Abstract
Copy number studies have led to an explosion in the discovery of new segmental duplication-mediated deletions and duplications. We have analyzed copy number changes in 2419 patients referred for clinical array comparative genomic hybridization studies. Twenty-three percent of the abnormal copy number changes we found are immediately flanked by segmental duplications ≥10 kb in size and ≥95% identical in direct orientation, consistent with deletions and duplications generated by non-allelic homologous recombination. Here, we describe copy number changes in five previously unreported loci with genomic organization characteristic of NAHR-mediated gains and losses; namely, 2q11.2, 7q36.1, 17q23, 2q13 and 7q11.21. Deletions and duplications of 2q11.2, deletions of 7q36.1 and deletions of 17q23 are interpreted as pathogenic based on their genomic size, gene content, de novo inheritance and absence from control populations. The clinical significance of 2q13 deletions and duplications is still emerging, as these imbalances are also found in phenotypically normal family members and control individuals. Deletion of 7q11.21 is a benign copy number change well represented in control populations and copy number variation databases. Here, we discuss the genetic factors that can modify the phenotypic expression of such gains and losses, which likely play a role in these and other recurrent genomic disorders.
INTRODUCTION
Segmental duplications are substrates of genomic instability that mediate deletions and duplications by unequal crossing-over between paralogous segments. This mechanism is responsible for the genomic changes underlying many classic genetic syndromes, including 22q11.2 deletion syndrome, Charcot-Marie-Tooth syndrome, hereditary neuropathy with liability to pressure palsies, Williams-Beuren syndrome, Prader-Willi syndrome, Angelman syndrome and Smith-Magenis syndrome (1). Recent whole-genome array comparative genomic hybridization (CGH) studies have led to the identification of new segmental duplication-mediated chromosome rearrangements that are emerging as distinct syndromes (2–11). Given that segmental duplications make up approximately 5% of the human genome (12), many more rearrangements await discovery. Copy number changes mediated by segmental duplications may be either pathogenic or exist as benign variants in the human population. Here, we describe five novel loci subject to copy number change flanked by highly homologous segmental duplications. As with other recurrent rearrangements generated by NAHR, we find reciprocal deletions and duplications.
RESULTS
We analyzed 2419 samples from patients referred to our clinical cytogenetics laboratory for array CGH testing. Clinical indications for testing were diverse, and included developmental delay, autism and birth defects among the most common features reported to us. Our array combines targeted and whole-genome coverage on a 44 000-oligonucleotide platform with a mean spacing of 75 kb between genomic probes (13). We identified 457 array cases representing clinically significant genomic abnormalities (19% abnormality rate) and compared these copy number changes to 130 rearrangement hotspots described by Sharp et al. (5). Rearrangement hotspots were defined as 50 kb to 10 Mb genomic regions flanked by segmental duplications that are ≥10 kb in size and ≥95% identical (5). We identified 98 patients with previously described genomic disorders mediated by segmental duplications (Supplementary Material, Table S1). Nine patients had deletions and/or duplications in four previously unreported loci flanked by segmental duplications (Fig. 1, Table 1, Supplementary Material, Table S2). Thus, 23% (107/457) of the clinically significant copy number changes identified by our array are flanked by segmental duplications. We also detected a benign deletion of chromosome 7q11.21 flanked by segmental duplications in one patient with no clinically significant copy number changes. All deletions and duplications were heterozygous (loss or gain of one copy) and were confirmed by fluorescence in situ hybridization (FISH), as described previously (13). To evaluate the inheritance of copy number changes, we performed FISH analysis on parental samples with probes specific to the regions of gain or loss. We then verified the de novo status of copy number changes not detected in parents by microsatellite analysis (15 loci evaluated, AmpFlSTR Identifiler PCR Amplification Kit no. 4322288, Applied Biosystems, Inc.), which confirmed familial relationships in all cases.
