Abstract
Nicotine dependence risk and lung cancer risk are associated with variants in a region of chromosome 15 encompassing genes encoding the nicotinic receptor subunits CHRNA5, CHRNA3 and CHRNB4. To identify potential biological mechanisms that underlie this risk, we tested for cis-acting eQTLs for CHRNA5, CHRNA3 and CHRNB4 in human brain. Using gene expression and disease association studies, we provide evidence that both nicotine-dependence risk and lung cancer risk are influenced by functional variation in CHRNA5. We demonstrated that the risk allele of rs16969968 primarily occurs on the low mRNA expression allele of CHRNA5. The non-risk allele at rs16969968 occurs on both high and low expression alleles tagged by rs588765 within CHRNA5. When the non-risk allele occurs on the background of low mRNA expression of CHRNA5, the risk for nicotine dependence and lung cancer is significantly lower compared to those with the higher mRNA expression. Together, these variants identify three levels of risk associated with CHRNA5. We conclude that there are at least two distinct mechanisms conferring risk for nicotine dependence and lung cancer: altered receptor function caused by a D398N amino acid variant in CHRNA5 (rs16969968) and variability in CHRNA5 mRNA expression.
INTRODUCTION
Cigarette smoking is a common addictive disorder. In 2007, an estimated 20% of Americans aged 18 years or older were current smokers (defined as those who reported that they smoked 100 cigarettes or more during their lifetime and were currently smoking every day or some days) (1). Among current cigarette smokers, 59% were nicotine-dependent (2). It is well established that smoking has detrimental effects on physical health, increasing risk for cancer, heart disease, stroke and chronic lung disease. According to the World Health Organization's report in 2006, tobacco use is responsible for about five million deaths annually, making it the largest cause of preventable mortality in the world (3). In the USA, tobacco use is the leading cause of morbidity and mortality; it accounts for 30% of all cancer deaths including 87% of all deaths from lung cancer (4,5).
Many aspects of cigarette smoking behavior cluster in families (6). Evidence from twin studies indicates that genetic factors contribute to the development of smoking, smoking persistence and nicotine dependence (7–9). Heritability estimates for nicotine dependence range from 60% to 72% (10), and the risk for the sibling of a nicotine-dependent individual of developing nicotine dependence is 2-fold increased over the general population rate.
Neuronal nicotinic acetylcholine receptors (nAChRs) are a family of pentameric (mostly hetero-pentameric) ligand-gated ion channels that can mediate fast signal transmission at synapses (11) as well as modulate the release of several neurotransmitters (12). Nicotine is an exogenous agonist of these receptors. Within a few seconds of smoking, nicotine produces physiological responses. The most abundantly expressed receptor subtype in the brain is the α4β2 subtype. Some α4β2 receptors also contain an α5 subunit (13). Inclusion of an α5 subunit significantly increases the rate of receptor desensitization and calcium permeability (14). Recent studies have shown that nicotine can also induce cell proliferation, tumor invasion and angiogenesis, and confer resistance to apoptosis, processes that are mediated through nAChRs (15–17). Thus, variations in nAChRs are strong candidate risk factors for nicotine-dependence and lung cancer.
Several genetic association studies involving addiction in humans have focused on genes encoding the major nAChR subunits expressed in the brain (α4 and β2) (18–20). Recently, our genome-wide association study (GWAS) and candidate gene study of nicotine dependence identified several variants in the gene cluster encoding the α5, α3 and β4 subunits on chromosome 15 that alter risk for nicotine dependence, including an amino acid substitution (a change from aspartic acid to asparagine at codon 398) in the α5 nicotinic receptor subunit gene (CHRNA5) and several non-coding variants across this gene cluster (21–23). These associations have now been replicated in independent smoking data sets (24–28). GWAS using lung cancer populations from Europe, the USA and Iceland have also demonstrated association between lung cancer susceptibility and the same variants or highly correlated variants in the CHRNA5/A3/B4 gene cluster (27,29–31).
There is extensive linkage disequilibrium across the CHRNA5/A3/B4 gene region. Two distinct LD bins (r2 > 0.8) are associated with both nicotine dependence and lung cancer (Supplementary Material, Fig. S1). One bin (bin 1) is tagged by rs16969968 and the minor allele is linked to increased risk for nicotine dependence (24–27,32) and lung cancer (27,29,30). The second bin (bin 2) is tagged by rs3743078 (or rs578776) and the minor allele is associated with a reduced risk for nicotine dependence (24,26,28) and lung cancer (30,31).
Rs16969968 is a missense variant that results in an amino acid substitution at codon 398 (D398N) of CHRNA5. Our in vitro functional study demonstrated that heterologously expressed nicotinic receptors containing the missense variant of CHRNA5, α4β2α5N398 exhibit reduced response to the nicotinic agonist epibatidine when compared with receptors containing the more common variant, α4β2α5D398 (25). No other obvious functional variants are in this LD bin. Using quantitative polymerase chain reaction (qPCR), we have previously shown that CHRNA5 mRNA levels in frontal cortex and in lymphocytes are highly variable between individuals and that some of this variability is explained by cis-acting polymorphisms (33). The goal of this study was to determine the influence of two potential biological mechanisms in the etiology of nicotine dependence and lung cancer: the influence of a missense mutation in a receptor subunit gene and the influence of subunit mRNA expression level. We find that both biological mechanisms play a role.
RESULTS
Postmortem interval, age, and smoking status are weakly associated with CHRNA5, CHRNA3 and CHRNB4 mRNA levels, respectively
We used linear regression to test for evidence of differential expression in samples of different genotype. First, we tested whether drinking status affected gene expression in the samples obtained from the Australian Brain Bank (ABDP). No significant differences in CHRNA5, CHRNA3 or CHRNB4 mRNA expression were observed between alcoholic and non-alcoholic subjects. We then combined brain tissues from both the Washington University Alzheimer's Disease Research Center (ADRC) and ABDP for further analysis. The site of brain bank, postmortem interval, age, gender and smoking history (ever smoke or never smoke) were tested for their influence on mRNA levels of CHRNA5, CHRNA3 and CHRNB4 (Supplementary Material, Table S1). For CHRNA5, only postmortem interval has a weak effect on mRNA expression levels (P = 0.02). Brain bank site, age and gender influence CHRNA3 mRNA expression. However, when theses variables were added to a linear regression model, only age remained as a significant covariate. For CHRNB4, site, gender and smoking history influence mRNA expression. When these variables were included in the logistic regression model, only smoking history has a significant effect on mRNA expression levels.
Variability in CHRNA5 mRNA levels is strongly associated with variants located upstream of the coding region of CHRNA5
Single variant analysis
To examine whether the polymorphisms associated with nicotine dependence in the chromosome 15q24-25.1 region are also associated with gene expression, we used real-time PCR to quantitate mRNA levels of CHRNA5, CHRNA3 and CHRNB4 in the frontal cortex in individuals of different genotype. We genotyped 104 brain samples of European descent with 44 variants spanning the CHRNA5/A3/B4 gene cluster (Table 1 and Supplementary Material, Table S2). These SNPs tag 79 of the 100 SNPs that have a minor allele frequency≥5% in the HapMap CEU reference sample in this gene cluster at an r2 of 0.8 or better (dbSNP build 129 and HapMap public release 23a); 94 are tagged at an r2 ≥ 0.6.
