Summary
The portal channel is a key component in the life cycle of bacteriophages and herpesviruses. The bacteriophage P22 portal is a 1 megadalton dodecameric oligomer of gp1 that plays key roles in capsid assembly, DNA packaging, assembly of the infection machinery, and DNA ejection. The portal is the nucleation site for the assembly of 39 additional subunits generated from multiple copies of four gene products (gp4, gp10, gp9, and gp26), which together form the multifunctional tail machine. These components are organized with a combination of 12- (gp1, gp4), 6- (gp10, trimers of gp9) and 3- (gp26, gp9) fold symmetry. Here we present the 3-dimensional structures of the P22 assembly-naïve portal formed from expressed subunits (gp1), and the intact tail machine purified from infectious virions. The assembly-naïve portal structure exhibits a striking structural similarity to the structures of the portal proteins of SPP1 and phi29 derived form X-ray crystallography. In the context of the tail machine the portal allows the precise definition of the boundary between gp1 and gp4, and leads to a proposal for the gene product constitution and mode of assembly. The detailed reconstruction facilitates a description of the subunit-subunit interactions that make up the tail machine, and the molecular events that occur during host cell attachment and DNA translocation.
Introduction
Formation of infectious dsDNA bacteriophages and herpesviruses occurs through a multistep process involving hundreds of specific protein-protein interactions. A crucial component that assembles along this pathway in double-stranded DNA (dsDNA) bacteriophages is a highly specialized nano machine that is responsible for adsorbing to and infecting host cells, commonly referred to as the tail (Ackermann, 1998). Discovered in 1952, the well-characterized Salmonella enterica-infecting bacteriophage P22, a Podoviridae within the order Caudovirales, has served as a model for virus assembly (Casjens & Weigele, 2005; Prevelige, 2005). The P22 tail is short, non-contractile, and incorporated into a pentameric opening at one of twelve icosahedral capsid shell vertices (Chang et al, 2006; Lander et al, 2006; Poteete, 1994; Tang et al, 2005). Able to withstand heating to 70°C in 2M urea and in the presence of detergent, the assembled tail machine is a remarkably stable macromolecule and lends itself well to structural studies (Tang et al, 2005). Despite its complexity, the P22 tail is significantly simpler than the infection machinery of other documented phages, some of which incorporate four times the number of gene products or hundreds of copies of protein subunits into their functional tails (Agirrezabala et al, 2005a; Chang et al, 2006; Katsura & Kuhl, 1975; Kondou et al, 2005; Kostyuchenko et al, 2005; Leiman et al, 2007; Mc Grath et al, 2006; Parker & Eiserling, 1983; Plisson et al, 2007; Tang et al, 2008; Xiang et al, 2006). An understanding of the P22 tail machine's architecture has provided a model for translocation and injection of DNA utilized by the more elaborate phage tails (Johnson & Chiu, 2007; Nemecek et al, 2008; Tang et al, 2005; Ziedaite et al, 2009).
Twelve copies of an 83kDa protein (gp1) form an oligomer, termed the portal, that is situated at a pentavalent opening in the capsid. In addition to its involvement in assembly of the immature phage (Moore & Prevelige, 2001), the P22 portal serves as the attachment site for an external dsDNA-bound complex with ATPase activity (Casjens & Weigele, 2005; Nemecek et al, 2008; Prevelige, 2005). This packaging motor pumps the ∼43.5kbp phage genome into the capsid through the portal's central channel, resulting in DNA concentrations of 500mg/ml (Earnshaw & Casjens, 1980). Upon completion of genome packaging, the terminase motor detaches from the portal structure, at which point the dodecameric oligomer functions as a nucleation site for the attachment of the additional tail machine subunits.
The tail machine is nearly three megadaltons and is comprised of gp1 (12 copies), gp4 (12), gp10 (6), gp9 (18), and gp26 (3). In addition to the symmetry mismatch arising from a twelve-fold symmetric portal oligomer embedded in a pentameric opening, the complete tail machine assembly itself is rife with symmetry mismatches, exhibiting a combination of twelve-, six-, and three-fold symmetrical substructures (Figure 1A). It is established that upon completion of DNA packaging, twelve copies of the 18.0kDa protein gp4 attach directly to the base of the portal (Olia et al, 2006), and that without initial attachment of this gene product no further components will assemble on the virion (Strauss & King, 1984). Upon formation of the gp1-gp4 complex, six copies of gp10 (52.5kDa) subsequently attaching at the base of gp4. As these two additional components alone are insufficient to prevent genome leakage from within the capsid interior, three copies of gp26 (24.7kDa) plug the distal end of the gp1-gp4-gp10 complex in the form of a thin needle-like trimeric coiled-coil. Stable assembly of all three gene products (gp4, gp10, and gp26) are necessary for retaining the genetic material housed within the capsid (Strauss & King, 1984). The gp26 needle seals the DNA exit tunnel of the tail machine and is hypothesized to penetrate the host cell envelope during infection by the phage (Olia et al, 2007; Olia et al, 2009).
Figure 1.
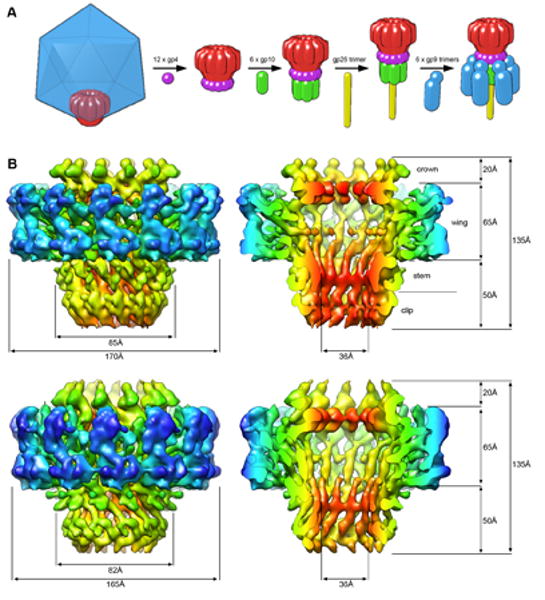
The tail machine assembly pathway and CryoEM structures of the 11-and 12-fold portals at sub nanometer resolution. A. The dodecameric portal (red) is shown at one pentameric vertex of the capsid (blue, not shown in subsequent steps). After DNA packaging, the tail assembles via sequential addition of multiple copies of gp4, gp10, gp26, and gp9 to the portal ring. B. Surface renderings of the reconstructed portal densities contoured at 1.5 sigma and colored radially from the central channel axis. The top two structures are of the 12-fold particles (8.6Å resolution), and the lower two of the 11-fold (8.8Å resolution). On the right are cutaway views of the portals, showing the arrangement of skewed helices in the stem region of the portals and a channel that is wide enough to accommodate double-stranded DNA. The same overall structural morphology is seen throughout both reconstructions.
