Abstract
Familial juvenile hyperuricaemic nephropathy (FJHN), an autosomal dominant disorder, is caused by mutations in the UMOD gene, which encodes Uromodulin, a glycosylphosphatidylinositol-anchored protein that is expressed in the thick ascending limb of the loop of Henle and excreted in the urine. Uromodulin contains three epidermal growth factor (EGF)-like domains, a cysteine-rich region which includes a domain of eight cysteines and a zona pellucida (ZP) domain. Over 90% of UMOD mutations are missense, and 62% alter a cysteine residue, implicating a role for protein misfolding in the disease. We investigated 20 northern European FJHN probands for UMOD mutations. Wild-type and mutant Uromodulins were functionally studied by expression in HeLa cells and by the use of western blot analysis and confocal microscopy. Six different UMOD missense mutations (Cys32Trp, Arg185Gly, Asp196Asn, Cys217Trp, Cys223Arg and Gly488Arg) were identified. Patients with UMOD mutations were phenotypically similar to those without UMOD mutations. The mutant Uromodulins had significantly delayed maturation, retention in the endoplasmic reticulum (ER) and reduced expression at the plasma membrane. However, Gly488Arg, which is the only mutation we identified in the ZP domain, was found to be associated with milder in vitro abnormalities and to be the only mutant Uromodulin detected in conditioned medium from transfected cells, indicating that the severity of the mutant phenotypes may depend on their location within the protein. Thus, FJHN-causing Uromodulin mutants are retained in the ER, with impaired intracellular maturation and trafficking, thereby indicating mechanisms whereby Uromodulin mutants may cause the phenotype of FJHN.
INTRODUCTION
Familial juvenile hyperuricaemic nephropathy (FJHN; OMIM 162000) is an autosomal dominant disorder characterized by gout, elevated serum urate concentrations, a low fractional renal urate excretion, interstitial nephropathy with basement membrane thickening and glomerulosclerosis and progression to end-stage renal failure (1). FJHN is genetically heterogeneous and caused by mutations in the UROMODULIN (UMOD) gene (2,3) located on chromosome 16p12.3–p13.11 (4), the hepatocyte nuclear factor 1β (HNF-1β) gene (5,6) located on chromosome 17cen–q21.3 and an unidentified gene located on chromosome 1q41 (7). UMOD mutations are also associated with medullary cystic kidney disease type 2 (MCKD2, OMIM: 603860), which is characterized by corticomedullary and medullary cysts, tubulointerstitial inflammation, progressive renal failure and variable hyperuricaemia (2), and MCKD2 and FJHN are considered to be allelic variants. In addition, UMOD mutations have been reported in glomerulocystic kidney disease (GCKD) (8,9).
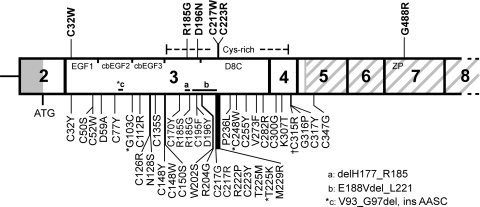
The UMOD gene, consisting of 11 exons, of which exons 2–11 are coding (10) (Fig. 1) encodes Uromodulin, also known as the Tamm–Horsfall glycoprotein (11), which is the most abundant protein in human urine (12). Uromodulin is a 640 amino acid protein with a 24 amino acid signal peptide at the N-terminus, 48 cysteines and 8 potential N-linked glycosylation sites (10), of which 7 appear to be occupied (13). Sequence homology indicates that Uromodulin is likely to contain three epidermal growth factor (EGF)-like domains, of which the second and third contain a calcium-binding (cb) motif (14); a domain of eight cysteines (D8C) (15) within a cysteine-rich region; and a zona pellucida (ZP) domain (16), which is responsible for the polymerization of extracellular proteins into helical filaments (17) (Fig. 1).
Figure 1.
Schematic representation of exons 2–8 of UMOD illustrating the locations of 46 mutations. The 40 previously described mutations and the 6 mutations described in this study are shown below and above the gene, respectively. The majority (>87%) of the mutations have been identified in FJHN patients, ∼10% in MCKD patients (*) and ∼2.5% in GCKD patients (†) (2,3,8,9,19–27). Missense substitutions are indicated by single-letter amino acid codes, and three inframe deletions are indicated by horizontal lines a–c. Most of the mutations cluster in exons 3 and 4, which encode the three epidermal growth factor (EGF)-like domains, and the cysteine-rich region, which includes the domain of eight cysteines (D8C). The G488R mutation is the first to be described in exon 7, which encodes part of the zona pellucida (ZP) domain (grey diagonal stripes).
Uromodulin is expressed exclusively in the thick ascending limb (TAL) of the loop of Henle and the early distal convoluted tubule (18). It is targeted mainly to the apical membrane by a glycosylphosphatidylinositol anchor, from where it is cleaved by a cellular protease into the tubule lumen (18) and excreted in the urine. Patients with Uromodulin mutations have a markedly reduced urinary Uromodulin, and only wild-type Uromodulin is excreted (19). The majority, i.e. >90%, of the reported 36 UMOD mutations associated with FJHN are missense mutations, and 61% of these alter a normal cysteine residue (2,3,8,9,19–27), implicating an important role for protein misfolding in the aetiology of the disease. Furthermore, formation of disulphide bonds is a rate-limiting step in the maturation of Uromodulin from an ∼84 kDa endoplasmic reticulum (ER)-resident precursor form to the ∼97 kDa mature form (28,29), and thus it has been postulated that mutant Uromodulin is retained in the ER. Indeed, this has been shown, by in vitro studies, for 19 UMOD mutants (8,25,30–32) and resulted in Uromodulin mutations being classified into two groups (I and II), defined on the basis of expression at the plasma membrane. Group I mutants represented those with reduced expression at the plasma membrane, whereas group II mutants represented those unable to reach the plasma membrane (25). Such studies of FJHN-causing UMOD mutations have yielded important insights, and in order to understand further the mechanisms underlying this renal disease, we ascertained patients with FJHN and investigated them for UMOD mutations and their functional consequences.
RESULTS
Segregation of FJHN and UMOD
The 20 FJHN probands (Table 1) of northern European origin that had been ascertained comprised 11 with a familial history and 9 without a family history. Three UMOD single-nucleotide polymorphisms (SNPs), which were located ∼3.6 kb 5′ of the ATG (rs13333226), within intron 6 (rs9928757) and ∼1.5 kb 3′ of the stop codon (rs4238595), were used for segregation studies in the 11 FJHN families. In three families, the SNPs did not co-segregate with FJHN (data not shown) and these were therefore not analysed for UMOD mutations. Thus, UMOD sequence analysis was undertaken in 17 probands (8 with familial disease and 9 without familial disease) (Table 1).
Table 1.