Table 1.
Association of CHRNA5 mRNA expression in human brain with variants in the CHRNA5–CHRNA3–CHRNB4 gene cluster
| Variant | Gene | Chromosome position | Allele 1 (Minor) | Allele 2 (Major) | Minor allele frequency | n | Regression coefficient | 95% confidence interval | P-value |
|---|---|---|---|---|---|---|---|---|---|
| rs880395 | Upstream of CHRNA5 transcription | 76631411 | A | G | 0.44 | 95 | 0.56 | 0.41, 0.71 | 1.10E−10 |
| rs7164030 | 76631716 | G | A | 0.46 | 91 | 0.56 | 0.40, 0.72 | 4.20E−10 | |
| rs905739 | 76632165 | G | A | 0.24 | 94 | −0.44 | −0.64, −0.24 | 4.21E−05 | |
| rs2036527 | 76638670 | A | G | 0.33 | 94 | −0.27 | −0.47, −0.07 | 8.42E−03 | |
| rs3841324 | 76644868 | S | L | 0.41 | 95 | 0.60 | 0.44, 0.75 | 6.36E−11 | |
| rs684513 | CHRNA5 | 76645455 | G | C | 0.22 | 91 | −0.37 | −0.59, −0.14 | 1.90E−03 |
| rs667282 | 76650527 | C | T | 0.25 | 94 | −0.38 | −0.58, −0.18 | 2.82E−04 | |
| rs588765 | 76652480 | T | C | 0.42 | 94 | 0.53 | 0.37, 0.68 | 1.11E−09 | |
| rs17486278 | 76654537 | C | A | 0.33 | 94 | −0.26 | −0.45, −0.06 | 1.30E−02 | |
| rs601079 | 76656634 | A | T | 0.45 | 92 | 0.54 | 0.38, 0.69 | 8.95E−10 | |
| rs680244 | 76658343 | T | C | 0.4 | 89 | 0.50 | 0.33, 0.67 | 1.51E−07 | |
| rs621849 | 76659916 | G | A | 0.41 | 90 | 0.52 | 0.36, 0.68 | 1.47E−08 | |
| rs569207 | 76660174 | T | C | 0.23 | 94 | −0.39 | −0.59, −0.20 | 1.69E−04 | |
| rs637137 | 76661031 | A | T | 0.23 | 94 | −0.38 | −0.58, −0.19 | 2.69E−04 | |
| rs692780 | 76663560 | G | C | 0.35 | 94 | 0.38 | 0.21, 0.55 | 3.57E−05 | |
| rs11637635 | 76664205 | A | G | 0.32 | 87 | 0.40 | 0.21, 0.58 | 7.18E−05 | |
| rs951266 | 76665596 | A | G | 0.34 | 94 | −0.24 | −0.43, −0.04 | 1.83E−02 | |
| rs555018 | 76666297 | G | A | 0.41 | 92 | 0.49 | 0.33, 0.65 | 3.51E−08 | |
| rs16969968** | 76669980 | A | G | 0.36 | 94 | −0.26 | −0.46, −0.06 | 1.26E−02 | |
| rs514743 | 76671282 | T | A | 0.34 | 94 | 0.38 | 0.21, 0.55 | 2.76E−05 | |
| rs615470 | 76673043 | T | C | 0.32 | 83 | 0.36 | 0.18, 0.55 | 2.32E−04 | |
| rs578776 | CHRNA3 | 76675455 | A | G | 0.29 | 93 | −0.21 | −0.41, 0.00 | 4.84E−02 |
| rs12910984 | 76678682 | G | A | 0.26 | 92 | −0.37 | −0.57, −0.18 | 2.76E−04 | |
| rs1051730* | 76681394 | A | G | 0.32 | 93 | −0.26 | −0.46, −0.06 | 1.13E−02 | |
| rs3743078 | 76681814 | G | C | 0.24 | 92 | −0.40 | −0.59, −0.20 | 1.80E−04 | |
| rs1317286 | 76683184 | G | A | 0.33 | 85 | −0.22 | −0.42, −0.01 | 3.83E−02 | |
| rs938682 | 76683602 | G | A | 0.26 | 92 | −0.39 | −0.59, −0.19 | 2.90E−04 | |
| rs11637630 | 76686774 | G | A | 0.23 | 93 | −0.39 | −0.59, −0.19 | 2.54E−04 | |
| rs6495308 | 76694711 | C | T | 0.24 | 94 | −0.38 | −0.58, −0.19 | 2.60E−04 | |
| rs8192479 | 76696453 | T | C | 0.01 | 92 | −0.11 | −0.85, 0.64 | 7.82E−01 | |
| rs3743075* | 76696507 | T | C | 0.33 | 94 | 0.37 | 0.20, 0.54 | 5.11E−05 | |
| rs3743074 | 76696535 | G | A | 0.35 | 91 | 0.36 | 0.19, 0.54 | 8.75E−05 | |
| rs8040868* | 76698236 | C | T | 0.4 | 94 | −0.11 | −0.30, 0.08 | 2.47E−01 | |
| rs8192475** | 76698285 | T | C | 0.08 | 95 | 0.54 | 0.15, 0.93 | 8.41E−03 | |
| rs1878399 | 76699058 | G | C | 0.44 | 94 | 0.49 | 0.33, 0.65 | 3.79E−08 | |
| rs12914008** | CHRNB4 | 76710560 | A | G | 0.06 | 94 | 0.61 | 0.18, 1.03 | 6.58E−03 |
| rs17487223 | 76711042 | T | C | 0.34 | 93 | −0.21 | −0.40, −0.02 | 3.64E−02 | |
| rs950776 | 76713073 | C | T | 0.35 | 94 | 0.44 | 0.26, 0.61 | 5.85E−06 | |
| rs12440014 | 76713781 | G | C | 0.26 | 93 | −0.38 | −0.58, −0.18 | 3.44E−04 | |
| rs11636605 | 76715933 | A | G | 0.23 | 95 | −0.41 | −0.61, −0.20 | 1.75E−04 | |
| rs3813567 | upstream of CHRNB4 transcription | 76721606 | G | A | 0.21 | 94 | −0.42 | −0.63, −0.20 | 2.47E−04 |
| rs4887075 | 76740186 | C | T | 0.06 | 93 | 0.04 | −0.35, 0.43 | 8.50E−01 | |
| rs1996371 | 76743861 | C | T | 0.41 | 95 | 0.02 | −0.16, 0.21 | 8.04E−01 | |
| rs16970006 | 76757314 | C | T | 0.07 | 92 | −0.01 | −0.42, 0.40 | 9.70E−01 |
The data shown here are from a linear regression including source of brain tissue as a covariate. Bold, italic and bold-italic representations denote highly correlated (r2 ≥ 0.6) variants in bins 1, 2 and 3, respectively. MAF, minor allele frequency. Asterisk (*) indicates a synonymous coding SNP; double asterisk (**) indicates a non-synonymous coding SNP.
