Abstract
Cellular differentiation is triggered by extracellular signals that cause target cells to adopt a particular fate. Differentiation in bacteria typically involves autocrine signaling in which all cells in the population produce and respond to the same signal. Here we present evidence for paracrine signaling in bacterial populations—some cells produce a signal to which only certain target cells respond. Biofilm formation in Bacillus involves two centrally important signaling molecules, ComX and surfactin. ComX triggers the production of surfactin. In turn, surfactin causes a subpopulation of cells to produce an extracellular matrix. Cells that produced surfactin were themselves unable to respond to it. Likewise, once surfactin-responsive cells commenced matrix production, they no longer responded to ComX and could not become surfactin producers. Insensitivity to ComX was the consequence of the extracellular matrix as mutant cells unable to make matrix responded to both ComX and surfactin. Our results demonstrate that extracellular signaling was unidirectional, with one subpopulation producing a signal and a different subpopulation responding to it. Paracrine signaling in a bacterial population ensures the maintenance, over generations, of particular cell types even in the presence of molecules that would otherwise cause those cells to differentiate into other cell types.
Keywords: Biofilm, signaling, cell differentiation, B. subtilis, development and quorum sensing
Developmental processes in multicellular organisms result in differentiation and specialization of the constituent cells. Hence, the progression of pluripotent stem cells through differentiation steps that define distinct coexisting cell types has been considered a defining feature of metazoans (Orkin and Zon 2008; Rossant 2008). In contrast, bacteria are often thought of as unable to differentiate into distinct coexisting cell types. Yet, bacteria can develop into morphologically complex multicellular communities composed of different subpopulations of specialized cell types (Aguilar et al. 2007). Examples of bacterial multicellularity are provided by the myxobacteria, which form elaborate fruiting bodies within which the specialized spore cells reside (Kaiser 2008), and the streptomycetes, which form complex colonies with substrate and aerial mycelia similar in appearance to some fungi (Flardh and Buttner 2009).
Surface-associated communities known as biofilms are another example of multicellularity in bacteria (Kolter and Greenberg 2006). The soil-dwelling bacterium Bacillus subtilis forms biofilms in which the constituent cells are encased in an extracellular matrix (Branda et al. 2005). These biofilms contain multiple coexisting cell types. For example, in a mature biofilm, some cells synthesize an extracellular matrix, while other cells are motile, and yet others go on to become spores (Vlamakis et al. 2008). The occurrence and participation of those different cell types in the process of biofilm development might be expected to require coordinating the physiologies of the different cell types through cell–cell communication (Lopez et al. 2009b).
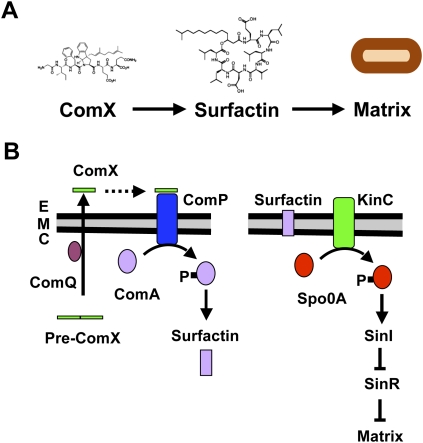
Cell–cell communication in bacteria is generally driven by the production and secretion of specific signaling molecules that can trigger diverse programs of cellular differentiation (Camilli and Bassler 2006). In the case of B. subtilis, much is known about the molecular mechanisms of several secreted signaling molecules that activate specific differentiation pathways. Germane to the studies presented here, the prenylated peptide ComX is secreted and from the outside of the cell activates the membrane histidine kinase ComP (Fig. 1). ComP in turn phosphorylates the transcriptional factor ComA, resulting in the transcription of a regulon that includes the srf operon, responsible for the synthesis of a surfactant, the cyclic lipopeptide surfactin (Dubnau 1991; Magnuson et al. 1994). Recently, surfactin was shown to function as an extracellular signaling molecule as well as a surfactant (Fig. 1; Lopez et al. 2009a). Surfactin indirectly activates the membrane histidine kinase KinC, which phosphorylates the master regulator Spo0A, eventually leading to the production of the biofilm's extracellular matrix via the SinI–SinR pathway (Kearns et al. 2005; Lopez et al. 2009a). While recent studies revealed that only a subpopulation of cells within a biofilm are matrix producers (Chai et al. 2008; Vlamakis et al. 2008), the dynamics of when and how ComX-producing cells trigger surfactin production and how surfactin triggers matrix production had not been explored. Here we present the finding that, while most cells in the population appear able to make ComX, only a small subset of the cells respond to the signal by becoming surfactin producers. Surprisingly, surfactin producers themselves do not go on to become matrix producers. Rather, a different subpopulation responds to surfactin. Strikingly, those matrix-producing cells become immune to the ComX signal and thus do not become surfactin producers themselves. The immunity to ComX is due to the presence of the extracellular matrix itself. Thus, surfactin is a paracrine signal that triggers other cells to produce matrix while preventing those cells from producing surfactin.
