Here, the crystal structure of an endoglucanase, Cel9A, from Alicyclobacillus acidocaldarius (Aa_Cel9A) is reported which displays a modular architecture composed of an N-terminal Ig-like domain connected to the catalytic domain. This paper describes the overall structure and the detailed contacts between the two modules.
Keywords: endoglucanases, thermoacidophiles, cellulases, biofuels
Abstract
The production of biofuels using biomass is an alternative route to support the growing global demand for energy and to also reduce the environmental problems caused by the burning of fossil fuels. Cellulases are likely to play an important role in the degradation of biomass and the production of sugars for subsequent fermentation to fuel. Here, the crystal structure of an endoglucanase, Cel9A, from Alicyclobacillus acidocaldarius (Aa_Cel9A) is reported which displays a modular architecture composed of an N-terminal Ig-like domain connected to the catalytic domain. This paper describes the overall structure and the detailed contacts between the two modules. Analysis suggests that the interaction involving the residues Gln13 (from the Ig-like module) and Phe439 (from the catalytic module) is important in maintaining the correct conformation of the catalytic module required for protein activity. Moreover, the Aa_Cel9A structure shows three metal-binding sites that are associated with the thermostability and/or substrate affinity of the enzyme.
1. Introduction
The next generation of biofuels will use cellulose derived from tailored crops as a source of fermentable sugars to produce fuels such as ethanol. The sugars present in this kind of biomass are located in the cell walls of plants, which are composed of lignin, hemicelluloses and cellulose (Parsiegla et al., 2008 ▶). Plants produce about 180 billion tons of cellulose per year globally, making this polysaccharide the largest organic carbon reservoir on earth (Festucci-Buselli et al., 2007 ▶). An efficient breakdown of cellulosic biomass is a prerequisite for the production of biofuels and cellulases are key enzymes in this process. The β-1,4-glucanase (EC 3.2.1.4) from Alicyclobacillus acidocaldarius (Aa_Cel9A), a thermoacidophilic Gram-positive bacterium, displays a temperature optimum of 343 K and a pH optimum of 5.5 (Eckert et al., 2002 ▶). Enzymes that can resist higher temperatures and a range of pHs are required since heat and/or chemical pretreatment processes are currently used to remove lignin to expose cellulose to cellulases (Sticklen, 2008 ▶).
Cellulases belong to a group of enzymes termed glycoside hydrolases (GHs). Several members of the GH family demonstrate a modular architecture composed of one or two catalytic modules connected to several kinds of accessory modules (Schubot et al., 2004 ▶). The accessory modules can be involved in numerous functions. For example, some cellulases contain carbohydrate-binding modules (CBMs), which enhance the association of the catalytic modules with insoluble carbohydrates. Currently, glycoside hydrolases have been grouped into 113 families (http://www.cazy.org).
Cellulases have been characterized as endo or exo and processive or nonprocessive cellulases according to their mode of action on the substrate (Parsiegla et al., 2002 ▶). The endocellulases cleave the cellulose chain at arbitrary points, while the exocellulases cleave at the terminus of a chain to start the degradation process. Nonprocessive enzymes become detached from their substrate after one step of substrate hydrolysis, while processive cellulases remain bound to the cellulose substrate and continue breaking down the polysaccharide. The nonprocessive endoglucanase Aa_Cel9A belongs to subfamily E1 of family 9 of glycoside hydrolases. Members of this group show an N-terminal immunoglobulin-like (Ig-like) domain followed by the catalytic domain. The function of the Ig-like module is still unclear; however, its deletion promotes complete loss of enzymatic activity in a related cellobiohydrolase, CbhA from Clostridium thermocellum (Kataeva et al., 2004 ▶).
