The crystal structure of the complex between ristocetin A and the cell-wall peptide mimetic N-acetyl-lysine-d-alanine-d-alanine has been solved. Structural details explaining the anticooperativity of the antibiotic have been identified.
Keywords: glycopeptide antibiotics, vancomycin, antimicrobial resistance
Abstract
Antimicrobial drug resistance is a serious public health problem and the development of new antibiotics has become an important priority. Ristocetin A is a class III glycopeptide antibiotic that is used in the diagnosis of von Willebrand disease and which has served as a lead compound for the development of new antimicrobial therapeutics. The 1.0 Å resolution crystal structure of the complex between ristocetin A and a bacterial cell-wall peptide has been determined. As is observed for most other glycopeptide antibiotics, it is shown that ristocetin A forms a back-to-back dimer containing concave binding pockets that recognize the cell-wall peptide. A comparison of the structure of ristocetin A with those of class I glycopeptide antibiotics such as vancomycin and balhimycin identifies differences in the details of dimerization and ligand binding. The structure of the ligand-binding site reveals a likely explanation for ristocetin A’s unique anticooperativity between dimerization and ligand binding.
1. Introduction
Glycopeptide antibiotics are used to treat serious infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA; Aoki & Kashiwagi, 1992 ▶; Foldes et al., 1983 ▶). However, the rise of antibiotic resistance is eroding the utility of many current therapeutic agents and new drugs must be developed to fight infection. A detailed understanding of the structural basis of target recognition should aid in the effort to design the next generation of antimicrobials.
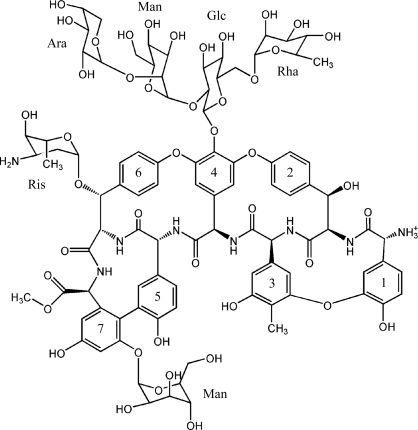
Vancomycin was the first glycopeptide antibiotic to be discovered and is still the best known member of this class of drugs (Kahne et al., 2005 ▶); however, many additional members of this family are known. All interfere with bacterial cell-wall biosynthesis by binding to the terminal d-Ala-d-Ala sequence of the muramyl pentapeptide intermediate formed during peptidoglycan biosynthesis, thus inhibiting transpeptidation and ultimately causing cell death (Kahne et al., 2005 ▶; Nieto & Perkins, 1971 ▶). The glycopeptide antibiotics all share a common structure based upon a heptapeptide backbone. The peptide side chains are oxidatively cross-linked to form macrocycles and carry sugar substituents at various positions (Loll & Axelsen, 2000 ▶). Glycopeptide antibiotics are divided into three classes depending on the type of residues found at positions 1 and 3 of the heptapeptide. Compounds bearing aliphatic residues at these positions (vancomycin, balhimycin and eremomycin) are designated as group I and compounds with individual aromatic residues at these positions (e.g. avoparcin) are assigned to group II, while group III compounds (ristocetin and teicoplanin) have aromatic residues at these positions that are covalently joined to each other (Fig. 1 ▶).
Figure 1.
Structure of the class III antibiotic ristocetin. The sugars are labeled as follows: Ara, arabinose; Man, mannose; Glc, glucose; Rha, rhamnose; Ris, ristosamine.
Ristocetin A is a group III glycopeptide antibiotic that was originally isolated from Nocardia lurida (later reclassified as Amycolatopsis orientalis) around the same time that vancomycin was discovered (Grundy et al., 1956 ▶). Although it was employed clinically to treat bacterial infections in the late 1950s and early 1960s, it was found to cause platelet aggregation, leading to its discontinuation as an antimicrobial. However, ristocetin-dependent platelet aggregation requires the presence of the plasma protein von Willebrand factor, making ristocetin a useful tool for diagnosing abnormalities in this protein (Macfarlane et al., 1975 ▶; Weiss et al., 1973 ▶). The aglycon of ristocetin A is free of the undesirable platelet side effect, but retains antimicrobial activity comparable to that of the parent compound (Herrin & Thomas, 1984 ▶), making it a useful lead compound for the development of new antibiotics directed against vancomycin-resistant bacteria (McComas et al., 2003 ▶).