Tail machine assembly is completed by attachment of six homotrimers of the 71.9kDa gp9 protein at the gp4-gp10 interface. Each tailspike trimer consists of an N-terminal head-binding domain and a C-terminal receptor-binding domain, and the crystal structure of each isolated domain has been solved (Steinbacher et al, 1996; Steinbacher et al, 1997). The head-binding domain (residues 5-108) serves as an anchor for the tailspike, binding at the cleft between gp4 and gp10, bridging the twelve- to six-fold symmetry mismatch (Chang et al, 2006; Lander et al, 2006; Maurides et al, 1990; Tang et al, 2005). The receptor-binding domain (residues 113-666), functions as both a viral adhesion protein, binding to the O-antigenic repeats of polysaccharides, and as an endoglycosidase, cleaving glycosidic bonds after attachment. There is a four-residue linker connecting these domains that are unaccounted for by the two crystal structures. The EM density suggests that the linker is flexible, allowing for the skewed orientation adopted by the head-binding domain relative to the receptor-binding domain upon attachment of the tailspike to the tail machine body (Chang et al, 2006; Lander et al, 2006; Tang et al, 2005).
Three-dimensional reconstructions of the isolated tail machine as well as within the context of the mature virion were achieved through the use of cryo-electron microscopy (cryoEM), allowing for putative assignments for the fifty-one different components (Chang et al, 2006; Lander et al, 2006; Tang et al, 2005), but the low resolution of these reconstructions prevented a detailed molecular description of the P22 tail machine anatomy. Here, we present the three-dimensional cryoEM structures of the isolated assembly-naïve portal complex, and of the intact tail machine complex released and purified from infectious virions. These reconstructions, in conjunction with existing x-ray structures, allowed segmentation of individual subunit density and the definition of all inter-subunit boundaries. The results presented here vary from those previously reported by Zheng et al. (2008) and they are discussed with regard to the dynamic character of the portal in its various states of interaction with tail machine components.
Results
Polymorphic oligomerization of the assembly-naïve P22 portal
Certain phage portal proteins, when expressed ectopically, assemble into rings that adopt different symmetries from those assembled in the context of the capsid, and may also display polymorphic oligomerization (Bazinet et al, 1988; Cingolani et al, 2002; Dube et al, 1993; Kocsis et al, 1995; Lander et al, 2006; Lurz et al, 2001; Newcomb et al, 2001; Orlova et al, 2003; Poliakov et al, 2007; Trus et al, 2004). This aberrant assembly of isolated portals points to a collaborative role of portal, scaffold, and capsid in proper assembly of the procapsid in vivo. P22 portal subunits assemble into a heterogeneous mixture of 11- and 12-fold rings in the absence of capsid and scaffolding proteins in solution (Lander et al, 2006; Poliakov et al, 2007; Zheng et al, 2008), which is problematic in reconstructing a three-dimensional structure by cryoEM, due to the lack of homogeneity required for the accurate reconstruction.
A purification protocol reported to yield a homogeneous population of 12-fold symmetric P22 portals was described by Zheng et al., which led to a reconstruction of a truncated portal at reportedly sub nanometer resolutions (Zheng et al, 2008). Following the same protocol, our attempts to reproduce the 12-fold homogeneity of full-length portals failed, as evidenced by reference-free classification of top-view particles in the frozen-hydrated state, which display a heterogeneous mixture of 11- and 12-fold symmetries (Supplementary Figure 2). Correspondence-analysis of the individual particles allowed three-dimensional reconstructions of the P22 portal structure in both its 11- and 12-mer states at sub nanometer resolutions (Figure 1b). Attempts to resolve a 10- or 13-mer structure by the same protocol failed, confirming a mixture of portals dominated by 11- and 12-fold symmetry.
P22 portal structure
The structures of the 11- and 12-fold P22 portal structure were refined from images of ice-embedded gp1 proteins expressed in the absence of other viral proteins (Supplementary Figure 1). The reported resolutions for the 11-fold particles were 7.1Å and 8.8Å using Fourier shell correlation (FSC) and Rmeasure at 0.5, respectively. The 12-fold portal was resolved to 6.9Å and 8.6Å by FSC and Rmeasure at 0.5, respectively (Sousa & Grigorieff, 2007; van Heel & Harauz, 1986). Observation of the reconstructed density confirmed that the structures are at sub nanometer resolutions, with alpha helices clearly visible. The structure resembles a hollow turbine, as reported for the previously solved P22 portal truncate and for other phage and herpesvirus portal structures (Agirrezabala et al, 2005b; Guasch et al, 2002; Lebedev et al, 2007; Orlova et al, 2003; Simpson et al, 2000; Trus et al, 2004; Zheng et al, 2008).
Surface renderings of both the oligomeric portal structures are shown in Figure 1; the dimensions for the 12-fold portal agree with the previously reported P22 truncate, whose channel is wide enough to accommodate dsDNA. As expected, the height of the 11-fold structure is identical to that of the 12-fold structure, although the lateral dimensions are smaller due to the absence of one gp1 monomer. Both reconstructions exhibit a similar domain morphology, which is defined following nomenclature introduced by Lebedev et al. as the (from top to bottom) crown, wing, stem, and clip domains (Figure 2B).
Figure 2.
