Abstract
Histone deacetylases (Hdacs) are transcriptional repressors with crucial roles in mammalian development. Here we provide evidence that Hdac8 specifically controls patterning of the skull by repressing a subset of transcription factors in cranial neural crest cells. Global deletion of Hdac8 in mice leads to perinatal lethality due to skull instability, and this is phenocopied by conditional deletion of Hdac8 in cranial neural crest cells. Hdac8 specifically represses the aberrant expression of homeobox transcription factors such as Otx2 and Lhx1. These findings reveal how the identity and patterning of vertebrate-specific portions of the skull are epigenetically controlled by a histone deacetylase.
Keywords: Histone deacetylation, craniofacial development, birth defects, homeobox, neura crest
Mammalian development is an intrinsically epigenetic process in which a zygote generates hundreds of different cell types, which show vast differences in their phenotype, behavior, and function, despite being genetically identical. How this “epigenetic landscape” (Goldberg et al. 2007) is modulated during development has been under intense study, as it has become clear that many aspects of developmental epigenetics are later recapitulated in disease (Haberland et al. 2009).
Epigenetic modifications, generally defined as the outcome of processes that lead to inheritance that is not reflected in the sequence of the nuclear DNA, are broadly divided into two categories: modifications of the DNA itself by methylation of CpG nucleotides and modifications of histones (Goldberg et al. 2007). Histone modifications are complex and varied, and dozens of enzymes that modify histones have been described. While many histone modifications such as phosphorylation, ubiquitination, sumoylation, propionylation, and butyrylation have been described, the most studied modifications so far are methylation and acetylation of histone tails (Kouzarides 2007). A large body of work has shown that these modifications are intricately linked with transcriptional regulation and epigenetic establishment of cell identity (Bernstein et al. 2007).
Acetylation of histones is mediated by histone acetyltransferases (HATs), which transfer an acetyl group to certain lysines in the tails of the core histones H2A, H2B, H3, and H4. Histone acetylation is generally coupled to transcriptional activation as a consequence of the relaxation of chromatin structure, as well as the creation of binding sites for bromodomain proteins, which function as transcriptional activators (Grunstein 1997). The stimulatory effect of histone acetylation on gene expression is counteracted by histone deacetylases (HDACs), which remove acetyl groups, thereby promoting chromatin compaction (Shahbazian and Grunstein 2007). There are 11 HDAC enzymes encoded in the mammalian genome, which are grouped by similarity and function into four different classes, class I (Hdac1, Hdac2, Hdac3, and Hdac8), IIa (Hdac4, Hdac5, Hdac7, and Hdac9), IIb (Hdac 6 and 10), and IV (Hdac11) (Yang and Seto 2008).
Gene knockout studies of three of the four class I Hdacs (Hdac1, Hdac2, and Hdac3) in mice have shown that these enzymes play specific roles in development. Deletion of Hdac1 leads to demise of the embryo early in embryogenesis (Lagger et al. 2002; Montgomery et al. 2007). Loss of Hdac2, on the other hand, leads to specific cardiac malformations and perinatal lethality (Montgomery et al. 2007; Trivedi et al. 2007). Global deletion of Hdac3 leads to early embryonic lethality, whereas tissue-specific deletion leads to perturbation of metabolic gene programs in heart and liver (Knutson et al. 2008; Montgomery et al. 2008). The in vivo functions of Hdac8 have not been described so far.
The evolution of vertebrates was associated with the invention of a “new head” structure, which allowed the transition from a passive to a more active predatory lifestyle (Gans and Northcutt 1983). The evolution of this “new head” was mainly possible because of another vertebrate invention, a cell type that sometimes has been called the fourth germ cell layer: the neural crest (Le Douarin et al. 2007). Among many other tissues and organs, these cells give rise to most of the mesenchymal components of the vertebrate head such as muscle, cartilage, and bone. Patterning of cranial neural crest cells (NCCs) is achieved by a complex pattern of homeobox transcription factors. Intriguingly, the most anterior domain of cranial NCCs, which serves as the source of progenitors of the anterior skull, does not express any Hox genes (Trainor and Krumlauf 2001; Le Douarin et al. 2007).