The endoglucanase Aa_Cel9A is most active against substrates containing β-1,4-linked glucans (including carboxymethylcellulose and lichenan), but also exhibits activity against β-1,4-xylans. The enzyme has been shown not to hydrolyze substrates such as starch (α-1,4) or laminarin (β-1,6) (Eckert et al., 2002 ▶). The crystallization and preliminary X-ray analysis of Aa_Cel9A have previously been reported (Eckert et al., 2003 ▶); however, the Aa_Cel9A structure was not subsequently described, most likely because anisotropic disorder in the crystal and weak diffraction thwarted structure solution. In the present paper, we describe the crystal structure of the endoglucanase Aa_Cel9A at 2.3 Å resolution. The Aa_Cel9A structure contains one zinc and two calcium ion-binding sites and the presence of these metals is associated with the temperature stability of the enzyme and/or substrate affinity. Moreover, the structure of Aa_Cel9A reveals the detailed contacts between the Ig-like and catalytic domains, providing new information about the interactions between them.
2. Materials and methods
2.1. Cloning, expression, purification and crystallization of Aa_Cel9A
Cloning, expression and purification have been reported elsewhere (Eckert et al., 2002 ▶). Aa_Cel9A was concentrated and dialyzed against 15 mM Tris–HCl buffer pH 7.5 containing 50 mM NaCl. The final concentration of Aa_Cel9A used for crystallization trials was 5 mg ml−1. The protein solution was brought to 5.0 mM CaCl2 and centrifuged prior to crystallization. Aa_Cel9A protein was screened using the sparse-matrix method (Jancarik & Kim, 1991 ▶) with a Phoenix Robot (Art Robbins Instruments, Sunnyvale, California, USA) using the following crystallization screens: Crystal Screen I and II, PEG/Ion, SaltRx and Index (Hampton Research, Aliso Viejo, California, USA). The optimum conditions for crystallization of Aa_Cel9A were found to be 0.1 M HEPES pH 7.3 and 55% 2-methyl-2,4-pentanediol (MPD). Crystals were obtained after 2 d by the sitting-drop vapor-diffusion method with the drops consisting of a mixture of 1.0 µl protein solution and 0.5 µl reservoir solution.
2.2. X-ray data collection and structure determination
Crystals were placed in a reservoir solution containing 55%(v/v) MPD and then flash-frozen in liquid nitrogen. A native data set for the endoglucanase Aa_Cel9A was collected on the Berkeley Center for Structural Biology beamline 5.0.1 of the Advanced Light Source at Lawrence Berkeley National Laboratory (LBNL). The diffraction data were recorded using an ADSC-Q210 detector. The data set was collected using 140° oscillation with Δϕ = 1° and a wavelength of 0.977 Å. The data were processed using the program HKL-2000 (Otwinowski & Minor, 1997 ▶).
The crystal structure of Aa_Cel9A was determined by the molecular-replacement method with the program Phaser (McCoy et al., 2007 ▶), using as a search model the structure of Cel9A (formerly CelD) from C. thermocellum (Ct_Cel9A; PDB code 1clc), which shows only 27% sequence identity with the target. The best solution was obtained with Euler angles α = 153.2, β = 122.4, γ = 103.1° and fractional coordinates Tx = 1.918, Ty = −0.237, Tz = 0.452. The atomic positions obtained from molecular replacement were used to initiate crystallographic refinement and model rebuilding. Structure refinement was performed using PHENIX (Adams et al., 2002 ▶). TLS refinement with both domains as a single TLS group was used in the process. Manual rebuilding using Coot (Emsley & Cowtan, 2004 ▶) and the addition of water molecules allowed construction of the final model. 5% of the data were randomly selected for cross-validation. The final model has an R factor of 19.6% and an R free of 23.3%.