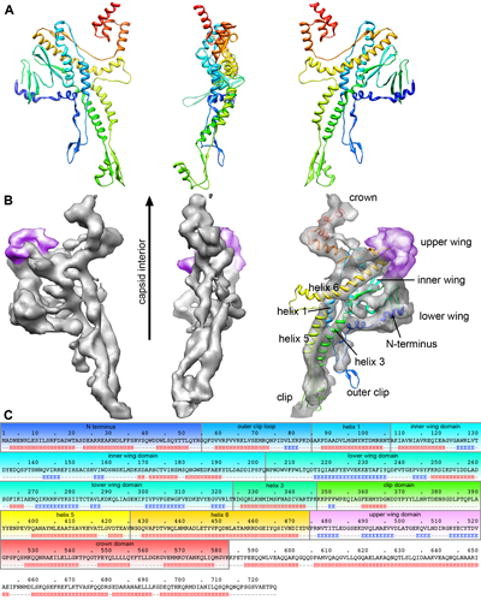
Comparison of the P22 portal monomeric subunit with the SPP1 portal monomer, and three-dimensional mapping of the P22 portal sequence. A. Side views of the SPP1 portal (PDB entry: 2JES) are represented as ribbons colored from the N-terminus (blue) to C-terminus (red). B. Contoured at 1.5 sigma, the details of the P22 subunit extracted from the 12-fold portal structure shows many similarities to the subunit structure of the SPP1 portal. Both have crown, wing, stem, and clip regions, with P22 densities that are positioned similarly to helices 3, 4, and 6 of SPP1 (helical numbers defined in (Simpson et al, 2000)). The SPP1 subunit has been refined into the P22 density and is shown in the rightmost P22 portal density. P22 has an additional domain for which SPP1 has no counterpart (colored in magenta). C. Hydrogen-deuterium exchange data (Kang et al, 2008) has been combined with secondary structure prediction (www.predictprotein.org) and information from the SPP1 crystal structure to assign the location of the entire P22 sequence into the segmented P22 density. Domains are colored in the sequence according to their corresponding locations in the SPP1 ribbon representation, with the exception of the upper wing domain (magenta), which has no SPP1 counterpart. Residues after 580 were determined to be disordered and likely continue above the crown domain.
Viewed in cross section from the side (Fig. 1b, 2), the crown domain is closest to the 12-fold portal axis and is likely to interact with the ejection proteins that form a channel within the intact virion (Chang et al, 2006; Lander et al, 2006). The wing domain is the largest of the four domains, extending radially from the portal channel, possibly interacting with spooling DNA during genome packaging. The lowermost funnel-like region, commonly referred to as the “stalk”, is divided into two parts - the alpha-helical stem, which spans the pentameric opening in the assembled capsid, and the clip domain. The clip domain is a multifunctional region that is involved in both binding to the packaging terminase in the viral procapsid, and upon completion of genome delivery, binding to the first of the tail machine components, gp4. The similarity of the 11- and 12-fold structures goes beyond discernible secondary structural elements, extending into tertiary topology. The fact that these densities were arrived upon without the introduction of any initial model bias provides additional confidence in the accuracy of the reconstruction.
Density corresponding to a single gp1 monomer was extracted from the symmetrized densities by first aligning the crown and wing region of the 11-fold density to the same regions of the 12-fold structure as a rigid body. Due to the difference in symmetry of the superimposed structures the inter-subunit boundaries were evident, and with minimal aid of the SPP1 crystal structure as a reference, monomeric densities were extracted for further comparisons and structural characterization.
Comparison of the P22 portal to crystal structures
Phage portal proteins range in size, from 37kDa in phi29 to 83kDa in P22, and have virtually no sequence homology. All portal proteins studied assemble into dodecameric oligomers in the virion (Agirrezabala et al, 2005b; Doan & Dokland, 2007; Guasch et al, 2002; Lebedev et al, 2007; Orlova et al, 2003; Simpson et al, 2000; Tang et al, 2008; Trus et al, 2004; Zheng et al, 2008). Portal structures, solved by either cryoEM or crystallography, exhibit a structurally conserved architecture, described above (Agirrezabala et al, 2005b; Guasch et al, 2002; Lebedev et al, 2007; Orlova et al, 2003; Simpson et al, 2000; Trus et al, 2004; Zheng et al, 2008). The wing regions vary widely in shape and size among phage, while the stalk is structurally conserved throughout dsDNA phages and herpesviruses.
The crystal structures of phi29 and SPP1 portal proteins are available (Guasch et al, 2002; Lebedev et al, 2007; Simpson et al, 2000), and the structural conservation inherent between these and P22 is evident. The stem regions, which in phi29 and SPP1 are comprised of two extended anti-parallel alpha helices (helices 3 and 5), both tilted between ∼30° and ∼50° relative to the tunnel axis, are strikingly similary to each other and the corresponding density in the segmented P22 portal density (Figure 2).
The conservation continues into the wing region, where in both phi29 and SPP1, alpha helix 6 is oriented perpendicular to alpha helices 3 and 4, and extends away from the central channel, along the bottom edge of the wing region, to its periphery. There is a beta-sheet located along the outermost rim of the wing domain in both phi29 and SPP1. Helix 6 and the beta sheet have corresponding densities in P22 portal including a 45 degree deviation from linearity of helix 6 near the central channel (Figure 2B). It is known that a mutation near this kink in helix 6 in SPP1 leaves the portal incapable of DNA packaging (Isidro et al, 2004a; Isidro et al, 2004b; Lebedev et al, 2007; Oliveira et al, 2005). Based on the conservation of these secondary structural elements in three different phages and the SPP1 mutagenesis, it is likely that this domain is involved with essential interactions necessary for DNA packaging.
The SPP1 portal protein is 21kDa larger than the phi29 portal protein, and exhibits a more complex architecture in the wing domain. Like P22, the SPP1 portal also has a crown domain that is absent in phi29. This structural elaboration in the SPP1 portal (and by analogy in P22) may facilitate “headful DNA packaging” utilized by SPP1 and P22. This requires sensing the equivalent of one genome-length of DNA during translocation, a role known to be associated with the portal in P22 (Casjens & Weigele, 2005).
The P22 portal is larger than SPP1 by 222 residues and there is a large density in the upper portion of the wing that has no SPP1 counterpart (colored purple in Figure 2). Combined with the assignments of the P22 secondary structure based on the SPP1 homology and additional prediction, this additional wing density appears to be the location for a large portion of the inserted residues. Residues 429 to 471 in the P22 portal are predicted to be a 40 residue alpha-helix. This extended alpha helix lines up with the long helix 6 that spans from the channel to the outermost portion of the wing in both phi29 and SPP1 (Figures 2 & 3). The phi29 portal structure terminates shortly after helix 6, while in SPP1 an abrupt turn links the C-terminal end of helix 6 to the helical crown domain. Secondary structure prediction (Figure 2C) shows a series of beta-strands that occur within a 50-residue region unique to P22 and immediately following helix 6. The EM density for this inserted upper wing domain connects to the C-terminus of helix 6, is distinctly flat and broad, and can accommodate the predicted insert of four beta strands. Following this insertion, the density again corresponds to the SPP1 structure and forms the crown domain. Combining information described above and from deuterium exchange studies of portal assembly (Kang et al, 2008), we believe that the P22 gp1 sequence 1-582 has been fully assigned to the cryoEM density (Figure 2). Residues 583 to 725 are predicted to be predominately helical, however there is no additional density to accommodate these residues. We conclude that they extend off the top of the crown and may make specific interactions with the ejection proteins when the portal is in the particle, but when free or in the tail machine they are highly dynamic and their density lost during reconstruction.