Using linear regression with postmortem interval as a covariate, 28 of 44 variants showed significant evidence for association (P < 0.001) with CHRNA5 mRNA levels (Table 1). The variant showing the strongest evidence for association is an insertion/deletion polymorphism (rs3841324) located upstream of the coding region of CHRNA5. However, several other variants in the upstream region show comparable levels of association and are in high linkage disequilibrium with each other (bin 3, Table 1). Subjects homozygous for the minor allele at rs3841324 (S, short allele) show a 2.9-fold increase in CHRNA5 mRNA levels in frontal cortex compared with major allele homozygotes (Supplementary Material, Fig. S2). This confirms our previous finding reported in a subset of the brain tissues used in this study (33). The same observation was seen in brain tissue from alcohol-dependent and non-dependent subjects. Under an additive model, 42% of the variation in CHRNA5 mRNA expression is explained by rs3841324 or highly correlated polymorphisms. The variants showing the strongest association with CHRNA5 mRNA expression are not associated with either nicotine dependence or lung cancer in single SNP association tests in European American samples (22,31).
Variants in the LD bin previously reported to be associated with reduced risk for nicotine dependence and tagged by rs3743078 are also associated with variability in CHRNA5 mRNA expression levels. We observed that 13% of the variability in CHRNA5 mRNA expression is explained by rs3743078 or highly correlated variants in an additive model. However, using stepwise discriminant analysis with rs3841324 (bin 3) and rs3743078 (bin 2), the association between rs3743078 and CHRNA5 mRNA expression is no longer significant. The D398N variant (rs16969968 in bin 1) is more weakly associated with CHRNA5 mRNA expression (P = 0.01) (Supplementary Material, Fig. S3), though the association is no longer significant after inclusion of rs3841324 in the model.
Among the 44 polymorphisms genotyped, 10 SNPs were not highly correlated with any variants in bins 1, 2 or 3. Using the same linear regression analysis, we observed that six of these SNPs, including two rare missense variants in CHRNA3 and CHRNB4, respectively (rs8192475 and rs12914008) are associated with CHRNA5 gene expression (Table 1). Using stepwise discriminant analysis conditioning on rs3841324, SNPs rs11636605 and rs3813567 (highly correlated with rs11636605) remain weakly associated with mRNA expression (P = 0.02 and 0.04, respectively). The other SNPs are no longer significantly associated.
Lower variability in mRNA expression was observed for CHRNA3 and CHRNB4. Using linear regression with age as a covariate, variants in bin 2 and bin 3 are weakly associated with CHRNA3 mRNA expression levels (0.02 ≤ P ≤ 0.05) (Supplementary Material, Table S2). Polymorphisms in bin 3 are also weakly associated with CHRNB4 mRNA expression (0.01 ≤ P ≤ 0.05) in human brains using smoking history as a covariate and dropping individuals with unknown smoking history (Supplementary Material, Table S2). The D398N variant (bin 1) is not associated with differences in mRNA expression of either CHRNA3 or CHRNB4 (Supplementary Material, Table S2).
Diplotype analysis
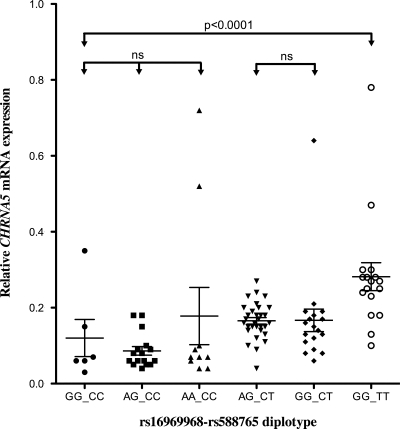
To determine which of the alleles of the D398N polymorphism exist as high- and low-expressing alleles, we performed haplotype and diplotype analyses using a variant associated with CHRNA5 expression levels. While the insertion/deletion variant, rs3841324, shows the strongest association with CHRNA5 mRNA expression, this variant has a lower genotyping success rate when compared with other variants in bin 3 and was not genotyped in all of the case–control series. Accordingly, we selected rs588765 for these analyses because it has a high genotyping call rate (>99%) and is genotyped in two of three case–control series. Haplotype and diplotype analysis with rs16969968 and rs588765 revealed three major haplotypes and six major diplotypes. The risk genotype (AA) at rs16969968 usually occurs with the low expression genotype (CC) at rs588765. However, the non-risk genotype (GG) at rs16969968 occurs with both the high expression genotype (TT at rs588765) and the low expression genotype (CC at rs588765) for CHRNA5 (Fig. 1, Supplementary Material, Table S3). By looking at diplotypes, we can examine the effect of each variant on a constant genotypic background for the other variant. For instance, among subjects with non-risk genotype at rs16969968, CHRNA5 mRNA expression levels are significantly associated with genotypic variants at rs588765 (GG_CC diplotype shows lower expression than diplotype GG_TT, P = 3.66 × 10−5) (Fig. 1 and Supplementary Material, Table S3). When examining the effect of rs16969968 on a constant genotypic background for rs588765 (see Fig. 1 and columns in Supplementary Material, Table S3), there are no differences in expression.
Figure 1.
Association of different rs16969968-rs588765 diplotypes with CHRNA5 mRNA expression (global test P < 0.0001). Diplotype analysis confirms that rs588765 alters CHRNA5 mRNA expression (GG_CC versus GG_TT; P = 3.66 × 10−5). It also demonstrates that rs16969968 does not influence CHRNA5 mRNA expression. The bars represent mean ± SD of CHRNA5 mRNA expression. We used SAS software to run t-test for pair-wise comparison of CHRNA5 mRNA expression with the specific genotype combination. The comparison of three diplotype groups (GGCC, AGCC and AACC) was analyzed with F-test.
Low levels of CHRNA5 mRNA are associated with lower risk for nicotine dependence
Based upon our biological evidence that the D398N variant in CHRNA5 alters receptor activity and that non-coding variation alters CHRNA5 mRNA expression, we performed a diplotype analysis in a large case–control series (ACS study) for nicotine dependence to test whether both the D398N variant and CHRNA5 mRNA expression levels influence risk. This analysis illustrates that both variants independently influence risk for nicotine dependence (Table 2). The non-risk genotype at rs16969968 occurs on both the high expression and low expression alleles of CHRNA5. When the non-risk genotype at rs16969968 occurs on the high-expression genotype of CHRNA5 (GG_TT diplotype), the risk for developing nicotine dependence is increased (OR = 1.72, CI 1.19–2.47) relative to GG_CC diplotype (Table 2, Fig. 1). Among the subjects heterozygous at rs16969968, subjects with higher expression (AG_CC versus AG_CT in Fig. 1) have higher risk for nicotine dependence (P = 3.59 × 10−6). The risk genotype of rs16969968 (AA) almost always occurs on the background of low expression. On this constant background, this SNP shows a dose-dependent increase in risk for nicotine dependence (Table 2), and subjects with the AA_CC diplotype have the highest risk for nicotine dependence (OR = 2.32, CI 1.58–3.43). Similar findings are seen in the COGEND data set (data not shown). While these findings indicate that the risk associated with the amino acid change outweighs the protective effect of low expression, risk for nicotine dependence is affected by both the D398N variant and variants that alter CHRNA5 mRNA expression.