Figure 1.
Signaling involved in extracellular matrix formation. (A) Flow scheme of the signaling cascade that leads to the production of the biofilm extracellular matrix in B. subtilis. The pheromone ComX is required for the production of surfactin. Surfactin in turn triggers the production of the extracellular matrix. Structures of the signaling molecules are shown. (B) Schematic representation of the respective signal transduction pathways leading to the production of surfactin (left) and the extracellular matrix (right). (Left) The quorum-sensing pheromone ComX triggers the production of surfactin by phosphorylation of the master regulator ComA, via activation of the membrane kinase ComP. (Right) The quorum-sensing molecule surfactin induces the phosphorylation of Spo0A via activation of the membrane kinase KinC. Spo0A induces the expression of SinI, which antagonizes the repressor SinR and causes derepression of genes involved in matrix synthesis.
Results
Surfactin-producing and matrix-producing cells are different subpopulations
We recently identified surfactin as a quorum-sensing molecule that induces a subpopulation of B. subtilis cells within a developing biofilm to become matrix producers (Lopez et al. 2009a). We were thus interested in studying the production of surfactin vis-à-vis the appearance of matrix-producing cells. Given that surfactin production is induced, in part, in response to ComX production, we decided to focus our attention on following the dynamics of subpopulations of cells that produced ComX, surfactin, and matrix within a growing biofilm.
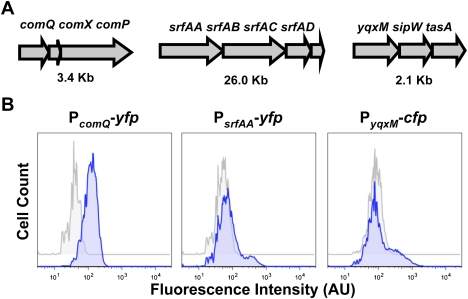
To identify individual cells producing ComX, surfactin, and matrix, we chose to monitor gene expression instead of directly assaying for the molecules, which are all extracellular and may not localize near the cells that produced them. Therefore, to most accurately monitor producing cells, we generated transcriptional fusions of promoters of relevant operons to genes encoding fluorescent proteins of different colors. Figure 2A shows three operons—comQXP, srfAA-AD, and yqxM-sipW-tasA—whose gene products are responsible for ComX, surfactin, and matrix protein (TasA) production, respectively (Nakano et al. 1991; Magnuson et al. 1994; Branda et al. 2004). We constructed strains harboring the transcriptional fusions PcomQ-yfp, PsrfAA-yfp, and PyqxM-cfp and determined the fluorescent cell populations in developing biofilms using flow cytometry (see Materials and Methods). We performed flow cytometry analyses of these three strains 72 h into biofilm development, when the biofilm has been shown to already contain several coexisting cell types (Vlamakis et al. 2008). The results are shown in Figure 2B. We assume for simplicity in what follows that cells expressing PcomQ-yfp, PsrfAA-yfp, or PyqxM-cfp were producing ComX, surfactin, or TasA, respectively, although it is possible that not all cells expressing a reporter may have been producing the corresponding peptide or protein.
Figure 2.
Expression of the operons responsible for the production of ComX, surfactin, and the matrix protein TasA. (A) Schematic representation of the three operons involved. Genes within each operon are drawn to scale. The overall size of each operon is indicated below. (B) Flow cytometry analyses to monitor the expression of each operon in developing biofilms using transcriptional fusions to reporter genes encoding fluorescent proteins. Colonies grown on MSgg for 72 h showed maximum heterogeneity in cell differentiation. (Left panel) In these conditions, expression of comX occurs in all the cells of the population, according to the shift observed in the expression profile of the strain labeled with the reporter PcomQ-cfp. Cells with no reporter construct were used as negative controls (gray peak in all panels). The expression of surfactin synthesis genes is limited to a small subpopulation of cells. (Center panel) The strain labeled with PsrfAA-yfp showed a shoulder of cells expressing the reporter at higher level than the background. The expression of the yqxM-sipW-tasA operon is limited to a larger subpopulation of cells. (Right panel) The profile of the strain labeled with the reporter PyqxM-cfp also showed a shoulder of cells expressing the reporter at a higher level than the background. Fluorescence intensity was measured in arbitrary units (AU).
