Abstract
Precise control of the timing and magnitude of Notch signaling is essential for the normal development of many tissues, but the feedback loops that regulate Notch are poorly understood. Developing T cells provide an excellent context to address this issue. Notch1 signals initiate T-cell development and increase in intensity during maturation of early T-cell progenitors (ETP) to the DN3 stage. As DN3 cells undergo β-selection, during which cells expressing functionally rearranged TCRβ proliferate and differentiate into CD4+CD8+ progeny, Notch1 signaling is abruptly down-regulated. In this report, we investigate the mechanisms that control Notch1 expression during thymopoiesis. We show that Notch1 and E2A directly regulate Notch1 transcription in pre-β-selected thymocytes. Following successful β-selection, pre-TCR signaling rapidly inhibits Notch1 transcription via signals that up-regulate Id3, an E2A inhibitor. Consistent with a regulatory role for Id3 in Notch1 down-regulation, post-β-selected Id3-deficient thymocytes maintain Notch1 transcription, whereas enforced Id3 expression decreases Notch1 expression and abrogates Notch1-dependent T-cell survival. These data provide new insights into Notch1 regulation in T-cell progenitors and reveal a direct link between pre-TCR signaling and Notch1 expression during thymocyte development. Our findings also suggest new strategies for inhibiting Notch1 signaling in pathologic conditions.
Keywords: Notch, T cell, lymphocytes, leukemia
Notch1 controls multiple essential functions during thymocyte development (Maillard et al. 2005; Ciofani et al. 2006). Notch1 signals initiate the generation of the earliest intrathymic T cells from multipotent hematopoietic progenitors (Radtke et al. 1999; Sambandam et al. 2005). Subsequently, Notch1 is required for αβ T-cell development through β-selection, an important checkpoint during which immature thymocytes expressing functionally rearranged TCRβ proliferate and then differentiate into quiescent CD4+CD8+ cells. Conditional inactivation of Notch1, Rbpj, or inhibition of Notch signaling by dominant-negative Mastermind-like 1 ((DNMAML) arrests T-cell development at the DN3 stage, prior to β-selection (Wolfer et al. 2002; Tanigaki et al. 2004; Maillard et al. 2006). In vitro studies using OP9 feeder cells have shown that both Notch1 and pre-TCR signals are required to traverse the β-selection checkpoint (Ciofani et al. 2004); Notch1 provides important differentiation, survival, proliferation, and metabolic signals during this juncture in T-cell development (Ciofani and Zuniga-Pflucker 2005; Campese et al. 2006).
Following β-selection, Notch signaling and Notch1 expression are abruptly down-regulated. CD27 expression can be used to separate DN3 cells into two distinct populations, DN3a and DN3b (Taghon et al. 2006). The pre-β-selection CD27−DN3a population is Notch-dependent, whereas post-β-selection CD27+DN3b cells do not require Notch signals for further intrathymic differentiation or survival. Significantly, Notch1 expression is high in DN3a cells and low in DN3b cells (Taghon et al. 2006; Weng et al. 2006).
Although the mechanism of Notch1 down-regulation in β-selected cells is poorly understood, high levels of Notch signaling post-β-selection may be oncogenic. For example, expression of the Notch1 intracellular domain (ICN1) driven by either a retroviral vector (Izon et al. 2002; Li et al. 2008) or a Lck transgene (Deftos et al. 2000; Fowlkes and Robey 2002) allows sustained Notch activity past the DN3 stage that is associated with increased proliferation and survival, a developmental block, and acute lymphoblastic T-cell leukemia (T-ALL). These findings emphasize the importance of precise control of Notch1 signaling at the β-selection checkpoint.
E-proteins, which include E12, E47, E2-2, and HEB in mammals, encode a class of widely expressed basic helix–loop–helix (bHLH) transcription factors that are critical for B-cell development and play important roles in thymocyte development (Murre 2005). E12 and E47 (collectively termed E2A) are encoded by one gene, Tcfe2a, and are generated through alternative splicing (Murre et al. 1989), whereas E2-2 and HEB are encoded by distinct genes (Murre 2005). The primary E-protein complex in thymocytes is a E47/HEB heterodimer (Sawada and Littman 1993; Barndt et al. 2000). The functions of E-proteins in thymocyte development have been revealed through several loss-of-function approaches. E2A knockout mice exhibit an incomplete block in early T-cell development at the DN1 stage (Bain et al. 1997), whereas HEB knockout mice display reduced thymic cellularity and increased immature single positive (ISP) cells (Barndt et al. 1999). Expression of a HEB dominant-negative protein causes a more severe decline in thymocyte numbers and an earlier block in T-cell development than HEB knockout mice, as this antagonist prevents compensation by other E-proteins (Barndt et al. 2000). Enforced expression of the E-protein antagonist Inhibitor of DNA binding 3 (Id3) in human T-lineage precursor cells blocks T-cell lineage differentiation from CD34+ progenitors (Heemskerk et al. 1997). Like Notch, E2A activity is dynamically regulated during thymocyte development. E2A is active prior to β-selection, whereupon pre-TCR signals up-regulate Id3 expression to reduce the DNA-binding activity of E2A in DP or DN thymocytes (Bain et al. 2001; Engel et al. 2001; Xi et al. 2006).
Emerging data suggest cross-talk between E2A and Notch signals during T-cell development (Ikawa et al. 2006; Rothenberg et al. 2008). Expression of several genes that are important in T-cell development, such as Hes1 and pTα, are coregulated by Notch and E2A, and both Notch1 and Notch3 mRNA levels are decreased in E47-deficient fetal thymocytes (Takeuchi et al. 2001; Ikawa et al. 2006). Furthermore, retroviral ICN1 expression in E2A−/− fetal thymocyte progenitors rescues the developmental arrest caused by E2A deficiency (Ikawa et al. 2006). Although they provide synergistic functions, the precise nature of the interactions between Notch and E2A have not been determined.
In this study, we investigate the mechanism underlying the dynamic regulation of Notch1 during β-selection. We show that prior to β-selection, Notch1 and E2A bind the Notch1 locus and promote Notch1 transcription in DN3 cells. At β-selection, MAPK-dependent pre-TCR signals up-regulate Id3 expression, which inhibits E2A binding to the Notch1 promoter and decreases Notch1 expression. Consistent with this model, loss of Id3 expression enhances Notch1 expression in post-β-selected thymocytes, whereas loss of E2A decreases Notch1 expression in pre-β-selected thymocytes in a dose-dependent manner. Furthermore, enforced Id3 expression inhibits Notch1 expression and Notch1-dependent cell survival in Notch1-dependent T-cell lines. Together, these data reveal a direct link between pre-TCR signaling and Notch1 expression during thymocyte development and provide new strategies to disable Notch1 expression and signaling.