Table 2.
Association of nicotine dependence with rs16969968-rs588765 diplotype in ACS study
| rs16969968 | rs588765 |
||
|---|---|---|---|
| CC (case/control) | CT (case/control) | TT (case/control) | |
| GG | 61/98 | 220/301 | 253/237 |
| OR (95% CI) | 1.00 (reference) | 1.17 (0.82 1.69) | 1.72 (1.19 2.47) |
| AG | 233/232 | 483/374 | 1/2 |
| OR (95% CI) | 1.61 (1.12 2.33) | 2.07 (1.47 2.94) | |
| AA | 191/132 | 0/2 | 0/0 |
| OR (95% CI) | 2.32 (1.58 3.43) | ||
Minor allele of rs3743078 tags the protective haplotype associated with lower CHRNA5 expression
Previous association studies have shown that rs3743078 and correlated SNPs in bin 2 (Table 1, Supplementary Material, Fig. S1) are associated with lower risk for nicotine dependence (22–24,26). In this study, we observe that these SNPs show moderate association with CHRNA5 mRNA levels, but are not strongly correlated with mRNA levels of CHRNA3 or CHRNB4 (Table 1, Supplementary Material, Table S1). This association is explained by LD between rs3743078 and rs588765 and correlated SNPs. To test whether the previously reported association with nicotine dependence also reflected differences in CHRNA5 mRNA expression levels, we performed a 2-SNP and 3-SNP haplotype analysis using rs16969968, rs588765 and rs3743078. Both analyses revealed only three major haplotypes (Tables 3 and 4). The risk variant (A allele) of rs16969968 primarily occurs with the major allele for rs588765 (C) and rs3743078 (C), which are correlated with low CHRNA5 mRNA expression. The major allele (G) of rs16969968 can pair with either major (C) (low expression) or minor (T) (high expression) alleles of rs588765. The GC haplotype (low expression) is associated with reduced risk for nicotine dependence (P = 5.27 × 10−9 in ACS data set; P = 1.82 × 10−3 in COGEND data set). Adding SNP rs3743078 to the haplotype analysis does not change the result indicating that the minor allele at rs3743078 is tagging the GC protective haplotype of rs16969968-rs588765 (Tables 3 and 4).
Table 3.
Association of nicotine dependence with 2-SNP haplotype of rs16969968-rs588765
| Data set | Haplotype | Frequency of affected subjects | Frequency of unaffected subjects | P-value | Effect on nicotine dependence | CHRNA5 variant D398N | CHRNA5 mRNA expression |
|---|---|---|---|---|---|---|---|
| ACSa (1422 cases, 1334 controls) | G_C | 0.20 | 0.27 | 5.27E−09 | protective | D | Low |
| G_T | 0.42 | 0.42 | 0.89 | Neutral | D | High | |
| A_C | 0.38 | 0.32 | 5.27E−07 | risk | N | Low | |
| COGENDb (797 cases, 813 controls) | G_C | 0.20 | 0.25 | 1.82E−03 | Protective | D | Low |
| G_T | 0.42 | 0.43 | 0.38 | Neutral | D | High | |
| A_C | 0.38 | 0.32 | 2.50E−04 | Risk | N | Low |
aGlobal test: P = 5.69E−10.
bGlobal test: P = 2.36E−04.
Table 4.
Association of nicotine dependence with 3-SNP haplotype of rs16969968-rs588765-rs3743078 in ACS data set
| Haplotype | Frequency of affected subjects | Frequency of unaffected subjects | P-value | Effect on nicotine dependence | CHRNA5 variant D398N | CHRNA5 mRNA expression |
|---|---|---|---|---|---|---|
| G_C_G | 0.20 | 0.26 | 3.20E−09 | Protective | D | Low |
| G_T_C | 0.42 | 0.42 | 0.80 | Neutral | D | High |
| A_C_C | 0.38 | 0.32 | 7.23E−07 | Risk | N | Low |
Global haplotype test: P = 4.68E−10.
Low levels of CHRNA5 mRNA reduce risk factor for lung cancer
To examine whether the same haplotypes are associated with lung cancer risk, we performed haplotype analysis with rs16969968 and rs6495306 in a case–control series for lung cancer (GELCC data set). We used rs6495306, which is highly correlated with rs588765 (r2 = 1 in COGEND data set) for haplotype analysis because rs588765 was not included in our lung cancer study. We observed the same three haplotypes that were seen in the case–control series for nicotine dependence. These haplotypes have similar effects on risk for lung cancer (Table 5). The risk allele of rs16969968 (A, coding for N398) for lung cancer is always associated with the major allele of rs6495306 (T), which is associated with lower mRNA expression of CHRNA5. The non-risk allele of rs16969968 (G, coding for D398) can be paired with either the major allele or the minor allele of rs6495306 (C). When D398 of rs16969968 occurs on the background of low mRNA expression of CHRNA5 (major allele of rs6495306, T), the haplotype is associated with lower risk for lung cancer (Table 5). In our data set over 86% of cases and 93% of controls used tobacco. Using pack years as a covariate, we observed similar association (global haplotype test P = 1.01 × 10−3). Because of the small size of this data set we were unable to separately analyze lung cancer individuals who have never smoked. Nonetheless, these data are consistent with part of the risk for lung cancer being explained by the same mechanism as for nicotine dependence.
Table 5.
Association of lung cancer with haplotype of rs16969968-rs6495306-rs3743078
| Data set | Haplotype | Frequency of affected subjects | Frequency of unaffected subjects | P-value | Effect on lung cancer | CHRNA5 Variant D398N | CHRNA5 mRNA expression |
|---|---|---|---|---|---|---|---|
| GELCC (194 cases, 219 controls) | G_T_G | 0.17 | 0.27 | 7.51E−04 | protective | D | Low |
| G_C_C | 0.43 | 0.43 | 0.96 | Neutral | D | High | |
| A_T_C | 0.40 | 0.30 | 3.90E−03 | Risk | N | Low |
Global haplotype test: P = 7.9E−04.
DISCUSSION
Further insights into the genetic basis of nicotine dependence and smoking have strong potential to inform ongoing lung cancer prevention and control efforts. The demonstration that variants in nAChRs are associated with nicotine dependence and lung cancer is an important step in understanding the pathogenesis of nicotine dependence and correlated medical illnesses.
In this study, we provide compelling evidence for at least two different mechanisms of action: both a coding variant that changes amino acid sequence in CHRNA5 (D398N) and non-coding variants that regulate CHRNA5 gene expression show association with risk for both nicotine dependence and lung cancer.