Clinical details of FJHN probands
| Family | Age | S.Cr, mg/dl (<1.5) | S.UAa, mg/dl | U.Cr, mg/dl (25–400) | U.UA, mg/dl (25–75) | FEUAb | Ultrasound or other scan | Remarks |
|---|---|---|---|---|---|---|---|---|
| 1c | 17 | 1.5 | 7.8↑↑ | NA | NA | 2.6%↓↓ | Small kidney size | |
| 2c | 39 | 2.98 | 4.67↑ | 26.1 | 6.36 | 7.0%↓ | Small kidneys, no cysts | Gout, subsequent NTX |
| 3d | 42 | 3.55 | 6.41- | 237.1 | 18.11 | 4.2%↓ | NA | Gout |
| 4d | 1.6 | 9.5↑ | NA | NA | 2.9%↓ | NA | Gout | |
| 5e | 16 | 1.5 | 12.3↑↑ | 53.2 | 24.3 | 5.6%↓↓ | Enhanced echogenicity | Treatment refused |
| 6e | 24 | 2.5 | 11.5↑↑ | NA | NA | NA | Bilateral small kidneys | Gout, HD, subsequent NTX |
| 7e | 50 | 7.55 | 14.4↑↑ | NA | NA | 8.4%- | Atrophic kidneys, no cysts | Gout, HD, NTX rejected, now on HD |
| 8d | 13 | 2.2 | 12.9↑↑ | 100 | 21.8 | 2.9%↓↓ | Echogenic, normal size | Gout, allopurinol, NTX |
| 9d | 45 | 2.3 | 4.8↑ | NA | NA | 4.8%↓↓ | Atrophic kidneys | Allopurinol, gout |
| 10c | 21 | 1.5 | 2.36- | 334.6 | 30.2 | 5.7%↓↓ | Normal | Allopurinol, no gout |
| 11d | 14 | 0.93 | 7.2↑ | 113.1 | 27.2 | 3.1%↓↓ | Normal | Allopurinol |
| 12f | 46 | 1.62 | 6.8↑ | NA | NA | NA | Normal | NTX after ESRF |
| 13f | 67 | 1.12 | 8.3↑ | 28.3 | 8.6 | 4.1%↓↓ | NA | NTX |
| 14f | 58 | 1.6 | 8.3↑ | 53.6 | 9.1 | 3.3%↓ | Normal | Allopurinol, NTX |
| 15f | 72 | 0.85 | 5.3↑ | 27.5 | 15.6 | 9.1%↓ | Normal | NTX after ESRF |
| 16f | 42 | 2.25 | 9.6↑ | 47.6 | 13.8 | 6.8%↓ | Normal | Allopurinol, NTX after ESRF |
| 17f | 57 | 1.5 | 6.3- | 93 | 5.7 | 1.4%↓ | Small right kidney | Allopurinol |
| 18f | 73 | 2.71 | 10.6↑↑ | 28.8 | 4.7 | 4.2%↓ | Cortical cysts, both kidneys | Proteinuria, Allopurinol |
| 19f | 47 | 1.5 | 5.9↑ | NA | 5.25 | NA | Small kidneys with cysts | Allopurinol, gout |
| 20f | 25 | 2.72 | 8.5↑↑ | NA | ↓ | ↓ | NA | Gout, 50% of glomeruli sclerosed, tubulointerstitial lesions |
NA, not available; S.Cr, serum creatinine (normal range); S.UA, serum uric acid; U.Cr, urinary creatinine (normal range); U.UA, urinary uric acid (normal range); FEUA, fractional excretion of uric acid; HD, haemodialysis; NTX, renal transplant; ESRF, end-stage renal failure.
aNormal S.UA values vary with sex and age (44); ↑ denotes value above upper limit of normal; ↑↑ denotes value above twice upper limit of normal; - denotes value in normal range.
bNormal FEUA values vary with sex and age (45); ↓ denotes value below lower limit of normal; ↓↓ denotes value below half-lower limit of normal; - denotes value in normal range.
cUMOD involvement excluded by UMOD SNP analysis.
dUninformative for UMOD SNPs.
eUMOD involvement could not be excluded by UMOD SNP analysis.
fOther family members not available.
UMOD mutations
DNA sequence analysis of the UMOD 11 exons and intron–exon boundaries, along with 1.5 kb upstream of exon 2 (including the non-coding exon 1), in 17 unrelated probands with FJHN (Table 1), revealed six missense mutations: c.96C>G (p.C32W); c.553C>G (p.R185G); c.586G>A (p.D196N); c.651C>G (p.C217W); c.667T>C (p.C223R); and c.1462G>C (p.G488R). Five of these UMOD mutations (C32W, D196N, C217W, C223R, G488R) are novel and one (R185G) has been previously reported in an unrelated FJHN patient (22). Three of the five novel mutations resulted in the substitution of a cysteine residue (C32W, C217W, C223R) (Table 2). In addition, the G488R mutation is the first to be reported in exon 7 and represents only one of two reported to occur within the ZP domain (Fig. 1). Each mutation was confirmed either by restriction digest analysis, amplification refractory mutation system (ARMS)-PCR (Fig. 2A–C) or repeat sequencing of independent PCR products. The absence of each of these six DNA sequence abnormalities in 110 alleles from 55 unrelated normal northern European individuals established that they are not common polymorphisms. Furthermore, all the mutations predicted significant alterations in conserved amino acid residues, consistent with likely functional changes.
Table 2.
UMOD mutations in FJHN families and patients
| Family/patient | Nucleotidea | Exon | Confirmation | Amino acid change | Uromodulin domain affected |
|---|---|---|---|---|---|
| 6 | c.96C>G | 3 | RE | Cys32Trp* (C32W) | EGF1 |
| 11 | c.553C>G | 3 | ARMS-PCR | Arg185Gly (R185G) | Cysteine-rich region |
| 5 | c.586G>A | 3 | ARMS-PCR | Asp196Asn* (D196N) | Cysteine-rich region |
| 15 | c.651C>G | 3 | RE | Cys217Trp* (C217W) | D8C |
| 9 | c.667T>C | 3 | RE | Cys223Arg* (C223R) | D8C |
| 20 | c.1462G>C | 7 | RS | Gly488Arg* (G488R) | ZP |
RE, restriction endonuclease; ARMS-PCR, amplification refractory mutation system-PCR; RS, repeat DNA sequence analysis using independent PCR products; novel mutations are marked by asterisks; EGF, epidermal growth factor-like; D8C, domain of eight cysteines; ZP, zona pellucida.
aNucleotide numbers refer to UMOD cDNA. 1 = A of ATG.
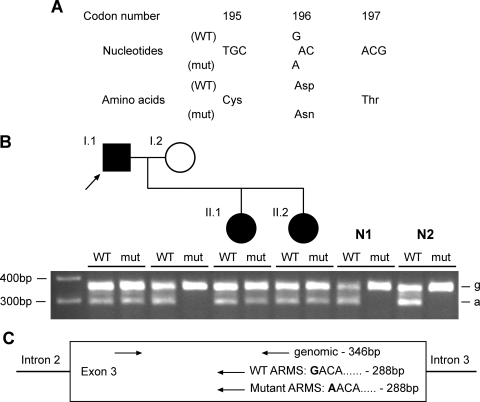
Figure 2.