Results
Notch1 directly autoregulates its own transcription
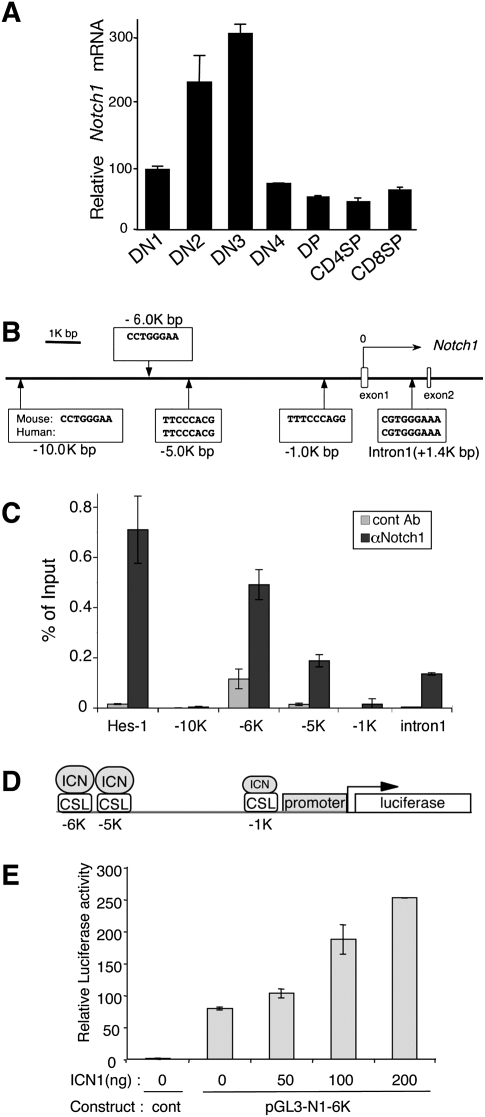
To investigate the regulation of Notch1 expression during thymocyte development, we quantified Notch1 mRNA in thymocyte subsets. A progressive increase in Notch1 mRNA occurred from the earliest intrathymic T-cell progenitors (ETP/DN1) to the DN3 stage (Fig. 1A). This was followed by an abrupt decrease in Notch1 mRNA after the DN3 stage, whereupon Notch1 mRNA remained low throughout intrathymic T-cell development (Fig. 1A). These data confirm previous observations (Huang et al. 2003; Taghon et al. 2006; Weng et al. 2006).
Figure 1.
Notch1 autoregulates its transcription via direct binding to CSL sites. (A) Notch1 expression in thymocyte subsets. Each fraction was sorted from C57BL/6 thymocytes. qPCR was performed and Notch1 expression relative to EF1α is shown as a mean of values from triplicate wells ±SD. Notch1 expression in DN1 cells was set at 100, and the expression in other subsets was set relative to this. Data are representative of three independent experiments. (B) Schematic representation of the 5′ region of the mouse Notch1 locus indicating potential CSL-binding sites. The CSL-binding sequences from mouse and human are shown. (C) ChIP analysis to identify Notch1 binding to CSL sites in Notch1. Rag2−/− DN3 thymocytes were used for ChIP analysis. qPCR was performed with primers flanking each putative CSL site. The Hes1 promoter region was the positive control. Values represent the mean of signal intensity relative to input DNA from triplicate wells ±SD. Data are representative of three independent experiments. (D) Schematic representation of the Notch1 promoter sequences used in the luciferase assays. The CSL sites are shown. (E) ICN1 enhances Notch1 promoter activity. NIH3T3 cells were transiently cotransfected with the Notch1 promoter construct containing the −6K, −5K, −1K CSL-binding sites, and with either empty vector or pcDNA3-ICN1. Luciferase activity relative to Renilla luciferase activity was normalized to the pGL3-basic alone. The data are shown as a mean of values from triplicate wells ±SD. Data are representative of at least three independent experiments.
To understand the basis for this dynamic pattern of Notch1 expression during early thymocyte development, we investigated how Notch1 transcription is regulated. We previously found an approximately fourfold decrease in endogenous mouse Notch1 mRNA when oncogenic Notch1 signals were blocked in T-cell leukemia cell lines (Weng et al. 2006), suggesting that Notch1 autoregulates its own transcription. To investigate this possibility, we searched for putative CSL (the protein encoded by Rbpj)-binding sites (CCTGGGAA or STGGGAR) using the VISTA program (Couronne et al. 2003). We found two CSL-binding sites that are conserved in mouse and human Notch1 in intron 1 and at −5 kb, and three nonconserved CSL-binding sites at −1 kb, −6 kb, and −10 kb (Fig. 1B). In a chromatin immunoprecipitation (ChIP) assay with DN3 cells from Rag2−/− thymocytes, we observed Notch1 binding at the −6-kb, −5-kb, and intron 1 sites (Fig. 1C). Little binding was found at the −1-kb and −10-kb sites (Fig. 1C).
To determine if these sites are important for Notch1 autoregulation, a construct containing the entire 6-kb upstream region of the Notch1 promoter fused to luciferase was studied for its responsiveness to Notch signals (Fig. 1D). Activated Notch1 produced a dose-dependent increase in reporter activity (Fig. 1E). These data demonstrate direct binding of endogenous Notch1 to Notch1 regulatory sequences in primary T cells and further show that Notch1 has the capacity to directly autoregulate its own promoter.
Notch1 mRNA expression is down-regulated by pre-TCR signals in DN3 cells
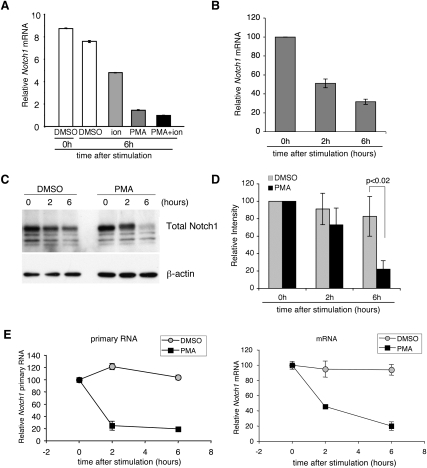
The marked decrease in Notch1 mRNA that occurs at the β-selection checkpoint suggests that pre-TCR signals down-regulate Notch1. Rag2−/− thymocytes are blocked at the DN3 stage due to the inability to rearrange a β-chain to form a pre-TCR, and therefore fail to undergo β-selection. To determine if down-regulation of Notch1 expression at the DN3 stage was associated with pre-TCR signals, DN3 cells from Rag2−/− mice were stimulated with PMA and/or ionomycin, which mimic signals generated through the pre-TCR. Notch1 expression was reduced with PMA and/or ionomycin treatment (Fig. 2A), but PMA stimulation alone reduced Notch1 expression by nearly the same amount as PMA/ionomycin stimulation, suggesting that signals downstream from PMA, which activates the PKC/Ras/MAPK pathways, are most important.