Here we show that deletion of Hdac8 in mice results in a highly specific deficiency of cranial NCCs, the loss of specific cranial skeletal elements, and consequent biomechanical instability of the skull, which results in perinatal death due to brain trauma. These severe abnormalities correlate with the derepression of a number of homeobox transcription factors specifically in NCCs, which have been implicated in patterning of the frontal skull. Our results reveal a unique role for Hdac8 in skull development and further demonstrate that each class I HDAC, despite being ubiquitously expressed, possesses highly specific functions in vivo.
Results and Discussion
Gene targeting of Hdac8
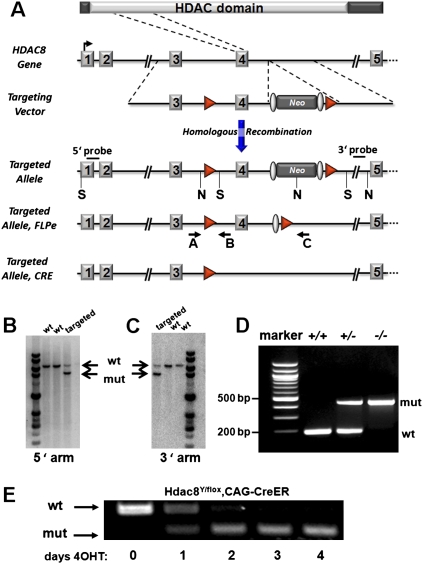
The mouse Hdac8 gene contains 11 exons and spans about 220 kilobases of DNA on the X chromosome. We generated a conditional loss-of-function allele of Hdac8 by introducing loxP sites around exon 4 through homologous recombination in ES cells (Fig. 1A). Deletion of exon 4 eliminates part of the catalytic domain of HDAC8 and creates a frame shift upon fusion of exon 3 to exon 5 (Supplemental Fig. 1). Mice heterozygous for this Hdac8neo-loxP allele were obtained by blastocyst injection and chimera germline transmission and then crossed to hACTB∷FLPe mice to delete the neomycin cassette, yielding the Hdac8loxP allele. We also bred mice with the Hdac8neo-loxP allele to mice expressing a CAG-Cre transgene, which expresses Cre in the germline, resulting in the Hdac8KO allele. Correct targeting and germline transmission was confirmed by Southern blot and PCR of genomic DNA (Fig. 1B–D). Generation of a conditional null allele was confirmed by induced deletion in mouse embryonic fibroblasts using a tamoxifen-inducible ubiquitous Cre (CAG-CreER) (Fig. 1E; Danielian et al. 1998).
Figure 1.
Generation of a conditional Hdac8 allele. (A) Strategy to generate a conditional Hdac8 allele. Protein domains, genomic structure, targeting vector, and targeted allele are shown. Exon 4 was flanked by loxP sites (red triangles), and the neomycin resistance cassette (Neo), flanked by FRT sites (gray ovals) was removed by crossing to FLPe transgenic mice. NcoI (N) and SacI (S) sites used for Southern blotting are indicated. Primers used for genotyping are depicted with arrows. (B,C) Representative Southern blots of genomic DNA showing correct targeting of the Hdac8 locus in ES cells. Genomic DNA was digested with NcoI and SacI and probed with 5′ and 3′ probes shown in A. (D) Genotyping of wild-type, heterozygous and homozygous null alleles from tail DNA by genomic PCR shows germline transmission and functionality of the conditional allele. (E) Deletion of Hdac8 in mouse embryonic fibroblasts using a tamoxifen-inducible Cre. Hdac8 mRNA levels were detected using primers spanning the deleted exon 4 at the indicated days after adding 4-hydroxy-tamoxifen (4OHT).