Root-mean-square deviation differences from ideal geometries for bond lengths, angles and dihedrals were calculated with PHENIX (Adams et al., 2002 ▶). The overall stereochemical quality of the final model for Aa_Cel9A was assessed by the program MOLPROBITY (Davis et al., 2007 ▶). Atomic models were superposed using the program LSQKAB from CCP4 (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
The crystal of Aa_Cel9A diffracted to 2.3 Å resolution and belonged to the orthorhombic space group P21212, with unit-cell parameters a = 49.06, b = 84.97, c = 129.48 Å. The crystallographic asymmetric unit contained one copy of the Aa_Cel9A protein. The statistics for the crystallographic data and refinement are summarized in Table 1 ▶. The electron-density map showed clear positions for the residues present in both the Ig-like and catalytic modules. The crystallization conditions and unit-cell parameters of Aa_Cel9A are very similar to those previously reported at 3.0 Å resolution (Eckert et al., 2003 ▶). We believe that the addition of 5 mM divalent ion (Ca2+) prior to crystallization experiments was essential in improving the quality of the crystal diffraction.
Table 1. Statistics for data collection and structure refinement of endoglucanase Cel9A.
| Data collection | |
| Wavelength (Å) | 0.979 |
| Resolution range (Å) | 50–2.3 (2.34–2.30) |
| Crystal-to-detector distance (mm) | 300 |
| Φ collected/ΔΦ (°) | 140/1.0 |
| Exposure time (s) | 3 |
| Temperature of data collection (K) | 100 |
| Data statistics | |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 49.06, b = 84.97, c = 129.48 |
| Total reflections | 248498 |
| Unique reflections | 23484 |
| Multiplicity | 5.6 (4.2) |
| Data completeness (%) | 94.8 (93.7) |
| I/σ(I) | 9.0 (1.70) |
| Rmerge† (%) | 0.140 (0.550) |
| Structure refinement | |
| Resolution range | 50–2.3 |
| R factor‡ (%) | 19.6 |
| Rfree§ (%) | 23.3 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.061 |
| Protein residues | 528 |
| Ca2+ | 2 |
| Zn2+ | 1 |
| Water molecules | 380 |
| Average isotropic B factors (Å2) | |
| Protein atoms | 27.8 |
| Solvent atoms | 35.6 |
| Ramachandran plot | |
| Favored region (%) | 97.3 |
| Outliers region (%) | 0.0 |
R
merge = 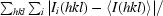
 , where
, where  denotes the sum over all reflections and
denotes the sum over all reflections and 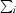 is the sum over all equivalent and symmetry-related reflections.
is the sum over all equivalent and symmetry-related reflections.
R factor = 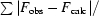
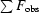 .
.
R free = R factor for 5% of the data that were not included during crystallographic refinement.
3.1. Overall structure of endoglucanase Aa_Cel9A
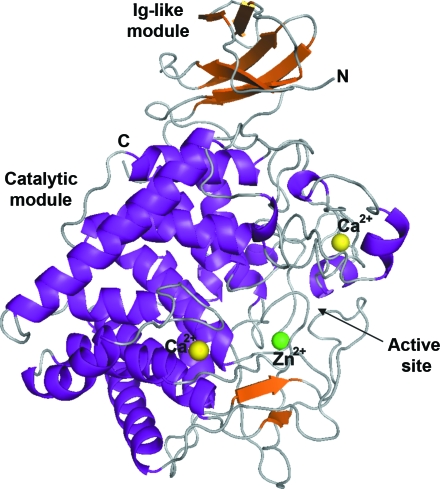
The 59 kDa endoglucanase Aa_Cel9A consists of two modules. The N-terminal Ig-like module is composed of 85 amino-acid residues and is linked to the catalytic module (residues 86–537). The overall fold of Aa_Cel9A is similar to the previously reported structures of endoglucanase Ct_Cel9A (Chauvaux et al., 1995 ▶) and the exoglucanase Ct_CbhA (Schubot et al., 2004 ▶), both from C. thermocellum. The r.m.s.d. between Cα positions for overlapping residues between Aa_Cel9A and Ct_Cel9A and Ct_CbhA are 1.622 and 1.923 Å, respectively.