Figure 3.
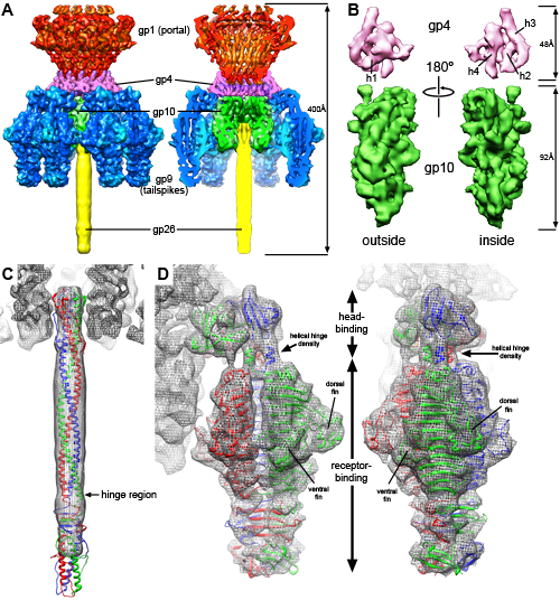
The P22 tail machine at 9.4Å resolution, as determined by cryoEM. A. Fifty-one subunits from five different gene products assemble to form the tail machine, and are represented in surface renderings at 1.5 sigma. The image on the right depicts a cutaway view to expose the tail machine interior. The gene products are colored as follows: gp1 are red, gp4 are magenta, gp10 are green, gp9 are blue, gp26 are yellow. B. The exterior and interior views of the segmented gp4 (above, magenta) and gp10 (below, green) monomeric densities. The gp4 density, which is predicted to consist of four alpha helices, exhibits four distinctly sausage-like densities that are characteristic of alpha helices. Two of these run laterally at the gp10 interface (h1 and h2), and the other two run parallel to the channel axis on the interior of the monomer (h3 and h4). These two interior helices appear to interact with gp1 and gp10 in the reconstructed density. The gp10 density is much more difficult to interpret that the gp4 density, and secondary structure prediction shows that this protein is mostly made up of beta strands. C. The gp26 tail needle (PDB entry: 2POH) is docked into the EM density, showing extensive interactions with gp10 at the N-terminal tip. Further from the tail machine body, the needle density becomes more disordered, becoming completely disordered shortly after passing the hinge domain. D. The crystal structures for the N-terminal head-binding domain and the C-terminal receptor-binding domain of gp9 (PDB entries: 1LKT, 1TYU) are docked into the EM density, fitting with high fidelity. The tail spike assumes an asymmetric organization upon binding to the tail machine with the head-binding domain tilted relative to the axis of the tail machine. Two of the head-binding subunits are involved in interactions that bridge gp4 and gp10, while the third subunit interacts with the three N-terminal helices of the receptor-binding domain.
The SPP1 portal crystal structure (PDB entry: 2JES) readily docks into the P22 segmented density. Several rounds of real-space flexible refinements show that a minimal amount of movement is necessary to improve the fit (Figure 2B). The Cα root-mean-square-deviation (RMSD) between the rigid body fitted and the flexible fitted SPP1 portal crystal structure is 3.7Å. The most discernible difference between the two models is a large ∼20Å movement of the SPP1 C-terminus to fit into the P22 electron density (Supplementary Figure 3).
P22 Tail Machine Structure
P22 tail machine particles were purified from disrupted mature virions. In a previous study, isolated tail machine particles adopted a preferred orientation in the frozen-hydrated state that required tilting of the microscope stage to perform the single-particle reconstruction (Tang et al, 2005) (Supplementary Figure 4A). In the current study, the particles were vitrified onto a holey grid with a thin overlaid layer of carbon in order to provide random orientations of the tail machines (Ruiz & Radermacher, 2006) (Supplementary Figure 4B). The density corresponding to the tail machine was extracted from an asymmetric reconstruction of P22 (Lander et al, 2006) and low-pass filtered to 30Å resolution for use as an initial model. Utilizing the same reconstruction scheme that was used for the free-portal reconstructions, over 350,000 automatically selected particle images were processed to arrive at the final 3D reconstruction.
The reconstructed density of the tail machine reveals a structure approximately 415Å in height with a maximal width of 250Å (Figure 3A). Salient features include the portal structure, which occupies the upper third of the tail machine, six tail spike trimers surrounding the central tube, and an extended needle-like structure plugging the central channel. The tail tube channel is devoid of density and varies in diameter from approximately 30Å to 45Å (if the larger portal channel is not considered).
Tail machine segmentation
Previous cryoEM reconstructions provided putative localizations for the five gene products that comprise this assembly, although the resolutions of these reconstructions did not provide sufficient detail to delineate the precise boundaries between the sub complexes or the monomeric subunits (Chang et al, 2006; Lander et al, 2006; Tang et al, 2005). The existing crystal structures for the tail fiber gp26 (Figure 3C) (Olia et al, 2007) and the tailspike gp9 (Figure 3D) (Steinbacher et al, 1996; Steinbacher et al, 1997) in conjunction with the known stoichiometry of the gene products (Tang et al, 2005) and the isolated 12-fold portal reconstruction, allowed determination of the inter-gene product boundaries and segmentation of monomeric subunits (Figure 3B).
The gp1 portal protein exhibits a very similar morphology in both the assembly-naïve state and in the context of the assembled tail machine. All secondary structural features in the wing and stalk domains are identical within the limits of the cryoEM resolutions, although the bottom-most domain of the clip region appears to have undergone a rearrangement to accommodate the binding of gp4. Conformational flexibility of this domain has also been shown for phage SPP1, although the change between the two P22 portal states observed here is not as dramatic.