Global deletion of Hdac8
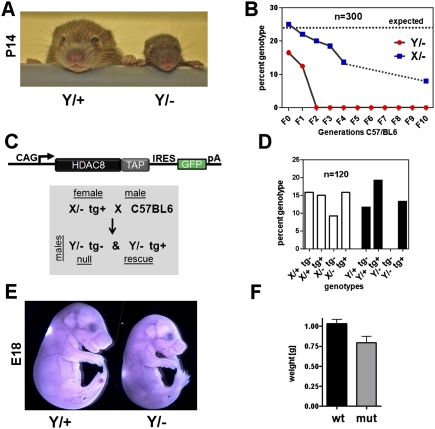
Conditional hemizygous male mice (Hdac8Y/loxP) and homozygous female Hdac8loxP/loxP animals were viable and fertile and obtained at Mendelian ratios. Hemizygous male Hdac8-null mice (Hdac8Y/−) on a mixed genetic background (129/C57BL6/CD1) were obtained at sub-Mendelian ratios at weaning and failed to thrive (Fig. 2A; Supplemental Fig. 2A). We backcrossed the mutants on a C57BL/6 background, and after two generations the observed phenotype became lethal with 100% penetrance at weaning (Fig. 2B). Somewhat surprisingly, heterozygous females were also recovered at sub-Mendelian ratios after backcrossing for several generations. We wondered whether the phenotype in heterozygous mutant female mice might result from random X inactivation of the wild-type allele, creating de facto null tissue in heterozygous females or, alternatively to the inadvertent creation of a dominant-negative allele encoding an N-terminal peptide from exons 1–3. To distinguish between these possibilities, we generated an Hdac8 transgenic model using the ubiquitous CAG (CMV immediate enhancer/β-actin) promoter (Niwa et al. 1991) to drive a bicistronic Hdac8-IRES-eGFP transgene. These mice were viable and without obvious phenotype, and the allele was subsequently bred into the Hdac8-null background (Fig. 2C). Expression of the Hdac8 transgene completely rescued the observed lethality of the hemizygous null animals (Fig. 2D), indicating that the partial lethality of females heterozygous for the Hdac8-null reflects random X-chromosome inactivation of the wild-type Hdac8 allele in a subset of tissues where Hdac8 is required for viability, rather than the generation of a dominant-negative allele.
Figure 2.
Global deletion of Hdac8. (A) Mice with a global deletion of Hdac8 are viable but show a runted phenotype with hypoplastic, dysmorphic skulls. Mice on a mixed genetic background are shown at the age of 2 wk. (B) Global deletion of Hdac8 is lethal on a pure background. Mice were backcrossed for several generations in a C57/BL6 background. No hemizygous null animals (male, Y/−) were found after the first generation of backcrosssing. Mice were genotyped between 14 and 21 d of age. Heterozygous females (X/−) were recovered at sub-Mendelian ratios. (C) Strategy for transgenic rescue experiment. The CAG promoter was used to drive ubiquitous expression of a bicistronic mRNA encoding TAP-tagged Hdac8 and an IRES-linked eGFP. The transgene was crossed into Hdac8 heterozygous females (X/− tg+). These were then crossed with wild-type C57/BL6 males to obtain hemizygous null males, which were either transgene-negative (null) or transgene-positive (rescue). (D) Genotypes obtained in the transgenic rescue experiment. No Hdac8-null animals (Y/− tg−) were recovered, but mice with transgenic re-expression of Hdac8 on the null background (Y/−, tg+) were recovered at expected frequencies. (E,F) Phenotype and weight of mutant animals at E18. Mutant animals were recovered at Mendelian frequencies before birth, although they were smaller and weighed less.
Perinatal lethality in Hdac8 mutant animals
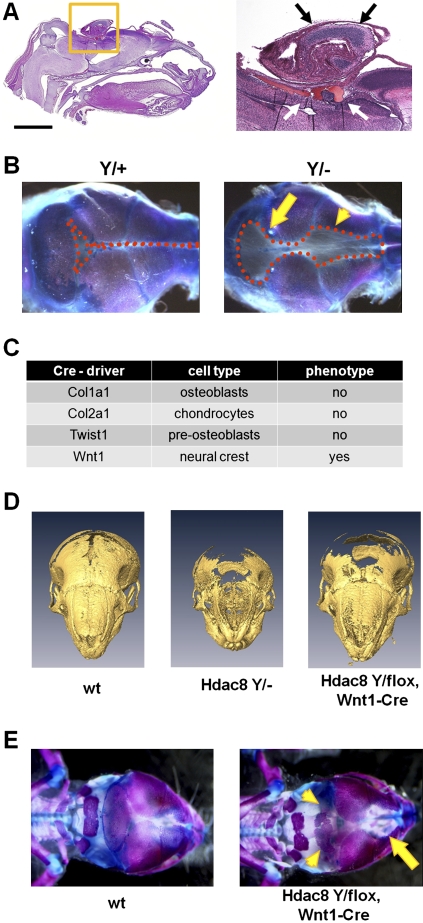
To determine the time of death of Hdac8-deficient mice, we analyzed offspring from a series of timed matings. Mutant animals were obtained at Mendelian ratios during embryogenesis and were viable until embryonic day 18.5 (E18.5), although they showed a small reduction in body size and weight (Fig. 2E,F). Immediately after birth, however, Hdac8-deficient animals showed deficiencies in movement, signs of hypoxia, were often neglected by their mothers and usually died within 4–6 h after birth (Supplemental Fig. 2B). Careful examination of the major organ systems in Hdac8 mutants did not reveal an obvious phenotype in the heart, lung, liver, kidney, intestine, and bladder (data not shown). However, we observed hemorrhage in the brains from mutant animals, and in some severe cases a herniation of brain and other soft tissue through the top of the skull was observed (Fig. 3A). Examination of alizarin red/alcian blue-stained skulls showed a distinct ossification defect with a wide foramen frontale and defects in the interparietal bone (Fig. 3B). No defects were observed in the rest of the skeleton (Supplemental Fig. 3). Corresponding to the genetic background-dependent lethality, the observed skull phenotype ranged from a small persistent anterior fontanelle in Hdac8 mutants on a mixed background to severe frontocranial dysplasia with obvious biomechanical instability in mice backcrossed to a C57BL/6 genetic background (Fig. 3B; Supplemental Fig. 4).
