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lappin SC, Dale TJ, Brown JT, Trezise DJ, Davies CH. Brain Res. 2005;1065:37. doi: 10.1016/j.brainres.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 230.Anderson NJ, Slough S, Watson WP. Eur. J. Pharmacol. 2006;546:48. doi: 10.1016/j.ejphar.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 231.Patel AJ, Honore E, Lesage F, Fink M, Romey G, Lazdunski M. Nat. Neurosci. 1999;2:422. doi: 10.1038/8084. [DOI] [PubMed] [Google Scholar]
- 232.Heurteaux C, Guy N, Laigle C, Blondeau N, Duprat F, Mazzuca M, Lang-Lazdunski L, Widmann C, Zanzouri M, Romey G, Lazdunski M. EMBO J. 2004;23:2684. doi: 10.1038/sj.emboj.7600234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Duprat F, Lesage F, Patel AJ, Fink M, Romey G, Lazdunski M. Mol. Pharmacol. 2000;57:906. [PubMed] [Google Scholar]
- 234.Heurteaux C, Laigle C, Blondeau N, Jarretou G, Lazdunski M. Neuroscience. 2006;137:241. doi: 10.1016/j.neuroscience.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 235.Sherratt RM, Bostock H, Sears TA. Nature. 1980;283:570. doi: 10.1038/283570a0. [DOI] [PubMed] [Google Scholar]
- 236.Smith KJ, Felts PA, John GR. Brain. 2000;123:171. doi: 10.1093/brain/123.1.171. [DOI] [PubMed] [Google Scholar]
- 237.Beeton C, Wulff H, Standifer NE, Azam P, Mullen KM, Pennington MW, Kolski-Andreaco A, Wei E, Grino A, Counts DR, Wang PH, Leehealey CJ, B SA, Sankaranarayanan A, Homerick D, Roeck WW, Tehranzadeh J, Stanhope KL, Zimin P, Havel PJ, Griffey S, Knaus HG, Nepom GT, Gutman GA, Calabresi PA, Chandy KG. Proc. Natl. Acad. Sci. USA. 2006;103:17414. doi: 10.1073/pnas.0605136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sidell N, Schlichter LC, Wright SC, Hagiwara S, Golub SH. J. Immunol. 1986;137:1650. [PubMed] [Google Scholar]
- 239.Schlichter L, Sidell N, Hagiwara S. Proc. Natl. Acad. Sci. USA. 1986;83:451. doi: 10.1073/pnas.83.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Duffy SM, Lawley WJ, Conley EC, Bradding P. J. Immunol. 2001;167:4261. doi: 10.4049/jimmunol.167.8.4261. [DOI] [PubMed] [Google Scholar]
- 241.Cruse G, Duffy SM, Brightling CE, Bradding P. Thorax. 2006;61:880. doi: 10.1136/thx.2006.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gallin EK. Biophys. J. 1984;46:821. doi: 10.1016/S0006-3495(84)84080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gallin EK, Sheehy PA. J. Physiol. (Lond.) 1985;369:475. doi: 10.1113/jphysiol.1985.sp015911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gallin EK, McKinney LC. J. Membr. Biol. 1988;103:55. doi: 10.1007/BF01871932. [DOI] [PubMed] [Google Scholar]
- 245.McKinney LC, Gallin EK. J. Membr. Biol. 1990;116:47. doi: 10.1007/BF01871671. [DOI] [PubMed] [Google Scholar]
- 246.Judge SI, Montcalm-Mazzilli E, Gallin EK. Am. J. Physiol. 1994;267:C1691. doi: 10.1152/ajpcell.1994.267.6.C1691. [DOI] [PubMed] [Google Scholar]
- 247.Ypey DL, Clapham DE. Proc. Natl. Acad. Sci. USA. 1984;81:3083. [Google Scholar]
- 248.Blunck R, Scheel O, Muller M, Brandenburg K, Seitzer U, Seydel U. J. Immunol. 2001;166:1009. doi: 10.4049/jimmunol.166.2.1009. [DOI] [PubMed] [Google Scholar]
- 249.DeCoursey TE, Kim SY, Silver MR, Quandt FN. J. Membr. Biol. 1996;152:141. doi: 10.1007/s002329900093. [DOI] [PubMed] [Google Scholar]
- 250.Eder C, Fischer HG. Naunyn Schmiedebergs Arch. Pharmacol. 1997;355:198. doi: 10.1007/pl00004932. [DOI] [PubMed] [Google Scholar]
- 251.Liu QH, Williams DA, McManus C, Baribaud F, Doms RW, Schols D, De Clercq E, Kotlikoff MI, Collman RG, Freedman BD. Proc. Natl. Acad. Sci. USA. 2000;97:4832. doi: 10.1073/pnas.090521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hanley PJ, Musset B, Renigunta V, Limberg SH, Dalpke AH, Sus R, Heeg KM, Preisig-Muller R, Daut J. Proc. Natl. Acad. Sci. USA. 2004;101:9479. doi: 10.1073/pnas.0400733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chung I, Zelivyanskaya M, Gendelman HE. J. Neuroimmunol. 2002;122:40. doi: 10.1016/s0165-5728(01)00462-3. [DOI] [PubMed] [Google Scholar]