The Ig-like module of Aa_Cel9A consists of six antiparallel strands forming two β-sheets. The first β-sheet (β-strands 1 and 4) packs in opposition to the second β-sheet (β-strands 2, 3, 5 and 6), forming a β-barrel structure (Fig. 1 ▶). The Ig-like module of Aa_Cel9A is more similar to the Ig-like module of Ct_Cel9A than that of Ct_CbhA, even though the Ig-like module of Ct_Cel9A contains seven antiparallel strands. The disulfide bridge between β-strands 2 and 6 conserved in immunoglobulin-domain structures is not observed in the Ig-like module of cellulases. Moreover, the structural similarity between the Ig-like module of cellulases and the immunoglobulin domains is significant despite low sequence homology (Juy et al., 1992 ▶).
Figure 1.
Modular architecture of Aa_Cel9A. The N-terminal Ig-like module consists of six β-strands, creating a β-barrel structure. The catalytic module has an (α/α)6-barrel motif formed by the inner α-helices. The metal ions bound to Aa_Cel9A are shown as large spheres. The zinc ion and the two calcium ions are shown in green and yellow, respectively. N, amino-terminus; C, carboxyl-terminus.
The catalytic module of members of GH family 9 shows an (α/α)6-barrel structure (Fig. 1 ▶). The 12 α-helices (α1–α12) display an alternating connection pattern between outer and inner helices, as is common in (α/α)6-barrel structures (Parsiegla et al., 1998 ▶). The barrel is formed by the parallel inner helices α2, α4, α6, α8, α10 and α12. Besides the 12 α-helices, the catalytic module of Aa_Cel9A shows two antiparallel β-strands and three short α-helices which are structurally conserved throughout the family 9 cellulases.
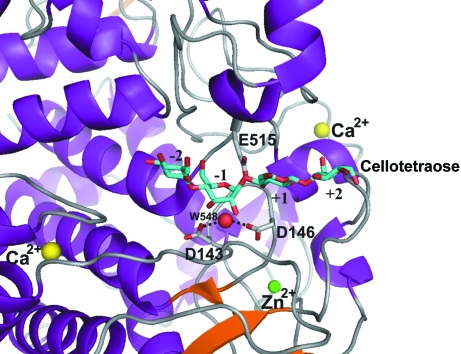
The active site of Aa_Cel9A is positioned at the N-terminal region of the inner helices of the barrel structure. The similarities between the active sites of Aa_Cel9A and of Ct_CbhA solved in complex with substrate (cellotetraose) permits inference of the residues involved in the catalytic mechanism. In Aa_Cel9A, residue Glu515 acts as a proton donor in the reaction. Asp143 and Asp146 are thought to deprotonate the water involved in nucleophilic attack. This catalytic water molecule is conserved in the Aa_Cel9A structure and is hydrogen bonded to Asp143 and Asp146 with distances of 2.71 and 2.96 Å, respectively (Fig. 2 ▶). A water chain connected to the catalytic Asp146 was observed in Aa_Cel9A for an efficient water supply. His461 and Arg463 are hydrogen bonded to a glucose unit at substrate subsite +1 (nomenclature according to Davies et al., 1997 ▶). Finally, the active site has residues Phe221, Tyr300, Trp343, Trp401, Tyr511, Tyr519 and Trp520 forming the substrate-binding cleft.
Figure 2.
Arrangement of the active site of Aa_Cel9A in complex with substrate (cellotetraose). Glu515 acts as a proton donor and Asp143 and Asp146 are thought to deprotonate the water (W548) involved in the nucleophilic attack. The modeling analyses and the docking of the cellotetraose molecule into the active site of Aa_Cel9A were performed based on the inactive mutant of the Ct_CbhA–cellotetraose complex (Schubot et al., 2004 ▶).
3.2. Metal-binding sites
Approximately 45% of the residues of the catalytic module of Aa_Cel9A are involved in α-helix formation. The presence of metal ions bound to the catalytic module enhances the intradomain interaction and compensates for the low composition of secondary structure (Juy et al., 1992 ▶). Any metal ions are bound to the Ig-like module.