Figure 2.
Pre-TCR signals down-regulate Notch1 transcription. (A) Notch1 mRNA was diminished by PMA treatment in DN3 cells. DN3 cells from Rag2−/− mice were treated with DMSO and/or PMA (20 ng/mL) and/or ionomycin (500 ng/mL) for 6 h, and qPCR was performed. Notch1 expression relative to EF1α is shown as the mean of values from triplicate wells ±SD. Notch1 expression in the PMA/ionomycin sample was set at 1, and the values in the other samples were normalized to this. The data are representative of three independent experiments. (B) Time course of Notch1 mRNA expression following PMA stimulation. DN3 cells from Rag2−/− mice were treated with PMA (20 ng/mL) for 0 h, 2 h, or 6 h, and qPCR was performed. Notch1 expression relative to EF1α is shown as the mean of values from triplicate wells ±SD. Data are relative to the 0-h sample, whose value was set at 100, and representative of three independent experiments. (C) Time course of Notch1 protein expression following PMA stimulation. Rag2−/− DN3 thymocytes were sorted and treated with DMSO or PMA (20 ng/mL) for 0 h, 2 h, or 6 h. Notch1 and β-actin expression were determined by Western blotting. Data are representative of three independent experiments. (D) Quantification of Notch1 protein expression after PMA stimulation. Protein amounts were measured by densitometer and quantified by Image Quant. The protein amounts were relative to unstimulated control, and error bars show SD from three independent experiments. (E) PMA-induced Notch1 down-regulation is regulated through transcriptional changes. Primary-transcript RT–PCR analysis was performed with DN3 cells from Rag1−/− mice. Cells were treated with either DMSO or PMA. Primers were designed to flank between Notch1 exon 27 and exon 28 for mRNA and exon 27 and intron 27 for primary RNA. Notch1 mRNA or primary RNA expression is relative to 18S rRNA and is shown as a mean of values from triplicate wells ±SD. Data are relative to the 0-h sample, whose value was set at 100 and is representative of two independent experiments.
To further dissect Notch1 down-regulation at the β-selection checkpoint, DN3 cells were stimulated with PMA for 1 h, 2 h, and 6 h. Notch1 mRNA expression was inhibited by ∼40% at 1 h, ∼50% at 2 h, and ∼70% at 6 h post-PMA stimulation (Fig. 2B). In contrast to the decrease in Notch1 mRNA, Notch1 protein amounts were not significantly decreased at 2 h post-PMA treatment (Fig. 2C,D). By 6 h post-PMA treatment, however, Notch1 protein decreased by ∼80% (Fig. 2C,D).
To determine whether pre-TCR signals influenced Notch1 mRNA or steady-state protein levels, we measured these parameters in the presence of proteasome and/or MEKK1/2 inhibitors. In the presence of the proteasome inhibitor MG132, Notch1 protein accumulated (Supplemental Fig. S1A, lanes 6–9), and Notch1 mRNA levels markedly increased (Supplemental Fig. S1B), consistent with Notch1 autoregulation. Even in the presence of elevated levels of Notch1 protein due to MG132 treatment, PMA stimulation decreased Notch1 mRNA levels (Supplemental Fig. S1B), showing that Notch1 mRNA is reduced in a proteasome-independent manner. Consistent with an important role for pre-TCR signals in the regulation of Notch1 mRNA, treatment of Rag2−/− DN3 cells with the MEKK1/2 inhibitor U-0126 prevented the PMA-induced decrease in Notch1 mRNA without affecting the steady-state amount of Notch1 protein (Supplemental Fig. S1). Together with the time-course studies, our data suggest that pre-TCR signaling decreased Notch1 mRNA expression before affecting steady-state protein levels.
To identify whether pre-TCR signals affected prespliced Notch1 transcripts, primary and spliced transcripts were compared in Rag2−/− DN3 cells. PMA stimulation for 2 h resulted in lower levels of both mature and immature Notch1 transcripts (Fig. 2E), a time at which Notch1 protein was not significantly decreased (Fig. 2C). Thus, our data demonstrate that pre-TCR signals down-regulate Notch1 mRNA, which leads to decreased Notch1 protein.
Pre-TCR signals block Notch1 transcription via Id3
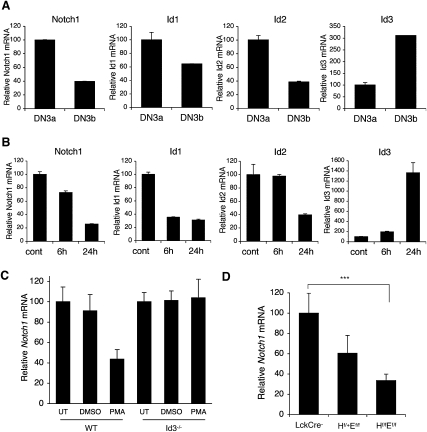
Previous work demonstrated that TCR and pre-TCR signals induce Id3 via ERK/MAPK signaling (Bain et al. 2001; Engel et al. 2001). These studies showed that TCR ligation reduced E2A DNA-binding activity as a consequence of up-regulated Id3 expression in CD4+CD8+ DP thymocytes and DN thymocytes. To determine whether pre-TCR stimulation induced Id3 expression in DN3 cells, we examined Id3 mRNA expression in both wild-type DN3a and DN3b cells (Fig. 3A) and thymocytes from Rag2−/− mice stimulated with α-CD3 in vivo (Fig. 3B). Like Notch1, the expression of Id1 and Id2, both reportedly Notch targets (Talora et al. 2003; Weng et al. 2006), decreased in DN3b cells (Fig. 3A). In contrast, Id3 expression was highly up-regulated in post-β-selected DN3b cells. Consistent with these observations, α-CD3-stimulated DN3 cells showed reduced Notch1, Id1, and Id2 expression, whereas Id3 expression was dramatically increased by 24 h (Fig. 3B).
Figure 3.