The smallest of the tail machine proteins, gp4, which is 18kDa, is composed of four distinct sausage-like densities that correspond to individual alpha helices at this resolution (Figure 3B). While there are no existing high-resolution structures of gp4 or homologous proteins, secondary structure prediction of the gp4 sequence indicates the presence of four alpha helices, ranging from 13 to 21 residues in length. Although we cannot discern the connectivity between these helices from the reconstructed density, this structure confirms that gp4 is composed of a four-helix bundle, two of which are oriented laterally relative to the central channel axis, and the other two in a perpendicular fashion. The segmented density shows that two gp4 subunits assemble on either side of a single portal clip domain, resulting in an interlocking set of interactions between gp1 and gp4.
Six copies of gp10 (52.5kDa) bind to the 12 copies of gp4 in the assembling tail machine. Secondary structure prediction indicates a structure that is mostly beta-sheet in composition, with no significant helical segments. Due to its size, a detailed analysis of the secondary structure constitution of the segmented gp10 monomer is difficult, but the lack of sausage-like densities is consistent with secondary structure prediction (Figure 3B). The symmetry mismatch that occurs between the dodecameric gp4 ring and the six copies of gp10 is accommodated by a pseudo-12-fold architecture of the uppermost domain of gp10 (Supplementary Figure 6A). Just as gp4 extends a helical segment between the clip regions of the portal gp1 proteins, gp4 similarly exhibits a helix that juts into gp4-gp10 interface. Six of these gp4 helices interact with a pocket formed between neighboring gp10 subunits, while the other six gp4 helices insert into a cavity within the gp10 monomer that structurally resembles the intra-gp10 pocket.
The gp26 trimeric needle, an extended trimeric coiled-coil, plugs into the central tail tube preventing genome leakage from the capsid. Due to the six-fold symmetry imposed on the tail machine density during EM processing, the three-fold details of this component are lost, although fitting of the crystal structure into the density shows a similar molecular envelope (Figure 3C) (Olia et al, 2007). The first 27 N-terminal residues of gp26 trimer fold down around the N-terminal tip of the coiled-coil to form an antiparallel six-stranded bundle roughly 30Å in diameter. Similar to the pseudo-12-fold symmetry exhibited by gp10 at the gp4 interface, the N-terminal domain of gp26 exhibits pseudo-6-fold symmetry in order to accommodate the interaction with gp10 (Supplementary Figure 6B). The first 27 residues that form this pseudo 6-fold symmetry in gp26 are known to be critical for binding of the needle to the tail machine, although it is not necessary for oligomerization of the needle structure (Bhardwaj et al, 2007). Fitting the crystal structure into the EM density shows that this domain is almost entirely enclosed within the tail machine's central channel, interacting extensively with gp10 (Figure 3C). The gp26 needle comes into very close proximity to the gp10 density at positions where the gp26 N-terminus exhibits alpha helical character, namely residues Asp3-Asn8 and Asp18-Leu22.
Two hundred angstroms away, at the opposite end of the gp26 needle, EM density for the C-terminal tip is disordered. This may be due in part to the fact that the gp26 needle is only held in place at its N-terminus to the gp10 subunits. The disorder may be exacerbated by the putative flexibility of the basic C-terminal domain (termed the lazo domain), thought to be involved with membrane penetration. This flexibility comes about due to a hinge-like region centered at Gln137 at the terminal end of the helical core, allowing rotation of this domain up to 18° away from the vertical axis (Figure 3C) (Olia et al, 2007; Olia et al, 2009).
The final addition to the tail machine is the gp9 tailspike, whose N-terminal head-binding domain attaches to the interface between the gp4 and gp10 subunits. From each of the trimeric head binding domains, two of the subunits interact with the interface, resulting in a pseudo-12 fold symmetric binding. The EM density provides sufficient resolution to distinguish residues Asp94-Ser98 as directly involved with binding to the gp4 subunit, although at different positions on the gp4 monomer. One gp9 head-binding domain appears to interact with the lowermost alpha helical segment of gp4, while the same residues on the neighboring gp9 monomer, along with Arg21 & Ser22, are situated to interact with the most peripheral region of a neighboring gp4 (Supplementary Figure 5A & B). The head binds such that the six tailspike trimers are in register with the six gp10 subunits, suggesting a specificity of the head-binding domain to gp10. One monomer from the head domain contains a short two-stranded beta sheet (Pro48-Pro64) that appears to create a bridge spanning two adjacent gp10 subunits (Supplementary Figure 5C). This bridging interaction would only be possible at the inter-gp10 crevasse, such that the 6-fold specificity may be set here.
A notable feature of the tailspike, as seen in earlier reconstructions, is an asymmetric arrangement of its two constituent domains. The head-binding domain, the smaller of the two domains and through which the tailspike attaches to the gp4-gp10 interface, assumes a skewed position relative to the larger receptor-binding domain, which is responsible for binding to and destroying the cell surface lipopolysaccharides (Figure 3D) (Chang et al, 2006; Lander et al, 2006; Tang et al, 2006). In the reconstructed tail machine presented here, details of the tailspike hinge are discerned. The uppermost portion of the receptor-binding domain is alpha helical before becoming disordered and these alpha helices are clearly seen in the EM density (Figure 3D). Furthermore, these three helical segments are situated directly underneath the peripheral monomer of the head-binding domain. The head-binding domain appears to be a multifaceted structure, able to interact with the gp4-gp10 interface and offering an interaction site for the uppermost helical portion of the receptor-binding domain.
The remarkable fit of the receptor-binding domain crystal structure into the EM density is evidence that lateral interactions exist between gp10 and the tailspike. If the tailspike receptor domains were mobile relative to the rest of the tail machine assembly, the density corresponding to the tail spikes would be an average of the receptor domain in many different positions. Since the density corresponds well with the crystal structure, and it is unlikely that the interaction between the head-binding domain and the receptor-binding domain alone would be sufficient to stabilize the orientation of the tailspike, there must be a gp10-tailspike interaction.
The tailspike contains an endo-rhamnosidase enzyme that is used by the phage to digest the polysaccharide layer that surrounds gram-negative bacteria (Iwashita & Kanegasaki, 1976). The binding site is situated in a 20Å cleft formed between two protruding domains of the tailspike, termed the ventral and dorsal “fins” (Steinbacher et al, 1997). From the EM reconstruction we observe that each of the tailspike trimers is positioned such that a receptor-binding domain is directed outward toward the solvent (Supplementary Figure 5D). It is also clear that the fins not only serve to form the binding cleft, but also are used to stabilize the receptor-binding domain in its position relative to the tail machine. The ventral fin of one of the tailspike subunits comes into close proximity to a gp10 subunit, although it is difficult to assess the stabilization mechanism without higher resolution detail of gp10 (Figure 3A & Supplementary Figure 5D). The remaining tailspike fins appear to undergo an interlocking lateral interaction around the circumference of the tail machine, where the dorsal fin of one tailspike is directed into the polysaccharide-binding cleft of the neighboring tailspike (Supplementary Figure 5D). It is likely that these interacting fins inhibit the enzyme activity of these gp9 monomers, leaving only one active site open per tail spike.