Detection and confirmation of novel Uromodulin mutation D196N in family 04/03 (Table 1). (A) DNA sequence analysis of individual I.1 revealed a heterozygous G-to-A transition at codon 196, thus altering the wild-type (WT) sequence GAC, encoding an aspartate (D), to the mutant (mut) sequence AAC, encoding an asparagine (N). (B) The D196N mutation was confirmed in the proband (arrowed) by ARMS-PCR, which also showed that the mutation was present in the other affected family members (II.1, II.2), but absent in a panel of 55 normal individuals, of which two (N1, N2) are shown. For each individual, ARMS-PCR products are loaded wild-type (WT)—mutant (mut). ‘g’ denotes genomic band, ‘a’ denotes ARMS band, as shown in the schematic diagram (C). Each ARMS primer was used in a reaction with both genomic primers.
Phenotypes of patients with UMOD mutations
Approximately 70% of the 20 FJHN probands analysed in our study did not have mutations in the UMOD gene, and this is consistent with previous reports (19,20,25), although these and our study have not searched for intronic mutations or exonic or whole gene UMOD deletions. In order to search for differences in the phenotypes of patients with and without UMOD mutations, we pooled the clinical and mutational data from this study and our previous reports (3,33). A comparison of the clinical features of FJHN between the patients with and without UMOD mutations revealed an absence of phenotypic differences (Table 3).
Table 3.
Clinical features of FJHN in patients subdivided into those with and without UMOD mutations
| Clinical feature | With UMOD mutation, n = 13 (n = 31)a | Without UMOD mutation, n = 25 | Two-tailed probabilityb |
|---|---|---|---|
| Gout | 30.8% (54.8%) | 28.0% | 1.00 (0.06) |
| Hyperuricaemia | 72.7% (75.9%) | 68.0% | 1.00 (0.56) |
| Low FEUA | 100% (92.0%) | 90.5% | 1.00 (1.00) |
| ESRF | 38.5% (29.0%) | 36.0% | 1.00 (0.77) |
| Renal insufficiency | 53.8% (71.0%) | 66.7% | 0.49 (0.77) |
| Cysts | 7.7% (12.9%) | 8.0% | 1.00 (0.68) |
| Small/atrophic kidney(s) | 23.1% (38.7%) | 24.0% | 1.00 (0.27) |
FEUA, fractional excretion of uric acid; ESRF, end-stage renal failure. Hyperuricaemia is defined as a serum urate concentration >1 standard deviation above the normal range for age and sex. Low FEUA is defined as an FEUA of <1 standard deviation below the normal range for age and sex. ESRF is defined by necessity for dialysis or a transplant. Renal insufficiency is defined as a serum creatinine >1.5 mg/dl.
aNumbers not in parentheses are from this study, and those in parentheses refer to the pooled data from this study and our previous reports (3,33).
bComparisons performed using two-tailed Fisher's exact test.
Uromodulin mutants result in maturation and trafficking defects
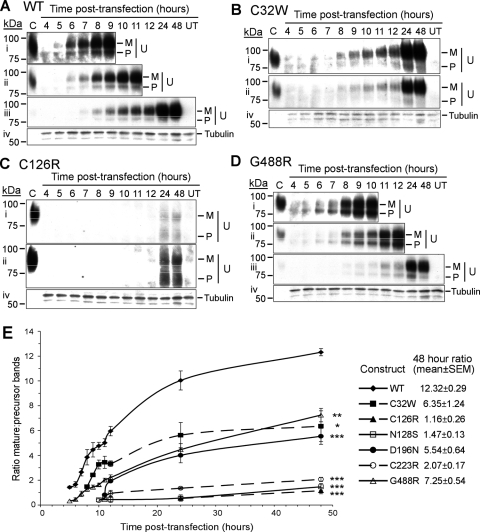
To study the effects of UMOD mutations, we chose six mutations (four novel and two previously reported) on the basis of their location and possible impact within different domains of Uromodulin. Thus, the four novel mutants and their locations were C32W (EGF1 domain), D196N (cysteine-rich region), C223R (D8C domain) and G488R (ZP domain) (Table 2), and the two previously reported mutations (3) were C126R and N128S, which are both in the cb-EGF3 domain. We did not investigate further the R185G and C217W mutants identified in this study, as they are located within the same domains as D196N and C223R which were included for further analysis. Wild-type and mutant UMOD constructs were transiently transfected into HeLa cells, and the maturation and trafficking of the expressed Uromodulin from the ∼84 kDa immature, ER-resident precursor protein, to the fully glycosylated post-ER ∼97 kDa mature protein (28,29) studied by western blot analysis and immunofluorescence. Both the precursor and mature forms of Uromodulin could be detected (Fig. 3A–D), and the ratio of mature:precursor Uromodulin was measured at several time points post-transfection (Fig. 3E). Wild-type Uromodulin could be detected after an initial lag time of ∼5 h, and both the precursor and mature forms were present throughout the time course (Fig. 3A). During the time course, the amount of mature wild-type Uromodulin was enriched ∼8.6-fold relative to the amount of precursor, indicating that the 84 kDa precursor form undergoes efficient folding and glycosylation to generate the 97 kDa mature form (Fig. 3E). This maturation appeared to slow after 24 h post-transfection; however, soon after this time, mature wild-type Uromodulin started to appear in the conditioned medium (see below). In contrast, all Uromodulin mutants had significantly delayed maturation when compared with wild-type Uromodulin. C32W, D196N and G488R showed a significant amount of maturation of the precursor form over the time course, with enrichment of the mature form to ∼50% of that of wild-type Uromodulin (Fig. 3E); whereas C126R, N128S and C223R showed significantly less maturation of the precursor form, with enrichments of the mature form to ∼25% of that of wild-type Uromodulin, and to ∼50% of the C32W, D196N and G488R mutants (Fig. 3E). This analysis of Uromodulin maturation by western blot analysis indicates the occurrence of two groups of Uromodulin mutants; group A contains Uromodulin mutants C32W, D196N and G488R, which had ∼50% maturation when compared with wild-type Uromodulin, and group B contains Uromodulin mutants C126R, N128S and C223R, which had ∼25% maturation when compared with wild-type Uromodulin. This subdivision of the Uromodulin mutants into two groups corresponds to that proposed by a previous study which subdivided Uromodulin mutants into two groups, group I and group II, on the basis of the presence or absence, respectively, of Uromodulin at the plasma membrane measured by FACS analysis (25). Among the group B Uromodulin mutants defined by our study, maturation of C223R Uromodulin was slightly less retarded than C126R and N128S Uromodulin (P < 0.05), suggesting that this may either represent a subclass of group B, or potentially another distinct group.
Figure 3.