Figure 3.
Skull dysmorphism in Hdac8 mutants. (A) H&E-stained sagittal sections of a P1 skull. The skin was removed for better fixation. Bar, 2 mm. Note the herniation of brain tissue (yellow square and black arrows) and hemorrhage (white arrows). (B) Skeletal prep of P3 mouse skull showing massive ossification defects. The dotted lines demarcate the boundaries of uncalcified soft tissue. Arrows point to the anterior and posterior fontanelle. (C) Tissue-specific deletion of Hdac8 with the indicated Cre deleter strains to determine the causative tissue for the observed phenotype of the global deletion. (D) Three-dimensional reconstruction of micro-CT scans of newborn mouse skulls. Note the severe ossification defects in Hdac8 mutant mice. (E) Neural crest deletion of Hdac8 leads to ossification defects in frontal (arrow) and interparietal bones (arrowheads) in E18 embryos.
Conditional deletion of Hdac8
To further characterize the observed phenotype, we used a variety of Cre drivers for tissue-specific deletion of Hdac8. We initially focused on Cre drivers directing deletion in cells of the skeletal system such as Col1a1-Cre, which deletes in osteoblasts (Dacquin et al. 2002), Col2a1-Cre for deletion in chondrocytes (Ovchinnikov et al. 2000), and Twist1-Cre, which deletes in preosteoblasts (Bialek et al. 2004). Surprisingly, none of these mutants recapitulated the skull phenotype observed with global deletion (Supplemental Fig. 5). As parts of the skull, and especially the frontal and interparietal bones most affected in the global knockout are derived from NCCs, we hypothesized that the skull abnormalities might be due to neural crest patterning defects. We thus used Wnt1-Cre (Echelard et al. 1994) to delete Hdac8 specifically in neural crest-derived tissue. This largely phenocopied the global deletion with ossification defects in frontal and interparietal bones (Fig. 3D,E). We did not observe any defects in other neural crest-derived tissues such as dorsal root ganglia, enteric nervous system, adrenal medulla, or melanocytes (data not shown).
We next tested if the cranial neural crest phenotype was specific for Hdac8. Deletion of conditional alleles for Hdac1 and Hdac2 with Wnt1-Cre deleter mice showed no phenotype in skeletal analysis (Supplemental Fig. 6). We conclude that Hdac8 exerts a unique function in NCCs to control the formation of specific cranial skeletal elements.