Surprisingly, the variants showing the strongest association with CHRNA5 expression are not associated with either nicotine dependence or lung cancer in single SNP association tests in European Americans. Only after diplotype analysis do we see that when subjects with the non-risk genotype for the D398N variant are associated with the low expression (GG_CC diplotype of rs16969968-rs588765 in ACS data set), the risk for developing nicotine dependence and lung cancer is significantly lower compared to those with the higher expression genotype (GG_TT diplotype of rs16969968-rs588765 in ACS data set). The variant CHRNA5 (D398N), which greatly increases the risk for both disorders, primarily occurs on the background of low mRNA expression of CHRNA5. A small number of individuals carry this risk allele on a high-expressing CHRNA5 allele. We speculate that the risk for developing nicotine dependence and lung cancer is further increased among these individuals, although the number of individuals with this haplotype was too small to formally test this hypothesis. The observation that lower mRNA expression of CHRNA5 with the non-risk genotype of rs16969968 is protective for nicotine dependence and lung cancer suggests that altered function of the N398 variant of α5 subunit likely does not fully explain the association between CHRNA5 and nicotine dependence and lung cancer. Genetic variants tagged by rs3740378 are associated with reduced risk for nicotine dependence and lung cancer in single SNP association tests. Our data show that this association reflects tagging of the protective haplotype associated with low mRNA expression of the normal CHRNA5 allele. Although we believe, based on our single SNP, haplotype and functional studies, that there are two distinct mechanisms associated with risk for nicotine dependence, we cannot completely rule out the possibility that there is a third untyped variant that could explain both the protective and risk effects detected here.
Neuronal nicotinic acetylcholine receptors form pentameric ligand-gated ion channels. In the brain, CHRNA5 is most commonly found in heteromeric receptors composed of α4β2α5 sub-units. In addition, the α5 subunit is expressed outside the brain, most prominently in postsynaptic receptors on ganglionic neurons (34). However, a number of studies have found that mRNA for the α5 subunit can be found in a number of non-neuronal cells (35). These observations indicate that genetic variation of α5 could have physiological effects at a number of sites of significance in terms of studies of behavior and addiction or carcinogenesis. Unfortunately, studies of the variants of the α5 subunit are at an early state, and only one previous functional study has been reported (25).
In speculating about possible connections, three aspects must be considered. The first is functional differences between receptors containing different variants. The second is the role of altered levels of protein expression of (either) α5 variant. Finally, there may be alterations in expression of other subunits in response to changes in α5 function or expression. Regarding the first point, our previous work has shown that α4β2* receptors containing the N398 CHRNA5 have a lower maximal response to acetylcholine than receptors containing α5 D398 (25). In addition, sorting and trafficking of nAChRs also depends strongly on the subunit composition and somatodendritic versus presynaptic localization of nAChRs in neurons will strongly affect neuronal responses to nicotine (36). Cellular trafficking (37) as well as interaction of α4β2* nAChRs with synaptic scaffolding proteins (13) is altered by the presence of the α5 subunit. The variant is located in the major cytoplasmic loop of the subunit, and might influence localization of receptors. Hence, there is reason to suggest that there will be differences in function and possibly in the location of receptors containing the two variants. Taking the second point, the specific subunit composition of nAChRs governs the acute responses to agonists, such as endogenous acetylcholine and exogenous nicotine. The presence of the α5 subunit affects the potency of agonists at α4β2* receptors, shifting the activation curve to lower concentrations of acetylcholine by 10-fold or more (37–39). Desensitization, which occurs at all nAChRs, describes the phenomenon that even in the maintained presence of agonist, the receptor has a closed channel. Kuryatov et al. (37) have shown that inclusion of the α5 subunit significantly increases the rate of desensitization of α4β2* nAChRs. In addition, calcium permeability varies markedly with subunit (40–42) composition, and inclusion of the α5 subunit in α4β2* nAChRs significantly increases calcium permeability (37,41). Since nicotine is much more slowly metabolized than acetylcholine, there may be a sustained Ca2+ influx which is thought to play a role in activating several signal transduction pathways that could lead to gene activation (in addiction) (43,44) or to cell proliferation (in cancer) (45). Finally, nicotine also acts as a pharmacological chaperone to assist in the folding and maturation of nAChRs (46–48). Selective pharmacological chaperoning of acetylcholine receptor number and stoichiometry is a general mechanism for the phenomenon first described in 1983 that chronic exposure to nicotine upregulates nAChRs (49–51). Such chaperoning by nicotine varies with detailed receptor stoichiometry, and in particular between α4β2 receptors and α4β2α5 receptors (although the reports differ in terms of the effect seen) (37,52). All of these studies indicate that the level of expression of the α5 subunit, in addition to functional differences between variants, will have an influence on the properties of α4β2* receptors. The final point, changes in the expression of other subunits depending on the level of expression of α5 protein, is suggested by studies of the α5-knockout mouse. Acetylcholine-induced dopamine release in striatum from α6 receptors was found to be inversely related to expression of the α5 subunit when wild-type mice, mice heterozygous for a null mutation in Chrna5 and mice homozygous for a null mutation in Chrna5 were compared (53).
Taking into account all of these observations, it is possible that the two biological factors are operating in two fashions. First, the risk for individuals having the D398 (lower risk) variant is lowered with low mRNA expression. This might suggest that a high level of α5 expression is a risk factor, per se, perhaps because of the functional consequences of incorporation of the α5 subunit into α4β2* receptors (discussed above). Second, the higher-risk, N398, variant is very strongly associated with low mRNA expression. This suggests that an additional property of this variant subunit confers an increased risk regardless of the expression level. It is not known what property this is, nor whether the risks for addiction or carcinogenesis will reflect the same property underlying this association.
Further studies are necessary to determine which of these mechanisms (desensitization, sorting, trafficking, permeability, chaperoning and potentially others) has the most significant impact on risk for nicotinic addiction.
Although investigators in the lung cancer field have suggested that the variants influencing risk for lung cancer may lie outside the nicotinic receptor gene cluster, gene expression data, functional data and genetic data point to the nicotinic receptor genes particularly for addiction and specifically CHRNA5 as the most likely candidate. Furthermore, a recent paper has reported that CHRNA5 mRNA expression is elevated in lung adenocarcinoma compared with normal lung tissue and that expression of CHRNA5 in normal lung tissue is associated with genotype at rs16969968 suggesting that CHRNA5 is also the most likely candidate gene for lung cancer (54). No other SNPs were tested for association with CHRNA5 mRNA expression in that study. However, based on our studies in brain and lymphocytes, we would anticipate that CHRNA5 mRNA expression in lung is more strongly associated with rs588765 and other highly correlated SNPs than with rs16969968.
This work provides a potential drug target for the treatment of nicotine addiction/lung cancer and will lead to a better understanding of the underlying biology of nicotine addiction and smoking-related illnesses.
MATERIALS AND METHODS
Quantitative gene expression analysis in human brain
Postmortem brain tissue derived from the frontal cortex of 44 unrelated, non-demented elderly European Americans was obtained from the Alzheimer's Disease Research Center (ADRC) of Washington University in St Louis (http://alzheimer.wustl.edu/). Smoking status (tobacco use) is available for these subjects. We have also obtained a second set of frontal cortex samples from the Australian Brain Donor Program (ABDP), Sydney, Australia. Thirty-four of these samples were derived from unrelated, non-alcohol-dependent subjects and 35 samples were from alcohol-dependent subjects. Fifty-nine samples were of European descent. Though the smoking history (ever smoke or never smoke) was not always recorded, 22 subjects who are alcohol-dependent were smokers, and two were non-smokers. Among the non-alcohol-dependent subjects 14 were smokers and four were non-smokers. Others are unknown.