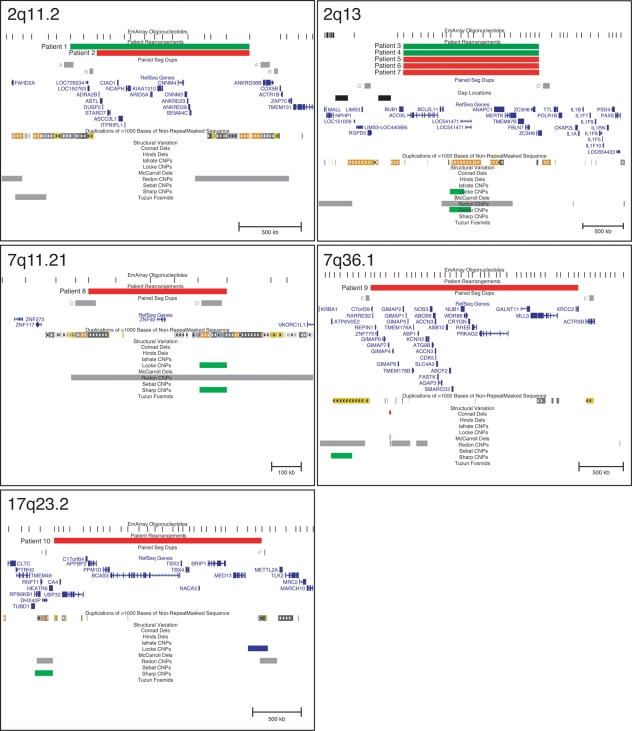
Figure 1.
Genome views of five loci flanked by segmental duplications, extracted from Build 36.1 (hg18) of the human genome assembly (44). Patient copy number changes are displayed as the minimal region of loss (red) or gain (green) detected by EmArray oligonucleotide probes (13). Flanking paired segmental duplications are labeled with letters corresponding to Table 2 and shown in gray. RefSeq genes, segmental duplications and structural variation are as described in http://www.genome.ucsc.edu/.
Table 1.
Summary of copy number changes flanked by segmental duplications
| Patient | Chr band | First oligo | Last oligo | Minimum size | Gain/loss | Genes | Inheritance |
|---|---|---|---|---|---|---|---|
| 1 | 2q11.2 | 95909077 | 97380005 | 1.47 Mb | Gain | 17 | De novo |
| 2 | 2q11.2 | 96130287 | 97380005 | 1.25 Mb | Loss | 17 | Unknown |
| 3 | 2q13 | 111158601 | 112782250 | 1.62 Mb | Gain | 10 | Paternal |
| 4 | 2q13 | 111158601 | 112782250 | 1.62 Mb | Gain | 10 | Paternal |
| 5 | 2q13 | 111158601 | 112782250 | 1.62 Mb | Loss | 10 | Unknown |
| 6 | 2q13 | 111158601 | 112782250 | 1.62 Mb | Loss | 10 | Paternal |
| 7 | 2q13 | 111158601 | 112782250 | 1.62 Mb | Loss | 10 | Unknown |
| 8 | 7q11.21 | 64247264 | 64708354 | 461 kb | Loss | 1 | Unknown |
| 9 | 7q36.1 | 149647886 | 152002289 | 2.35 Mb | Loss | 35 | De novo |
| 10 | 17q23 | 55527482 | 57670085 | 2.14 Mb | Loss | 11 | De novo |
We report genomic size as the minimum region of copy number change detected by array CGH. The boundaries of the first and last oligonucleotides for a given copy number change are listed. Genomic coordinates correspond to Build 36.1 (hg18) of the human genome assembly. Unknown inheritance indicates cases where parents were unavailable for testing.
Next we analyzed the genomic architecture of the five regions relative to the reference genome assembly (Build 36.1, hg18). Segmental duplications were identified using the duplication track on the UCSC browser (14,15) and confirmed using the cross_match alignment program (http://www.phrap.org/). We identified six highly homologous segmental duplication pairs in direct orientation flanking the deletion and duplication regions (Table 2). Such conformations are capable of mediating reciprocal gains and losses via non-allelic homologous recombination (NAHR) (1).
Table 2.