Expression of Notch1 inversely correlates with Id3. (A) DN3a cells and DN3b cells were sorted from thymocytes of C57BL/6 mice. Notch1, Id1, Id2, and Id3 expression relative to EF1α is shown as the mean of values from triplicate wells ±SD. Data are representative of three independent experiments. (B) Rag2−/− mice were injected with α-CD3 or PBS. Thy1.2+ cells were sorted from the thymocytes 6 h or 24 h post-injection. The transcript quantities relative to EF1α are shown as the mean of values from triplicate wells ±SD. Data are representative of three independent experiments. (C) DN3a cells were sorted from C57BL/6 or Id3−/− thymocytes and stimulated with DMSO or PMA for 2 h. Notch1 expression relative to EF1α was determined by qPCR. Data are relative to untreated samples (UT) of each mouse and represent the mean of three independent experiments ±SD. (D) Dose-dependent Notch1 expression in E2A/HEB double-deficient thymocytes. Notch1 expression relative to EF1α was determined from DN3a cells of Tcf12f/fTcfe2af/fLckCre− (LckCre−), Tcf12f/+Tcfe2af/fLckCre+ (Hf/+Ef/f), Tcf12f/fTcfe2af/fLckCre+ (Hf/fEf/f) thymocytes. Data are from triplicates of two independent experiments (n = 3) and relative to the LckCre− sample. (***) P < 0.05, Student's t-test, two-tailed. Graphed results are means with error bars representing the standard error of the mean (SEM).
The inverse relationship between Id3 and Notch1 expression suggested that Id3 mediates pre-TCR-induced inhibition of Notch1 transcription. To directly test this model, we measured Notch1 mRNA in PMA-treated Id3−/− DN3a thymocytes (Fig. 3C). PMA treatment of Id3 knockout DN3a cells failed to down-regulate Notch1, whereas Notch1 expression decreased by 60% in PMA-treated DN3a cells from littermate controls. These data show that Id3 has an important role in mediating the decrease in Notch1 transcription that occurs at β-selection.
Id3 functions by inhibiting E-protein-mediated transcriptional activation. Thus, if loss of Id3 activity prevented the decrease in Notch1 transcription, loss of E2A activity should result in decreased Notch1 transcription. HEB and E47, encoded by the genes Tcf12 and Tcfe2a, form the major E2A heterodimer in developing T cells (Barndt et al. 2000). We tested the association between E2A signaling and Notch1 transcription by measuring Notch1 mRNA in DN3a thymocytes lacking E47 and/or HEB. DN3a thymocytes were purified from conditional knockout mice in which both E47 and HEB were deleted with an Lck-Cre transgene (Tcf12f/fTcfe2af/fLckCre+). These mice exhibit a developmental block at the DN3 stage (Wojciechowski et al. 2007). The genotypes of the floxed mice were either Tcf12f/+Tcfe2af/fLckCre+ or Tcf12f/fTcfe2af/fLckCre+, which provided the opportunity to compare the effects of different E-protein levels on Notch1 expression. The amount of Notch1 expression correlated with the amount of E2A (Fig. 3D). Thus, the complementary results provided by the Id3 and E2A loss-of-function studies suggest that E2A is an important regulator of Notch1 transcription.
E2A directly activates Notch1 transcription
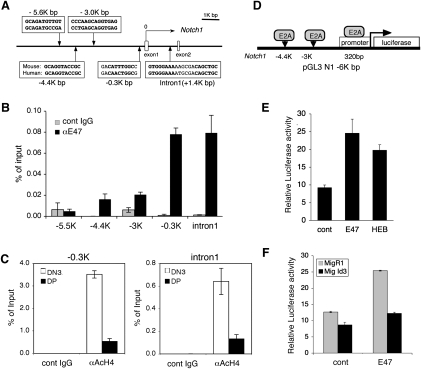
To investigate whether E2A directly regulates Notch1 transcription, we searched for canonical E-box sites (CANNTG) in the Notch1 locus. Five binding sites conserved between human and mouse were found in a region spanning from 6 kb upstream of the transcriptional start to intron 1 of the Notch1 gene (Fig. 4A). We performed ChIP assays with DN3 cells from Rag2−/− thymocytes and observed strong E47 binding to the −0.3-kb and intron 1 sites (Fig. 4B). Binding was minimal at the −4.4-kb and −3-kb sites and absent at the −5.5-kb site of Notch1. Histone 4 (H4) was hyperacetylated at both the −0.3-kb and intron 1 sites in DN3 cells but not in DP cells, indicating that these are likely to be regions of open chromatin in DN3 cells but not in DP cells (Fig. 4C).
Figure 4.
E47 directly up-regulates Notch1 transcription. (A) Schematic representation of the 5′ region of the Notch1 locus with putative E2A-binding sites. The translational start site is labeled “0.” Only the first two Notch1 exons are shown. (B) E47 binds to E2A-binding sites of Notch1. Rag2−/− DN3 thymocytes were subjected to ChIP analysis. qPCR was performed with primers flanking putative E2A-binding sites at 5.5, 4.4, 3, and 0.3 kb upstream of the transcriptional start site as well as intron 1 of Notch1. Graphs represent the mean of the ratio of the amount of immunoprecipitated DNA/input from values from triplicate wells ±SD. Data are representative of two independent experiments. (C) Differential binding activity of acetylated histone 4 to the Notch1 locus in DN3a and DP cells. ChIP analysis to identify acetylated Histone 4 was performed on DN3a and DP cells from wild-type B6 mice. qPCR was performed to detect E2A-binding sites at −0.3 kb and intron 1 of Notch1. Data are representative of two independent experiments. (D) Schematic of the Notch1 promoter luciferase construct including the E2A-binding sites. (E) E47 and HEB transactivate the Notch1 promoter. NIH3T3 cells were transfected with the Notch1 promoter construct containing the indicated E2A-binding sites and with the empty vector, E47, or HEB. Luciferase activity relative to Renilla luciferase activity is shown as the mean of values from triplicate wells ±SD. Data are representative of two independent experiments. (F) Id3 inhibits E47-induced transactivation of the Notch1 promoter. NIH3T3 cells were transduced with either MigR1 or Id3. At 18 h, cells were transiently cotransfected with the Notch1 promoter construct with empty vector or E47. Luciferase activity relative to Renilla luciferase activity is shown as a mean of values from triplicate wells ±SD. Data are representative of three independent experiments.
To evaluate the importance of these regions in Notch1 promoter activity, we investigated whether E47 up-regulated luciferase activity in our Notch1 reporter that contains ∼6 kb of the Notch1 promoter region, which includes the E-box sites (Fig. 4D). Both E47 and HEB increased Notch1 reporter activity (Fig. 4E). Furthermore, the E47-mediated increase in Notch reporter activity was abrogated by Id3 (Fig. 4F). These data demonstrate that E47 and HEB directly activate Notch1 transcription.