Analysis of cells harboring the PcomQ-yfp reporter demonstrated that at the time of sampling, the distribution of cells expressing fluorescence was unimodal (Fig. 2B, left panel). Thus, most of the cells in the biofilm were expressing the genes involved in ComX production to a greater or lesser extent. In contrast, cells harboring PsrfAA-yfp expressed fluorescence in a bimodal pattern, with most cells exhibiting little or no expression of the reporter and a subpopulation of cells exhibiting high fluorescence (Fig. 2B, center panel). [The bimodal expression of the surfactin operon observed in B. subtilis strain 3610 used here is not observed in the domesticated laboratory strain 168. See Supplemental Figure S1.] This result showed that only a small fraction of the population responded to ComX by producing surfactin (or at least expressing the PsrfAA-yfp reporter). In turn, surfactin was sensed and responded to by expression of the matrix protein operon in only a subpopulation of cells because when we monitored the PyqxM-cfp reporter, we observed that the distribution of fluorescent cells was also bimodal (Fig. 2B, right panel).
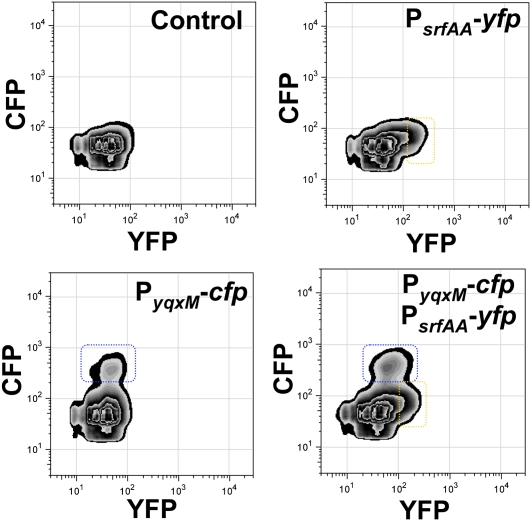
The significant difference between the percent of the population expressing the reporter for the surfactin operon (∼10%) compared with the percent expressing the reporter for matrix production operon (∼35%) led us to hypothesize that the processes might not be occurring in the same cells. To test this hypothesis, we double-labeled a strain with the transcriptional reporters PsrfAA-yfp and PyqxM-cfp, and we monitored YFP and CFP fluorescence simultaneously. Cells were harvested after 72 h of growth on biofilm-inducing medium. The results were plotted in graphs in which YFP fluorescence was on the X-axis and CFP fluorescence was on the Y-axis. The number of cells expressing different levels of fluorescence was then plotted on a Z-axis perpendicular to the plane of the X- and Y-axes, as shown by the isolines (Fig. 3). The top left panel of Figure 3 shows the control of background fluorescence for both YFP and CFP in a strain harboring no fluorescent protein genes. We first distinguished each subpopulation of cells (surfactin-producing and matrix-producing cells) individually in each channel (YFP and CFP, respectively), using single-labeled strains as controls (PsrfAA-yfp and PyqxM-cfp). Indeed, the single-labeled control strains showed a subpopulation of ∼12% of the cells expressing surfactin (Fig. 3, top right panel, dashed yellow rectangle) and ∼33% of the cells expressing matrix genes (Fig. 3, bottom left panel, dashed blue rectangle). Importantly, when the double-labeled strain PsrfAA-yfp, PyqxM-cfp was monitored, both subpopulations still appeared as separated peaks of isolines in each channel with little signal overlap (Fig. 3, bottom right panel, dashed blue and yellow rectangles). These results indicate that the surfactin-producing cells and matrix-producing cells are distinct subpopulations.
Figure 3.
Cells expressing surfactin and matrix reporters are two different subpopulations. Flow cytometry monitoring the expression of the reporter PsrfAA-yfp (YFP fluorescence on the X-axis) and PyqxM-cfp (CFP fluorescence on the Y-axis) from 72-h colonies grown on MSgg medium. The number of cells is represented by isolines on the Z-axis coming out of the plane of the paper. The top left panel shows the control of background fluorescence for both CPF and YFP in a strain harboring no fluorescent protein genes. (Top right panel) The strain harboring PsrfAA-yfp showed a subpopulation of cells (framed in yellow) highly expressing the reporter. (Bottom left panel) The strain harboring PyqxM-cfp showed a subpopulation of cells (framed in blue) highly expressing the reporter. (Bottom right panel) The strain harboring both reporters, PsrfAA-yfp and PyqxM-cfp, showed two subpopulations of highly expressing cells. Each population is framed, showing there is no overlap.