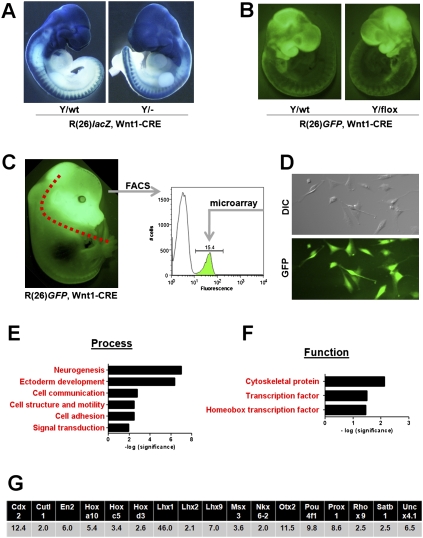
To further delineate the observed phenotype, we performed lineage tracing experiments, using the double-transgenic ROSA26, Wnt1-CRE system crossed into the Hdac8 mutant background to label NCCs (Soriano 1999). In this system, the expression of Cre deletes the Hdac8 allele and at the same time activates a marker gene—i.e., lacZ or GFP—facilitating the visualization of mutant NCCs. Analysis of neural crest migration at various embryonic time points and using different markers did not reveal any major differences in NCC migration between wild-type and Hdac8 mutant animals (Fig. 4A,B; Supplemental Figure 7).
Figure 4.
Analysis of Hdac8-null NCCs. (A) Lineage trace of Hdac8-null NCCs using a double-transgenic ROSA26(lacZ), Wnt1-Cre system. Wnt-1 Cre marks the delaminating NCCs by activating lacZ expression. All the descendant cells are marked blue. (B) Lineage trace of Hdac8 using the ROSA26(GFP), Wnt-Cre system. In this system, NCCs are marked by GFP expression. (C) Strategy for microarray analysis of FACS sorted cranial NCCs. The anterior part of the skull was dissected from E15.5 embryos that expressed GFP in NCCs as indicated. Cells were dissociated and used for FACS analysis. (D) Cranial NCCs in culture. (Top panel) DIC. (Bottom panel) GFP (E,F) Gene ontology analysis of microarray analysis showing molecular process and function of up-regulated genes in Hdac8 mutant cranial NCCs. (G) Fold changes of up-regulated homeobox transcription factors.
Transcriptional profiling of Hdac8-null cranial NCCs
In order to determine the mechanistic basis of the observed phenotype in Hdac8 mutant mice, we sought to purify cranial NCCs for transcriptional profiling. We again used the ROSA26, Wnt1-CRE compound genetic system to label NCCs with eGFP. We then dissected the frontal skull of wild-type and Hdac8 neural crest-deleted animals, dissociated the cells, and purified the eGFP-positive fraction via FACS sorting (Fig. 4C). As the skeletal phenotype is first observed around E16.5 (Supplemental Fig. 8), we chose E15.5 as the time point to detect transcriptional changes. Plating of the purified cells indicated that they were close to 100% pure; i.e., GFP-positive (Fig. 4D). Transcriptional analysis was then performed directly on the sorted cells using Affymetrix microarrays. Using a cutoff of twofold regulation, we found 838 genes (3.9% of interrogated transcripts) up-regulated and 928 (4.3%) down-regulated (Supplemental Table 1).
We compared the dysregulated genes for enrichments in certain groups of molecular function using gene ontology (GO) annotations provided by PANTHER (Thomas et al. 2003). Given that HDACs usually act as transcriptional repressors (and direct targets thus should be derepressed in mutant cells), we focused on up-regulated genes. PANTHER analysis indicated that the most significantly enriched biological process in mutant cells was neurogenesis, whereas cytoskeletal proteins as well as homeobox transcription factors were significantly enriched when up-regulated transcripts were analyzed by biological function (Fig. 4E,F).
Transgenic expression of Otx2 and Lhx1 in NCCs causes cranial dysplasia
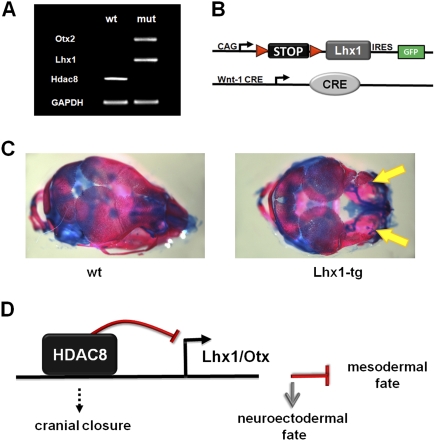
We found 17 homeobox genes up-regulated in the HDAC8-null cranial NCCs (Fig. 4G). To investigate whether the dysregulation of these factors might be causative for the observed phenotype, we attempted to overexpress some of these factors in cranial NCCs. We focused on Otx2 and Lhx1, as genetic loss-of-function analysis in mice had indicated these factors as being crucial for patterning of the anterior skull (Matsuo et al. 1995; Shawlot and Behringer 1995). We validated the dysregulation of these two genes in isolated cranial NCCs by RT–PCR (Fig. 5A). As no promoter driving robust and continuous expression in cranial NCCs has been described so far, we used a conditional transgenic approach for the overexpression of these cDNAs. In this model, the transgene is separated from a ubiquitous promoter (CAG) by a loxP flanked STOP cassette. Upon crossing mice carrying this transgene with Wnt1-CRE-expressing mice, the STOP cassette is deleted leading to continuous expression of the transgene (Fig. 5B). Expression of the transgenes in the different lines can be verified by eGFP expression. Overexpression of both Lhx1 (Fig. 5C) and Otx2 (Supplemetal Fig. 9) caused a severe cranial phenotype with dysmorphism of the frontal skull resembling the Hdac8-null phenotype.