- 254.Vicente R, Escalada A, Coma M, Fuster G, Sanchez-Tillo E, Lopez-Iglesias C, Soler C, Solsona C, Celada A, Felipe A. J. Biol. Chem. 2003;278:46307. doi: 10.1074/jbc.M304388200. [DOI] [PubMed] [Google Scholar]
- 255.Vicente R, Escalada A, Villalonga N, Texido L, Roura-Ferrer M, Martin-Satue M, Lopez-Iglesias C, Soler C, Solsona C, Tamkun MM, Felipe A. J. Biol. Chem. 2006;281:37675. doi: 10.1074/jbc.M605617200. [DOI] [PubMed] [Google Scholar]
- 256.Villalonga N, Escalada A, Vicente R, Sanchez-Tillo E, Celada A, Solsona C, Felipe A. Biochem. Biophys. Res. Commun. 2007;352:913. doi: 10.1016/j.bbrc.2006.11.120. [DOI] [PubMed] [Google Scholar]
- 257.Eder C, Fischer HG, Hadding U, Heinemann U. J. Membr. Biol. 1995;147:137. doi: 10.1007/BF00233542. [DOI] [PubMed] [Google Scholar]
- 258.Schlichter LC, Sakellaropoulos G, Ballyk B, Pennefather PS, Phipps D. J. Glia. 1996;17:225. doi: 10.1002/(SICI)1098-1136(199607)17:3<225::AID-GLIA5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 259.Eder C. Am. J. Physiol. 1998;275:C327. doi: 10.1152/ajpcell.1998.275.2.C327. [DOI] [PubMed] [Google Scholar]
- 260.Kotecha SA, Schlichter LC. J. Neurosci. 1999;19:10680. doi: 10.1523/JNEUROSCI.19-24-10680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Khanna R, Roy L, Zhu X, Schlichter LC. Am. J. Physiol. Cell. Physiol. 2001;280:C796. doi: 10.1152/ajpcell.2001.280.4.C796. [DOI] [PubMed] [Google Scholar]
- 262.Schilling T, Stock C, Schwab A, Eder C. Eur. J. Neurosci. 2004;19:1469. doi: 10.1111/j.1460-9568.2004.03265.x. [DOI] [PubMed] [Google Scholar]
- 263.Fay AJ, Qian X, Jan YN, Jan LY. Proc. Natl. Acad. Sci. USA. 2006;103:17548. doi: 10.1073/pnas.0607914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ghanshani S, Wulff H, Miller MJ, Rohm H, Neben A, Gutman GA, Cahalan MD, Chandy KG. J. Biol. Chem. 2000;275:37137. doi: 10.1074/jbc.M003941200. [DOI] [PubMed] [Google Scholar]
- 265.Wulff H, Calabresi PA, Allie R, Yun S, Pennington M, Beeton C, Chandy KG. J. Clin. Invest. 2003;111:1703. doi: 10.1172/JCI16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wulff H, Knaus HG, Pennington M, Chandy KG. J. Immunol. 2004;173:776. doi: 10.4049/jimmunol.173.2.776. [DOI] [PubMed] [Google Scholar]
- 267.Lewis RS, Cahalan MD. Annu. Rev. Immunol. 1995;13:623. doi: 10.1146/annurev.iy.13.040195.003203. [DOI] [PubMed] [Google Scholar]
- 268.Cahalan MD, Wulff H, Chandy KG. J. Clin. Immunol. 2001;21:235. doi: 10.1023/a:1010958907271. [DOI] [PubMed] [Google Scholar]
- 269.Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. Science. 2006;312:1220. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. Nature. 2005;437:902. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Peinelt C, Vig M, Koomoa DL, Beck A, Nadler MJ, Koblan-Huberson M, Lis A, Fleig A, Penner R, Kinet JP. Nat. Cell. Biol. 2006;8:771. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lewis RS. Ann. Rev. Immunol. 2001;19:497. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 273.Feske S. Nat. Rev. Immunol. 2007;7:690. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 274.Wulff H, Beeton C, Chandy KG. Curr. Opin. Drug Discov. Devel. 2003;6:640. [PubMed] [Google Scholar]
- 275.Wulff H, Pennington M. Curr. Opin. Drug Discov. Devel. 2007;10:438. [PubMed] [Google Scholar]