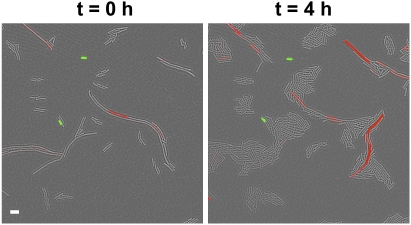
Direct microscopic observation of cells corroborated the flow cytometry results. We disrupted developing biofilms of the double-labeled strain (PsrfAA-yfp, PyqxM-cfp) after 72 h of growth and observed individual cells using light and fluorescence microscopy. This allowed us to determine the fate of numerous individual cells. As expected from the results shown in Figure 3, most cells were not fluorescent. But, among the fluorescent cells, we never observed a cell expressing both YFP and CFP. These cell fates did not change even after several hours of following individual cells. A representative field of view followed over a subsequent 4 h is shown in Figure 4. Fluorescent cells were falsely colored green if they were expressing YFP (surfacing producers) and red if they were expressing CFP (matrix producers). Superimposing images captured for both fluorescence wavelengths onto the same field captured in light microscopy yielded only green and red fluorescent cells, along with nonfluorescent cells. We never observed yellow cells, which would have been indicative of cells expressing both fluorescent proteins. These microscopic observations supported our flow cytometry data and confirmed that surfactin-producing and matrix-producing cells are two different subpopulations within developing biofilms. This finding, coupled to the fact that surfactin is itself required for the induction of matrix production, led us to conclude that cell–cell signaling is unidirectional; cells producing surfactin do not appear able to respond to it. Rather, it is a subpopulation from among the nonsurfactin producers, which senses and responds to this signaling molecule. It is interesting to note that after 4 h of observation, a time period during which the nonfluorescent cells and the matrix producers underwent several cell divisions, the surfactin-producing cells did not grow nor divide. The mechanisms that render surfactin producers insensitive to surfactin and unable to grow remain unknown.
Figure 4.
Subpopulations of surfactin and matrix producers are distinct under the microscope. Micrographs taken 4 h apart of the same field of cells of the strain harboring both reporter fusions, PsrfAA-yfp and PyqxM-cfp, growing in MSgg medium. The image is an overlay of fluorescence and transmitted light images. Colonies were grown for 72 h, and individual cells were separated by mild sonication and placed on a thin MSgg agar layer on a microscope slide and covered with a coverslip to allow observation of individual cells for several hours. The two subpopulations of cells expressing PsrfAA-yfp (artificially colored green) and PyqxM-cfp (artificially colored red) were distinct and did not overlap. Bar, 6 μm.
Extracellular matrix prevents cells from becoming surfactin producers
Thus far we have presented results consistent with the following scenario: In a developing biofilm, many cells produce ComX (or at least express the operon responsible for its production). Only a subpopulation responds to ComX and differentiates into surfactin producers (or at least into cells that express the operon for surfactin production). Surfactin producers themselves do not respond to surfactin, but other cells do respond to it and become matrix producers. Once surfactin producers and matrix producers are generated, they remain stably differentiated for prolonged periods—individual cells simultaneously expressing both surfactin and matrix synthesis genes were never observed.
The question immediately arises as to the reason why, with the passage of time, the matrix producers (as well as the other cells) do not become surfactin producers. If ComX is made by many cells, then it stands to reason that eventually more cells should sense ComX and respond to it, differentiating into surfactin producers. Yet, this did not occur. The hallmark that distinguishes the subpopulation of matrix producers is matrix itself. This prompted us to explore whether the extracellular matrix might be involved in preventing ComX action on matrix producers during biofilm development.