Solution NMR studies of ristocetin A in complex with the di-N-acetyl-Lys-d-Ala-d-Ala peptide revealed that like other glycopeptide antibiotics, ristocetin A forms a back-to-back homodimer (Gerhard et al., 1993 ▶; Waltho & Williams, 1989 ▶; Groves, Searle, Mackay et al., 1994 ▶; Groves et al., 1995 ▶; Kaplan et al., 2001 ▶; Mackay, Gerhard, Beauregard, Westwell et al., 1994 ▶; Schäfer et al., 1998 ▶). This dimer is asymmetric, with the conformation of the two monomeric units differing in the orientation of the tetrasaccharide. Solution studies have revealed that unlike other glycopeptide antibiotics, ristocetin A does not dimerize and bind ligand cooperatively; it is the only antibiotic for which such behaviour has been described (Mackay, Gerhard, Beauregard, Maplestone et al., 1994 ▶; Mackay, Gerhard, Beauregard, Westwell et al., 1994 ▶).
We have determined the crystal structure of the complex of ristocetin A with the cell-wall mimetic N-acetyl-Lys-d-Ala-d-Ala. As observed previously, ristocetin forms an asymmetric homodimer essentially similar to those seen in other glycopeptide structures. However, ristocetin differs from other antibiotics in the details of dimerization, ligand binding and oligomerization. The structure reveals how asymmetry in the tetrasaccharide orientation creates different ligand-binding environments in the two halves of the dimer and provides an explanation for ristocetin’s anticooperative behaviour.
2. Materials and methods
2.1. Crystallization
Pharmaceutical grade ristocetin and the N-acetyl-lysine-d-alanine-d-alanine peptide were obtained from Sigma–Aldrich (St Louis, USA) and were used without further purification. Crystals were prepared by the hanging-drop vapor-diffusion method at 291 K. Prior to crystallization, ristocetin was mixed with 1.5 equivalents of the N-acetyl-lysine-d-alanine-d-alanine peptide to form a 20 mg ml−1 solution of the complex in water. 2 µl drops of this solution were mixed with equal volumes of reservoir buffer. A large number of conditions were screened; the best crystals were found using a reservoir solution containing 2.7 M ammonium sulfate and 8%(v/v) 2-propanol. Hexagonal prisms formed within 1 d, growing to maximum dimensions of 0.1 × 0.1 × 0.2 mm.
2.2. Data collection and processing
Diffraction data were collected from one cryocooled crystal on beamline X6A at the National Synchrotron Light Source (NSLS). Prior to data collection, the crystal was transferred into Fomblin Y oil (Sigma–Aldrich), mounted on a cryoloop and flash-cooled by plunging into liquid nitrogen. The crystal was maintained at 100 K in a stream of N2 gas during data collection. To minimize overloads, two data-collection passes were made, using different exposure times for the low- and the high-resolution data sets. Integration and scaling were accomplished using XDS (Kabsch, 1993 ▶). Data-collection and processing statistics are presented in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength (Å) | 0.8984 |
| Space group | P3121 |
| Unit-cell parameters (Å) | a = b = 22.65, c = 64.15 |
| Resolution range (Å) | 25–1.00 (1.03–1.00) |
| No. of observations | 91815 (1655) |
| No. of unique reflections | 10422 (589) |
| Completeness (%) | 95.5 (75.4) |
| Multiplicity | 8.8 (2.8) |
| Mean I/σ(I) | 39.7 (3.8) |
| Rmerge | 0.027 (0.337) |
| Rmeas† | 0.028 (0.396) |
| Refinement | |
| Resolution range (Å) | 20–1.00 |
| No. of reflections used | 10233 |
| No. of antibiotic atoms | 188 |
| No. of peptide atoms | 23 |
| No. of water molecules | 45 |
| Mean B values (Å2) | |
| Antibiotic | 10.6 |
| Peptide | 12.0 |
| Water | 28.2 |
| R.m.s. deviations from ideal geometry | |
| Bond distances (Å) | 0.024 |
| Bond angles (°) | 3.15 |
| Rcryst/Rfree | 0.172/0.192 |
The redundancy-independent residual on intensities R meas is defined according to Diederichs & Karplus (1997 ▶).