Discussion
The ability of portals within headful packaging bacteriophages to sense the building pressure within a capsid has long been observed, and portal structures have been shown to be structurally dynamic (Casjens et al, 1992; Isidro et al, 2004a; Lander et al, 2006; Tavares et al, 1992; Xiang et al, 2006; Zheng et al, 2008). In the present study we compared the structures of the assembly-naïve P22 portal to its conformation in the assembled tail machine and it is evident that they are virtually identical. In particular our results are inconsistent with a large conformational reorganization of the portal gp1 proteins upon binding gp4 (Zheng et al, 2008).
Zheng et al. describe conformation changes upon binding of gp4 that include a widening of the central channel, the appearance of a DNA scaffolding “dome” directly above the central channel, and a dramatic reorientation of the stem domain from a skewed to a vertical morphology. While we note that Zheng et al. were working with a truncation mutant that is missing the 123 C-terminal residues, we are not convinced that such a truncation would result in the assembly of an entirely novel structure that maintains functional binding of gp4, but that behaves in a disparate manner to the wild type.
The portal structures reported here, with and without gp4 association, are virtually identical in dimension and morphology. The central channel does not widen, although the portal appears to undergo a subtle conformational switch in the lowermost clip region in order to accommodate the binding of gp4. A similar transition is observed in the SPP1 portal structure, where the clip domain widens in order to accommodate the binding of the subsequent tail components. It is logical that a conformational switch would occur in this domain, as it is the binding site of two very different components. Following assembly of the P22 procapsid, the terminase complex binds to the portal at this clip domain, and at the conclusion of headful packaging, this site becomes the new binding site for gp4. The headful sensing mechanism utilized by the portal likely changes the conformation of the clip domain, lowering the binding affinity for the terminase, and increasing its ability to bind gp4. In this manner, the tail machine would not assemble prematurely before the genome is packaged. We cannot describe the specifics of the headful packaging mechanism based on the reconstructions shown here, but it may involve a subtle structural signal that passes from the portal's crown or wing to the clip domain through the stem.
Our results also differ from Zheng et al. regarding the location of the C-terminal 123 residues in their truncation mutant. They propose that this C-terminal portion of the structure localizes to the upper wing domain of the portal structure, but we believe that it is dynamic, not obeying rigid 12-fold symmetry, and is therefore disordered in the reconstruction. Although predicted to be high in helical content, the P22 portal protein after residues 577 were shown not be necessary for portal oligomerization by hydrogen-deuterium exchange (Kang et al, 2008). These findings suggest the C-terminus to be a flexible domain that would not be resolved by single-particle processing. Based on the similarity of the SPP1 crystal structure and the P22 portal density, we believe that the upper wing domain described by Zheng et al. as C-terminus to be in fact a beta-sheet-rich domain from residues 472-526.
The assembled tail machine structure additionally questions the validity of the dome-like structure observed by Zheng et al. in their reconstructed gp1-gp4 complex. No significant density appears above the central channel in the tail machine reconstruction, although Zheng et al. state that this dome-like density is among the strongest densities in their reconstructed complex.
The portal and tail machine structures of various other Podoviridae have been described at moderate resolutions in asymmetric cryoEM reconstructions of phages T7, phi29, epsilon15, K1E, and K1-5 (Agirrezabala et al, 2005a; Jiang et al, 2006; Leiman et al, 2007; Tang et al, 2008; Xiang et al, 2006). Despite the marked structural differences in the overall shapes of the tail machines described by this collection of densities, it is possible to pinpoint common features that are shared by them all. Almost all of the P22 tail components presented in this work have direct structural analogs in these other virions, such that interactions described here likely serve as a model for these other systems. In all the Podoviridae reconstructions the portal is always evident within the capsid interior, and although they vary in size and complexity, they all appear to contain a homologous core domain. The utilization of differing packaging mechanisms and a lack of portal sequence similarity between these phage points to this core domain performing a common ancestral role in DNA translocation. The tail machine assembles on the virion through attachment of a smaller gene product at the base of the portal. This attachment protein will likely have 12-fold symmetry, matching the symmetry of the portal. Podoviridae all have enzymatic tail spikes, although the numbers and arrangements can differ between them. The P22 structure presented here clearly shows the attachment of six tailspikes in a symmetric manner around the tail machine. Epsilon15 also contains six tailspikes, although they deviate from a symmetric arrangement around the tail, each tailspike adopting a slightly different orientation (Jiang et al, 2006). The K1E and K1-5 phages contain a six-fold symmetric arrangement of their tailspike enzymes, although the tailspike content differs between the two. K1E contains six endosialidase tailspikes while K1-5, which also contains six endosialidases, additionally incorporates six P22-like endorhamonidase tailspikes into its tail structure (Leiman et al, 2007). The phi29 tail machine contains twelve tailspike appendages surrounding the tailspike that, similarly to epsilon15, deviates from the azimuthal symmetry (Tang et al, 2008; Xiang et al, 2006). The symmetry of the remainder of the tail machine is likely dictated by the arrangement of tailspikes around the tail, since they attach to the tail machine at the interface of two subunits (gp4 and gp10 in P22). While the P22 gp26 needle does not have structural homologs in the other Podoviridae reconstructions, it is likely that the gene products present at the distal end of the tail machine play a similar dual role of closing the tail machine channel to secure the genome within the capsid, and in infection upon arrival at the host surface.
The P22 tail machine architecture is a testament to nature's ability to multitask structurally. The portal, which occupies the upper third of the structure, is used for DNA translocation, pressure sensing, and genome injection. Using electron microscopy, we have shown that the dynamic nature of this macromolecule must function at a level that is much more subtle than previously proposed. The base of the portal additionally sets the stage for attachment of components that assemble via a series of symmetry mismatches and asymmetries. Once assembled, the tail machine is an incredibly stable structure, yet we know that it must also be dynamic in order to act in signaling for genome delivery upon attachment to a host cell. In conjunction with the density presented in this work, future crystallographic studies of gp1, gp4 and gp10 will reveal the specific molecular mechanisms that define maturation, assembly, and infection events. Given the structural similarities observed here between P22, SPP1, and phi29, these mechanisms will more than likely introduce common motifs employed by all dsDNA phage and herpesviruses.