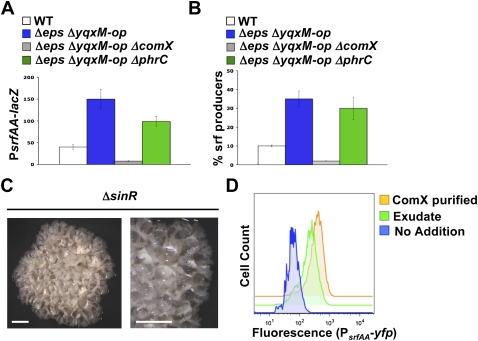
We began by monitoring the levels of surfactin gene expression as a reporter for the activation of ComX in the presence or absence of the extracellular matrix. We constructed a lacZ transcriptional fusion to the surfactin operon promoter (PsrfAA-lacZ) and assayed its level of expression in different genetic backgrounds (Fig. 5A). The observed levels of transcription were increased more than threefold in the mutant lacking matrix (Δeps ΔyqxM-op) compared with the wild type. [The designation ΔyqxM-op is used to indicate deletion of the entire yqxM-sipW-tasA operon.] Transcription was abolished, as expected, when a ΔcomX mutation was introduced into the Δeps ΔyqxM-op strain. Mutating a second extracellular signal that modulates surfactin gene expression, CSF (competence and sporulation stimulating factor) (Lazazzera et al. 1997), only slightly reduced the elevated expression levels observed in the matrix mutant, indicating that most of the effect was due to increased ComX signaling. Control experiments monitoring the expression of the genes responsible for ComX and CSF synthesis indicated that the observed increase in surfactin gene expression was not due to a corresponding increase in the synthesis of the signaling molecules in the mutant strains (Supplemental Fig. S2).
Figure 5.
Extracellular matrix represses expression of surfactin by blocking the action of the ComX pheromone. (A) β-Galactosidase assays monitoring the expression of a surfactin reporter fusion (PsrfAA-lacZ) in various mutant backgrounds. The absence of extracellular matrix (Δeps ΔyqxM-op) increased the expression of the reporter. The increase was abrogated by the absence of the pheromone ComX (ΔcomX), but not the signaling peptide CSF (ΔphrC). (B) Flow cytometry monitoring the subpopulation of surfactin producers as reported by the PsrfAA-yfp fusion. The absence of extracellular matrix caused an increase in the subpopulation of surfactin producers. The increase is eliminated by the absence of the pheromone ComX (ΔcomX), but not the signaling peptide CSF (ΔphrC). (C) Hyperwrinkled colony morphology of the ΔsinR mutant. The overproduction of extracellular matrix showed consequent hyperwrinkling and accumulation of droplets of an aqueous exudate (detailed picture on the right). Image taken after 3 d of growth on MSgg agar. Bars, 1 mm. (D) Flow cytometry of the tester strain (168 ΔcomQXP, amyE∷comP, lacA∷PsrfAA-yfp) to assay for ComX activity in the droplets produced by the ΔsinR mutant. The tester strain responded to the exudate in a similar fashion as when using purified ComX.
In the wild-type strain, only a subpopulation of cells expresses the surfactin genes (see Fig. 2B). Thus, the increase in surfactin gene expression observed in the absence of matrix using the lacZ fusion could either be due to an increase in expression per cell in the same subpopulation of cells or due to an increase in the number of cells expressing the surfactin genes. Flow cytometric analyses of cells expressing PsrfAA-yfp demonstrated that there was an increase in the number of cells expressing surfactin genes in the absence of matrix (Fig. 5B). We reasoned that the matrix might be the reason why the laboratory strain, unable to produce matrix, has a large subpopulation of surfactin-expressing cells. Indeed, artificial production of matrix in this strain inhibited surfactin expression to levels comparable with the undomesticated strain (Supplemental Fig. S3). The foregoing results are thus all consistent with the hypothesis that the matrix is somehow interfering with the ability of ComX to activate ComP.
The production of active ComX involves not only transcription of the structural genes, but also processing of the peptide and its export from the cell. The possibility thus remained that in the presence of matrix, the processing and/or export of ComX were somehow reduced, and elimination of matrix production led to more ComX being produced. To test this possibility, we exploited the phenotype of mutants lacking the master regulator of matrix gene expression, SinR. As described in Figure 1B, SinR functions to repress matrix gene expression, and SinR mutant strains therefore express matrix genes constitutively, resulting in a hyperwrinkled colony morphology (Fig. 5C). Consistent with the matrix interfering with surfactin gene expression, SinR mutants did not detectably produce surfactin (Supplemental Fig. S4); because all of the cells produce matrix in these mutant strains, ComX signaling is always blocked. Close inspection of these ΔsinR colonies revealed droplets of liquid exudate (Fig. 5C). We collected this exudate and assayed it for ComX activity. Using a PsrfAA-yfp fusion as a reporter of ComX activity in a strain background that is unable to make ComX but is able to respond to it, we were able to detect active ComX in the exudate (Fig. 5D). This strain depends on exogenous ComX to activate ComP and transcribe the surfactin genes. Thus, even when all the cells were making matrix, functional ComX was being synthesized. These results indicate that, rather than interfering with ComX production, the matrix somehow interferes with ComX's ability to activate ComP.