We used Qiagen's DNeasy Blood and Tissue Kit and RNeasy Lipid Tissue kit (http://www.qiagen.com) to extract DNA and total RNA from brain tissues, respectively. A cDNA library was prepared from total RNA using the High Capacity cDNA Archive Kit (http://www.appliedbiosystems.com). Genomic DNA from all subjects was genotyped for 44 polymorphisms in the CHRNA5/A3/B4 gene cluster. We used the Sequenom MassArray platform for genotyping. A detailed genotyping protocol using MassArray technology is described elsewhere (55).
TaqMan assays (Applied Biosystems, CA, USA) were used to quantify the expression levels of CHRNA5 (Hs00181248_m1), CHRNA3 (Hs01088199_m1) and CHRNB4 (Hs00609520_m1) mRNAs in human frontal cortex. Gene expression levels were analyzed by real-time PCR using an ABI-7500 real-time PCR system. The program, Primer Express 3 (ABI) was used to design primers and a TaqMan probe for the GAPDH gene. Each real-time PCR run included within-plate duplicates and each experiment was performed twice for each sample. Correction for sample-to-sample variation was done by simultaneously amplifying GAPDH as a reference. Real-time data was analyzed using the comparative Ct method (56).
We used linear regression to test for evidence of differential expression in samples of different genotypes. To minimize possible effects of sample heterogeneity, we performed our association analyses in subjects of European descent only. The origin of the sample (brain bank site), postmortem interval, age, gender, drinking status and smoking history were used as covariates. For diplotype analysis, we first log-transformed relative mRNA expression to obtain a normal distribution and then used a t-test to run pair-wise comparisons of CHRNA5 mRNA expression with the specific genotype combination. A comparison of three diplotype groups (GGCC, AGCC and AACC) was made using F-test in the SAS software release 9.1 (SAS Institute, Cary, NC, USA).
COGEND study
The Collaborative Genetic Study of Nicotine Dependence recruited subjects from three urban areas in the USA: St Louis, Detroit and Minneapolis. A community-based telephone-screening interview was used to identify individuals who had smoked at least 100 cigarettes in their lifetime (smokers). These individuals then completed the Fagerstorm Test of Nicotine Dependence (FTND) questionnaire. Case subjects were current smokers with a score of four or more on the FTND, which defines nicotine dependence. Control subjects smoked 100 cigarettes but never exhibited symptoms of nicotine dependence (FTND = 0), even during the heaviest period of smoking. Details of this sample and genotyping process have been reported elsewhere (21,23,57).
ACS study
The smokers used in this study were participants in the American Cancer Society CPS-II Cohort, a prospective study of cancer mortality begun in 1982, and the CPS-II Nutrition Cohort, a prospective study of cancer incidence formed in 1992 using a subset of CPS-II participants. Details regarding recruitment into these studies are described elsewhere (26). Information on smoking behavior from questionnaires administered in 1982, 1992 and 1997 was used to identify light and heavy smokers among participants. Individuals who reported smoking at least 30 cigarettes/day for at least 5 years were defined as heavy smokers. This phenotype is strongly correlated with nicotine dependence (26). We randomly selected 750 heavy smoking men and 750 heavy smoking women for this study (total n = 1500). Light smokers were defined as individuals who reported smoking for at least 1 year during their lifetime and in 1982 and 1992, reported always smoking fewer than 10 cigarettes/day. This was a rare phenotype especially among men. Among participants with DNA, 461 men and 1482 women met these criteria. We included all men defined as light smokers as well as a random sample of 1039 light smoking women to give a total of 1500 light smokers. A detailed description of the genotyping process was described elsewhere (26).
Lung cancer study subjects
We genotyped 194 cases with familial lung cancer and 219 cancer-free control subjects of European descent from the Genetic Epidemiology of Lung Cancer Consortium (GELCC) using Affymetrix 500K or Affymetrix Genome-Wide Human SNP Array 6.0 (Santa Clara, CA, USA). A detailed description of the genotyping process is described elsewhere (31). A sample of unrelated case individuals was identified by selecting one case from each high-risk lung cancer family. Among 194 cases, 186 individuals were smokers. Non-cancer control subjects were recruited from a combination of unaffected spouses from GELCC families (n = 36), unaffected individuals from the Coriell Institute for Medical Research (Camden, NJ, USA) (n = 11) and the Fernald Medical Monitoring Program (Fernald, OH, USA) (n = 172). These control subjects had no blood relationship with any selected case patients. Among 219 controls, 205 individuals used tobacco. Basic characteristics of the GELCC subjects are presented elsewhere (31).
Data analysis
To minimize possible effects of sample heterogeneity, we performed our association analyses in subjects of European descent only. We used the program PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) to generate haplotypes and used linear regression to test the association of different haplotypes with nicotine dependence and lung cancer. The SAS software release 9.1 (SAS Institute, Cary, NC, USA) was used to test the association of specific genotype combinations with nicotine dependence.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
COGEND project
In memory of Theodore Reich, founding Principal Investigator of Collaborative Genetic Study of Nicotine Dependence (COGEND), we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. The COGEND project is a collaborative research group and part of the NIDA Genetics Consortium. Subject collection was supported by NIH grant CA89392 (PI-L Bierut) from the National Cancer Institute. Lead investigators directing data collection are Laura Bierut, Naomi Breslau, Dorothy Hatsukami and Eric Johnson. The authors thank Heidi Kromrei and Tracey Richmond for their assistance in data collection. Genotyping work at Perlegen Sciences was performed under NIDA Contract HHSN271200477471C. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C (PIs J Tischfield and J Rice). Genotyping services were also provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096.
The following authors are included under the COGEND collaborators: N. Breslau1, R. Culverhouse2, D. Hatsukami3, A. Hinrichs2 and Eric Johnson4.
1Department of Epidemiology, Michigan State University, East Lansing, MI 48824, USA.
2Department of Psychiatry, Washington University, St Louis, MO, USA.
3Department of Psychiatry, University of Minnesota, Minneapolis, MN 55454, USA.
4Research Triangle Institute International, Research Triangle Park, NC 27709, USA.
GELCC project
The Genetic Epidemiology of Lung Cancer Consortium (GELCC) was formed by scientists from several US universities, plus the National Cancer Institute and the Human Genome Research Institute to identify lung cancer susceptibility genes in familial lung cancer populations. The Principal Investigator is Dr Marshall Anderson, and the members of GELCC are: University of Cincinnati (M Anderson, SM Pinney, J Lee, E Kupert), Washington University (M You, Y Wang, P Liu, H Vikis, Y Lu), Mayo Foundation & Clinic (M de Andrade, P Yang, GM Petersen), Karmanos Cancer Center (AG Schwartz), University of Colorado Health Science (PR Fain), University of Toledo College of Medicine (C Gaba), University of Texas Southwestern Medical Center (J Minna, A Gazdar), Louisiana State University (Diptasri Mandal), National Cancer Institute (D Seminara), National Human Genome Research Institute (JE Bailey-Wilson), MD Anderson Cancer Center (CI Amos). This work was supported by the NIH grant U01CA076293 from the National Cancer Institute.