Pre-TCR signals inhibit binding of E47 and Notch1 to the Notch1 promoter
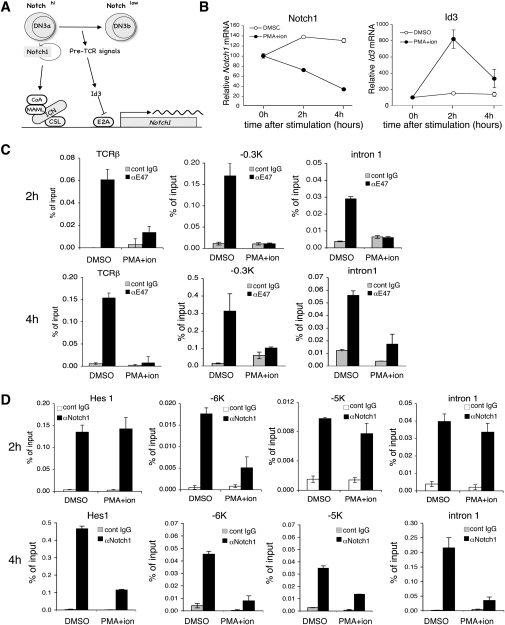
Our data demonstrate that both E2A and Notch1 are direct positive regulators of Notch1 transcription. At the β-selection checkpoint, pre-TCR signals induce Id3, which blocks E2A, leading to decreased Notch1 transcription and therefore decreased Notch1 protein resulting in a block in the positive feedback loop. Thus, we propose that pre-TCR signaling is the switch that extinguishes Notch1 transcription (Fig. 5A). Since PMA and/or PMA/ionomycin treatment of primary DN3 cells markedly reduced Notch1 mRNA amounts within several hours (Fig. 2A), the events proposed in our model should occur within this time frame. In order to obtain sufficient protein for these studies, we tested this hypothesis in Scid-adh cells. Scid-adh cells phenotypically resemble DN3 thymocytes and respond to stimuli mimicking pre-TCR signals by undergoing molecular and cellular changes resembling the DN3-to-DN4 transition (Carleton et al. 1999). Thus, these cells serve as an in vitro model to study the role of pre-TCR signaling in driving T-cell differentiation. Notch1 mRNA amounts were reduced by 80% at 4 h post-treatment of Scid-adh cells with PMA/ionomycin (Fig. 5B). The effects on Notch1 mRNA were independent of Scid-adh differentiation, as these cells did not show phenotypic evidence of differentiation until after 6 h of PMA treatment (Supplemental Fig. S2). PMA/ionomycin treatment of Scid-adh cells increased the amount of Id3 mRNA by fourfold within 2 h of treatment (Fig. 5B). ChIP was performed to detect binding of E47 and Notch1 to the sites we identified in the Notch1 promoter and intron 1 (Figs. 1B, 4A). By 2 h following PMA/ionomycin stimulation, the interaction of E47 with the previously identified E2A target gene TCRβ (Vβ5.1) (Agata et al. 2007) was significantly reduced, showing that E2A DNA-binding activity was reduced by 2 h of PMA/ionomycin stimulation. The interaction of E47 with both of the strong E2A-binding sites on Notch1 in DN3 cells (−0.3 kb and intron 1) was also abrogated (Fig. 5C). In contrast, Notch1 binding was significantly reduced at the −6-kb binding site but was unaffected at the −5-kb and intron 1-binding sites (Fig. 5D). Furthermore, Notch1 binding to the Hes1 promoter region was unaffected 2 h after treatment with PMA/ionomycin (Fig. 5D); whereas by 4 h post-PMA/ionomycin treatment when Notch1 mRNA levels were maximally reduced, Notch1 binding to all three binding sites in Notch1 as well as the Hes1 promoter was markedly reduced (Fig. 5D). E47 binding to the Notch1 sites remained low at this time (Fig. 5B). Taken together, these data demonstrate that pre-TCR signals inhibit Notch1 transcription via a mechanism that rapidly abrogates the ability of both E2A and Notch1 to stimulate Notch1 transcription.
Figure 5.
Id3 down-regulates Notch1 expression. (A) Model for Notch1 and E2A transcriptional regulation of Notch1. Both Notch and E2A are positive regulators of Notch1 transcription. Following β-selection, pre-TCR signaling induces Id3 expression. This inhibits transcriptional activation of Notch1 by E2A leading to lower Notch1 protein, which extinguishes the initial positive autoregulatory loops. (B) Time course of Notch1 and Id3 mRNA down-regulation by PMA stimulation in Scid-adh cells. qPCR was performed on Scid-adh cells that were stimulated with PMA (20 ng/mL) for 0 h, 2 h, and 6 h. Notch1 and Id3 expression relative to EF1α is shown as the mean of values from triplicate wells ±SD. Data are relative to the 0-h sample, which was set at 100, and are representative of three independent experiments. (C,D) E2A binding (C) and Notch1 binding (D) to Notch1 is inhibited by pre-TCR stimulation. Scid-adh cells were treated with DMSO or PMA/ionomycin for 2 h or 4 h and fixed for ChIP. qPCR was performed with primers flanking either the E2A-binding sites or CSL-binding sites on the Notch1 locus. E47 and Notch1 binding were also measured at the TCRβ locus or at the Hes1 locus as a control of E47 or Notch1 signaling activity. Values represent the mean of the ratio of the amount of immunoprecipitated DNA/input from values from triplicate wells ±SD. Data are representative of two independent experiments.
Id3 down-regulates Notch1 expression in multiple Notch1 T-ALL cell lines
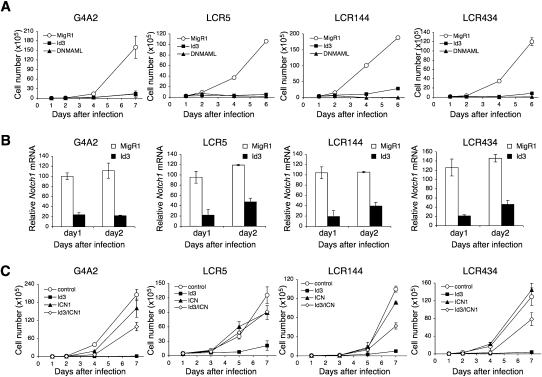
The ability of pre-TCR signaling and Id3 to inhibit Notch1 transcription suggested that this signaling axis might inhibit Notch1 transcription in contexts outside of the down-regulation of Notch1 transcription that normally occurs following successful β-selection. We were particularly interested in Notch1-dependent T-ALLs, which contain activating point and insertion/deletion mutations that do not appear to affect the regulation of Notch1 transcription. We thus asked whether ectopic Id3 expression could diminish Notch1 expression and thereby block Notch-dependent growth of these cells. For these studies, we used four different Notch-dependent T-ALL cell lines: a BCR-ABL-transformed T-ALL, G4A2 (Pear et al. 1998), and three different Kras-induced leukemia lines—LCR5, LCR144, and LCR434—that have mutations in the Notch1 PEST domains (Chiang et al. 2008). Retroviral expression of the pan-Notch inhibitor, DNMAML, or GSI treatment blocked the growth of all four T-ALL cell lines, showing Notch dependency of these tumor cell lines (Fig. 6A; data not shown). Retroviral expression of Id3 also blocked the growth of each of these Notch-dependent cell lines to an extent that was comparable with DNMAML expression (Fig. 6A). Id3-induced growth inhibition was accompanied by markedly decreased Notch1 expression by day 1 following Id3 expression (Fig. 6B).