2.3. Structure determination and refinement
The structure of ristocetin A was determined by ab initio methods using SnB v.2.3 (Miller et al., 2007 ▶). 5000 trial structures were evaluated using the full 1.00 Å data set. A bimodal distribution of R min values was obtained, indicating that approximately 35 of the 5000 trials were presumptive solutions. About 50% of the atoms of the aglycon could be identified from the peaks in the initial E map of the best solution and the full initial model of the aglycon was easily completed using iterative cycles of refinement with REFMAC5 (Murshudov et al., 1997 ▶) and rebuilding with Coot (Emsley & Cowtan, 2004 ▶). Two alternative conformations were built for the tetrasaccharide. Geometric restraints for ristocetin were derived using PRODRG (Schüttelkopf & van Aalten, 2004 ▶). Water molecules were added with the combined criteria of a peak of greater than 2.5σ in the (F o − F c) difference map and reasonable intermolecular interactions. 5% of the data were used for the R free calculation. The atomic coordinates have been deposited with the Cambridge Crystallographic Data Centre under deposition number CCDC 718620 (these data can be obtained free of charge via http://www.ccdc.cam.ac.uk/data_request/cif). The solvent-accessible surface was calculated using PyMOL with a probe radius of 1.4 Å (DeLano, 2002 ▶).
3. Results and discussion
3.1. Description of the overall structure
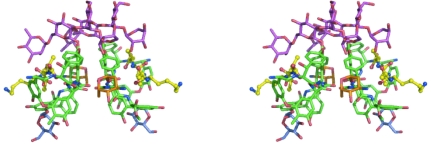
The crystal structure of ristocetin A represents the first crystal structure of a group III glycopeptide antibiotic. The crystal asymmetric unit contains one monomer of ristocetin A and one ligand molecule bound to the antibiotic. This monomer dimerizes with a second ristocetin molecule related to the first by a crystallographic twofold symmetry axis. The dimer interface lies at the convex surface (‘back’) of the molecule. The interactions that stabilize the back-to-back dimer are similar to those observed in the structures of other glycopeptide antibiotics (Groves et al., 1995 ▶; Groves, Searle, Mackay et al., 1994 ▶; Kaplan et al., 2001 ▶; Schäfer et al., 1998 ▶; Loll et al., 1997 ▶) and include aromatic ring stacking involving the side chains of residues 4 and 6 and four antiparallel β-type hydrogen bonds between the two peptide backbones. The N- and C-terminal ends of the glycopeptide bend outward away from the dimer interface, conferring a crescent shape to the aglycon portion of the monomer. The concave face of the antibiotic forms a shallow cleft that binds the cell-wall peptide. The carbohydrate portions of the dimer project outward above this concave surface and form part of the ligand-recognition pocket (Fig. 2 ▶).
Figure 2.
Stereoview of the back-to-back ristocetin A dimer. The aglycon is shown in green, the tetrasaccharide sugars in magenta, the ristosamine in orange, the mannose attached to residue 7 in blue and the N-acetyl-Lys-d-Ala-d-Ala peptide in yellow. Figs. 2–6 were created using PyMOL (DeLano, 2002 ▶).
Superposition of ristocetin A with group I antibiotics such as vancomycin and bahimycin reveals both similarities and differences in the structures of both the monomer and the dimer. Firstly, superposition of the aglycon monomers shows that the backbones of the antibiotics adopt very similar conformations, with r.m.s. differences of 0.26 Å in Cα positions between ristocetin and either vancomycin or balhimycin. Despite this similarity in backbone configuration, there are significant differences between ristocetin and the group I antibiotics, most notably in the vicinity of residues 1, 3 and 7. In ristocetin, the side chains of residues 1 and 3 contain covalently linked aromatic rings and residue 7 is mannosylated. The mannose sugar and the aromatic ring of residue 1 form two arms that extend outward from either end of the ristocetin molecule, wrapping around and partially covering the ligand. These two arms are linked by two hydrogen bonds between the mannose and residue 1.