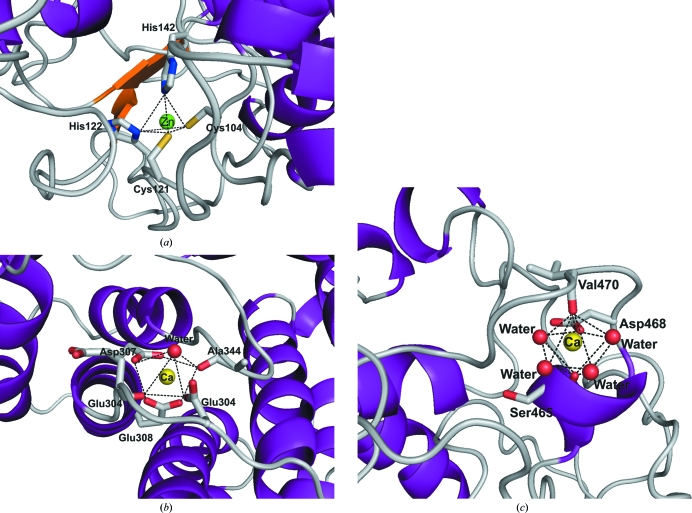
The identity of the metal ions was determined based on highly conserved binding sites among homologous structures and confirmed by their coordination spheres. The Aa_Cel9A structure shows one zinc-binding site and two calcium-binding sites. The type and quantity of metal ions bound to the Aa_Cel9A structure (this work) are supported biochemically by X-ray fluorescence analysis (Eckert et al., 2002 ▶). The zinc-binding site shows a general tetrahedral coordination involving two cysteines (Cys104 and Cys121) and two histidines (His122 and His142). The Zn2+-binding site is formed by the residues in a coil structure between α-helix 1 and α-helix 2 of the catalytic module. Of particular interest is the interaction via His142, which is located in the loop that contains the catalytic residues Asp143 and Asp146. The zinc-binding site is conserved in several cellulases of family 9, but not in the Ct_CbhA structure, in which the residues Cys121 and His142 (Aa_Cel9A numbering) are replaced by a glycine and a tyrosine residue, respectively. Therefore, the presence of a Zn2+-binding site in the catalytic module is not crucial for the enzyme reaction mechanism, but rather is associated with the thermal stability of cellulases (Chauvaux et al., 1995 ▶).
The Ct_Cel9A structure has been shown to possess three calcium-binding sites; however, only two of them are conserved in Aa_Cel9A. The Aa_Cel9A calcium-binding sites are characterized by seven or eight O atoms. In contrast to the zinc-binding site, where the coordination is formed exclusively by the side chains of protein residues, the calcium polyhedral coordination is from side chains of aspartic or glutamic acid residues, carbonyl groups of the main chain and water molecules. The first Ca2+ is located close to the active site of Aa_Cel9A (the nonreducing end), with a coordination formed by the residues Asp302, Glu304, Asp307, Glu308, Ala344 and one water molecule. This calcium-binding site is highly conserved in members of GH family 9.
The second Ca2+ is also positioned close to the active site, but at its opposite end (the reducing end). The coordination is formed by residues Ile465, Asp468 and Val470 and four water molecules. Functional analysis of Ct_Cel9A has shown that Ca2+ bound to this site is able to increase the substrate-binding affinity (Chauvaux et al., 1995 ▶). This is because the Ca2+ coordination residues Ile465, Asp468 and Val470 are located in same loop region as the substrate-binding residues His461 and Arg463. The coordination sphere of all metal ions bound to Aa_Cel9A is shown in Fig. 3 ▶.
Figure 3.
The coordination spheres of the metal-binding sites in Aa_Cel9A. (a) The zinc-binding site shows a tetrahedral coordination. (b, c) The two calcium-binding sites show polyhedral coordination formed exclusively from O atoms. For a complete discussion, see the main text.