Western blot and densitometry analyses of Uromodulin maturation. (A–D) Representative western blots of wild-type and mutant Uromodulin in HeLa cell lysates are shown: blots i—iii probed for Uromodulin [(i) 30 min exposure, (ii) 5 min exposure, (iii) 30 s exposure]; C, control (human urine); M, mature; P, precursor; U, Uromodulin. Blot iv is a control probed for Tubulin and exposed for 30 min. (A) wild-type (WT) Uromodulin, (B) C32W Uromodulin, (C) C126R Uromodulin, (D) G488R Uromodulin. Blots for N128S Uromodulin, D196N Uromodulin and C223R Uromodulin (not shown) were similar to those for C126R Uromodulin, with dispersed precursor bands and no detectable expression before 11 h post-transfection. (E) Densitometric analysis of blots for wild-type Uromodulin and all the mutants studied. Data (mean ± SEM) from four independent transfections showing mature:precursor ratios are shown. *P < 0.005, **P < 0.0002, ***P < 0.0001 at 48 h post-transfection, with respect to wild-type Uromodulin, calculated by unpaired, two-tailed Student's t-test. Mutants could be divided into group A (C32W, D196N, G488R) with reduced maturation, and group B (C126R, N128S, C223R) with almost no maturation.
Analysis of Uromodulin secretion
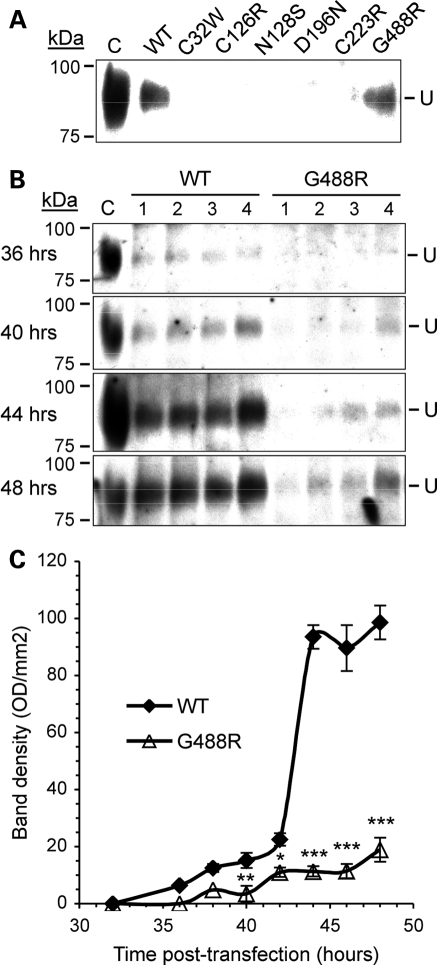
In order to study the secretion of Uromodulin, samples of conditioned media from HeLa cells transfected with wild-type or mutant (C32W, C126R, N128S, D196N, C223R and G488R) Uromodulin were collected 48 h post-transfection for western blot analysis. Wild-type Uromodulin could be detected in the conditioned medium of HeLa cells, and this is consistent with a previous report (29). The G488R Uromodulin was the only mutant to be detected in the conditioned medium (Fig. 4A). Western blot (Fig. 4B) and densitometry (Fig. 4C) of conditioned media samples from cells expressing wild-type or G488R Uromodulin, between 24 and 48 h post-transfection, revealed that wild-type Uromodulin was detectable by 36 h post-transfection, after which it rapidly increased to a maximum at 42 h post-transfection. However, the G488R mutant Uromodulin was detectable only at low levels; for example, at 48 h post-transfection, the level of the G488R mutant was significantly reduced at <20% of the wild-type Uromodulin level (G488R mutant mean ± SEM: 18.93 ± 6.88 versus wild-type: 98.59 ± 6.48, P < 0.0002). This significantly reduced level of G488R Uromodulin in the conditioned medium could be due to inhibition of the mechanism of secretion of Uromodulin by the G488R mutation, or because of less G488R Uromodulin being present at the plasma membrane. In order to investigate this, the subcellular location of wild-type and mutant Uromodulins was examined.
Figure 4.
Western blot and densitometric analyses of Uromodulin secretion. (A) Conditioned media from HeLa cells transfected with wild-type (WT) and mutant Uromodulin were collected at 48 h post-transfection and analysed by western blot. Only wild-type and G488R Uromodulin could be detected in conditioned media. (B) Conditioned medium samples from HeLa cells transfected with wild-type or G488R Uromodulin were collected at several time points between 24 and 48 h post-transfection and analysed by western blot. Western blots for samples collected at 36, 40, 44 and 48 h post-transfection are shown. Samples were collected from four independent transfections (1– 4) at each time point and analysed on the same blot. C, control (human urine). (C) Densitometric analysis of wild-type and G488R bands from western blots (data are shown as mean ± SEM). *P < 0.05, **P < 0.01, ***P < 0.0002 with respect to wild-type Uromodulin calculated by unpaired, two-tailed Student's t-test.
Subcellular location of Uromodulin mutants
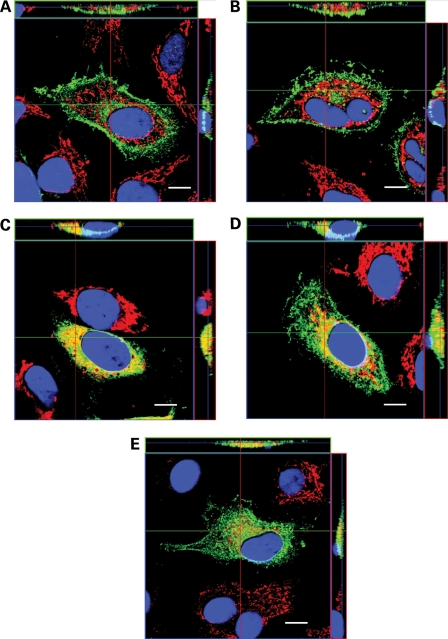
The results of western blot analysis showed that the Uromodulin mutants could be subdivided into two groups, with group A (C32W, D196N and G488R) containing mutants that had delayed and reduced maturation of ∼50% when compared with wild-type Uromodulin, and group B (C126R, N128S and C223R) containing mutants with maturation of ∼25% when compared with wild-type Uromodulin (Fig. 3). Confocal microscopy of transiently transfected HeLa cells that were fixed and co-immunostained with antibodies against Uromodulin (green) and the ER-resident protein Calnexin (red) revealed the subcellular location of wild-type and mutant Uromodulins (Fig. 5A–E). Wild-type Uromodulin was almost exclusively present at the plasma membrane at 12 h post-transfection (Fig. 5A), except for a small amount of intracellular Uromodulin, which did not co-localize with the ER. The group A mutants (C32W, D196N, G488R), as expected, had some Uromodulin at or near the plasma membrane at 24 h post-transfection (Fig. 5B and E). However, some Uromodulin was also present intracellularly, where it was localized within the ER and cytoplasm, which is well illustrated by the several large punctae of C32W Uromodulin (Fig. 5B). These results indicate that the reduced level of mutant G488R Uromodulin in the conditioned medium (Fig. 4) is likely to be due to an inhibition of secretion rather than a lack of mutant Uromodulin at the plasma membrane. The group B mutants C126R (Fig. 5C) and N128S (data not shown), as expected, were retained exclusively within the ER at 24 h post-transfection and were not trafficked to the plasma membrane. However, the group B mutant C223R co-localized with Calnexin to a lesser extent than C126R Uromodulin at 24 h post-transfection, and a small amount of C223R Uromodulin was localized at or near the plasma membrane. The remaining intracellular C223R Uromodulin had a broad cytoplasmic pattern with some punctate Uromodulin staining, the majority of which did not co-localize with Calnexin (Fig. 5D). The punctate staining of the C223R mutant Uromodulin, which was also observed with the other Uromodulin mutants (C32W, C126R, N128S, D196N and G488R), suggested that these mutant Uromodulins may be co-localized with lysosomes, which are vesicles that have been reported to yield such staining (34). Furthermore, mutant proteins retained in the ER may undergo degradation via the lysosomal, proteasomal and autophagic pathways (35–37). Given the punctate immunostaining pattern, we performed co-immunostaining using lysosome-associated membrane protein 1 (LAMP-1), which is a lysosomal resident protein (38). This revealed an absence of co-localization between the mutant (C32W, C126R, N128S, D196N, C223R and G488R) Uromodulins and LAMP-1 (Supplementary Material, Fig. S1), thereby indicating that the mutant Uromodulins (C32W, C126R, N128S, D196N, C223R and G488R) are not located within lysosomes but at other cytoplasmic sites in addition to the ER. These other cytoplasmic sites may be associated with proteasomes or autophagosomes, which are part of the degradative pathways for misfolded proteins that are retained in the ER (35–37).