- 276.Grissmer S, Dethlefs B, Wasmuth JJ, Goldin AL, Gutman GA, Cahalan MD, Chandy KG. Proc. Natl. Acad. Sci. USA. 1990;87:9411. doi: 10.1073/pnas.87.23.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Douglass J, Osborne PB, Cai YC, Wilkinson M, Christie MJ, Adelman JP. J. Immunol. 1990;144:4841. [PubMed] [Google Scholar]
- 278.DeCoursey TE, Chandy KG, Gupta S, Cahalan MD. Nature. 1984;307:465. doi: 10.1038/307465a0. [DOI] [PubMed] [Google Scholar]
- 279.Matteson DR, Deutsch C. Nature. 1984;307:468. doi: 10.1038/307468a0. [DOI] [PubMed] [Google Scholar]
- 280.Mackenzie AB, Chirakkal H, North RA. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L862. doi: 10.1152/ajplung.00095.2003. [DOI] [PubMed] [Google Scholar]
- 281.Pappone PA, Ortiz-Miranda SI. Am. J. Physiol. 1993;264:C1014. doi: 10.1152/ajpcell.1993.264.4.C1014. [DOI] [PubMed] [Google Scholar]
- 282.Chittajallu R, Chen Y, Wang H, Yuan X, Ghiani CA, Heckman T, C.J M, Gallo V. Proc. Natl. Acad. Sci. USA. 2002;99:2350. doi: 10.1073/pnas.042698399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Fadool DA, Levitan IB. J. Neurosci. 1998;18:6126. doi: 10.1523/JNEUROSCI.18-16-06126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Fadool DA, Tucker K, Perkins R, Fasciani G, Thompson RN, Parsons AD, Overton JM, Koni PA, Flavell RA, Kaczmarek LK. Neuron. 2004;41:389. doi: 10.1016/s0896-6273(03)00844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Helms LM, Felix JP, Bugianesi RM, Garcia ML, Stevens S, Leonard RJ, Knaus HG, Koch R, Wanner SG, Kaczorowski GJ, Slaughter RS. Biochemistry. 1997;36:3737. doi: 10.1021/bi962351p. [DOI] [PubMed] [Google Scholar]
- 286.Koo GC, Blake JT, Talento A, Nguyen M, Lin S, Sirotina A, Shah K, Mulvany K, Hora D, Jr., Cunningham P, Wunderler DL, McManus OB, Slaughter R, Bugianesi R, Felix J, Garcia M, Williamson J, Kaczorowski G, Sigal NH, Springer MS, Feeney W. J. Immunol. 1997;158:5120. [PubMed] [Google Scholar]
- 287.Cahalan MD, Chandy KG. Curr. Opin. Biotechnol. 1997;8:749. doi: 10.1016/s0958-1669(97)80130-9. [DOI] [PubMed] [Google Scholar]
- 288.Beeton C, Wulff H, Barbaria J, Clot-Faybesse O, Pennington M, Bernard D, Cahalan MD, Chandy KG, Beraud E. Proc. Natl. Acad. Sci. USA. 2001;98:13942. doi: 10.1073/pnas.241497298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pereira LE, Villinger F, Wulff H, Sankaranarayanan A, Raman G, Ansari AA. Exp. Biol. Med. (Maywood) 2007;232:1338. doi: 10.3181/0705-RM-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Beeton C, Chandy KG. Neuroscientist. 2005;11:550. doi: 10.1177/1073858405278016. [DOI] [PubMed] [Google Scholar]
- 291.Freedman BD, Fleischmann BK, Punt JA, Gaulton G, Hashimoto Y, Kotlikoff MI. J. Biol. Chem. 1995;270:22406. doi: 10.1074/jbc.270.38.22406. [DOI] [PubMed] [Google Scholar]
- 292.Lewis RS, Cahalan MD. Science. 1988;239:771. doi: 10.1126/science.2448877. [DOI] [PubMed] [Google Scholar]
- 293.Koni PA, Khanna R, Chang MC, Tang MD, Kaczmarek LK, Schlichter LC, Flavella RA. J. Biol. Chem. 2003;278:39443. doi: 10.1074/jbc.M304879200. [DOI] [PubMed] [Google Scholar]
- 294.Xu J, Koni PA, Wang P, Li G, Kaczmarek L, Wu Y, Li Y, Flavell RA, Desir GV. Hum. Mol. Genet. 2003;12:551. doi: 10.1093/hmg/ddg049. [DOI] [PubMed] [Google Scholar]
- 295.Xu J, Wang P, Li Y, Li G, Kaczmarek LK, Wu Y, Koni PA, Flavell RA, Desir GV. Proc. Natl. Acad. Sci. USA. 2004;101:3112. doi: 10.1073/pnas.0308450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Lucero MT, Pappone PA. J. Gen. Physiol. 1989;93:451. doi: 10.1085/jgp.93.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ramirez-Ponce MP, Mateos JC, Carrion N, Bellido JA. Biochem. Biophys. Res. Commun. 1996;223:250. doi: 10.1006/bbrc.1996.0880. [DOI] [PubMed] [Google Scholar]
- 298.Ramirez-Ponce MP, Mateos JC, Bellido JA. J. Membr. Biol. 2003;196:129. doi: 10.1007/s00232-003-0631-1. [DOI] [PubMed] [Google Scholar]