3.3. Interactions between the Ig-like module and catalytic module
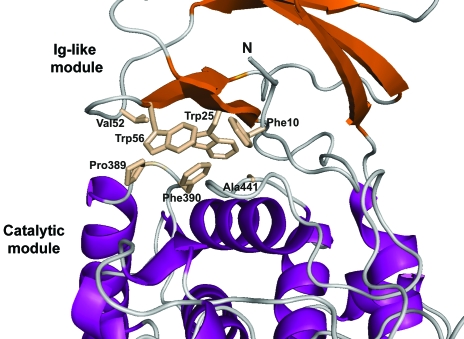
The Aa_Cel9A structure displays practically autonomous folding of the Ig-like module and catalytic module. However, we observe many direct or water-mediated hydrogen bonds and hydrophobic contacts between the two modules. All of the residues and waters involved in hydrogen bonds are listed in Table 2 ▶. Analysis of hydrophobic interactions shows that 15 residues of the Ig-like module are involved in van der Waals contacts with 23 residues of the catalytic module. A highly hydrophobic region is created by residues Phe10, Trp25, Val52 and Trp56 of the Ig-like module and residues Pro389, Phe390 and Ala441 of the catalytic module region (Fig. 4 ▶).
Table 2. Direct and water-mediated hydrogen bonds between the Ig-like and catalytic modules.
(a).
Direct hydrogen bonds.
| Ig-like module | Catalytic module | Distance (Å) |
|---|---|---|
| Tyr11 O | Ala441 N | 3.33 |
| Tyr11 OH | Gln453 OE1 | 3.38 |
| Asn12 ND2 | Tyr88 OH | 3.13 |
| Asn12 ND2 | Asp436 OD1 | 2.78 |
| Gln13 N | Phe439 O | 3.14 |
| Gln13 O | Gln454 OE1 | 3.25 |
| Gln13 OE1 | Gln454 N | 3.04 |
| Gln13 OE1 | Arg455 N | 3.18 |
| Gln13 NE2 | Gly445 O | 2.97 |
| Gly15 O | Tyr88 OH | 2.75 |
| Trp25 NE1 | Pro389 O | 2.86 |
| His84 O | Arg86 N | 2.92 |
| His84 O | Ala87 N | 3.25 |
| His84 ND1 | Arg86 N | 2.96 |
(b).
Water-mediated hydrogen bonds.
| Ig-like module | Distance (Å) | Water | Distance (Å) | Catalytic module |
|---|---|---|---|---|
| Tyr11 N | 3.26 | H2O 563 | 3.03 | Gly440 O |
| Leu17 N | 2.94 | H2O 551 | 3.06 | Gln432 OE1 |
| Asp21 O | 2.95 | H2O 551 | 3.06 | Gln432 OE1 |
| Asp21 O | 2.81 | H2O 552 | 2.89 | Asp436 OD2 |
| Asp21 O | 2.81 | H2O 552 | 3.20 | Arg433 NH1 |
| Arg23 N | 2.87 | H2O 552 | 2.89 | Asp436 OD2 |
| Arg23 N | 2.87 | H2O 552 | 3.20 | Arg433 NH1 |
| Asp60 OD2 | 3.03 | H2O 687 | 3.16 | Arg433 NH2 |
| Arg80 O | 2.77 | H2O 276 | 2.34 | Gln454 NE2 |
Figure 4.
Position of the nonpolar residues Phe10, Trp25, Trp26 and Val52 (Ig-like module) and Pro389, Phe390 and Ala441 (catalytic module) at the intermodular region forming a hydrophobic pocket. In addition to polar interactions, Aa_Cel9A has a large number of residues involved in hydrophobic contacts between the Ig-like and catalytic modules.
Even though 14 direct hydrogen bonds are present at the interface of the Ig-like and catalytic modules of Aa_Cel9A, only two are conserved in both the Ct_Cel9A and Ct_CbhA structures. The two conserved hydrogen bonds involve residues Gln13 with Phe439 and Trp25 with Pro389 of endoglucanase Aa_Cel9A. These hydrogen bonds are equivalent to residues Ser58 with Phe494 and Thr170 with Gly446 in the Ct_Cel9A structure and residues Gln218 with Leu715 and Thr230 with Gly661 in the Ct_CbhA structure. The role of the second conserved hydrogen bond has been established by the creation of the T230A mutant of Ct_CbhA (Kataeva et al., 2004 ▶). The mutant form T230A Ct_CbhA showed the same catalytic efficiency as the wild-type enzyme, confirming the hydrogen bond to be inessential for enzymatic activity. However, the interaction of Gln13 with Phe439 (Aa_Cel9A numbering) seems structurally important because this interaction between atoms of the main chain of nonconserved residues stabilizes the position of helix 11 and consequently the overall folding of the catalytic module. In addition, helix 11 of the catalytic module is connected to the loop region containing the two residues His461 and Arg463 involved in substrate binding.