Experimental Procedures
Production and Purification of Samples
Cloning and purification of full length His-tagged P22 gp1 protein was purified as described previously (Zheng et al, 2008). Intact P22 tail machines were purified from mature virions as described previously (Tang et al, 2005).
Specimen preparation
Portals and tail machine particles were prepared for cryoEM analyses by preservation in vitreous ice over a holey carbon substrate via rapid freeze plunging. Holey carbon films consisted of a layer of pure carbon fenestrated by two-micron holes spaced four microns apart overlaid on a 400 mesh copper grid. These grids were developed at NRAMM and are currently available from Protochips Inc. as “C-flats”. A plasma cleaner (Gatan, Inc.) was used to clean the grid surface prior to freezing using a mixture of 75% argon and 25% oxygen for 10 seconds. A 3 μl aliquot of sample was applied to the grids, which were loaded into an FEI Vitrobot (FEI company) that was set to 4°C, 100% humidity, and a blot offset of -2. Grids were blotted for six seconds and immediately plunged into liquid ethane. Grids were stored at liquid nitrogen temperatures until loaded into the microscope for data collection.
Assembly-naïve portal data collection and reconstruction
Data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 keV, using a dose of ∼18 e-/Å2 and a nominal underfocus ranging from -2.5 to 3.4 μm. 1075 images were automatically collected at a nominal magnification of 80,000X at a pixel size of 0.098 nm at the specimen level. All images were recorded with a Tietz F415 4K × 4K pixel CCD camera (15 μm pixel) utilizing the Leginon data collection software (Suloway et al, 2005). Experimental data were processed by the Appion software package, which interfaces with the Leginon database infrastructure (Lander et al, 2009). The contrast transfer function (CTF) for each micrograph was estimated using the Automated CTF Estimation (ACE) package (Mallick et al, 2005). 101,244 particles were automatically selected from the micrographs using a template-based particle picker (Roseman, 2004) and extracted at a box size of 288 pixels. Only particles whose CTF estimation had an ACE confidence of 80% or better were extracted. Phase correction of the single particles was carried out within EMAN during creation of the particle stack, and the particles were subsequently centered using the EMAN program “cenalignint”. Stacked particles were binned by a factor of 2 for the final reconstruction. The final stack contained 73,940 particles.
Due to the heterogeneous symmetry of the sample, it was necessary to avoid any symmetry bias during the reconstruction, so the portal density was extracted from the low-resolution asymmetric P22 virion reconstruction (Lander et al, 2006) and radially averaged to remove all symmetry. This radially averaged model was used as the starting point for both the 11- and 12-fold reconstructions. The 3D reconstruction was carried out using a combination of both the SPIDER and EMAN reconstruction packages (Frank et al, 1996; Ludtke et al, 1999). The same stack, initial model, and parameters were used for both 11- and 12-fold reconstructions, except for the imposed symmetry during the reconstruction. Creation of projections of the 3D model and subsequent classification of the particles was performed by EMAN, after which a SPIDER script was employed to perform a reference-free hierarchical clustering analysis of the particles in each class. The resulting SPIDER class that exhibited the highest cross-correlation value to the corresponding model projection was used in the creation of the 3D density for the following iteration. With this methodology, 30.6% of the initial stacked particles contributed to the final 11-fold reconstructed density, and 32% contributed to the 12-fold density. Resolution was assessed by calculating the Fourier Shell Correlation (FSC) at a cutoff of 0.5, which provided a value of 7.1 and 6.9Å resolution for the 11- and 12-fold particles, respectively (van Heel & Harauz, 1986). Calculation of the resolution by rmeasure (Sousa & Grigorieff, 2007) at a 0.5 cutoff yielded a resolution of 8.8 and 8.6Å for the 11- and 12-fold particles, respectively.
Tail machine data collection and reconstruction
Data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 keV, using a dose of ∼18 e-/Å2 and a nominal underfocus ranging from 1.2 to 2.9 μm. 595, 1512, and 1683 images were automatically collected during three sessions at a nominal magnification of 50,000X at a pixel size of 0.163 nm at the specimen level. All images were recorded and processed using the same techniques as for the portal particles, except that particle selection was performed using an algorithm based on a difference of gaussians (Voss et al, 2009). 358,893 particles were extracted at a box size of 384 pixels and binned by a factor of 2 for the final reconstruction. The tail machine density was extracted from the asymmetric P22 reconstruction (Lander et al, 2006) and low-pass filtered to 30Å resolution for use as an initial model. The 3D reconstruction was carried out using the same methodology as the portal reconstructions, enforcing 6-fold symmetry instead of 12 or 11. No additional symmetry was imposed on any of the components in the final density. Resolution was assessed by calculating the Fourier Shell Correlation (FSC) at a cutoff of 0.5, which provided a value of 9.8 Å resolution. Calculation of the resolution by rmeasure (Sousa and Gridgorieff 2007) at a 0.5 cutoff yielded a resolution of 9.4Å.
The density Fourier amplitudes of all final reconstructions were adjusted to fit an experimental 1D low angle X-ray scattering curve using the SPIDER software package (Frank et al, 1996). UCSF Chimera was utilized for segmentation of the individual subunits from the reconstructed density and creation of figure images (Goddard et al, 2007).
SPP1 modeling
The crystal structure of the SPP1 portal (PDB entry 2JES) subunit was manually docked into the segmented P22 EM density and refined in real-space using the fitting tool of UCSF Chimera (Goddard et al, 2007). The helices of the fitted model were subjected to real space refinement using the program Coot (Emsley & Cowtan, 2004), with the torsion, planar, ramachandran, and alpha helix restraints enabled. The C-terminus of the SPP1 structure was manually fitted into the EM density as a rigid body. The entire model was relaxed in centroid followed by full-atom mode with the program Rosetta (Rohl et al, 2004). Constraints were used during the Rosetta run to limit the movement of the Cα atoms to 0.25Å from the original position. The resulting model was then subjected to flexible fitting using the Flex-em module of Modeller (Topf et al, 2008). This procedure was repeated until the Modeller score refrained from improving. The cross-correlation value for the docked models in Chimera before and after modeling into the SPP1 crystal structure EM density increased from 0.18 to 0.43.