- 299.Panyi G, Sheng Z, Deutsch C. Biophys. J. 1995;69:896. doi: 10.1016/S0006-3495(95)79963-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Levy DI, Deutsch C. Biophys. J. 1996;70:798. doi: 10.1016/S0006-3495(96)79619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sands SB, Lewis RS, Cahalan MD. J. Gen. Physiol. 1989;93:1061. doi: 10.1085/jgp.93.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Leonard R, Garcia M, Slaughter R, Reuben J. Proc. Natl. Acad. Sci. USA. 1992;89:10094. doi: 10.1073/pnas.89.21.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Crest M, Jacquet G, Gola M, Zerrouk H, Benslimane A, Rochat H, Mansuelle P, Martin-Eauclaire MF. J. Biol. Chem. 1992;267:1640. [PubMed] [Google Scholar]
- 304.Garcia-Calvo M, Leonard RJ, Novick J, Stevens SP, Schmalhofer W, Kaczorowski GJ, Garcia ML. J. Biol. Chem. 1993;268:18866. [PubMed] [Google Scholar]
- 305.Bednarek M, Bugianesi R, Leonard R, Felix J. Biochem. Biophys. Res. Commun. 1994;198:619. doi: 10.1006/bbrc.1994.1090. [DOI] [PubMed] [Google Scholar]
- 306.Dauplais M, Gilquin B, Possani L, Gurrola-Briones G, Roumestand C, Menez A. Biochemistry. 1995;34:16563. doi: 10.1021/bi00051a004. [DOI] [PubMed] [Google Scholar]
- 307.Lebrun B, Romi-Lebrun R, Martin-Eauclaire MF, Yasuda A, Ishiguro M, Oyama Y, Pongs O, Nakajima T. Biochem. J. 1997;328:321. doi: 10.1042/bj3280321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Koschak A, Bugianesi RM, Mitterdorfer J, Kaczorowski GJ, Garcia ML, Knaus HG. J. Biol. Chem. 1998;273:2639. doi: 10.1074/jbc.273.5.2639. [DOI] [PubMed] [Google Scholar]
- 309.Peter M, Jr., Varga Z, Hajdu P, Gaspar R, Jr., Damjanovich S, Horjales E, Possani LD, Panyi G. J. Membr. Biol. 2001;179:13. doi: 10.1007/s002320010033. [DOI] [PubMed] [Google Scholar]
- 310.Bagdany M, Batista CV, Valdez-Cruz NA, Somodi S, Rodriguez de la Vega RC, Licea AF, Varga Z, Gaspar R, Possani LD, Panyi G. Mol. Pharmacol. 2005;67:1034. doi: 10.1124/mol.104.007187. [DOI] [PubMed] [Google Scholar]
- 311.Mouhat S, Visan V, Ananthakrishnan S, Wulff H, Andreotti N, Grissmer S, Darbon H, De Waard M, Sabatier JM. Biochem. J. 2005;385:95. doi: 10.1042/BJ20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Durell S, Guy H. Biophys. J. 1992;62:238. doi: 10.1016/S0006-3495(92)81809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Nature. 2006;440:959. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 314.Lange A, Becker S, Seidel K, Giller K, Pongs O, Baldus M. Angew. Chem. Int. Ed. Engl. 2005;44:2089. doi: 10.1002/anie.200462516. [DOI] [PubMed] [Google Scholar]
- 315.Lange A, Giller K, Pongs O, Becker S, Baldus M. J. Recept. Signa.l Transduct. Res. 2006;26:379. doi: 10.1080/10799890600932188. [DOI] [PubMed] [Google Scholar]
- 316.Aneiros A, Garcia I, Martinez JR, Harvey AL, Anderson AJ, Marshall DL, Engstrom A, Hellman U, Karlsson E. Biochim. Biophys. Acta. 1993;1157:86. doi: 10.1016/0304-4165(93)90082-j. [DOI] [PubMed] [Google Scholar]
- 317.Castaneda O, Sotolongo V, Amor AM, Stocklin R, Anderson AJ, Harvey AL, Engstrom A, Wernstedt C, Karlsson E. Toxicon. 1995;33:603. doi: 10.1016/0041-0101(95)00013-c. [DOI] [PubMed] [Google Scholar]
- 318.Pennington M, Mahnir V, Khaytin I, Zaydenberg I, Byrnes M, Kem W. Biochemistry. 1996;35:16407. doi: 10.1021/bi962463g. [DOI] [PubMed] [Google Scholar]
- 319.Dauplais M, Lecoq A, Song J, Cotton J, Jamin N, Gilquin B, Roumestand C, Vita C, de Medeiros CLC, Rowan EG, Harvey AL, Menez A. J. Biol. Chem. 1997;272:4302. doi: 10.1074/jbc.272.7.4302. [DOI] [PubMed] [Google Scholar]
- 320.Kalman K, Pennington MW, Lanigan MD, Nguyen A, Rauer H, Mahnir V, Paschetto K, Kem WR, Grissmer S, Gutman GA, Christian EP, Cahalan MD, Norton RS, Chandy KG. J. Biol. Chem. 1998;273:32697. doi: 10.1074/jbc.273.49.32697. [DOI] [PubMed] [Google Scholar]