Figure 5.
Dysregulated expression of homeobox transcription factors causes skull dysmorphism. (A) Semiquantitative RT–PCR showing loss of Hdac8 and up-regulation of Otx2 and Lhx1 in NCCs. (B) Strategy for conditional, transgenic overexpression in NCCs. A ubiquitous promoter is separated by a STOP cassette from a bicistronic cDNA-IRES-GFP expression cassette. Wnt1-Cre deletes the STOP cassette, leading to transgenic expression in NCCs. (C) Phenotype of transgenic overexpression of Lhx1 in NCCs. Skeletal prep of a P1 skull is shown. Note the severe skeletal dysmorphism in the transgenic animal (arrows). (D) Model of Hdac8 function in cranial NCCs. Hdac8 represses the expression of several homeobox transcription factors in cranial NCCs, which thus adopt a mesodermal fate. Loss of Hdac8 leads to up-regulation of these factors with cranial NCCs consequently adopting a neuroectodermal fate.
Nonredundant roles for class I HDACs in development
Class I HDACs are thought to be broadly expressed transcriptional repressors that act mainly by deacetylating histones in the nucleus. Although they clearly exist in different repression complexes, no substrate specificity has been demonstrated so far for the different isoforms (Yang and Seto 2008). Recent analyses of class I HDAC mutant mice have shown that global loss of each class I HDAC is invariably lethal. Mice lacking either Hdac1 or Hdac3 die very early in embryogenesis due to proliferation and gastrulation defects (Lagger et al. 2002; Montgomery et al. 2007, 2008; Knutson et al. 2008). Hdac2, on the other hand, is not required for embryogenesis per se, but mutant animals succumb to severe cardiac malformations in the first 24 h after birth (Montgomery et al. 2007). Together with the results of this study, we conclude that the complete loss of any class I HDAC is invariably lethal. Thus, despite sharing a high degree of homology and a ubiquitous expression pattern, individual class I HDACs cannot compensate for each other during mouse development. However, for Hdac1, Hdac2, and Hdac3, no specific cell type has been identified that autonomously mediates lethality; e.g., all the tissue-specific deletions described so far are viable and do not recapitulate the phenotypes of the global deletion. Although Hdac8 will most likely have additional functions in different cell types or organ systems, deletion in NCCs phenocopies the global deletion closely, indicating that a primary function of Hdac8 is to act cell-autonomously within cranial NCCs to control the transcriptional network that mediates the formation of specific cranial skeletal elements.
Epigenetic establishment of cranial NCC identity by Hdac8
NCCs are specified at the neural plate border from which they migrate to form a variety of different cell types and tissues such as neurons and glia cells, medullary secretory cells, smooth muscle cells, and melanocytes, as well as osteoblasts and chondrocytes (LaBonne and Bronner-Fraser 1999). Neural crest-derived skeletogenic cells eventually develop into most of the facial skull. Although the gene regulatory networks that control the initial steps of neural crest induction, epithelial-to-mesenchymal transformation as well as migration have been well established, little is known about the mechanisms that regulate NCC fate determination and differentiation (Sauka-Spengler and Bronner-Fraser 2008). One component of this fate determination cascade is the positional homeobox code along the anterior–posterior axis. NCCs usually express Hox genes equivalent to the rhombomeres from which they migrate; however, the NCCs from which the anterior skull develops are Hox-negative (Trainor and Krumlauf 2001). The caudal domain expresses the homeobox transcription factors Otx2 and Lhx1, whereas this expression is repressed in the rostral domain (Matsuo et al. 1995; Creuzet et al. 2002). The derepression of these genes in the Hdac8 mutant animals suggests that Hdac8 is involved in transcriptional repression in the anterior parts of the skull. Deletion of Hdac8 leads to up-regulation of these genes as well as a variety of other homeobox genes that are normally repressed in cranial NCCs. This presumably leads to mispatterning of the skull elements, possibly by influencing the expression patterns of structural genes, as revealed by microarray analysis (Fig. 4 E–G). Interestingly, Otx2 and Lhx1 are crucially important for the formation of the frontal skull, as deletion of either gene leads to embryos lacking all structures rostral to the otic vesicles despite normal trunk structures (Matsuo et al. 1995; Shawlot and Behringer 1995). This indicates that the expression levels and expression domains of these genes must be exquisitely fine tuned for correct patterning to occur.