The ristosamine amino sugar attached to the side chain of residue 6 also contributes to the hydrogen-bonding network that stabilizes the dimer, forming two hydrogen bonds to the carbonyl O atom of residue 3 of its dimer partner. One of these hydrogen bonds was also found in the NMR structure of ristocetin (Groves et al., 1995 ▶) and similar hydrogen bonds have been identified for balhimycin (Lehmann et al., 2002 ▶). The presence of a sugar at position 6 also promotes dimerization for both eremomycin and A82846B (Mackay, Gerhard, Beauregard, Westwell et al., 1994 ▶), probably by creating the same hydrogen bonds. The ristosamine sugar also affects the geometry of the back-to-back dimer, a fact that can be appreciated by superimposing the ristocetin A dimer upon dimers of other glycopeptide antibiotics. In the case of balhimycin, which also contains a sugar at position 6, the r.m.s. difference in Cα positions obtained upon superimposing dimers is 0.36 Å, only slightly higher than the value obtained when superimposing monomers. However, when superimposing the ristocetin A dimer upon vancomycin, which does not contain a sugar at position 6, the r.m.s. difference increases to 1.16 Å, which is much greater than the difference seen when superimposing monomers and reveals that the dimers are assembled differently for ristocetin and vancomycin. If one of the two molecules forming the ristocetin dimer (call it molecule 1) is superimposed on the corresponding molecule 1 of a vancomycin dimer, then molecule 2 in the ristocetin dimer is rotated by about 20° with respect to molecule 2 of the vancomycin dimer. It seems clear that this difference arises from the sugar in position 6, since modeling a sugar at position 6 in the vancomycin dimer leads to steric clashes between the two halves of the dimer. In ristocetin A (and other antibiotics containing a sugar in position 6), the two dimer partners rotate with respect to each other (compared with vancomycin) in order to prevent this steric clash and establish favorable hydrogen bonding. This slight reorientation may also serve to optimize the geometry at the dimer interface, since ristocetin, eremomycin and balhimycin show higher dimerization constants than vancomycin (Williams et al., 1993 ▶).
3.2. The ristocetin tetrasaccharide
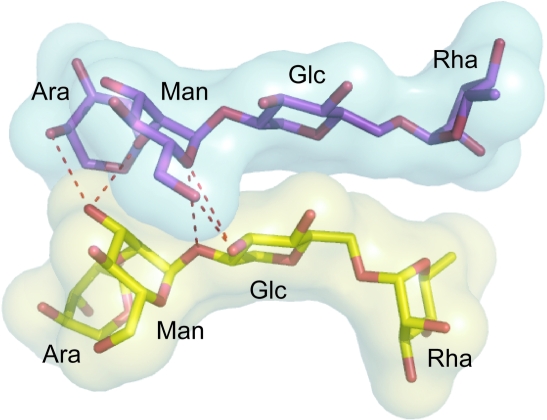
Apart from the monosaccharides found at positions 6 and 7, ristocetin A also contains a tetrasaccharide attached to the aromatic ring at position 4. The tetrasaccharide was not as well defined as the aglycon in the initial electron-density map, with none of the sugar atoms being identified by SnB. Using alternating cycles of rebuilding and refinement, we were able to build two alternate conformations (a and b) for the tetrasaccharide, assigning an occupancy of 0.5 to each conformer. Conformations a and b are related by a 180° rotation of the tetrasaccharide about the phenolic hydroxyl bond on the side chain of residue 4. In conformation a the central glucose turns away from the ligand-binding site, placing the rhamnose above the peptide ligand, whereas in conformation b the glucose turns towards the ligand-binding site, placing the arabinose over the ligand-binding site. If both monomers in the dimer adopted the b conformation, steric clashes would result, while if both assumed the a conformation no contacts would be formed between the two sugar moieties, which is not consistent with the observation that sugar–sugar interactions contribute to dimer stabilization (Gerhard et al., 1993 ▶; Groves et al., 1995 ▶; Mackay, Gerhard, Beauregard, Maplestone et al., 1994 ▶). Therefore, we expect that any given ristocetin dimer is asymmetric, containing one monomer with the tetrasaccharide in the a conformation and one in the b conformation. In this asymmetric dimer the two tetrasaccharide groups line up side by side. At one end of the tetrasaccharide pair, a network of hydrogen bonds connects the glucose and mannose of conformer a and the mannose and arabinose of conformer b (Fig. 3 ▶). The other end of the tetrasaccharide pair shows little surface complementarity between the sugars and is probably more important in ligand binding than dimerization. Thus, our crystallographic results agree with solution studies indicating that ristocetin A forms asymmetric homodimers in which each monomer displays a different orientation of the tetrasaccharide (Groves et al., 1995 ▶).