Figure 6.
Id3 down-regulates Notch1 expression and inhibits the growth of Notch1-dependent T-ALL cells. (A) Id3 inhibits T-ALL cell growth. G4A2, LCR5, LCR144, and LCR434 were transduced with either GFP (MigR1), Mig-DNMAML, or Mig-Id3. GFP+ cells were sorted at day 1 post-transduction (day 1) and cultured for the indicated days. Extrapolated GFP+ cell numbers are shown as a mean of values from triplicate wells ±SD. Data are representative of three independent experiments. (B) Id3 represses Notch1 mRNA expression. T-ALL cells were transduced with either MigR1 or Mig-Id3. GFP-expressing cells were sorted at day 1 post-transduction and cultured for 1 d (day 2). Notch1 expression relative to EF1α was determined at days 1 and 2, and is shown as a mean of values from triplicate wells ±SD. Data are representative of three independent experiments. (C) ICN1 rescues Id3-mediated growth arrest of T-ALL cells. T-ALL cells were cotransduced with the GFP (MigR1), Mig-ICN1, NGFR, and NGFR-Id3 retroviruses; and GFP+NGFR+ cells were sorted at day 1 post-transduction. GFP+NGFR+ cells were cultured for the indicated days. The extrapolated cell numbers of the GFP+NGFR+ fraction are shown as a mean of values from triplicate wells ±SD. Data are representative of three independent experiments.
To confirm that the effects of Id3 on proliferation of the Notch-dependent cells were Notch-specific, we used retroviruses to coexpress Id3 and a constitutively active form of Notch1 (ICN1). Because retrovirally expressed ICN1 lacks the Id3-dependent regulatory sequences, we reasoned that activated Notch1 expressed in this fashion would resist the effects of Id3. Consistent with this prediction, ICN1 maintained the proliferation of Id3-sensitive cell lines in the presence of Id3 expression (Fig. 6C), which (as expected) suppressed the expression of the endogenous murine Notch1 alleles (Supplemental Fig. S3). Id3 suppression of Notch1 mRNA was lower when ICN was coexpressed, likely due to the stimulatory effects of activated Notch on Notch1 expression (Supplemental Fig. S3). We also investigated the possibility that Id3 inhibited ICN1 directly by assaying the ability of Id3 to suppress ICN1-mediated activation of a Notch1-dependent Hes1 luciferase reporter (Supplemental Fig. S4). Id3 failed to suppress the ability of ICN1 to stimulate the Hes1 reporter; however, it did suppress a control E2A-luciferase reporter in the presence of E2A. Together, these data show that Id3 antagonizes the growth of Notch-dependent tumor cell lines by inhibiting Notch1 expression.
Discussion
Notch receptor expression is regulated by both transcriptional and post-transcriptional mechanisms. Important post-transcriptional mechanisms include the events that regulate Notch cleavage and trafficking prior to its appearance at the cell surface (Gordon et al. 2008). In contrast, the mechanisms that regulate Notch transcription are poorly understood. T-cell development provides a particularly good context to study Notch transcription as multiple developmental stages are known and Notch transcription changes during development. The initial steps of thymocyte development are Notch1-dependent, and Notch1 mRNA progressively increases during the early steps of thymocyte development up to and including the β-selection checkpoint. The level of Notch1 mRNA plummets following β-selection, and this is associated with stages of T-cell differentiation where the requirement for Notch signaling is no longer absolute. Despite this dynamic expression of Notch1 mRNA, very little is known about its regulation.
In this study, we describe a Notch1-dependent autoregulatory loop that is an important contributor to the progressive increase in Notch1 expression that occurs during the earliest stages of T-cell development and further show how pre-TCR signals disrupt this loop to down-regulate Notch1 transcription following β-selection. Thus, our data directly link pre-TCR signaling and Notch1 expression. We propose that the ability of pre-TCR signals to down-regulate Notch1 expression occurs via pre-TCR-mediated induction of the bHLH inhibitor Id3. Id3 then prevents E2A-dependent activation of Notch1 transcription, leading to a decrease in both Notch1 mRNA and protein. In this model, the loss of E2A-mediated transcriptional activation of Notch1 would initiate a progressive decrease in Notch1 transcription due to the loss of transcriptional activation by both E2A and Notch1. Our model also provides a mechanistic explanation for the observation of Murre and coworkers (Ikawa et al. 2006), who showed that ectopic E2A signals could up-regulate Notch1 mRNA. In this context, E2A provides requisite signals for the initial Notch1 up-regulation, which then leads to Notch1-driven positive autoregulation. In fact, our reporter studies suggest that E2A exerts a larger effect on Notch1 transcription than Notch1 itself.
Previous studies showed that pre-TCR and TCR signals induced expression of the E-protein inhibitor Id3, thereby blocking E-protein transcriptional activation (Bain et al. 2001; Engel et al. 2001; Xi et al. 2006). We have now linked Id3 expression directly to Notch, where it caused a decrease in Notch1 reporter activity, a decrease in Notch1 mRNA, and blocked growth of Notch-dependent T-cell lines, which is recovered by retrovirally expressing ICN1. Thus, E2A regulation appears to be a critical link between pre-TCR signaling and Notch1 transcriptional regulation.
Although it was known that Notch1 mRNA expression undergoes wide fluctuations in developing T cells, the mechanism behind this dynamic regulation had not been described. We identified Notch1 regulatory sequences that are directly controlled by Notch1 itself and E-proteins. Our data show that Notch1 directly binds autoregulatory CSL sites in the Notch1 locus to increase its own transcription; thus providing the mechanism to explain how Notch1 influences its own transcription (Huang et al. 2004; Weng et al. 2006). It will be interesting to determine whether these CSL-binding sites are exclusive to Notch1 or may be occupied by other Notch family members. Binding of E2A and Notch1 to the Notch1 regulatory sequences was accompanied by H4 hyperacetylation, showing that these interactions influence the chromatin structure of Notch1. This suggests that the pre-TCR-initiated down-regulation of Notch1 transcription ultimately leads to epigenetic changes that result in low levels of Notch1 transcription during the subsequent stages of thymocyte development.