- 321.Rauer H, Pennington M, Cahalan M, Chandy KG. J. Biol. Chem. 1999;274:21885. doi: 10.1074/jbc.274.31.21885. [DOI] [PubMed] [Google Scholar]
- 322.Middleton RE, Sanchez M, Linde AR, Bugianesi RM, Dai G, Felix JP, Koprak SL, Staruch MJ, Bruguera M, Cox R, Ghosh A, Hwang J, Jones S, Kohler M, Slaughter RS, McManus OB, Kaczorowski GJ, Garcia ML. Biochemistry. 2003;42:13698. doi: 10.1021/bi035209e. [DOI] [PubMed] [Google Scholar]
- 323.Yan L, Herrington J, Goldberg E, Dulski PM, Bugianesi RM, Slaughter RS, Banerjee P, Brochu RM, Priest BT, Kaczorowski GJ, Rudy B, Garcia ML. Mol. Pharmacol. 2005;67:1513. doi: 10.1124/mol.105.011064. [DOI] [PubMed] [Google Scholar]
- 324.Beeton C, Pennington MW, Wulff H, Singh S, Nugent D, Crossley G, Khaytin I, Calabresi PA, Chen CY, Gutman GA, Chandy KG. Mol. Pharmacol. 2005;67:1369. doi: 10.1124/mol.104.008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Tudor JE, Pallaghy PK, Pennington MW, Norton RS. Nat. Struct. Biol. 1996;3:317. doi: 10.1038/nsb0496-317. [DOI] [PubMed] [Google Scholar]
- 326.Lanigan MD, Kalman K, Lefievre Y, Pennington MW, Chandy KG, Norton RS. Biochemistry. 2002;41:11963. doi: 10.1021/bi026400b. [DOI] [PubMed] [Google Scholar]
- 327.Baell JB, Harvey AJ, Norton RS. J. Comput. Aided. Mol. Des. 2002;16:245. doi: 10.1023/a:1020214720560. [DOI] [PubMed] [Google Scholar]
- 328.Harvey AJ, Gable RW, Baell JB. Bioorg. Med. Chem. Lett. 2005;15:3193. doi: 10.1016/j.bmcl.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 329.Beeton C, Smith BJ, Sabo JK, Crossley G, Nugent D, Khaytin I, Chi V, Chandy KG, Pennington MW, Norton RS. J. Biol. Chem. 2008;283:988. doi: 10.1074/jbc.M706008200. [DOI] [PubMed] [Google Scholar]
- 330.Beeton C, Wulff H, Singh S, Bosko S, Crossley G, Gutman GA, Cahalan MD, Pennington M, Chandy KG. J. Biol. Chem. 2003;278:9928. doi: 10.1074/jbc.M212868200. [DOI] [PubMed] [Google Scholar]
- 331.Pragl B, Koschak A, Trieb M, Obermair G, Kaufmann WA, Gerster U, Blanc E, Hahn C, Prinz H, Schutz G, Darbon H, Gruber HJ, Knaus HG. Bioconjug. Chem. 2002;13:416. doi: 10.1021/bc015543s. [DOI] [PubMed] [Google Scholar]
- 332.Chandy KG, Wulff H, Beeton C, Calabresi PA, Gutman GA, Pennington MW. In: Voltage-Gated Ion Channels as Drug Targets. Triggle DJ, Gopalakrishnan M, Rampe D, Zheng W, editors. Vol. 29. Weinheim: Wiley-VCH; 2006. p. 214. [Google Scholar]
- 333.Panyi G, Possani LD, Rodriguez de la Vega RC, Gaspar R, Varga Z. Curr. Pharm. Des. 2006;12:2199. doi: 10.2174/138161206777585120. [DOI] [PubMed] [Google Scholar]
- 334.Goetz MA, Hensens OD, Zink DL, Borris RP, Morales F, Tamayo-Castillo G, Slaughter RS, Felix J, Ball RG. Tetrahedron Lett. 1998;39:2895. [Google Scholar]
- 335.Felix JP, Bugianesi RM, Schmalhofer WA, Borris R, Goetz MA, Hensens OD, Bao JM, Kayser F, Parsons WH, Rupprecht K, Garcia ML, Kaczorowski GJ, Slaughter RS. Biochemistry. 1999;38:4922. doi: 10.1021/bi982954w. [DOI] [PubMed] [Google Scholar]
- 336.Koo GC, Blake JT, Shah K, Staruch MJ, Dumont F, Wunderler D, Sanchez M, McManus OB, Sirotina-Meisher A, Fischer P, Boltz RC, Goetz MA, Baker R, Bao J, Kayser F, Rupprecht KM, Parsons WH, Tong XC, Ita IE, Pivnichny J, Vincent S, Cunningham P, Hora D, Jr., Feeney W, Kaczorowski G, et al. Cell. Immunol. 1999;197:99. doi: 10.1006/cimm.1999.1569. [DOI] [PubMed] [Google Scholar]
- 337.Hanner M, Schmalhofer WA, Green B, Bordallo C, Liu J, Slaughter RS, Kaczorowski GJ, Garcia ML. J. Biol. Chem. 1999;274:25237. doi: 10.1074/jbc.274.36.25237. [DOI] [PubMed] [Google Scholar]