Figure 3.
Asymmetry of the tetrasaccharide in the ristocetin A back-to-back dimer. This is a view looking down on what would be the top of the dimer in Figs. 1 ▶ and 2 ▶; the viewer is looking approximately down the twofold axis that relates the two halves of the aglycon dimer. The aglycon (not shown) would lie beyond the sugars in this view. The tetrasaccharide sugars are colored yellow (conformation a) and purple (conformation b). Hydrogen bonds between the two tetrasaccharides are represented by red dashed lines. The semi-transparent surfaces represent the solvent-accessible surfaces of the sugars and demonstrate how the two tetrasaccharide groups are tightly packed at only one end.
We see no evidence that the tetrasaccharide conformation of any given dimer is correlated with those of neighboring dimers in the crystal lattice. The tetrasaccharide moieties on neighboring dimers are not in close contact, nor is there any evidence in the diffraction images for the presence of a superlattice. Therefore, we believe that the conformations of the tetrasaccharide pairs are randomly distributed throughout the lattice (i.e. each back-to-back homodimer in the crystal has a 50:50 chance of having either the ab conformation or the ba conformation).
3.3. Ligand-binding pocket
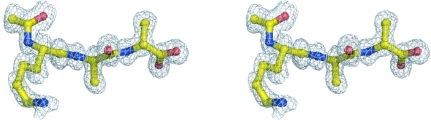
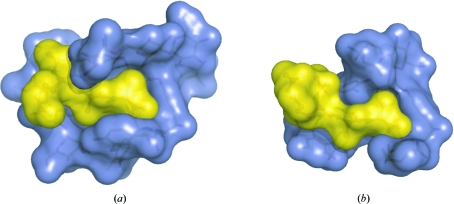
Clear electron density was observed for the peptide ligand, N-acetyl-Lys-d-Ala-d-Ala, allowing it to be built without any ambiguity (Fig. 4 ▶). The ligand lies inside the curved face of the ristocetin A monomer, making a network of interactions with the antibiotic. At its C-terminal extremity, the peptide is deeply embedded in the curved structure formed by the aromatic side chains of residues 1 and 3 of the antibiotic. As observed for other glycopeptide antibiotic–ligand complexes, the C-terminal carboxylate of the peptide makes strong hydrogen bonds to the backbone amides of residues 2, 3 and 4, while the amide N atom of the C-terminal d-Ala and the carbonyl O atom of the lysine form hydrogen bonds to the amide N atom and the carbonyl of residue 5 of ristocetin, respectively. The arms that extend from either end of the antibiotic monomer and engulf the ligand cause the peptide to be more buried in ristocetin than in vancomycin or balhimycin (Fig. 5 ▶). For ristocetin A the peptide buried area is 314 Å2 for conformer a and 337 Å2 for conformer b, corresponding to 50% and 53% of the ligand’s total area, respectively. In contrast, in the complex of vancomycin with di-acetyl-Lys-d-Ala-d-Ala 44% of the ligand’s surface area is buried (PDB structure 1fvm; Y. Nitanai, K. Kakoi & K. Aoki, unpublished work), while in the complex of balhimycin with Lys-d-Ala-d-Ala the ligand is 42% buried (PDB structure 1go6; Lehmann et al., 2002 ▶).
Figure 4.
Final 2F o − F c electron-density map of the N-acetyl-Lys-d-Ala-d-Ala peptide in the ligand-binding site.
Figure 5.