In addition to Notch1 and E2A, the only other protein shown to directly bind and regulate Notch1 transcription is p53, which induces Notch1 transcription in epithelial cells (Lefort et al. 2007; Yugawa et al. 2007). The extent to which p53 regulates Notch1 transcription in T cells remains to be determined, and current data suggest that p53 regulation of Notch1 in T cells may be post-transcriptional (Laws and Osborne 2004).
Synergistic signaling by Notch and E2A is evolutionarily conserved. The enhancer of split complex is an important mediator of Drosophila sensory organ development whose transcription is coregulated by Notch and bHLH activity (Bailey and Posakony 1995). Several members of this complex contain both CSL and E-box sites, which are synergistically activated by Notch and daughterless, an E2A ortholog (Bailey and Posakony 1995). Some of the enhancers of split loci, such as m8, contain paired CSL-binding sites (SPS) with an adjacent E-box (Bailey and Posakony 1995; Cave et al. 2005). Caudy and coworkers (Cave et al. 2005) suggest that the paired binding site/E-box configuration is necessary for transcriptional activation of this locus and that this activation may depend on physical interactions between Notch and daughterless. Recently, Blacklow and colleagues (Nam et al. 2007) showed that the SPS sites allow for Notch dimerization, which is critical for Notch-induced transcriptional activation at these sites. Thus, specific configurations of Notch- and E2A-binding sites may be required to activate a subset of Notch targets. Our initial analysis of the Notch1 locus has not identified canonical SPS sites, but it is certainly possible that there are long-range interactions between proteins bound at regulatory sites distant from each other.
Notch1-activating mutations are frequent in human and mouse T-ALLs, and proliferation of cell lines derived from these tumors requires persistent Notch signaling (Weng et al. 2004; Aster et al. 2008). This provided the rationale for treating patients with agents that block Notch receptor activation, such as γ secretase inhibitors (Aster et al. 2008). Our studies show that inhibiting Notch1 transcription with Id3 can also be used to block Notch-dependent growth of T-ALL cell lines. Ectopic Id3 expression inhibited Notch1 transcription and blocked growth of four Notch-dependent T-ALL cell lines as effectively as dominant-negative mastermind, a potent Notch inhibitor. This effect was Notch-specific as retroviral expression of ICN1, which was resistant to Id3 inhibition, rescued growth of these cell lines. These data suggest that therapeutic strategies that block Notch1 transcription may be beneficial in treating Notch1-dependent tumors. Even though enforced Id3 expression inhibited T-ALL growth, manipulation of E2A for therapeutic purposes will require a better understanding of the effects of inhibiting E2A. For example, inactivating E2A in the germline of mice is frequently associated with T-ALL (Bain et al. 1997), whereas tumors are rare when E2A is conditionally inactivated in thymocytes (Pan et al. 2002).
In summary, we identified a novel regulatory mechanism for Notch1 expression, which involves Notch1 itself and E-proteins. Our data explain the rapid diminution in Notch1 mRNA expression that occurs after β-selection. In our model, E2A and Notch1 are important transcriptional regulators of Notch expression in pre-β-selected thymocytes. With the onset of β-selection, pre-TCR signals lead to Id3 expression, which inhibits E2A activity leading to decreased Notch1 transcription and decreased Notch1 protein; thus extinguishing the positive feedback loop. Not only do these findings provide new insights into the control of Notch signaling in T-cell development, but they also suggest new strategies for inhibiting Notch1 transcription in pathologic conditions.
Materials and methods
Mice
Eight-week-old to 12-wk-old C57BL/6 mice (National Cancer Institute, Frederick, MD) and 4- to 6-wk-old Rag1−/− or Rag2−/− mice (Taconic Laboratories) were purchased. Id3−/−, Hf/fEf/fLckCre+ mice were generated as described (Pan et al. 1999; Zhang et al. 1999; Wojciechowski et al. 2007). All animal experiments were performed according to guidelines from the National Institutes of Health and with an approved protocol from the University of Pennsylvania Animal Care and Use Committee.
Isolation of thymocyte subsets
CD4−CD8− DN thymocytes were negatively selected with α-CD4 and α-CD8 MACs beads by MACS-LD columns (Militenyi Biotec) from thymocytes of C57BL/6 mice and were stained with labeled antibodies against lineage markers (TCRβ, TCRγ, CD3ɛ, CD4, CD8, NK1.1, Gr-1, CD11b, CD11c, B220, ter119), CD25 and CD44, or c-Kit antigen (Pharmingen). DN1 (CD44hi or c-Kithi, CD25−/lo Lin−), DN2 (CD44hi or c-Kithi, CD25hi, Lin−), DN3 (CD44lo or c-Kitlo, CD25hi, Lin−) and DN4 (CD44lo or c-Kitlo, CD25−/lo, Lin−) subsets were purified by cell sorting on a FACS Moflo (Cytomation). DN3a (CD44lo CD25hi, Lin−, CD27lo) and DN3b (CD44lo CD25hi, Lin−, CD27hi) cells were purified by sorting after staining with α-CD27 antibody (eBioscience) as described (Taghon et al. 2006). Thymocytes of C57BL/6 mice were stained with antibodies against CD4, CD8, and TCR-β (Pharmingen) and purified by cell sorting to obtain CD4+CD8+ (DP), CD4+CD8− (CD4SP), and CD4−CD8+ (CD8SP) cells and CD4−CD8+ TCR-β− cells (ISP). DN3 cells from Rag2−/− mice were negatively selected with biotinylated antibodies against NK1.1, CD11b, CD11c, B220, Ter119, and CD44 with streptavidin beads (Militenyi Biotec) and confirmed to be >98% pure.
In vivo stimulation
Rag2−/− mice were injected with either PBS or 50 μg of purified α-CD3 (Pharmingen). Thymocytes were obtained at 6 h or 24 h, purified using Thy1.2 beads (Militenyi Biotec), and used for quantitative real-time PCR (qPCR). Three mice were used at each time point, and data are representative of three independent experiments.
qPCR
RNA was purified with the RNeasy Mini Kit or micro kit (Qiagen), digested with DNase I, and used for reverse transcription according to the manufacturer's instructions (SuperScript II kit; Invitrogen). Genes were validated with the primer sets indicated in Supplemental Table S1, and relative quantities were determined against EF1α, whose expression was stable in our experiments. Transcripts were amplified with Sybr Green PCR Master Mix (Applied Biosystems) on the ABI 7900HT sequence detection system (Applied Biosystems). All qPCR reactions were performed in triplicate.