Segmental duplications (SDs) flanking patient copy number gains and losses
| Patient | Chr band | SDs | SD size | Percent ID | Distance between SDs |
|---|---|---|---|---|---|
| 1 | 2q11.2 | A/A′ | 74 kb | 95.0 | 1.53 Mb |
| 2 | 2q11.2 | B/B′ | 31 kb | 98.3 | 1.48 Mb |
| 3–7 | 2q13 | C/C′ | 76 kb | 99.6 | 2.06 Mb |
| 8 | 7q11.21 | D/D′ | 96 kb | 99.4 | 415 kb |
| 9 | 7q36.1 | E/E′ | 43 kb | 98.8 | 2.50 Mb |
| 10 | 17q23 | F/F′ | 15 kb | 98.7 | 2.24 Mb |
The size, percent identity and distance between paired SDs are listed. See Figure 1 for the genomic organization of SDs.
We interpreted the clinical significance of deletions and duplications based on the size of gain/loss, gene content, inheritance pattern and frequency in control populations (Fig. 1, Table 1). Given the relatively few individuals carrying any particular copy number change, comprehensive genotype–phenotype correlations were not possible at this time. The major phenotypic features of the patients in our study may be found in the supplementary data (Supplementary Material, Table S3).
To determine the frequency of copy number changes at our five loci in the general population, we searched previously reported copy number variation (CNV) data sets ascertained from apparently normal individuals (16–25) and consulted CNVs reported in online databases (http://www.genome.ucsc.edu/; http://projects.tcag.ca/variation/). We also screened a control population of 876 individuals for gains and losses of the five loci (Affymetrix Genome-Wide Human SNP Array 6.0: n = 347 unaffected parents of schizophrenia probands, plus n = 529 Ashkenazi Jewish patients with Crohn's disease, dystonia or Parkinson's disease; J.G.M., unpublished data).
We identified two patients with overlapping 1.5-Mb copy number changes in 2q11.2 flanked by clusters of segmental duplications. Patient 1's duplication is slightly larger than Patient 2's deletion, corresponding to neighboring segmental duplication pairs A/A′ and B/B′ (Fig. 1). Segmental duplications A and B are not homologous, rather they are part of large duplication clusters flanking the rearrangements. The deletion and duplication span a minimum of 17 genes, none of which have been associated with human disease. Parental studies demonstrated that Patient 1's duplication was de novo; however, Patient 2's parents were unavailable for study. Neither the duplication nor the deletion was found in our control population or in CNV databases, consistent with the interpretation of these imbalances as pathogenic copy number changes.
The 2.35 Mb deletion of 7q36.1 spans 35 genes, including two genes involved in heart function, PRKAG2 and KCNH2 (Fig. 1). Heterozygous mutations in PRKAG2 cause hypertrophic cardiomyopathy with Wolff–Parkinson–White syndrome (MIM 602743) (26–28). KCNH2 encodes a voltage-gated potassium channel, and mutations in this gene cause long QT syndrome type 2 (MIM 152427) (29,30) and short QT syndrome type 1 (MIM 609620) (31). Mutations in both PRKAG2 and KCNH2 are inherited in an autosomal dominant manner, exhibiting variable expressivity and incomplete penetrance (26,32–34). Patient 9 had a normal EKG at 19 months of age, suggesting incomplete penetrance of the cardiac defects associated with loss of function of PRKAG2 and KCNH2. However, heterozygous mutations in KCNH2 have been found in patients with epilepsy (35,36), and Patient 9 has a history of seizures. The 7q36.1 deletion occurred de novo and was not present in either our control population or CNV databases.
The 2.14-Mb deletion of 17q23 removes 11 genes, including TBX4. Notably, heterozygous loss-of-function mutations in TBX4 have been identified in families with autosomal dominant small patella syndrome (MIM 147891) (37); nevertheless, Patient 14 shows no patellar, pelvic or foot anomalies typical of the syndrome. The 17q23 deletion occurred de novo and is not found in our control population or other CNV databases. The absence of a skeletal phenotype in Patient 14 may be owing to incomplete penetrance of small patella syndrome.