Figure 5.
Confocal imaging of HeLa cells expressing wild-type and mutant Uromodulin. Orthogonal reconstructions of Z-stacks of transfected cells fixed and stained for Uromodulin (green) and Calnexin (red). Co-localization of Uromodulin and Calnexin yields a yellow signal. Nuclei are stained with DAPI (blue). Scale bars: 10 µm. (A) Wild-type, 12 h post-transfection; (B) C32W Uromodulin, 24 h post-transfection; (C) C126R Uromodulin, 24 h post-transfection; (D) C223R Uromodulin, 24 h post-transfection; (E) G488R Uromodulin, 12 h post-transfection. N128S Uromodulin showed an identical pattern to that of C126R Uromodulin, and D196N Uromodulin showed a similar pattern to that of C223R Uromodulin (data not shown).
DISCUSSION
Our results, which identified six UMOD mutations (C32W, R185G, D196N, C217W, C223R and G488R) in a cohort of 20 patients with FJHN (Fig. 1, Tables 1 and 2), expand the spectrum of such mutations associated with this renal tubular disorder. The C32W, D196N, C217W, C223R and G488R (Fig. 1, Table 2) represent novel UMOD mutations, whereas the R185G has been previously reported in an unrelated patient with FJHN (22). The total number of different UMOD mutations now reported, including the results of our present study, is 46, and >95% of these are located in exons 3 and 4, which encode the three EGF-like domains, and the D8C within a cysteine-rich region (Fig. 1) (2,3,8,9,19–27). In addition, 93% of these 46 UMOD mutations are missense substitutions, and the remaining 7% are inframe deletions. Thus, all UMOD mutations are predicted to result in a full-length protein; therefore, FJHN due to these UMOD mutations is likely to result from abnormalities of protein folding, maturation and trafficking, rather than a Uromodulin deficiency. Indeed, our functional characterization, using western blot analysis to study maturation of Uromodulin over a time course, has revealed defects in maturation and trafficking of Uromodulin mutants (Fig. 3). Moreover, these abnormalities in Uromodulin maturation could be subdivided into two groups, with group A mutants (C32W, D196N and G488R) showing a significantly reduced maturation of Uromodulin of ∼50% when compared with wild-type, and the group B mutants (C126R, N128S and C223R) showing ∼25% maturation of precursor Uromodulin when compared with wild-type. These results using western blot analysis are in broad agreement with our results from confocal microscopy, which revealed the subcellular localization of these mutants (Fig. 5). Thus, the group A mutants localized to the plasma membrane, whereas the group B mutants did not localize to the plasma membrane but were retained in the ER. These findings sit well with the previously proposed classification of UMOD mutations into two groups (group I and group II) on the basis of their presence or absence, respectively, at the plasma membrane that was determined using FACS analysis (25). However, the C223R mutant Uromodulin appears to be an exception, as our results showed a significant reduction in maturation to ∼25% that of wild-type, similar to the other group B mutants C126R and N128S, by western blot analysis (Fig. 3E), yet a small amount appeared to be present at the plasma membrane (Fig. 5D). These results also indicated that mutations which substitute a cysteine residue do not necessarily lead to a more severely retarded phenotype, as demonstrated particularly by C32W, and that non-cysteine substituting mutations may also affect protein folding and maturation, as illustrated by N128S. A further analysis of the locations of these UMOD mutations revealed potentially interesting insights about the structure–function relationships of the domains.
Thus, the C126R and N128S mutant Uromodulins, which are located in cbEGF3 (Fig. 1), showed the most severely retarded maturation defects (Fig. 3E). N128 is one of the residues which co-ordinate with the Ca2+ ion (3), and thus may also have an effect on the folding of this domain in the ER. Furthermore, cbEGF domains are strongly recognized by ER chaperones (39), thus any mutation which potentially disrupts the structure of this domain is likely to result in ER retention. Unsurprisingly, given the lack of expression at the plasma membrane, neither C126R Uromodulin or N128S Uromodulin was detected in the conditioned medium.
The C223R mutant Uromodulin, which is located within the D8C (Fig. 1), was almost as severely retarded as C126R and N128S. The function of the D8C is unknown but it is conserved in several proteins that also contain ZP domains (e.g. liver-specific ZP domain-containing protein) (15). The eight cysteines of the D8C are predicted to form four intra-domain disulphide bonds within a structure of seven β-strands (15), again suggesting a highly structured domain which would be disrupted by substitution of one of the cysteine residues. The importance of the D8C is underscored by the D196N mutant Uromodulin, which is slightly N-terminal of the D8C domain (Fig. 1), yet has a faster maturation rate when compared with C223R Uromodulin (Fig. 3E). Neither the D196N or C223R mutant Uromodulins were detected in conditioned medium, and this is likely to be a consequence of the reduced expression of D196N and C223R Uromodulin at the plasma membrane.