Comparison of the ligand-buried surface in the structures of ristocetin A (a) and vancomycin (b). The structures of the complexes of the two antibiotics with N-acetyl-Lys-d-Ala-d-Ala are shown in equivalent orientations. The solvent-accessible surfaces are shown in blue for the antibiotics and in yellow for the peptide ligands.
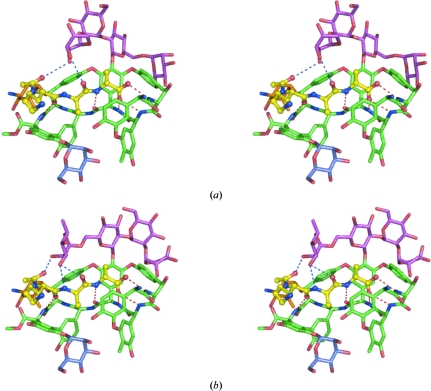
The peptide interacts with both conformations of the tetrasaccharide. Because different sugars cap the ligand-binding site for the two tetrasaccharide conformers, each conformer leads to a different network of interactions between the ligand and the antibiotic (Fig. 6 ▶). In conformer a the arabinose sugar overhangs the ligand-binding site. The electron density is not as well defined for this sugar as it is for the other sugars of the tetrasaccharide; nonetheless, it can be seen that the arabinose sits atop the lysine residue of the ligand and makes weak hydrogen bonds to the peptide, one to the carbonyl of the second d-Ala (3.32 Å) and one to the carbonyl of the N-terminal acetate (3.56 Å). Together with residue 7 of ristocetin, which packs against the opposite side of the ligand, the arabinose in conformer a creates a ‘grip-like’ structure that holds the peptide in position. This result is consistent with the solution NMR structure, in which this sugar was also seen to cap the ligand-binding site and to form a single hydrogen bond to the peptide’s N-terminal acetyl group. For conformer b of the tetrasaccharide the rhamnose sugar lies above the ligand-binding site, making strong contacts with the peptide. While the rhamnose creates a similar kind of ‘grip-like’ structure around the lysine residue as does the arabinose, it appears to grip the ligand more strongly, closing more tightly around the peptide and making three strong hydrogen bonds to the ligand, one with the carbonyl of the acetate and two with the amide and the carbonyl of the second d-Ala. In addition to the rhamnose, the glucose also interacts with the peptide in this conformer, packing about 4 Å above the C-terminal d-Ala. Similar interactions with the sugars have also been observed in the NMR structure (Groves et al., 1995 ▶).
Figure 6.
Close-up stereoviews of the ligand-binding sites in conformers a (a) and b (b). The aglycon is in green, the tetrasaccharide sugars in magenta, the ristosamine in orange, the mannose attached to residue 7 in blue and the N-acetyl-Lys-d-Ala-d-Ala peptide in yellow. The five hydrogen bonds between the peptide and the aglycon are represented as red dashed lines, while hydrogen bonds between the peptide and the tetrasaccharide are shown as blue dashed lines.
3.4. Dimerization and cooperative ligand binding
The two halves of the ristocetin A asymmetric homodimer are known to bind ligand with different affinities (Cho et al., 1996 ▶). This can be explained in terms of the structure. In conformer b, stronger hydrogen bonds mediate peptide recognition than in conformer a. This asymmetry in ligand binding gives rise to an unusual anticooperativity between ligand binding and dimerization in ristocetin. For all dimeric glycopeptide antibiotics except ristocetin A, ligand binding and dimerization are cooperative (Groves, Searle, Chicarelli-Robinson et al., 1994 ▶); ristocetin is unique in that ligand binding decreases the proportion of dimer present in solution (Mackay, Gerhard, Beauregard, Maplestone et al., 1994 ▶). Solution studies have shown that binding of ligand to one high-affinity site in ristocetin is cooperative with dimerization (in the sense that binding to the dimer is tighter than binding to the monomer); binding of a second ligand to a lower affinity site is weaker than binding to the monomer and hence is anticooperative with dimerization (Cho et al., 1996 ▶). We believe that the sugar conformer b corresponds to the high-affinity ligand-binding site, while conformer a represents the lower affinity site. In monomeric ristocetin, the tetrasaccharide is likely to be highly mobile and can oscillate between the two conformers. Ligand will preferentially bind to conformer b and dimerization will stabilize this conformer (and hence the ristocetin–ligand complex). Stabilization will occur through the network of hydrogen bonds connecting the two tetrasaccharide groups of the dimer and perhaps also via vibrational effects (Jusuf et al., 2003 ▶).