- 338.Vianna-Jorge R, Oliveira C, Garcia M, Kaczorowski G, Suarez-Kurtz G. Br. J. Pharmacol. 2000;131:772. doi: 10.1038/sj.bjp.0703620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Vianna-Jorge R, Oliveira CF, Garcia ML, Kaczorowski GJ, Suarez-Kurtz G. Br. J. Pharmacol. 2003;138:57. doi: 10.1038/sj.bjp.0705023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Bao J, Miao S, Kayser F, Kotliar AJ, Baker RK, Doss GA, Felix JP, Bugianesi RM, Slaughter RS, Kaczorowski GJ, Garcia ML, Ha SN, Castonguay L, Koo GC, Shah K, Springer MS, Staruch MJ, Parsons WH, Rupprecht KM. Bioorg. Med. Chem. Lett. 2005;15:447. doi: 10.1016/j.bmcl.2004.10.058. [DOI] [PubMed] [Google Scholar]
- 341.Singh SB, Zink DL, Dombrowski AW, Dezeny G, Bills GF, Felix JP, Slaughter RS, Goetz M. A. Org. Lett. 2001;3:247. doi: 10.1021/ol006891x. [DOI] [PubMed] [Google Scholar]
- 342.Ding S, Ingleby L, Ahern CA, Horn R. J. Gen. Physiol. 2005;126:213. doi: 10.1085/jgp.200509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Bruhova I, Zhorov BS. B.M.C. Struct. Biol. 2007;7:5. doi: 10.1186/1472-6807-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zhorov BS, Govyrin VA. Dokl. Akad. Nauk. SSSR. 1983;273:497. [PubMed] [Google Scholar]
- 345.Ananthanarayanan VS. Biochem. Cell. Biol. 1991;69:93. doi: 10.1139/o91-014. [DOI] [PubMed] [Google Scholar]
- 346.Zhorov BS. J. Membr. Biol. 1993;135:119. doi: 10.1007/BF00231437. [DOI] [PubMed] [Google Scholar]
- 347.Ananthanarayanan VS, Tetreault S, Saint-Jean A. J. Med. Chem. 1993;36:1324. doi: 10.1021/jm00062a004. [DOI] [PubMed] [Google Scholar]
- 348.Belciug MP, Ananthanarayanan VS. J. Med. Chem. 1994;37:4392. doi: 10.1021/jm00051a017. [DOI] [PubMed] [Google Scholar]
- 349.Tetreault S, Ananthanarayanan VS. J. Med. Chem. 1993;36:1017. doi: 10.1021/jm00060a009. [DOI] [PubMed] [Google Scholar]
- 350.Zhorov BS, Ananthanarayanan VS. J. Biomol. Struct. Dyn. 1996;14:173. doi: 10.1080/07391102.1996.10508106. [DOI] [PubMed] [Google Scholar]
- 351.Zhorov BS, Folkman EV, Ananthanarayanan VS. Arch. Biochem. Biophys. 2001;393:22. doi: 10.1006/abbi.2001.2484. [DOI] [PubMed] [Google Scholar]
- 352.Yamaguchi S, Zhorov BS, Yoshioka K, Nagao T, Ichijo H, Adachi-Akahane S. Mol. Pharmacol. 2003;64:235. doi: 10.1124/mol.64.2.235. [DOI] [PubMed] [Google Scholar]
- 353.Tikhonov DB, Zhorov BS. FEBS Lett. 2005;579:4207. doi: 10.1016/j.febslet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 354.Tikhonov DB, Bruhova I, Zhorov BS. FEBS Lett. 2006;580:6027. doi: 10.1016/j.febslet.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 355.Hille B. Ion channels of excitable membranes. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 356.Wang SY, Mitchell J, Tikhonov DB, Zhorov BS, Wang GK. Mol. Pharmacol. 2006;69:788. doi: 10.1124/mol.105.018200. [DOI] [PubMed] [Google Scholar]
- 357.Wang SY, Tikhonov DB, Zhorov BS, Mitchell J, Wang GK. Pflugers Arch. 2007;454:277. doi: 10.1007/s00424-006-0202-2. [DOI] [PubMed] [Google Scholar]
- 358.Wang SY, Tikhonov DB, Zhorov BS, Mitchell J, Wang GK. Channels. 2007;1:179. doi: 10.4161/chan.4437. [DOI] [PubMed] [Google Scholar]
- 359.Bohuslavizki HK, Hinck-Kneip C, Kneip A, Koppenhofer E, Reimers A. Neuroopthalmol. 1993;13:191. [Google Scholar]
- 360.Wulff H, Rauer H, During T, Hanselmann C, Ruff K, Wrisch A, Grissmer S, Haensel W. J. Med. Chem. 1998;41:4542. doi: 10.1021/jm981032o. [DOI] [PubMed] [Google Scholar]
- 361.Vennekamp J, Wulff H, Beeton C, Calabresi PA, Grissmer S, Hansel W, Chandy KG. Mol. Pharmacol. 2004;65:1364. doi: 10.1124/mol.65.6.1364. [DOI] [PubMed] [Google Scholar]
- 362.Azam P, Sankaranarayanan A, Homerick D, Griffey S, Wulff H. J Invest Dermatol. 2007;127:1419. doi: 10.1038/sj.jid.5700717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Baell JB, Gable RW, Harvey AJ, Toovey N, Herzog T, Hansel W, Wulff H. J. Med. Chem. 2004;47:2326. doi: 10.1021/jm030523s. [DOI] [PubMed] [Google Scholar]