The C32W mutant Uromodulin, which is located in EGF1 (Fig. 1), had a significantly higher maturation rate of 50% when compared with the other cysteine-altering mutants (C126R and C223R) which had a maturation rate of ∼25% when compared with wild-type (Fig. 5E). This may reflect a lesser importance of EGF1 than cbEGF3, which is the location of C126R, and the D8C, which is the location of C223R. Alternatively, since C32 is the most N-terminal cysteine residue in Uromodulin, EGF1 may be able to adopt an alternative conformation in isolation, thus allowing the rest of the protein to fold correctly. Our results for C32W are in close agreement with those reported for the C32Y mutant, which was found to be largely expressed at the plasma membrane (25). Given the relatively high plasma membrane expression of C32W Uromodulin (Fig. 5B), it was surprising that no C32W mutant Uromodulin was detected in the conditioned medium (Fig. 4A). However, this finding accords well with the almost undetectable urinary levels of Uromodulin in patients with the C32Y and C50S mutations, which are located in EGF1 (25,26). Furthermore, the patient with the C50S mutation did not have the intracellular globular aggregates of Uromodulin, which are usually present in tubular cells of patients with other Uromodulin mutations (8,25). This corresponds well with our findings for the C32W mutant Uromodulin, which was the only mutant that did not have significant amounts of intracellular Uromodulin (Fig. 5B).
The G488R mutation is only the second mutation to have been described within the ZP domain of Uromodulin, which mediates polymerization of Uromodulin (40). The G488R mutant Uromodulin had more similarities in its maturation rates and presence in conditioned media, to wild-type Uromodulin than any of the other mutants (Figs 3–5); however, its maturation and trafficking to the plasma membrane were still significantly slower than that of wild-type Uromodulin, although it was the only mutant to be detected in the conditioned medium, but at greatly reduced levels and at a slower rate than wild-type Uromodulin. In addition to intracellular retention, the G488R mutation may have consequences for Uromodulin at the plasma membrane, or after its release into the tubule and urinary tract, by altering the polymerization of the mutant Uromodulin.
There appear to be no correlations between: the results of altered Uromodulin maturation and subcellular localization and the FJHN phenotype; the UMOD mutations and the FJHN phenotypes (Table 1), consistent with the results of other studies (8,25,31); or the presence or absence of a UMOD mutation and the FJHN phenotype (Table 3). A comparison of clinical features in patients with group A mutations compared with those with group B mutations also revealed no significant differences (data not shown). Finally, 68% of patients with FJHN in our study did not have UMOD mutations, even though they were clinically indistinguishable from those who had UMOD mutations (Table 3). This finding is consistent with those of other studies which have reported an absence of UMOD abnormalities in 17–84% of FJHN patients. This reflects the genetic heterogeneity of FJHN, which may also be caused by HNF-1β mutations (5,6), and an as-yet-unidentified gene on chromosome 1q41 (7).
In conclusion, we have identified five novel UMOD mutations in patients with FJHN and shown that UMOD mutants are associated with ER retention, and delayed maturation and trafficking of the mutant Uromodulin, which is likely to be due to protein misfolding. Accumulation of misfolded proteins within the ER may be associated with apoptosis, and this may be a possible mechanism for the pathogenesis of FJHN due to Uromodulin mutations.
MATERIALS AND METHODS
Patients
Twenty unrelated northern European probands with FJHN and their relatives were examined (Table 1). Informed consent was obtained from all individuals, using guidelines approved by the local ethical committee.
SNP analysis of the UMOD gene
Venous blood samples were obtained and leucocyte DNA extracted as described previously (41) and utilized for SNP analysis by the use of three SNPs: rs13333226, rs9928757 and rs4238595. The location of rs9928757 is intragenic and within intron 6, whereas those of rs13333226 and rs4238595 are ∼3.6 kb 5′ of the ATG and ∼1.5 kb 3′ of the stop codon, respectively. The surrounding DNA sequence was amplified by PCR using specific primers (details available upon request), and the SNPs detected by restriction endonuclease analysis and agarose gel electrophoresis (42).
DNA sequence analysis of the UMOD gene
Leucocyte DNA was used with UMOD gene-specific genomic primers (details available on request) for PCR amplification of the exons, intron–exon boundaries and the promoter region of the UMOD gene, as described previously (41). The DNA sequences of the PCR products were determined using DYEnamic ET Terminators (Amersham Biosciences) and resolved by electrophoresis using the ABI377 system (Applied Biosciences Inc.), as described (41). DNA sequence abnormalities were confirmed by restriction endonuclease analysis, or ARMS-PCR, or by repeat DNA sequence analyses of independently obtained PCR products as described (41). The DNA sequence abnormalities were demonstrated to be absent as common polymorphisms in the DNA from 55 unaffected, unrelated individuals (31 males, 24 females, of northern European origin) as described previously (41).
Uromodulin expression constructs
A pDNR-LIB plasmid containing wild-type Uromodulin cDNA (IMAGE clone 4611787, Clontech) was altered by site-directed mutagenesis (QuikChange® XL, Stratagene) to generate mutant Uromodulin cDNAs. The sequences of all cDNAs were verified by DNA sequence analysis. Wild-type and mutant Uromodulin cDNAs were subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen) using ApaI and EcoRI restriction endonucleases (43).
Cell culture and transfection
HeLa cells (ATCC CCL-2) were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% FCS, 2 mm l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2/95% air. Cells were seeded at a density of 2 × 105 cells per cm2 onto 35 mm dishes (Nunc) for protein preparations and collection of conditioned media, or onto glass coverslips for immunofluorescence studies. Twenty-four hours later, cells were transiently transfected with 1 µg Uromodulin expression construct using FuGene 6 reagent (Roche) (43). After 3 h, transfection reagent was removed and replaced with fresh, warm growth medium. Untransfected controls were included in all experiments. Transfection efficiency for wild-type and mutant constructs was quantified by assessing immunofluorescence of Uromodulin-expressing cells.
Collection and preparation of conditioned medium and cell lysates
Nine 15 µl collections of conditioned medium were made from the same 35 mm dish between 24 and 48 h post-transfection. The HeLa cells in the dish were then lysed to obtain protein. Media samples were centrifuged to remove any cells and stored at −80°C until analysed by western blotting. Cells growing in 35 mm dishes were harvested every hour from 4 to 12 h post-transfection, and at 24 and 48 h post-transfection. Cells were incubated in 450 µl lysis buffer [60 mm β-octylglucoside (Sigma), 5 mm EDTA, in PBS] (29) with a protease inhibitor cocktail (Roche) for 10 min at 4°C, harvested, sonicated and incubated at 4°C for a further 1 h. The lysate was centrifuged to pellet insoluble material, and the total protein concentration of the soluble fraction was measured using the Micro BCA Protein Assay (Pierce).
Western blotting and densitometry
Cell lysate and conditioned media samples, containing equal amounts of total protein for all cell lysates or conditioned media, were mixed 1:1 with Laemmli buffer, incubated at 95°C for 4 min, quenched on ice and resolved on 8% denaturing SDS–PAGE gels. Proteins were transferred to PVDF membrane (PerkinElmer) and hybridized with sheep anti-Uromucoid antibody (Biodesign International), followed by horseradish peroxidase (HRP)-conjugated mouse anti-sheep antibody (Sigma) and visualized using enhanced chemiluminescence (GE Healthcare). For cell lysates, identical sister gels were run and membranes hybridized with mouse anti-α-Tubulin (Santa Cruz), followed by HRP-conjugated goat anti-mouse (Bio-Rad) (43). Densitometry of western blots was performed using a GS-710 densitometer and Quantity One software (Bio-Rad). Band densities were measured on western blots from four independent transfections. Statistical analysis was performed using unpaired, two-tailed Student's t-test.