Binding of a second ligand molecule to the conformer a site can occur, but will do so with lower affinity because of the less favorable interactions between the ligand and the arabinose sugar.
Alternatively, the ristocetin dimer can dissociate, allowing the tetrasaccharide to rotate and adopt the higher affinity conformer b, explaining why binding the second ligand molecule is actually anticooperative with dimerization (Cho et al., 1996 ▶). This model predicts that anticooperativity is linked to the rhamnose sugar and is consistent with the observation that forms of ristocetin lacking this rhamnose only display positive cooperativity between dimerization and ligand binding (Mackay, Gerhard, Beauregard, Westwell et al., 1994 ▶; Bardsley & Williams, 1998 ▶).
3.5. Higher order oligomerization
Vancomycin has been observed to form supramolecular structures in which two back-to-back dimers associate in a ‘face-to-face’ assembly mediated by antiparallel β-type hydrogen bonds between the peptide ligands on facing dimers (Loll et al., 2009). Since the formation of such supramolecular assemblies may increase the avidity of binding for multivalent cell-wall structures on bacterial targets, it is of interest to ask whether ristocetin can form similar structures. Unlike vancomycin and balhimycin, where intimate face-to-face interactions occur as a result of crystal packing, ristocetin does not form tight face-to-face interactions in the crystal lattice. Two symmetry-related monomers can be seen to face one another, but the two ligands bound to these two molecules do not approach sufficiently closely to hydrogen bond to each other and overall the contacts between the two complexes are quite limited. In fact, modeling suggests that face-to-face interactions of the sort seen with vancomycin are not possible for ristocetin; the bulky macrocycle formed by residues 1 and 3, the large tetrasaccharide attached to residue 4 and the mannose sugar attached to residue 7 all wrap extensively around the ligand, making it much less accessible than it would be when bound to vancomycin. Attempts to build vancomycin-like supramolecular complexes with ristocetin lead to extensive steric clashes between residue 1 of one monomer and residue 7 on the facing monomer, as well as between the tetrasaccharide groups, making it impossible to bring the complexes sufficiently close for the two peptide ligands to hydrogen bond to one another. Such steric considerations suggest that the formation of supramolecular complexes may be limited to group I glycopeptide antibiotics.
4. Conclusion
Here, we have investigated the crystal structure of the complex between ristocetin A and a cell-wall peptide mimetic. We have shown that the antibiotic forms an asymmetric dimer that is essentially similar to those observed for other glycopeptides; however, slight differences in the structure of ristocetin lead to significant differences in dimerization, ligand binding and oligomerization. Furthermore, the structure that is presented in this paper clearly reveals the basis for ristocetin’s unique anticooperative binding and dimerization behaviour: it has two different ligand-binding sites, with different ligand coordination and hence different binding affinities.
Acknowledgments
This work was supported by grant GM079508 from the National Institutes of Health (PJL). Diffraction data were collected on the X6A beamline of the NSLS, funded by the National Institute of General Medical Sciences, National Institutes of Health under agreement GM-0080. The NSLS at Brookhaven National Laboratory is supported by the US Department of Energy under contract No. DE-AC02-98CH10886.
References
- Aoki, Y. & Kashiwagi, H. (1992). Chemotherapy (Tokyo), 40, 997–1004.
- Bardsley, B. & Williams, D. H. (1998). J. Chem. Soc. Perkin Trans. 2, pp. 1925–1928.
- Cho, Y. R., Maguire, A. J., Try, A. C., Westwell, M. S., Groves, P. & Williams, D. H. (1996). Chem. Biol.3, 207–215. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific LLC, Palo Alto, California, USA.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Foldes, M., Munro, R., Sorrell, T. C., Shanker, S. & Toohey, M. (1983). J. Antimicrob. Chemother.11, 21–26. [DOI] [PubMed]
- Gerhard, U., Mackay, J. P., Maplestone, R. A. & Williams, D. H. (1993). J. Am. Chem. Soc.115, 232–237.