The most frequent site of deletion/duplication in our study is a 1.62-Mb region of 2q13 that includes 10 genes. There is an assembly gap in the interval of gain/loss; thus, the region may be larger or smaller than 1.62 Mb (Fig. 1). We identified two duplications (both paternally inherited) and three reciprocal deletions (one paternally inherited, two of unknown inheritance) (Table 1). Patient 7 also carries an unbalanced translocation, complicating our interpretation of how the 2q13 deletion contributes to his phenotype. Thus, we compared the phenotypes of Patients 3–6 to discern common features of the deletion and duplication. Most phenotypic findings were non-specific (Supplementary Material, Table S3); however, tooth abnormalities were noted in three patients. Patients 3 and 4 (duplication carriers) have dental crowding, and Patient 6 (deletion carrier) has widely spaced teeth. Interestingly, the FBLN7 gene is located in the 2q13 region of gain/loss. FBLN7 is a cell adhesion molecule that plays a critical role in the differentiation and maintenance of odontoblasts and in dentin formation (38). To date, there are no known loss-of-function mutations in FBLN7; however, it is tempting to speculate that loss and gain of the gene may affect tooth development.
An overlapping 2q13 deletion has been reported previously, though the flanking segmental duplications and mechanism of rearrangement were not described (39). Brothers with the deletion had developmental delay and dysmorphic features not present in their unaffected mother who transmitted the deletion. The more affected proband had hypotonia and epilepsy, features found in a subset of our patients with 2q13 deletions (Supplementary Material, Table S3). Tooth abnormalities were not described in this family.
We detected one individual with the same 2q13 duplication in our control set. The reciprocal deletion, on the other hand, was not detected in our controls, but the deletion was transmitted from an apparently normal parent in at least one family in our study and another in the literature (39). Approximately 850 kb of the 1.62-Mb 2q13 region has been reported in CNV databases; however, many genes lie outside of this region, including FBLN7 (Fig. 1). Thus, there may be phenotypic differences between those carrying gains and/or losses of the common 850 kb CNV and the 1.6 Mb region we describe. Like several other genomic changes (7,9,11,40–42), deletions or duplications of this region could be pathogenic, with variable expressivity and/or incomplete penetrance in some individuals. Conversely, this could be a copy number change that affects tooth development but does not contribute to cognitive disabilities.
One deletion in our study falls clearly into the benign variant category. The 461-kb loss of 7q11.21 deletes a single zinc finger gene, ZNF92 (Fig. 1), a member of a large and copy number variable gene family (43). The inheritance of Patient 8's deletion is unknown; however, we found three losses and one gain of this region in our control population. Further, gains and losses overlapping this entire region have been reported in multiple population-wide CNV studies with a frequency similar to that in our control population (16,24). It is important to note that the same recombination mechanism that generates recurrent pathogenic deletions and duplications can also give rise to normal CNV in the human genome.
DISCUSSION
We found that 23% of the clinically significant copy number changes detected by array CGH testing are flanked by highly homologous segmental duplications. Owing to the genome-wide coverage of our array CGH design, we were able to detect previously described as well as novel NAHR events. We identified five regions of copy number change in affected patients. Similar to the genomic architecture that mediates deletions and duplications underlying other NAHR-driven genomic disorders (1,5), the segmental duplication pairs in our study are ≥10 kb in length and ≥95% identical (Table 2). Within the regions of gain or loss, we find genes in which heterozygous loss-of-function is disease-causing, as well as genes with putative roles in patient phenotypes.
In three out of five loci, we detected more than one individual with the deletion and/or reciprocal duplication, consistent with a recurrent mechanism of chromosome rearrangement. Deletions of 7q36.1 and 17q23.2 were singleton events, rare in our patient population but likely to occur again given the organization of segmental duplications. We expect that further study of other affected populations will reveal more individuals with the same genomic changes, leading to a better understanding of the phenotypic spectrum associated with particular recurrent deletions and duplications. The discovery of recurrent genomic imbalances is critical to the burgeoning field of CNV. Deletions and duplications associated with the 130 segmental duplication hotspots identified by Sharp et al. (5) may be embryonic lethal, pathogenic in liveborn individuals, or benign copy number changes (25). In this study we have characterized three loci associated with pathogenic copy number changes and one benign copy number change.