Discussion
Here we addressed the question of how different cell types arise and coexist within a multicellular community. Two prior sets of observations served as a starting point for the experiments presented here. First, several extracellular signals that serve to determine specific cell fates have been characterized (Pottathil and Lazazzera 2003). Of direct relevance to our work, ComX is required for the generation of surfactin-producing cells (Magnuson et al. 1994), and surfactin is involved in the production of cells that synthesize an extracellular matrix (Lopez et al. 2009a). Second, in a developing biofilm, different cell types coexist and display a high degree of spatiotemporal organization (Vlamakis et al. 2008). These observations led us to investigate which cells in a biofilm make ComX, surfactin, and matrix, using as surrogates fluorescent reporters for the expression of operons involved in the production of the two signaling molecules and the matrix protein TasA. We found that while many cells expressed the genes for ComX production, only a subpopulation made surfactin, and another, different subpopulation made matrix, and that these two subpopulations were able to coexist. These two cell types do not comprise the entire population; we presume that the rest of the cells that do not differentiate as surfactin producers or responders probably represent other cell types that B. subtilis is known to differentiate into (Veening et al. 2008; Lopez et al. 2009b). Ultimately, each differentiation process sets the stage for a subsequent cell type. For example, in biofilms, it has been shown that matrix-producing cells are the predominant cells that differentiate to become spores (Vlamakis et al. 2008). Additionally, surfactin gene expression is a prerequisite for competence (D'Souza et al. 1994). Therefore, differentiation of cells early in biofilm development could serve to dictate what the final population constituents are. Several questions arise regarding the mechanisms that underlie the fact that only subpopulations respond to the extracellular signals. We discuss these in the following paragraphs.
That only a subpopulation of cells initially responds to ComX and begins transcribing the surfactin synthesis genes might be due to stochasticity and a positive feedback loop that could give rise to a bimodal population (Dubnau and Losick 2006). ComX is only one component of a complex regulatory circuitry that controls surfactin gene expression (Lazazzera et al. 1999). When an individual cell senses ComX via ComP activation, ComA can be phosphorylated and the surfactin genes are transcribed (see Fig. 1). ComA∼P also activates transcription of the gene (phrC) for a second extracellular signaling molecule (CSF) that, at least at low concentrations, has an autostimulatory effect on ComA∼P (Lazazzera et al. 1997). Thus, the first cells to sense ComX could quickly accumulate a level of ComA∼P above a threshold and differentiate into surfactin producers.
Bacterial cell–cell communication via extracellular signals is most often referred to by the general term of “quorum sensing” (Fuqua et al. 1994; Camilli and Bassler 2006). This term was initially adopted because bacterial extracellular signaling was first discovered as the phenomenon of “autoinduction” wherein cells could use a self-generated signal to induce changes in gene expression (Nealson et al. 1970). These signaling mechanisms were thus seen as a way for cells to measure their population density. As quorum-sensing mechanisms have been investigated in more detail, it has become apparent that the extracellular signaling can involve much more than just sensing population density. For example, by having multiple extracellular signaling molecules where the synthesis of one depends on the prior synthesis of the other, Pseudomonas aeruginosa appears to convert its quorum-sensing systems into a timing device (Schuster et al. 2003). Quite recently, the production of one extracellular signal, p-coumaroyl-homoserinelactone by Rhodopseudomonas palustris, was shown to depend on coumaric acid being present in the medium (Schaefer et al. 2008). Thus, quorum sensing in this case is directly connected to the substrate present in the environment. Yet, in all these cases of quorum sensing, it has been generally assumed that all of the cells in a population are physiologically similar and thus able to produce the signal and to respond to it. In this sense, quorum sensing is an example of autocrine signaling.
Here we present evidence for unidirectional or paracrine extracellular signaling in a population of genetically identical bacteria. The subpopulation producing surfactin does not respond to the molecule; the responders are a different subpopulation. An indication that quorum sensing can involve directed signaling was provided in the discovery of a pentapeptide from Escherichia coli that triggers programmed cell death (Kolodkin-Gal et al. 2007). That signaling peptide was assumed to be the product of dead or dying cells but the subpopulation of producer cells was not directly identified as physiologically different from the responding cells.
In order to have unidirectional extracellular signaling and different cell types coexisting for prolonged periods, cells must have a way to develop selective immunity to the extracellular signals. We showed here that in a developing biofilm, B. subtilis cells that are producing surfactin develop immunity to surfactin and, similarly, matrix-producing cells develop immunity to ComX. In the following paragraphs we discuss possible mechanisms that might mediate these two distinct immunity processes.