Implication for human disease
HDAC inhibitors have been approved recently for the treatment of human disease (Duvic and Vu 2007; Mann et al. 2007), and they have shown benefit in a wide variety of preclinical disease models (Haberland et al. 2009). The data from this and other class I Hdac knockout studies indicate that HDAC inhibitors might be teratogenic, and indeed, this has been demonstrated in rodent models (Menegola et al. 2006). Craniofacial abnormalities have also been described in children born from mothers treated with the HDAC inhibitor valproic acid (Alsdorf and Wyszynski 2005).
Interestingly, Hdac8 has recently been implicated in tumorigenesis of neuroblastoma (Oehme et al. 2009), a common and deadly childhood tumor of neural crest origin (Brodeur 2003). Moreover, high Hdac8 expression is correlated with markers of poor outcome, advanced stage disease, and poor survival in neuroblastoma patients (Oehme et al. 2009). Thus, it is tempting to speculate that low Hdac8 expression, analogous to the results observed in this study, leads to an induction of neuroectodermal markers, and thus to a more differentiated and benign phenotype in neuroblastoma.
Materials and methods
Hdac8 mutant mice were generated by flanking exon 4 with loxP sites using homologous recombination and pGKNEO-F2L2DTA as a targeting vector (Addgene plasmid 13445). Germline transmission was achieved for two of three injected clones, which were kept as independent lines until identical phenotypes could be confirmed for both lines. Otx2 and Lhx1 transgenic expression constructs were generated by using the ubiquitously expressed CAG promoter followed by a loxP-STOP-loxP cassette fused to the murine cDNAs and an IRES-linked eGFP-polyA cassette. The Hdac8 transgenic expression construct was generated by fusing a CAG promoter with a tagged Hdac8 cDNA and an IRES-linked eGFP-polyA cassette. Skeletal preps were stained with alizarin red and alcian blue using standard techniques. μCT analysis was performed using an eXplore Locus scanner and images were rendered using Amira. GFP-based analysis and FACS sorting was performed essentially as described (Singh et al. 2005). Further details and other methods are in the Supplemental Material.
Acknowledgments
Work in the laboratory of E.N.O. was supported by grants from the NIH, the Donald W. Reynolds Foundation for Clinical Cardiovascular Research, the Robert A. Welch Foundation, and the Sandler Foundation for Asthma Research. M.H was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG, HA 3335/2-1).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1809209.
Supplemental material is available at http://www.genesdev.org.
References
- Alsdorf R, Wyszynski DF. Teratogenicity of sodium valproate. Expert Opin Drug Saf. 2005;4:345–353. doi: 10.1517/14740338.4.2.345. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Duvic M, Vu J. Vorinostat: A new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural crest and the origin of vertebrates: A new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Brain Res Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes & Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- Menegola E, Di Renzo F, Broccia ML, Giavini E. Inhibition of histone deacetylase as a new mechanism of teratogenesis. Birth Defects Res C Embryo Today. 2006;78:345–353. doi: 10.1002/bdrc.20082. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von Deimling A, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Singh S, Bhattacherjee V, Mukhopadhyay P, Worth CA, Wellhausen SR, Warner CP, Greene RM, Pisano MM. Fluorescence-activated cell sorting of EGFP-labeled neural crest cells from murine embryonic craniofacial tissue. J Biomed Biotechnol. 2005;2005:232–237. doi: 10.1155/JBB.2005.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol. 2001;13:698–705. doi: 10.1016/s0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 β activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]