- 364.Harvey AJ, Baell JB, Toovey N, Homerick D, Wulff H. J. Med. Chem. 2006;49:1433. doi: 10.1021/jm050839v. [DOI] [PubMed] [Google Scholar]
- 365.Schmalhofer WA, Slaugther RS, Matyskiela M, Felix JP, Tang YS, Rupprecht K, Kaczorowski GJ, Garcia ML. Biochemistry. 2003;42:4733. doi: 10.1021/bi034122q. [DOI] [PubMed] [Google Scholar]
- 366.Gradl SN, Felix JP, Isacoff EY, Garcia ML, Trauner D. J. Am. Chem. Soc. 2003;125:12668. doi: 10.1021/ja036155z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Gardos G. Biochem. Biophys. Acta. 1958;30:653. doi: 10.1016/0006-3002(58)90124-0. [DOI] [PubMed] [Google Scholar]
- 368.Joiner WJ, Wang LY, Tang MD, Kaczmarek LK. Proc. Natl. Acad. Sci. USA. 1997;94:11013. doi: 10.1073/pnas.94.20.11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ishii TM, Silvia C, Hirschberg B, Bond CT, Adelman JP, Maylie J. Proc. Natl. Acad. Sci. USA. 1997;94:11651. doi: 10.1073/pnas.94.21.11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Logsdon NJ, Kang J, Togo JA, Christian EP, Aiyar J. J. Biol. Chem. 1997;272:32723. doi: 10.1074/jbc.272.52.32723. [DOI] [PubMed] [Google Scholar]
- 371.Fanger CM, Ghanshani S, Logsdon NJ, Rauer H, Kalman K, Zhou J, Beckingham K, Chandy KG, Cahalan MD, Aiyar J. J. Biol. Chem. 1999;274:5746. doi: 10.1074/jbc.274.9.5746. [DOI] [PubMed] [Google Scholar]
- 372.Chen MX, Gorman SA, Benson B, Singh K, Hieble JP, Michel MC, Tate SN, Trezise DJ. Naunyn Schmiedebergs Arch. Pharmacol. 2004;369:602. doi: 10.1007/s00210-004-0934-5. [DOI] [PubMed] [Google Scholar]
- 373.Pena TL, Rane SG. J. Membr. Biol. 1999;172:249. doi: 10.1007/s002329900601. [DOI] [PubMed] [Google Scholar]
- 374.Neylon CB, Lang RJ, Fu Y, Bobik A, Reinhart PH. Circ Res. 1999;85:e33. doi: 10.1161/01.res.85.9.e33. [DOI] [PubMed] [Google Scholar]
- 375.Grgic I, Eichler I, Heinau P, Si H, Brakemeier S, Hoyer J, Kohler R. Arterioscler. Thromb. Vasc. Biol. 2005;25:704. doi: 10.1161/01.ATV.0000156399.12787.5c. [DOI] [PubMed] [Google Scholar]
- 376.Pena TL, Chen SH, Konieczny SF, Rane SG. J. Biol. Chem. 2000;275:13677. doi: 10.1074/jbc.275.18.13677. [DOI] [PubMed] [Google Scholar]
- 377.Rufo PA, Jiang L, Moe SJ, Brugnara C, Alper SL, Lencer WI. J. Clin. Invest. 1996;98:2066. doi: 10.1172/JCI119012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Devor DC, Singh AK, Lambert LC, DeLuca A, Frizzel RA, Bridges RJ. J. Gen. Physiol. 1999;113:743. doi: 10.1085/jgp.113.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Eichler I, Wibawa J, Grgic I, Knorr A, Brakemeier S, Pries AR, Hoyer J, Kohler R. Br. J. Pharmacol. 2003;138:594. doi: 10.1038/sj.bjp.0705075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Feletou M, Vanhoutte PM. Pharmacol. Res. 2004;49:565. doi: 10.1016/j.phrs.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 381.Tharp DL, Casati J, Turk JR, Bowles DK. FASEB J. 2005;19:A1628. [Google Scholar]
- 382.Tharp DL, Wamhoff BR, Turk JR, Bowles DK. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H2493. doi: 10.1152/ajpheart.01254.2005. [DOI] [PubMed] [Google Scholar]
- 383.Kaushal V, Koeberle PD, Wang Y, Schlichter LC. J. Neurosci. 2007;27:234. doi: 10.1523/JNEUROSCI.3593-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Begenisich T, Nakamoto T, Ovitt CE, Nehrke K, Brugnara C, Alper SL, Melvin JE. J. Biol. Chem. 2004;279:47681. doi: 10.1074/jbc.M409627200. [DOI] [PubMed] [Google Scholar]
- 385.Si H, Heyken WT, Wolfle SE, Tysiac M, Schubert R, Grgic I, Vilianovich L, Giebing G, Maier T, Gross V, Bader M, de Wit C, Hoyer J, Kohler R. Circ. Res. 2006;99:537. doi: 10.1161/01.RES.0000238377.08219.0c. [DOI] [PubMed] [Google Scholar]