We do not know how surfactin producers become immune to the action of the lipoprotein signaling molecule, but we speculate that the mechanism involves a protein called ComS. ComS is the product of a gene located within the srf operon and hence the gene is transcribed in all surfactin-producing cells (D'Souza et al. 1994). ComS triggers the entry of cells into a nondividing and stress-resistant state known as the “K-state” (Berka et al. 2002; Maamar and Dubnau 2005; Leisner et al. 2007). It does so by effecting the release of the activator for genes expressed in the K-state, ComK, from a second protein called MecA (Turgay et al. 1998). MecA is an adaptor protein that causes ComK to be degraded. Hence, the ComS-induced release of ComK from MecA allows ComK to accumulate. In addition, however, the gene for ComK is subject to a mechanism of bistable control that ensures that only a subset of cells accumulate high levels of the regulatory protein and enter the K-state (Suel et al. 2006). Thus, the K-state is not likely the basis for the immunity of surfactin producers to surfactin action; only a minority of surfactin producers is in the K-state. Instead, we suspect that ComS—via its interaction with MecA—might be involved in inactivating Spo0A, which is needed to respond to surfactin (see Fig. 1B; Lopez et al. 2009a). An important recent finding consistent with this possibility is the discovery that MecA binds Spo0A (M Defrancesco, P Prepiak, and D Dubnau, pers. comm.). Thus, by freeing MecA from ComK, ComS might allow MecA to bind to and inhibit Spo0A. While much remains to be done before we understand the molecular basis for the surfactin immunity of surfactin-producing cells, it will be interesting to see whether ComS plays a dual role in triggering the K-state and blocking responsiveness to the lipoprotein.
In contrast to the case of immunity to surfactin, the molecular mechanism whereby matrix-producing cells become immune to ComX and thus do not become surfactin producers appears to be clear. In that case, we were able to show directly that the presence of the extracellular matrix was responsible for preventing the ComX-dependent activation of the ComP histidine kinase. The extracellular matrix did not affect the synthesis of ComX, so the simplest explanation for the observed effect is that the interaction between ComX and ComP is somehow inhibited. This might result from physical occlusion of the extracellular sensing domain of ComP. However, since the precise molecular details of the ComX–ComP interaction have not been completely elucidated, any discussion of how the matrix might physically interfere is purely speculative. Nonetheless, from the images provided in Figure 4, the fact that none of the cells under observation became surfactin producers suggests that once matrix begins to be synthesized by some cells, it is able to block ComX signaling even in the nonmatrix producers. Thus, matrix serves not only to hold all of these cell types together but also as a timing mechanism. Once cells begin to make matrix as the result of surfactin signaling, no new surfactin producers are generated. Remarkably, similar to what we propose here, blockage of signaling molecules caused by extracellular matrix has been reported in eukaryotes to define the distinct cell fates during morphogenesis. For instance, Drosophila melanogaster requires specific signaling molecules to dictate cell fate during embryonic development. Although these signaling molecules are uniformly expressed, they are heterogeneously redistributed in a concentration gradient, which defines polarity during development of the embryo. This unequal distribution of the signals relies on interaction of the signaling molecules with type IV collagen, a major component of the extracellular matrix, and is critical for proper Drosophila development (Wang et al. 2008).
The results presented in this study provide additional support to the growing body of evidence indicating that bacteria often display attributes of multicellular organisms (Shapiro 1998; Aguilar et al. 2007). Clearly, the large amount of work on B. subtilis differentiation that has been previously published made it possible to follow the different cell types as described here. It will be extremely interesting to determine if similar mechanisms to induce and maintain different cell types are a common feature of bacteria during biofilm formation.
Materials and methods
Strains, media, culture conditions, strain construction, and reporters
B. subtilis NCIB3610 and B. subtilis 168 and their derivatives were used throughout this study and are listed in Supplemental Table S1. B. subtilis and E. coli were routinely grown on LB medium. When necessary, antibiotics were used at the following concentrations: MLS (1 μg mL−1 erythromycin, 25 μg mL−1 lincomycin); spectinomycin (100 μg mL−1); tetracycline (10 μg mL−1); kanamycin (10 μg mL−1). For biofilm formation experiments 3 μL of a cell suspension was spotted onto minimal MSgg medium supplemented with 1.5% Bacto agar and incubated at 30°C for 3 d (Branda et al. 2001).