Immunofluorescence
Transiently transfected HeLa cells grown on glass coverslips were fixed in ice-cold 4% paraformaldehyde for 15 min, permeablized in 0.1% Triton X-100 for 10 min and blocked in 10% pre-immune donkey serum (Sigma) for 30 min. Coverslips were incubated in sheep anti-Uromucoid antibody (Biodesign International) diluted 1:500, and mouse anti-Calnexin antibody (Abcam) diluted 1:200, or rabbit anti-LAMP1 (Abcam) diluted 1:200, for 1 h at room temperature. Secondary antibodies were AlexaFluor®488 donkey anti-sheep (Uromodulin), and AlexaFluor®594 donkey anti-mouse (Calnexin) or AlexaFluor®594 anti-rabbit (LAMP-1) (Molecular Probes), diluted 1:500 and incubated with coverslips for 1 h at room temperature. Coverslips were mounted onto slides in VECTASHIELD® mounting medium with DAPI (Vector Laboratories) (43). LAMP-1 fluorescence was viewed by UV light microscopy using an Eclipse E400 microscope under a ×100 oil-immersion lens (both Nikon), and images were captured using a DXM1200C digital camera and NIS-Elements BR2.30 software (both Nikon).
Confocal microscopy
Fixed and immunostained HeLa cells were visualized by confocal imaging using an LSM 10 META confocal laser scanning module arranged on an Axiovert 200 microscope, and a Plan-Apochromat ×63/1.4 oil immersion objective (all from Carl Zeiss, Germany). An argon laser was used to excite AlexaFluor 488 at λ = 488 nm; a HeNe laser was used to excite AlexaFluor 594 at λ = 594 nm. The DNA marker DAPI was excited in the two-photon mode using the 740 nm line of an infrared light Chameleon laser (Coherent, USA). The emission of fluophores used was detected in META channels of the scanning module in the multitrack mode within the following spectral ranges: 511–544 nm for AlexaFluor 488; 608–630 nm for AlexaFluor 594 and 447–480 nm for DAPI. Vertical sectioning of samples for obtaining Z-stackes of images was done at 0.5 µm intervals.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by Kidney Research UK (S.E.W. and R.V.T.); the Wellcome Trust (A.A.C.R., J.G., P.R. and R.V.T.); the Medical Research Council (A.A.C.R. and R.V.T.); and EuReGene, a Framework 6 programme grant by the European Commission (05085) (S.E.W., A.A.C.R., C.A. and R.V.T.).
ACKNOWLEDGEMENTS
We would like to thank Dr Juergen Strehlau for kindly providing two of the families in this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Duncan H., Dixon A.S. Gout, familial hypericaemia, and renal disease. Q. J. Med. 1960;29:127–135. [PubMed] [Google Scholar]
- 2.Hart T.C., Gorry M.C., Hart P.S., Woodard A.S., Shihabi Z., Sandhu J., Shirts B., Xu L., Zhu H., Barmada M.M., et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J. Med. Genet. 2002;39:882–892. doi: 10.1136/jmg.39.12.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner J.J., Stacey J.M., Harding B., Kotanko P., Lhotta K., Puig J.G., Roberts I., Torres R.J., Thakker R.V. UROMODULIN mutations cause familial juvenile hyperuricemic nephropathy. J. Clin. Endocrinol. Metab. 2003;88:1398–1401. doi: 10.1210/jc.2002-021973. [DOI] [PubMed] [Google Scholar]
- 4.Pook M.A., Jeremiah S., Scheinman S.J., Povey S., Thakker R.V. Localization of the Tamm–Horsfall glycoprotein (uromodulin) gene to chromosome 16p12.3–16p13.11. Ann. Hum. Genet. 1993;57:285–290. doi: 10.1111/j.1469-1809.1993.tb00902.x. [DOI] [PubMed] [Google Scholar]
- 5.Bingham C., Ellard S., van't Hoff W.G., Simmonds H.A., Marinaki A.M., Badman M.K., Winocour P.H., Stride A., Lockwood C.R., Nicholls A.J., et al. Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1beta gene mutation. Kidney Int. 2003;63:1645–1651. doi: 10.1046/j.1523-1755.2003.00903.x. [DOI] [PubMed] [Google Scholar]
- 6.Edghill E.L., Bingham C., Ellard S., Hattersley A.T. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J. Med. Genet. 2006;43:84–90. doi: 10.1136/jmg.2005.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodanova K., Majewski J., Kublova M., Vyletal P., Kalbacova M., Stiburkova B., Hulkova H., Chagnon Y.C., Lanouette C.M., Marinaki A., et al. Mapping of a new candidate locus for uromodulin-associated kidney disease (UAKD) to chromosome 1q41. Kidney Int. 2005;68:1472–1482. doi: 10.1111/j.1523-1755.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 8.Rampoldi L., Caridi G., Santon D., Boaretto F., Bernascone I., Lamorte G., Tardanico R., Dagnino M., Colussi G., Scolari F., et al. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum. Mol. Genet. 2003;12:3369–3384. doi: 10.1093/hmg/ddg353. [DOI] [PubMed] [Google Scholar]
- 9.Lens X.M., Banet J.F., Outeda P., Barrio-Lucia V. A novel pattern of mutation in uromodulin disorders: autosomal dominant medullary cystic kidney disease type 2, familial juvenile hyperuricemic nephropathy, and autosomal dominant glomerulocystic kidney disease. Am. J. Kidney Dis. 2005;46:52–57. doi: 10.1053/j.ajkd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Pennica D., Kohr W.J., Kuang W.J., Glaister D., Aggarwal B.B., Chen E.Y., Goeddel D.V. Identification of human uromodulin as the Tamm–Horsfall urinary glycoprotein. Science. 1987;236:83–88. doi: 10.1126/science.3453112. [DOI] [PubMed] [Google Scholar]
- 11.Tamm I., Horsfall F.L., Jr Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc. Soc. Exp. Biol. Med. 1950;74:108–114. [PubMed] [Google Scholar]
- 12.Kumar S., Muchmore A. Tamm–Horsfall protein–uromodulin (1950–1990) Kidney Int. 1990;37:1395–1401. doi: 10.1038/ki.1990.128. [DOI] [PubMed] [Google Scholar]
- 13.van Rooijen J.J., Voskamp A.F., Kamerling J.P., Vliegenthart J.F. Glycosylation sites and site-specific glycosylation in human Tamm–Horsfall glycoprotein. Glycobiology. 1999;9:21–30. doi: 10.1093/glycob/9.1.21. [DOI] [PubMed] [Google Scholar]