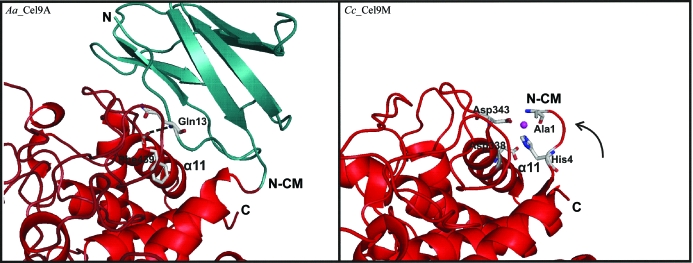
The deletion of the entire Ig-like module promotes the complete loss of enzymatic activity of Ct_CbhA (Kataeva et al., 2004 ▶). However, in some other cellulases the sequence lacks the Ig-like module and the question arises as to why their catalytic activity is not influenced by the absence of the Ig-like module. The cellulase Cel9M from C. cellulolyticum (Cc_Cel9M; Parsiegla et al., 2002 ▶) which, besides a short N-terminal dockerin module (30 amino acids), only contains the catalytic module, was compared with Aa_Cel9A. Analysis of the catalytic module of the Cc_Cel9M structure shows a large shift of the N-terminal region compared with the catalytic module of the Aa_Cel9A structure (Fig. 5 ▶). The N-terminal region of the catalytic module of Cc_Cel9M is located close to the region of Gln13 in the Ig-like module of Aa_Cel9A. The conserved hydrogen bond (Gln13–Phe439) observed in Aa_Cel9A is replaced by a metal-binding site in the Cc_Cel9M structure. The metal-ion site of the catalytic module of Cc_Cel9M is coordinated by the N-terminal residues Ala1 and His4 and residues Asp343 and Asp338 from helix 11 (Fig. 5 ▶). A similar shift of the N-terminus is observed in Cel9A (formerly called E4) from Thermonospora fusca (Tf_Cel9A), which in addition to the catalytic module has two C-terminal cellulose-binding modules and lacks the Ig-like module. However, the metal-binding site is not conserved in the Tf_Cel9A structure because the His4 observed in the Cc_Cel9M is replaced by a phenylalanine residue. Phe4 of Tf_Cel9A makes several hydrophobic contacts with the residue Leu354 (equivalent to residue Phe439 of Aa_Cel9A), Gly355 and Asp356. This observation supports our hypothesis that the essential interaction involving the residues Gln13 (Ig-like module) and Phe439 (catalytic module) of Aa_Cel9A is critical to the maintenance of enzymatic activity.
Figure 5.
Highly conserved polar interactions between the Ig-like module (Gln13) and the catalytic module (Phe439) of Aa_Cel9A. The Cc_Cel9M structure lacks the Ig-like module and there is a large shift of the N-terminal region of the catalytic module (N-CM) to help stabilize the region of helix 11 (α11). A metal ion is observed in an equivalent position to the conserved polar interaction in Aa_Cel9A. The Ig-like module and catalytic module are shown in green and red, respectively; the metal ion bound to Cc_Cel9M is represented by a small sphere (magenta) and the arrow indicates the shift direction of N-CM of Cc_Cel9M; N, amino-terminus; C, carboxyl-terminus.