The following authors are included under the GELCC collaborators: Haris Vikis1, Yan Lu1, Yian Wang1, Ping Yang2, Susan M. Pinney3, Gloria M. Petersen2, Mariza de Andrade2, Ann G. Schwartz4, Adi Gazdar5, Colette Gaba6, Diptasri Mandal7, Elena Kupert3, Juwon Lee3, Daniela Seminara8, Pamela R. Fain9, John Minna5, Joan E. Bailey-Wilson10, Yafang Li11, Christopher I. Amos11
1Department of Surgery, Washington University, St Louis, MO, USA.
2Mayo Clinic, Rochester, MN, USA.
3University of Cincinnati, Cincinnati, OH, USA.
4Karmanos Cancer Institute, Detroit, MI, USA.
5University of Texas Southwestern Medical Center, Dallas, TX, USA.
6University of Toledo College of Medicine, Toledo, OH, USA.
7Louisiana State University Health Science Center from Louisiana State University, New Orleans, LA, USA.
8National Cancer Institute, Bethesda, MD, USA.
9University of Colorado, Denver, CO, USA.
10National Human Genome Research Institute, Bethesda, MD, USA.
11M. D. Anderson Cancer Center, Houston, TX, USA.
COGA Project
This study is partially supported by the COGA project. The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis and storage take place. The nine sites and Principal Investigators and Co-investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr, T. Foroud); University of Iowa (S. Kuperman); SUNY Downstate (B. Porjesz); Washington University in St Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Q. Max Guo is the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field.
Conflict of Interest statement. L.J.B., A.M.G., A.J. Hinrichs, J.P.R., S.F.S. and J.C.W. are listed as inventors on a patent (US 20070258898) held by Perlegen Sciences, Inc., covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. N.L.S. is the spouse of S.F.S., who is listed as an inventor on the above-mentioned patent. Bierut has acted as a consultant for Pfizer, Inc. in 2008.
ACKNOWLEDGEMENT
We are grateful to the families for their participation in the studies at the Washington University Alzheimer's Disease Research Center (ADRC) and at the Australian Brain Donor Program (ABDP), Sydney, Australia. Funding for the research at the ADRC was provided by grants from the National Institute on Aging: P50 AG05681 and P01 AG03991 to J.C.M. We thank Dr Henry Lester for helpful discussions.
REFERENCES
- 1.CDC. Tobacco use among adults–United States. Morb. Mortal. Wkly. Rep. 2007;55:1145–1148. [PubMed] [Google Scholar]
- 2.SAMHSA. Results from the 2005 National Survey on Drug Use and Health: National Findings. Substance abuse and mental health services administration: NSDUH Series H-30. 2006 DHHS Publication No. SMA 06-4194. [Google Scholar]
- 3.World Health Organization. Regulation urgently needed to control growing list of deadly tobacco products. 2006 www.who.int/entity/mediacentre/news/releases/2006/pr28/en/ [Google Scholar]
- 4.CDC. Tobacco and alcohol use. Morb. Mortal. Wkly. Rep. 2004;53:19–32. [Google Scholar]
- 5.Mokdad A., Marks J., Stroup D., Gerberding J. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 6.Bierut L., Dinwiddie S., Begleiter H., Crowe R., Hesselbrock V., Nurnberger J., Jr, Porjesz B., Schuckit M., Reich T. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch. Gen. Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 7.Kendler K., Neale M., Sullivan P., Corey L., Gardner C., Prescott C. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol. Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- 8.Lessov C., Martin N., Statham D., Todorov A., Slutske W., Bucholz K., Heath A., Madden P. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol. Med. 2004;34:865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- 9.True W., Xian H., Scherrer J., Madden P., Bucholz K., Heath A., Eisen S., Lyons M., Goldberg J., Tsuang M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch. Gen. Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- 10.Carmelli D., Swan G., Robinette D., Fabsitz R. Genetic influence on smoking–a study of male twins. N. Engl. J. Med. 1992;327:829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- 11.Gotti C., Zoli M., Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Dani J., Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- 13.Conroy W., Berg D. Nicotinic receptor subtypes in the developing chick brain: appearance of a species containing the alpha4, beta2 and alpha5 gene products. Mol. Pharmacol. 1998;53:392–401. doi: 10.1124/mol.53.3.392. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Latorre J., Yu C., Qu X., Perin F., Karlin A., Role L. Functional contributions of alpha5 subunit to neuronal acetylcholine receptor channels. Nature. 1996;380:347–351. doi: 10.1038/380347a0. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta P., Chellappan S. Nicotine-mediated cell proliferation and angiogenesis: new twists to an old story. Cell Cycle. 2006;5:2324–2328. doi: 10.4161/cc.5.20.3366. [DOI] [PubMed] [Google Scholar]
- 16.Dasgupta P., Kinkade R., Joshi B., Decook C., Haura E., Chellappan S. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc. Natl Acad. Sci. USA. 2006;103:6332–6337. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta P., Rizwani W., Pillai S., Kinkade R., Kovacs M., Rastogi S., Banerjee S., Carless M., Kim E., Coppola D., et al. Nicotine induces cell proliferation, invasion and epithelial-mesenchymal transition in a variety of human cancer cell lines. Int. J. Cancer. 2009;124:36–45. doi: 10.1002/ijc.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehringer M., Clegg H., Collins A., Corley R., Crowley T., Hewitt J., Hopfer C., Krauter K., Lessem J., Rhee S., et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2007;144:596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y., Niu T., Xing H., Xu X., Chen C., Peng S., Wang L., Laird N., Xu X. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am. J. Hum. Genet. 2004;75:112–121. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., Beuten J., Ma J., Payne T., Lou X., Garcia V., Duenes A., Crews K., Elston R. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum. Mol. Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 21.Bierut L., Madden P., Breslau N., Johnson E., Hatsukami D., Pomerleau O., Swan G., Rutter J., Bertelsen S., Fox L., et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saccone N., Saccone S.F., Hinrichs A.L., Stitzel J.A., Duan W., Pergadia M.L., Agrawal A., Breslau N., Grucza R.A., Hatsukami D., et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saccone S., Hinrichs A., Saccone N., Chase G., Konvicka K., Madden P., Breslau N., Johnson E., Hatsukami D., Pomerleau O., et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berrettini W., Yuan X., Tozzi F., Song K., Francks C., Chilcoat H., Waterworth D., Muglia P., Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bierut L., Stitzel J., Wang J., Hinrichs A., Grucza R., Xuei X., Saccone N., Saccone S., Bertelsen S., Fox L., et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens V., Bierut L.J., Talbot J.T., Wang J.C., Sun J., Hinrichs A.L., Thun M.J., Goate A., Calle E.E. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol. Biomarkers Prev. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorgeirsson T., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K., Manolescu A., Thorleifsson G., Stefansson H., Ingason A., et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss R., Baker T., Cannon D., von Niederhausern A., Dunn D., Matsunami N., Singh N., Baird L., Coon H., McMahon W., et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amos C., Wu X., Broderick P., Gorlov I., Gu J., Eisen T., Dong Q., Zhang Q., Gu X., Vijayakrishnan J., et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung R., McKay J., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P., et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 31.Liu P., Vikis H.G., Wang D., Lu Y., Wang Y., Schwartz A.G., Pinney S.M., Yang P., de Andrade M., Petersen G.M., et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherva R., Wilhelmsen K., Pomerleau C.S., Chasse S.A., Rice J.P., Snedecor S.M., Bierut L.J., Neuman R.J., Pomerleau O.F. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103:1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Grucza R., Cruchaga C., Hinrichs A., Bertelsen S., Budde J., Fox L., Goldstein E., Reyes O., Saccone N., et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao D., Yasuda R.P., Fan H., Wolfe B.B., Kellar K.J. Heterogeneity of nicotinic cholinergic receptors in rat superior cervical and nodose Ganglia. Mol. Pharmacol. 2006;70:1693–1699. doi: 10.1124/mol.106.027458. [DOI] [PubMed] [Google Scholar]
- 35.Chini B., Hukovic F., Hukovic N., Sher E. Neuronal-type alpha-bungarotoxin receptors and the alpha 5-nicotinic receptor subunit gene are expressed in neuronal and nonneuronal human cell lines. Proc. Natl Acad. Sci. USA. 1992;89:1572–1576. doi: 10.1073/pnas.89.5.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams B., Temburni M., Levey M., Bertrand S., Bertrand D., Jacob M. The long internal loop of the alpha 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nat. Neurosci. 1998;1:557–562. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- 37.Kuryatov A., Onksen J., Lindstrom J. Roles of accessory subunits in alpha4beta2(*) nicotinic receptors. Mol. Pharmacol. 2008;74:132–143. doi: 10.1124/mol.108.046789. [DOI] [PubMed] [Google Scholar]
- 38.Brown R., Collins A.C., Lindstrom J.M., Whiteaker P. Nicotinic alpha5 subunit deletion locally reduces high-affinity agonist activation without altering nicotinic receptor numbers. J. Neurochem. 2007;103:204–215. doi: 10.1111/j.1471-4159.2007.04700.x. [DOI] [PubMed] [Google Scholar]
- 39.Kassam S., Herman P.M., Goodfellow N.M., Alves N.C., Lambe E.K. Developmental excitation of corticothalamic neurons by nicotinic acetylcholine receptors. J. Neurosci. 2008;28:8756–8764. doi: 10.1523/JNEUROSCI.2645-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sands S., Barish M.E. Neuronal nicotinic acetylcholine receptor currents in phaeochromocytoma (PC12) cells: dual mechanisms of rectification. J. Physiol. 1992;447:467–487. doi: 10.1113/jphysiol.1992.sp019012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapia L., Kuryatov A., Lindstrom J. Ca2+ permeability of the (alpha4)3(beta2)2 stoichiometry greatly exceeds that of (alpha4)2(beta2)3 human acetylcholine receptors. Mol. Pharmacol. 2007;71:769–776. doi: 10.1124/mol.106.030445. [DOI] [PubMed] [Google Scholar]
- 42.Vernino S., Amador M., Luetje C.W., Patrick J., Dani J.A. Calcium modulation and high calcium permeability of neuronal nicotinic acetylcholine receptors. Neuron. 1992;8:127–134. doi: 10.1016/0896-6273(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 43.Brunzell D., Russell D.S., Picciotto M.R. In vivo nicotine treatment regulates mesocorticolimbic CREB and ERK signaling in C57Bl/6J mice. J. Neurochem. 2003;84:1431–1441. doi: 10.1046/j.1471-4159.2003.01640.x. [DOI] [PubMed] [Google Scholar]
- 44.Shaw S., Bencherif M., Marrero M.B. Janus kinase 2, an early target of alpha 7 nicotinic acetylcholine receptor-mediated neuroprotection against Abeta-(1–42) amyloid. J. Biol. Chem. 2002;277:44920–44924. doi: 10.1074/jbc.M204610200. [DOI] [PubMed] [Google Scholar]
- 45.Catassi A., Servent D., Paleari L., Cesario A., Russo P. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat. Res. 2008;659:221–231. doi: 10.1016/j.mrrev.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Kuryatov A., Luo J., Cooper J., Lindstrom J. Nicotine acts as a pharmacological chaperone to up-regulate human alpha4beta2 acetylcholine receptors. Mol. Pharmacol. 2005;68:1839–1851. doi: 10.1124/mol.105.012419. [DOI] [PubMed] [Google Scholar]
- 47.Nashmi R., Lester H. Cell autonomy, receptor autonomy, and thermodynamics in nicotine receptor up-regulation. Biochem. Pharmacol. 2007;74:1145–1154. doi: 10.1016/j.bcp.2007.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sallette J., Pons S., Devillers-Thiery A., Soudant M., Prado de Carvalho L., Changeux J.P., Corringer P.J. Nicotine upregulates its own receptors through enhanced intracellular maturation. Neuron. 2005;46:595–607. doi: 10.1016/j.neuron.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 49.Lester H., Xiao C., Srinivasan R., Son C., Miwa J., Pantoja R., Dougherty D., Banghart M., Goate A., Wang J.C. Nicotine is a selective pharmacological chaperone of acetylcholine receptor number and stoichiometry. Implications for drug discovery. Am. Assoc. Pharma. Sci. 2009;11:167–177. doi: 10.1208/s12248-009-9090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks M., Burch J., Collins A. Effects of chronic nicotine infusion on tolerance development and nicotinic receptors. J. Pharmacol. Exp. Ther. 1983;226:817–825. [PubMed] [Google Scholar]
- 51.Schwartz R., Kellar K.J. Nicotinic cholinergic receptor binding sites in the brain: regulation in vivo. Science. 1983;220:214–216. doi: 10.1126/science.6828889. [DOI] [PubMed] [Google Scholar]
- 52.Mao D., Perry D.C., Yasuda R.P., Wolfe B.B., Kellar K.J. The alpha4beta2alpha5 nicotinic cholinergic receptor in rat brain is resistant to up-regulation by nicotine in vivo. J. Neurochem. 2008;104:446–456. doi: 10.1111/j.1471-4159.2007.05011.x. [DOI] [PubMed] [Google Scholar]
- 53.Salminen O., Murphy K., McIntosh J., Drago J., Marks M., Collins A., Grady S. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol. Pharmacol. 2004;65:1526–1535. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- 54.Falvella F., Galvan A., Frullanti E., Spinola M., Calabrò E., Carbone A., Incarbone M., Santambrogio L., Pastorino U., Dragani T. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin. Cancer Res. 2009;15:1837–1842. doi: 10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- 55.Wang J., Hinrichs A.L., Stock H., Budde J., Allen R., Bertelsen S., Kwon J.M., Wu W., Dick D.M., Rice J., et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum. Mol. Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- 56.Muller P., Janovjak H., Miserez A., Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. 1376, 1378–1379. [PubMed] [Google Scholar]
- 57.Saccone N., Saccone S., Goate A., Grucza R., Hinrichs A., Rice J., Bierut L. In search of causal variants: refining disease association signals using cross-population contrasts. BMC Genet. 2008;9:58. doi: 10.1186/1471-2156-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.