Supplementary Material
Acknowledgments
We would like to thank Dr. William Young for providing extensive computational support throughout the reconstruction processes. Electron microscopic imaging and reconstruction was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the National Center for Research Resources' P41 program (RR17573). This work was also supported by the NIH grants R01 GM054076 to J.E.J., R01 GM47980 to P.E.P, and a fellowship from the ARCS foundation to G.C.L.
Footnotes
Accession Numbers: Density maps of the 11- and 12-fold assembly-naïve portals have been deposited at the Electron Microscopy Data Bank (EMDB) and assigned reference codes EMDB-5050 and EMDB-5049, respectively. The assembled tail machine density has been deposited with the accession code EMDB-5051.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann HW. Tailed bacteriophages: the order caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005a;24(21):3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. Journal of molecular biology. 2005b;347(5):895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Bazinet C, Benbasat J, King J, Carazo JM, Carrascosa JL. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27(6):1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Olia AS, Walker-Kopp N, Cingolani G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. Journal of molecular biology. 2007;371(2):374–387. doi: 10.1016/j.jmb.2007.05.051. [DOI] [PubMed] [Google Scholar]
- Casjens S, Weigele PR. Headful DNA packaging by bacteriophage P22. In: Catalano CE, editor. Viral genome packaging machines genetics, structure, and mechanism. Georgetown, Tex. New York, N.Y.: Landes Bioscience/Eurekah.com; Kluwer Academic/Plenum Publishers; 2005. pp. 80–88. [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. Journal of molecular biology. 1992;224(4):1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14(6):1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Moore SD, Prevelige PE, Jr, Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139(1):46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- Doan DN, Dokland T. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J Struct Biol. 2007;157(2):432–436. doi: 10.1016/j.jsb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12(4):1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21(2):319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. Journal of structural biology. 2007;157(1):281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. Journal of molecular biology. 2002;315(4):663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004a;322(2):253–263. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Isidro A, Santos MA, Henriques AO, Tavares P. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol Microbiol. 2004b;51(4):949–962. doi: 10.1046/j.1365-2958.2003.03880.x. [DOI] [PubMed] [Google Scholar]
- Iwashita S, Kanegasaki S. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur J Biochem. 1976;65(1):87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17(2):237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kang S, Poliakov A, Sexton J, Renfrow MB, Prevelige PE., Jr Probing conserved helical modules of portal complexes by mass spectrometry-based hydrogen/deuterium exchange. Journal of molecular biology. 2008;381(3):772–784. doi: 10.1016/j.jmb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura I, Kuhl PW. Morphogenesis of the tail of bacteriophage lambda. III. Morphogenetic pathway. Journal of molecular biology. 1975;91(3):257–273. doi: 10.1016/0022-2836(75)90379-4. [DOI] [PubMed] [Google Scholar]
- Kocsis E, Cerritelli ME, Trus BL, Cheng N, Steven AC. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy. 1995;60(2):219–228. doi: 10.1016/0304-3991(95)00070-2. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Kitazawa D, Takeda S, Tsuchiya Y, Yamashita E, Mizuguchi M, Kawano K, Tsukihara T. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. Journal of molecular biology. 2005;352(4):976–985. doi: 10.1016/j.jmb.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12(9):810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg S, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009 doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. Embo J. 2007;26(7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. Journal of molecular biology. 2007;371(3):836–849. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P. Structural organisation of the head-to-tail interface of a bacterial virus. Journal of molecular biology. 2001;310(5):1027–1037. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104(1):8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Maurides PA, Schwarz JJ, Berget PB. Intragenic suppression of a capsid assembly-defective P22 tailspike mutation. Genetics. 1990;125(4):673–681. doi: 10.1093/genetics/125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Grath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brondsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D. Anatomy of a lactococcal phage tail. J Bacteriol. 2006;188(11):3972–3982. doi: 10.1128/JB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Prevelige PE., Jr Structural transformations accompanying the assembly of bacteriophage P22 portal protein rings in vitro. J Biol Chem. 2001;276(9):6779–6788. doi: 10.1074/jbc.M007702200. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. Journal of molecular biology. 2008;383(3):494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75(22):10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Al-Bassam J, Winn-Stapley DA, Joss L, Casjens SR, Cingolani G. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. Journal of molecular biology. 2006;363(2):558–576. doi: 10.1016/j.jmb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 2009;18(3):537–548. doi: 10.1002/pro.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L, Alonso JC, Tavares P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. Journal of molecular biology. 2005;353(3):529–539. doi: 10.1016/j.jmb.2005.08.063. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22(6):1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ML, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. J Virol. 1983;46(1):239–249. doi: 10.1128/jvi.46.1.239-249.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, Tavares P, Orlova EV. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. Embo J. 2007;26(15):3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, van Duijn E, Lander G, Fu CY, Johnson JE, Prevelige PE, Jr, Heck AJ. Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J Struct Biol. 2007;157(2):371–383. doi: 10.1016/j.jsb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Poteete AR. P22 bacteriophage. In: Webster RG, Granoff A, editors. Encyclopedia of virology. London; San Diego: Academic Press; 1994. pp. 1009–1013. [Google Scholar]
- Prevelige PE. Phage P22. In: Calendar R, editor. The Bacteriophages. 2nd. Oxford; New York: Oxford University Press; 2005. pp. 457–468. [Google Scholar]
- Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- Roseman AM. FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145(12):91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Ruiz T, Radermacher M. Three-Dimensional Analysis of Single Particles by Electron Microscopy. Totowa, NJ: Humana Press; 2006. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157(1):201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(20):10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. Journal of molecular biology. 1997;267(4):865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. Journal of molecular biology. 1984;172(4):523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16(6):935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure. 2006;14(5):837–845. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. Embo J. 2005;24(12):2087–2095. doi: 10.1038/sj.emboj.7600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. Journal of molecular biology. 1992;225(1):81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16(2):295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78(22):12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G. Resolution criteria for three dimensional reconstruction. Optik. 1986;73:119–122. [Google Scholar]
- Voss NR, Yoshioka C, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009 doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage phi29 upon DNA packaging and release. Embo J. 2006;25(21):5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Olia AS, Gonen M, Andrews S, Cingolani G, Gonen T. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol Cell. 2008;29(3):376–383. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziedaite G, Kivela HM, Bamford JK, Bamford DH. Purified Membrane-Containing Procapsids of Bacteriophage PRD1 Package the Viral Genome. Journal of molecular biology. 2009 doi: 10.1016/j.jmb.2008.12.068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