- Groves, P., Searle, M. S., Chicarelli-Robinson, I. & Williams, D. H. (1994). J. Chem. Soc. Perkin Trans. 1, pp. 659–665.
- Groves, P., Searle, M. S., Mackay, J. P. & Williams, D. H. (1994). Structure, 2, 747–754. [DOI] [PubMed]
- Groves, P., Searle, M. S., Waltho, J. P. & Williams, D. H. (1995). J. Am. Chem. Soc.117, 7958–7964.
- Grundy, W. E., Sinclair, A. C., Theriault, R. J., Goldstein, A. W., Rickher, C. J., Warren, H. B. Jr, Oliver, T. J. & Sylvester, J. C. (1956). Antibiot. Annu., pp. 687–692. [PubMed]
- Herrin, T. R. & Thomas, A. M. A. L. (1984). Chem. Abstr.101, 210855.
- Jusuf, S., Loll, P. J. & Axelsen, P. H. (2003). J. Am. Chem. Soc.125, 3988–3994. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kahne, D., Leimkuhler, C., Lu, W. & Walsh, C. (2005). Chem. Rev.105, 425–448. [DOI] [PubMed]
- Kaplan, J., Korty, B. D., Axelsen, P. H. & Loll, P. J. (2001). J. Med. Chem.44, 1837–1840. [DOI] [PubMed]
- Lehmann, C., Bunkoczi, G., Vertesy, L. & Sheldrick, G. M. (2002). J. Mol. Biol.318, 723–732. [DOI] [PubMed]
- Loll, P. J. & Axelsen, P. H. (2000). Annu. Rev. Biophys. Biomol. Struct.29, 265–289. [DOI] [PubMed]
- Loll, P. J., Bevivino, A. E., Korty, B. D. & Axelsen, P. H. (1997). J. Am. Chem. Soc.119, 1516–1522.
- Loll, P. J., Derhovanessian, A., Shapovalov, M. V., Kaplan, J., Yang, L. & Axelsen, P. H. (2009). J. Mol. Biol.385, 200–211. [DOI] [PMC free article] [PubMed]
- Macfarlane, D. E., Stibbe, J., Kirby, E. P., Zucker, M. B., Grant, R. A. & McPherson, J. (1975). Thromb. Diath. Haemorrh.34, 306–308. [PubMed]
- Mackay, J. P., Gerhard, U., Beauregard, D. A., Maplestone, R. A. & Williams, D. H. (1994). J. Am. Chem. Soc.116, 4573–4580.
- Mackay, J. P., Gerhard, U., Beauregard, D. A., Westwell, M. S., Searle, M. S. & Williams, D. H. (1994). J. Am. Chem. Soc.116, 4581–4590.
- McComas, C. C., Crowley, B. M., Hwang, I. & Boger, D. L. (2003). Bioorg. Med. Chem. Lett.13, 2933–2936. [DOI] [PubMed]
- Miller, R., Shah, N., Green, M. L., Furey, W. & Weeks, C. M. (2007). J. Appl. Cryst.40, 938–944.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nieto, M. & Perkins, H. R. (1971). Biochem. J.124, 845–852. [DOI] [PMC free article] [PubMed]
- Schäfer, M., Sheldrick, G. M., Schneider, T. R. & Vértesy, L. (1998). Acta Cryst. D54, 175–183. [DOI] [PubMed]
- Schüttelkopf, A. W. & van Aalten, D. M. F. (2004). Acta Cryst. D60, 1355–1363. [DOI] [PubMed]
- Waltho, J. P. & Williams, D. H. (1989). J. Am. Chem. Soc.111, 2475–2480.
- Weiss, H. J., Rogers, J. & Brand, H. (1973). J. Clin. Invest.52, 2697–2707. [DOI] [PMC free article] [PubMed]
- Williams, D. H., Searle, M. S., Mackay, J. P., Gerhard, U. & Maplestone, R. A. (1993). Proc. Natl Acad. Sci. USA, 90, 1172–1178. [DOI] [PMC free article] [PubMed]