ChIP assays
ChIP was performed with the ChIP assay kit (Millipore, #17-295). DN3 cells, purified from Rag2−/− mice, and Scid-adh cells were treated with either DMSO or PMA (20 ng/mL) for 4 h. All procedures have been described (Weng et al. 2006). Briefly, 2.5 × 106 cells for each immunoprecipitation sample were fixed and immunoprecipitated with control antibody (rabbit IgG; Santa Cruz Biotechnologies, Inc., #sc-3888), Notch1 TAD/PEST-specific antiserum (Aster et al. 2000), α-E47 (Santa Cruz Biotechnologies, #sc-763), or anti-acetylated histone 4 (Upstate Biotechnologies, #06-866). After DNA purification, SYBR Green qPCR was performed with primers flanking the CSL-binding sites on the Hes1 promoter (330 base pairs [bp] upstream of the first exon of Hes1) or the Notch1 locus (Fig. 1A) and for E2A-binding sites on the Notch1 locus (Fig. 5A). The DNA quantity recovered from each ChIP sample is shown as the relative value to the DNA input sample that was not immunoprecipitated. Primer sequences are provided in Supplemental Table S1. Values are from triplicate wells ±SD. Data are representative of three independent experiments.
Luciferase reporter assay
Approximately 6.3 kb of Notch1 sequences upstream of the translational start site was cloned (Notch1-6K) into the pGL3-basic vector (Promega). The primer sequences are provided in Supplemental Table S1. Id3, E47, and HEB cDNAs were cloned into the MigR1 retroviral vector. NIH3T3 cells were transduced with MigR1 or Id3-MigR1 retroviral supernatants 18 h prior to the transfection for Figure 4F. Cells were transfected with pGL3-Notch1-6K (200 ng) and pRL-TK (25 ng) by Lipofectamin-2000 (Invitrogen). MigR1, ICN1-MigR1, E47-MigR1, or HEB-MigR1 (400 ng) were cotransfected as indicated. Cells were collected at 30 h post-transfection, and luciferase activity was measured with the Dual-Luciferase reporter assay kit (Promega).
Cell preparation and Western blotting
Rag2−/− DN3 cells (5 × 105) were stimulated with PMA (20 ng/mL) or DMSO. Cells were collected at 0, 2, or 6 h after treatment, and 20 μg of protein lysate was subjected to Western blotting. Antibodies against the Notch1 TAD/PEST domain and β-actin (Sigma) were used.
Retroviral transduction and proliferation assay
Mig-ICN1 and Mig-DNMAML have been described (Weng et al. 2003; Maillard et al. 2004). Id3 was also cloned into MSCV-tNGFR, which coexpresses the truncated nerve growth factor receptor (tNGFR) as a surrogate marker (Izon et al. 2002). T-ALL cells (2 × 106) were centrifuged with the appropriate amount of viral supernatant and 8 μg/mL polybrene (Sigma) for 90 min at 2500 rpm (day 0). GFP+ or NGFR+ cells were purified by cell sorting on a FACS MoFlo (Cytomation). Cells were cultured for up to 7 d, and the total cell number was determined using a hemocytometer. Each experiment was carried out at least three times.
Real-time, primary-transcript RT–PCR
All procedures have been described (Murray 2005). Briefly, total RNA was isolated from DN3a cells from Rag1−/− mice using TRIzol (Invitrogen). RNA was resuspended at a final concentration of 0.1 μg/μL in 100 μL and treated with 10 U of RNase-free DNase I (Promega) for 30 min, then purified with acid phenol/chloroform. RNA (0.4 μg) was reverse-transcribed using SuperScript II (Invitrogen). Primers were designed to amplify the region exon 27 and exon 28 for mRNA and exon 27 and intron 27 for primary RNA on Notch1. All samples were processed in triplicate, and relative quantities were determined against a diluted standard of 18S rRNA.
Statistical analysis
Statistical analysis was done using an unpaired Student's t-test.
Acknowledgments
We thank John Choi, Yueming Li, Zissimos Mourelatos, Marianti Kiriakido, and Hong Sai for providing advice and reagents. We are grateful to Jon Aster, Avinash Bhandoola, Tom Kadesch, Karen Keeshan, Ivan Maillard, Al Singer, and members of the Pear laboratory for sage advice and critical reading of the manuscript. We thank William Demuth for cell sorting. This work was supported by grants from the National Institutes of Health to S.C.B., Y.Z., and W.S.P., and a LLS SCOR Award to S.C.B. and W.S.P.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1793709.
Supplemental material is available at http://www.genesdev.org.
References
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix–loop–helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix–loop–helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campese AF, Garbe AI, Zhang F, Grassi F, Screpanti I, von Boehmer H. Notch1-dependent lymphomagenesis is assisted by but does not essentially require pre-TCR signaling. Blood. 2006;108:305–310. doi: 10.1182/blood-2006-01-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Ruetsch NR, Berger MA, Rhodes M, Kaptik S, Wiest DL. Signals transduced by CD3ɛ, but not by surface pre-TCR complexes, are able to induce maturation of an early thymic lymphoma in vitro. J Immunol. 1999;163:2576–2585. [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C, Childs ME, Gimotty PA, Aster JC, Pear WS. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zuniga-Pflucker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the αβ and γδ T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes BJ, Robey EA. A reassessment of the effect of activated Notch1 on CD4 and CD8 T cell development. J Immunol. 2002;169:1817–1821. doi: 10.4049/jimmunol.169.4.1817. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling–a structural and biochemical perspective. J Cell Sci. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk MH, Blom B, Nolan G, Stegmann AP, Bakker AQ, Weijer K, Res PC, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: Correlation with the double-negative to double-positive transition. J Immunol. 2003;171:2296–2304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- Huang YH, Li D, Winoto A, Robey EA. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc Natl Acad Sci. 2004;101:4936–4941. doi: 10.1073/pnas.0401133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- Laws AM, Osborne BA. p53 regulates thymic Notch1 activation. Eur J Immunol. 2004;34:726–734. doi: 10.1002/eji.200324772. [DOI] [PubMed] [Google Scholar]
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes & Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205:2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc Natl Acad Sci. 2005;102:8686–8691. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Helix–loop–helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Takeuchi A, Yamasaki S, Takase K, Nakatsu F, Arase H, Onodera M, Saito T. E2A and HEB activate the pre-TCR α promoter during immature T cell development. J Immunol. 2001;167:2157–2163. doi: 10.4049/jimmunol.167.4.2157. [DOI] [PubMed] [Google Scholar]
- Talora C, Campese AF, Bellavia D, Pascucci M, Checquolo S, Groppioni M, Frati L, von Boehmer H, Gulino A, Screpanti I. Pre-TCR-triggered ERK signalling-dependent downregulation of E2A activity in Notch3-induced T-cell lymphoma. EMBO Rep. 2003;4:1067–1072. doi: 10.1038/sj.embor.7400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPT, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORγt, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, et al. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]