Deletion mutants were generated using long flanking homology PCR (Wach 1996) (using primers listed in Supplemental Table S2). Transcriptional fusions PsrfAA-yfp, PyqxM-cfp, and PcomQ-yfp and the cassette PcomQ-comP in the tester strain were inserted by double recombination into neutral integration sites (amyE or lacA) in the genome of B. subtilis by inducing natural competence (Hardwood and Cutting 1990). The vectors used to construct the transcriptional fusions were pKM008 for fusions expressing yfp and pKM003 for constructions expressing cfp for the insertions in the amyE locus, kindly provided by Professor D.Z. Rudner (Harvard Medical School), and pDR183 for insertions in lacA locus (Doan et al. 2005). Transcriptional fusions to lacZ, PcomQ-lacZ, and Pcsf-lacZ were cloned in pDG1663 vector using the primers listed in Supplemental Table S2 (Guerout-Fleury et al. 1996) and inserted into the amyE locus by natural competence. Constructs were transferred to NCIB3610 when required by SPP1 phage transduction (Yasbin and Young 1974), and β-galactosidase assays were performed according to Branda et al. (2006).
Image capture and analysis
Colonies were photographed using a Zeiss Stemi SV6 Stereoscope connected to a color AxioCam. Microscopy images were taken on a Nikon Eclipse TE2000-U microscope with X-cite 120 illumination system attached, using a Hamamatsu digital camera model ORCA-ER. YFP/CFP fluorescence signal was taken using a dual-band filter set (Ex436/500). The exposure times were 800 and 300 msec, respectively. Image processing was done using MetaMorph Software and Photoshop.
Flow cytometry
Biofilms were harvested, centrifuged, and resuspended in 1 mL of PBS buffer and dispersed with three pulses of mild sonication. Cells were fixed in 4% paraformaldehyde for 7 min. After fixation, cells were washed with PBS, resuspended in GTE buffer (50 mM Glucose, 10 mM EDTA at pH 8, 20 mM Tris-HCl at pH 8), and stored at 4°C for further analysis. Mild sonication of the samples was required to obtain single cells prior to flow cytometry (Branda et al. 2006).
For flow cytometric analysis, cells were diluted 1:100 in PBS and measured on a BD LSR II flow cytometer (BD Biosciences). For YFP fluorescence, we used a laser excitation of 488 nm coupled with 530/30 and 505LP sequential filters. For CFP fluorescence, we used laser excitation at 405 nm coupled with 408/40 and 460LP sequential filters. The photomultiplier voltage was set between 400 and 500 V. Every sample was analyzed measuring 50,000 events using FACS Diva (BD Biosciences) software to capture the data. Further analysis was performed using FlowJo 8.7.2 software (http://www.flowjo.com).
ComX purification
Partial purification was performed as described previously (Magnuson et al. 1994). The supernatant of 500 mL of culture in MC medium grown overnight was loaded into a C-18 Sep-Pak cartridge (Waters) previously stabilized with aqueous 20% CH3CN and 0.1% trifluoroacetic acid. ComX was eluted from the cartridge with 10 mL of aqueous 50% CH3CN and 0.1% trifluoroacetic acid and neutralized with ammonium.
Acknowledgments
We thank the core facilities of the Harvard FAS Center for Systems Biology and P. Rogers and B. Tilton for their expert guidance in flow cytometry. We thank D. Dubnau for helpful discussions and critical reading of this manuscript. We thank A. Earl, Y. Chai, F. Chu, A. McLoon, D. Romero, and C. Aguilar for helpful discussions. This work was funded by NIH grants GM58213 to R.K. and GM18568 to R.L. D.L. was the recipient of a post-doctoral fellowship from the Fundación Séneca, Comunidad Autónoma de la Región de Murcia (Spain).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1813709.
Supplemental material is available at http://www.genesdev.org.
References
- Aguilar C, Vlamakis H, Losick R, Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: Genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- D'Souza C, Nakano MM, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Flardh K, Buttner MJ. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: The LuxR–LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier NP. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hardwood CR, Cutting SM. Wiley; New York: 1990. Molecular biological methods for Bacillus. [Google Scholar]
- Kaiser D. Myxococcus—From single-cell polarity to complex multicellular patterns. Annu Rev Genet. 2008;42:109–130. doi: 10.1146/annurev.genet.42.110807.091615. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Lazazzera BA, Kurtser IG, McQuade RS, Grossman AD. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J Bacteriol. 1999;181:5193–5200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a ‘switching-window’ into the K-state of Bacillus subtilis. Mol Microbiol. 2007;63:1806–1816. doi: 10.1111/j.1365-2958.2007.05628.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci. 2009a;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009b;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Magnuson R, Myers A, Curry J, Grossman AD, Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991;173:1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottathil M, Lazazzera BA. The extracellular Phr peptide–Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci. 2003;8:d32–d45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes & Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]