- 386.Castle NA, Lodon DO, Creech C, Fajloun Z, Stocker JW, Sabatier J-M. Mol. Pharmacol. 2003;63:409. doi: 10.1124/mol.63.2.409. [DOI] [PubMed] [Google Scholar]
- 387.Visan V, Fajloun Z, Sabatier JM, Grissmer S. Mol. Pharmacol. 2004;66:1103. doi: 10.1124/mol.104.002774. [DOI] [PubMed] [Google Scholar]
- 388.Kharrat R, Mabrouk K, Crest M, Darbon H, Oughideni R, Martin-Eauclaire MF, Jacquet G, el Ayeb M, Van Rietschoten J, Rochat H, Sabatier JM. Eur. J. Biochem. 1996;242:491. doi: 10.1111/j.1432-1033.1996.0491r.x. [DOI] [PubMed] [Google Scholar]
- 389.Rauer H, Lanigan MD, Pennington MW, Aiyar J, Ghanshani S, Cahalan MD, Norton RS, Chandy KG. J. Biol. Chem. 2000;275:1201. doi: 10.1074/jbc.275.2.1201. [DOI] [PubMed] [Google Scholar]
- 390.Alvarez J, Montero M, Garcia-Sancho J. J. Biol. Chem. 1992;267:11789. [PubMed] [Google Scholar]
- 391.Brugnara C, Armsby CC, Sakamoto M, Rifai N, Alper SL, Platt OJ. Pharmacol. Exp. Ther. 1995;273:266. [PubMed] [Google Scholar]
- 392.De Franceschi L, Saadane N, Trudel M, Alper SL, Brugnara C, Beuzard Y. J. Clin. Invest. 1994;93:1670. doi: 10.1172/JCI117149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Brugnara C, Gee B, Armsby CC, Kurth S, Sakamoto M, Rifai N, Alper SL, Platt OS. J. Clin. Invest. 1996;97:1227. doi: 10.1172/JCI118537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Wulff H, Gutman GA, Cahalan MD, Chandy KG. J. Biol. Chem. 2001;276:32040. doi: 10.1074/jbc.M105231200. [DOI] [PubMed] [Google Scholar]
- 395.Stocker JW, De Franceschi L, McNaughton-Smith GA, Corrocher R, Beuzard Y, Brugnara C. Blood. 2003;101:2412. doi: 10.1182/blood-2002-05-1433. [DOI] [PubMed] [Google Scholar]
- 396.Swerdlow P, Ataga KI, Smith W, Saunthararajah Y, Stocker JW. Blood. 2006;108 Abstract 685. [Google Scholar]
- 397.Urbahns K, Horvath E, Stasch JP, Mauler F. Bioorg. Med. Chem. Lett. 2003;13:2637. doi: 10.1016/s0960-894x(03)00560-2. [DOI] [PubMed] [Google Scholar]
- 398.Urbahns K, Goldmann S, Kruger J, Horvath E, Schuhmacher J, Grosser R, Hinz V, Mauler F. Bioorg. Med. Chem. Lett. 2005;15:401. doi: 10.1016/j.bmcl.2004.10.063. [DOI] [PubMed] [Google Scholar]
- 399.Mauler F, Hinz V, Horvath E, Schuhmacher J, Hofmann HA, Wirtz S, Hahn MG, Urbahns K. Eur. J. Neurosci. 2004;20:1761. doi: 10.1111/j.1460-9568.2004.03615.x. [DOI] [PubMed] [Google Scholar]
- 400.Reich EP, Cui L, Yang L, Pugliese-Sivo C, Golovko A, Petro M, Vassileva G, Chu I, Nomeir AA, Zhang LK, Liang X, Kozlowski JA, Narula SK, Zavodny PJ, Chou CC. Eur. J. Immunol. 2005;35:1027. doi: 10.1002/eji.200425954. [DOI] [PubMed] [Google Scholar]
- 401.Kivisakk P, Mahad DJ, Callahan MK, Sikora K, Trebst C, Tucky B, Wujek J, Ravid R, Staugaitis SM, Lassmann H, Ransohoff RM. Ann. Neurol. 2004;55:627. doi: 10.1002/ana.20049. [DOI] [PubMed] [Google Scholar]
- 402.Rus H, Pardo CA, Hu L, Darrah E, Cudrici C, Niculescu T, Niculescu F, Mullen KM, Allie R, Guo L, Wulff H, Beeton C, Judge SI, Kerr DA, Knaus HG, Chandy KG, Calabresi PA. Proc. Natl. Acad. Sci. USA. 2005;102:11094. doi: 10.1073/pnas.0501770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Wojtulewski JA, Gow PJ, Walter J, Grahame R, Gibson T, Panayi GS, Mason J. Ann. Rheum. Dis. 1980;39:469. doi: 10.1136/ard.39.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Moritoki Y, Lian Z, Wulff H, Yang G, Chuang Y, Ueno Y, Ansari AA, Ross L, Mackay IR, Gershwin ME. Hepatology. 2006;45:314. doi: 10.1002/hep.21522. [DOI] [PubMed] [Google Scholar]
- 405.Neylon CB. Vascul. Pharmacol. 2002;38:35. doi: 10.1016/s1537-1891(02)00124-6. [DOI] [PubMed] [Google Scholar]
- 406.Wermuth CG. J. Med. Chem. 2004;47:1303. doi: 10.1021/jm030480f. [DOI] [PubMed] [Google Scholar]
- 407.Singh S, Syme CA, Singh AK, Devor DC, Bridges RJ. J. Pharmacol. Exp. Ther. 2001;296:600. [PubMed] [Google Scholar]