4. Conclusions
Thermally resistant cellulases are required for the production of second-generation biofuels and metal-binding sites play an essential role in the thermal stability of these enzymes. Based on the structural information we have obtained for Aa_Cel9A, protein bioengineering can be used to produce mutant forms that are more catalytically efficient and/or resistant to high temperatures. The presence of direct hydrogen bonds at the interface between the Ig-like and catalytic modules, based on other cellulase structures, is a potential mechanism for the thermostabilization of Aa_Cel9A. Addition of a third calcium-binding site only previously observed in Ct_Cel9A and Ct_CbhA and absent in the Aa_Cel9A structure is also a possible approach to increasing temperature stability.
Finally, the Aa_Cel9A structure suggests that the Ig-like module has a role in stabilizing the correct folding of the catalytic module. We base this hypothesis on the following observations: (i) the Ig-like module is not directly involved in catalytic function because several cellulases lack this module, (ii) some related cellulases possess other stabilizing mechanisms, such as metal coordination, in the same region of the Ig-like module interact with the catalytic module and (iii) the addition of the extra domain (the Ig-like module) may create a more rigid architecture conferred by a large number of polar interactions and hydrophobic contacts and thus could be important in stabilizing the ‘active form’ of the catalytic module.
Supplementary Material
PDB reference: Aa_Cel9A, 3ez8, 3ez8sf
Acknowledgments
This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. We are grateful to the staff of the Berkeley Center for Structural Biology at the Advanced Light Source of Lawrence Berkeley National Laboratory. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231. We also would like to thank Professor E. Schneider at Humboldt-Universität zu Berlin for the gift of the original gene construct of Cel9A that was used to subclone the gene for expression of the protein.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Chauvaux, S., Souchon, H., Alzari, P. M., Chariot, P. & Beguin, P. (1995). J. Biol. Chem.270, 9757–9762. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davies, G. J., Wilson, K. S. & Henrissat, B. (1997). Biochem. J.321, 557–559. [DOI] [PMC free article] [PubMed]
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Soeyink, J., Richardson, J. C. & Richardson, D. C. (2007). Nucleic Acids Res.35, 375–383. [DOI] [PMC free article] [PubMed]
- Eckert, K., Ernst, H. A., Schneider, E., Larsen, S. & Lo Leggio, L. (2003). Acta Cryst. D59, 139–141. [DOI] [PubMed]
- Eckert, K., Zielinski, F., Lo Leggio, L. & Schneider, E. (2002). Appl. Microbiol. Biotechnol.60, 428–436. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Festucci-Buselli, R. A., Otoni, W. C. & Joshi, C. P. (2007). Braz. J. Plant Physiol.19, 1–13.
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Juy, M., Amit, A. G., Alzari, P. M., Poljak, R. J., Claeyssens, M., Beguin, P. & Aubert, J.-P. (1992). Nature (London), 357, 89–91.
- Kataeva, I. A., Uversky, V. N., Brewer, J. M., Schubot, F., Rose, J. P., Wang, B.-C. & Ljungdahl, G. L. (2004). Protein Eng. Des. Sel.17, 759–769. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parsiegla, G., Belaich, A., Belaich, J. P. & Haser, R. (2002). Biochemistry, 41, 11134–11142. [DOI] [PubMed]
- Parsiegla, G., Juy, M., Reverbel-Leroy, C., Tardif, C., Belaich, J.-P., Driguez, H. & Haser, R. (1998). EMBO J.17, 5551–5562. [DOI] [PMC free article] [PubMed]
- Parsiegla, G., Reverbel, C., Tardif, C., Driguez, H. & Haser, R. (2008). J. Mol. Biol.375, 499–510. [DOI] [PubMed]
- Schubot, F. D., Kataeva, I. A., Chang, J., Shah, A. K., Ljungdahl, L. G., Rose, J. P. & Wang, B.-C. (2004). Biochemistry, 43, 1163–1170. [DOI] [PubMed]
- Sticklen, M. B. (2008). Nature (London), 9, 433–443. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Aa_Cel9A, 3ez8, 3ez8sf
PDB reference: Aa_Cel9A, 3ez8, 3ez8sf