We have also identified copy number changes of uncertain clinical significance. We found deletions and duplications of 2q13 in some apparently normal individuals; however, copy number changes in these regions include several genes and are not common copy number variants in control populations (25). This type of inheritance has been described in other recurrent genomic disorders (7,9,11,40–42), and complicates any interpretation of the clinical significance of such findings.
Our data highlight the importance of other genetic factors in modifying the expression of genomic imbalances. Loss-of-function mutations in PRKAG2, KCNH2 and TBX4 cause autosomal dominant syndromes, but do not produce the associated phenotypes in some patients with heterozygous deletions encompassing those genes. Further, large deletions and duplications may be inherited from a phenotypically normal parent and exist as a rare copy number change (<1%) in cohorts of normal individuals. Genetic effects of other loci modify phenotypic expression and penetrance of copy number changes. In the case of deletions, loss of one gene copy may unmask a recessive allele on the intact chromosome. Both deletions and duplications could be subject to genomic imprinting, whereby the parental origin of the transmitted copy number change determines phenotype. All these factors are critical when considering newly described deletions and duplications in a relatively small group of patients. Genomic regions identified in this study are subject to recurrent deletion and duplication; thus we expect more individuals with the same copy number changes to emerge, which will lead to a more refined phenotype, as has been the case with other genomic disorders.
MATERIALS AND METHODS
Patient samples
Patient samples were collected as previously described (13). This study was approved by the Emory University Institutional Review Board. Informed consent was obtained as per the study protocol.
Array comparative genomic hybridization
Our oligonucleotide microarray combines targeted and genome-wide coverage in a 4 X 44K format (Agilent Technologies, Santa Clara, CA, USA) (13). Mean backbone spacing is approximately 75 kb between genomic probes. Array CGH was performed following the manufacturer's protocol as described previously (13). Genomic DNA was extracted from peripheral blood from patients and control individuals. Patient DNA was co-hybridized with a pool of five sex-mismatched control DNA samples. Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA) and signal intensities were evaluated using Feature Extraction Version 9.5.1.1 software (Agilent Technologies). We used DNA Analytics Version 4.0 software (Agilent Technologies) to analyze the array data and determine copy number gains and losses. Deletions and duplications were confirmed by FISH, as described previously (13).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health (R01 MH074090 to D.H.L. and C.L.M.).
ACKNOWLEDGEMENTS
We thank Daniel Moreno De Luca for helpful discussions and critical reading of the manuscript. Andres Moreno De Luca made the initial observation of the recurrent 2q11.2 copy number change. We also thank Cheryl Strauss for editorial assistance. The Emory Cytogenetics Laboratory performed the array CGH experiments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 2.Willatt L., Cox J., Barber J., Cabanas E.D., Collins A., Donnai D., FitzPatrick D.R., Maher E., Martin H., Parnau J., et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am. J. Hum. Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 4.Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 5.Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 6.Mefford H.C., Clauin S., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 8.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr, Dobyns W.B., et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 9.Hannes F.D., Sharp A.J., Mefford H.C., de Ravel T., Ruivenkamp C.A., Breuning M.H., Fryns J.P., Devriendt K., Van Buggenhout G., Vogels A., et al. Recurrent reciprocal deletions and duplications of 16p13.11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J. Med. Genet. 2008 doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D., et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin E.L., Lee J.Y., Blake D.M., Bunke B.P., Alexander C.R., Kogan A.L., Ledbetter D.H., Martin C.L. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet. Med. 2008;10:415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 14.Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 17.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 18.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 20.Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 21.Hinds D.A., Kloek A.P., Jen M., Chen X., Frazer K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 22.Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 24.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itsara A., Cooper G.M., Baker C., Girirajan S., Li J., Absher D., Krauss R.M., Myers R.M., Ridker P.M., Chasman D.I., et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gollob M.H., Green M.S., Tang A.S., Gollob T., Karibe A., Ali Hassan A.S., Ahmad F., Lozado R., Shah G., Fananapazir L., et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N. Engl. J. Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 27.Burwinkel B., Scott J.W., Buhrer C., van Landeghem F.K., Cox G.F., Wilson C.J., Grahame Hardie D., Kilimann M.W. Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the gamma 2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am. J. Hum. Genet. 2005;76:1034–1049. doi: 10.1086/430840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arad M., Maron B.J., Gorham J.M., Johnson W.H., Jr, Saul J.P., Perez-Atayde A.R., Spirito P., Wright G.B., Kanter R.J., Seidman C.E., et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N. Engl. J. Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 29.Priori S.G., Napolitano C., Schwartz P.J. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 30.Westenskow P., Splawski I., Timothy K.W., Keating M.T., Sanguinetti M.C. Compound mutations: a common cause of severe long-QT syndrome. Circulation. 2004;109:1834–1841. doi: 10.1161/01.CIR.0000125524.34234.13. [DOI] [PubMed] [Google Scholar]
- 31.Brugada R., Hong K., Dumaine R., Cordeiro J., Gaita F., Borggrefe M., Menendez T.M., Brugada J., Pollevick G.D., Wolpert C., et al. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation. 2004;109:30–35. doi: 10.1161/01.CIR.0000109482.92774.3A. [DOI] [PubMed] [Google Scholar]
- 32.Splawski I., Shen J., Timothy K.W., Lehmann M.H., Priori S., Robinson J.L., Moss A.J., Schwartz P.J., Towbin J.A., Vincent G.M., et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1 and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 33.Moss A.J. Long QT syndrome. JAMA. 2003;289:2041–2044. doi: 10.1001/jama.289.16.2041. [DOI] [PubMed] [Google Scholar]
- 34.Goldenberg I., Zareba W., Moss A.J. Long QT syndrome. Curr. Probl. Cardiol. 2008;33:629–694. doi: 10.1016/j.cpcardiol.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Vincent G.M., Timothy K.W., Leppert M., Keating M. The spectrum of symptoms and QT intervals in carriers of the gene for the long-QT syndrome. N. Engl. J. Med. 1992;327:846–852. doi: 10.1056/NEJM199209173271204. [DOI] [PubMed] [Google Scholar]
- 36.Johnson J.N., Hofman N., Haglund C.M., Cascino G.D., Wilde A.A., Ackerman M.J. Identification of a possible pathogenic link between congenital long QT syndrome and epilepsy. Neurology. 2008 doi: 10.1212/01.wnl.0000335760.02995.ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bongers E.M., Duijf P.H., van Beersum S.E., Schoots J., Van Kampen A., Burckhardt A., Hamel B.C., Losan F., Hoefsloot L.H., Yntema H.G., et al. Mutations in the human TBX4 gene cause small patella syndrome. Am. J. Hum. Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vega S., Iwamoto T., Nakamura T., Hozumi K., McKnight D.A., Fisher L.W., Fukumoto S., Yamada Y. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J. Biol. Chem. 2007;282:30878–30888. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 39.Bisgaard A.M., Kirchhoff M., Nielsen J.E., Brandt C., Hove H., Jepsen B., Jensen T., Ullmann R., Skovby F. Transmitted cytogenetic abnormalities in patients with mental retardation: pathogenic or normal variants? Eur. J. Med. Genet. 2007;50:243–255. doi: 10.1016/j.ejmg.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 40.McDonald-McGinn D.M., Tonnesen M.K., Laufer-Cahana A., Finucane B., Driscoll D.A., Emanuel B.S., Zackai E.H. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet. Med. 2001;3:23–29. doi: 10.1097/00125817-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Ballif B.C., Theisen A., Coppinger J., Gowans G.C., Hersh J.H., Madan-Khetarpal S., Schmidt K.R., Tervo R., Escobar L.F., Friedrich C.A., et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol. Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helbig I., Mefford H.C., Sharp A.J., Guipponi M., Fichera M., Franke A., Muhle H., de Kovel C., Baker C., von Spiczak S., et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009 doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoovers J.M., Mannens M., John R., Bliek J., van Heyningen V., Porteous D.J., Leschot N.J., Westerveld A., Little P.F. High-resolution localization of 69 potential human zinc finger protein genes: a number are clustered. Genomics. 1992;12:254–263. doi: 10.1016/0888-7543(92)90372-y. [DOI] [PubMed] [Google Scholar]
- 44.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.