- 14.Serafini-Cessi F., Malagolini N., Cavallone D. Tamm–Horsfall glycoprotein: biology and clinical relevance. Am. J. Kidney Dis. 2003;42:658–676. doi: 10.1016/s0272-6386(03)00829-1. [DOI] [PubMed] [Google Scholar]
- 15.Yang H., Wu C., Zhao S., Guo J. Identification and characterization of D8C, a novel domain present in liver-specific LZP, uromodulin and glycoprotein 2, mutated in familial juvenile hyperuricaemic nephropathy. FEBS Lett. 2004;578:236–238. doi: 10.1016/j.febslet.2004.10.092. [DOI] [PubMed] [Google Scholar]
- 16.Prasadan K., Bates J., Badgett A., Dell M., Sukhatme V., Yu H., Kumar S. Nucleotide sequence and peptide motifs of mouse uromodulin (Tamm–Horsfall protein)—the most abundant protein in mammalian urine. Biochim. Biophys. Acta. 1995;1260:328–332. doi: 10.1016/0167-4781(94)00240-4. [DOI] [PubMed] [Google Scholar]
- 17.Jovine L., Qi H., Williams Z., Litscher E., Wassarman P.M. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat. Cell Biol. 2002;4:457–461. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- 18.Cavallone D., Malagolini N., Serafini-Cessi F. Mechanism of release of urinary Tamm–Horsfall glycoprotein from the kidney GPI-anchored counterpart. Biochem. Biophys. Res. Commun. 2001;280:110–114. doi: 10.1006/bbrc.2000.4090. [DOI] [PubMed] [Google Scholar]
- 19.Dahan K., Devuyst O., Smaers M., Vertommen D., Loute G., Poux J.M., Viron B., Jacquot C., Gagnadoux M.F., Chauveau D., et al. A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J. Am. Soc. Nephrol. 2003;14:2883–2893. doi: 10.1097/01.asn.0000092147.83480.b5. [DOI] [PubMed] [Google Scholar]
- 20.Wolf M.T., Mucha B.E., Attanasio M., Zalewski I., Karle S.M., Neumann H.P., Rahman N., Bader B., Baldamus C.A., Otto E., et al. Mutations of the Uromodulin gene in MCKD type 2 patients cluster in exon 4, which encodes three EGF-like domains. Kidney Int. 2003;64:1580–1587. doi: 10.1046/j.1523-1755.2003.00269.x. [DOI] [PubMed] [Google Scholar]
- 21.Kudo E., Kamatani N., Tezuka O., Taniguchi A., Yamanaka H., Yabe S., Osabe D., Shinohara S., Nomura K., Segawa M., et al. Familial juvenile hyperuricemic nephropathy: detection of mutations in the uromodulin gene in five Japanese families. Kidney Int. 2004;65:1589–1597. doi: 10.1111/j.1523-1755.2004.00559.x. [DOI] [PubMed] [Google Scholar]
- 22.Bleyer A.J., Hart T.C., Shihabi Z., Robins V., Hoyer J.R. Mutations in the uromodulin gene decrease urinary excretion of Tamm–Horsfall protein. Kidney Int. 2004;66:974–977. doi: 10.1111/j.1523-1755.2004.00845.x. [DOI] [PubMed] [Google Scholar]
- 23.Tinschert S., Ruf N., Bernascone I., Sacherer K., Lamorte G., Neumayer H.H., Nurnberg P., Luft F.C., Rampoldi L. Functional consequences of a novel uromodulin mutation in a family with familial juvenile hyperuricaemic nephropathy. Nephrol. Dial. Transplant. 2004;19:3150–3154. doi: 10.1093/ndt/gfh524. [DOI] [PubMed] [Google Scholar]
- 24.Calado J., Gaspar A., Clemente C., Rueff J. A novel heterozygous missense mutation in the UMOD gene responsible for familial juvenile hyperuricemic nephropathy. BMC Med. Genet. 2005;6:5. doi: 10.1186/1471-2350-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vylet'al P., Kublova M., Kalbacova M., Hodanova K., Baresova V., Stiburkova B., Sikora J., Hulkova H., Zivny J., Majewski J., et al. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 26.Benetti E., Caridi G., Vella M.D., Rampoldi L., Ghiggeri G.M., Artifoni L., Murer L. Immature renal structures associated with a novel UMOD sequence variant. Am. J. Kidney Dis. 2009;53:327–331. doi: 10.1053/j.ajkd.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Lhotta K., Gehringer A., Jennings P., Kronenberg F., Brezinka C., Andersone I., Strazdins V. Familial juvenile hyperuricemic nephropathy: report on a new mutation and a pregnancy. Clin. Nephrol. 2009;71:80–83. doi: 10.5414/cnp71080. [DOI] [PubMed] [Google Scholar]
- 28.Serafini-Cessi F., Malagolini N., Hoops T.C., Rindler M.J. Biosynthesis and oligosaccharide processing of human Tamm–Horsfall glycoprotein permanently expressed in HeLa cells. Biochem. Biophys. Res. Commun. 1993;194:784–790. doi: 10.1006/bbrc.1993.1890. [DOI] [PubMed] [Google Scholar]
- 29.Malagolini N., Cavallone D., Serafini-Cessi F. Intracellular transport, cell-surface exposure and release of recombinant Tamm–Horsfall glycoprotein. Kidney Int. 1997;52:1340–1350. doi: 10.1038/ki.1997.459. [DOI] [PubMed] [Google Scholar]
- 30.Choi S.W., Ryu O.H., Choi S.J., Song I.S., Bleyer A.J., Hart T.C. Mutant Tamm–Horsfall glycoprotein accumulation in endoplasmic reticulum induces apoptosis reversed by colchicine and sodium 4-phenylbutyrate. J. Am. Soc. Nephrol. 2005;16:3006–3014. doi: 10.1681/ASN.2005050461. [DOI] [PubMed] [Google Scholar]
- 31.Bernascone I., Vavassori S., Di Pentima A., Santambrogio S., Lamorte G., Amoroso A., Scolari F., Ghiggeri G.M., Casari G., Polishchuk R., et al. Defective intracellular trafficking of uromodulin mutant isoforms. Traffic. 2006;7:1567–1579. doi: 10.1111/j.1600-0854.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 32.Jennings P., Aydin S., Kotanko P., Lechner J., Lhotta K., Williams S., Thakker R.V., Pfaller W. Membrane targeting and secretion of mutant uromodulin in familial juvenile hyperuricemic nephropathy. J. Am. Soc. Nephrol. 2007;18:264–273. doi: 10.1681/ASN.2006020158. [DOI] [PubMed] [Google Scholar]
- 33.Stacey J.M., Turner J.J., Harding B., Nesbit M.A., Kotanko P., Lhotta K., Puig J.G., Torres R.J., Thakker R.V. Genetic mapping studies of familial juvenile hyperuricemic nephropathy on chromosome 16p11–p13. J. Clin. Endocrinol. Metab. 2003;88:464–470. doi: 10.1210/jc.2002-021268. [DOI] [PubMed] [Google Scholar]
- 34.Cantalupo G., Alifano P., Roberti V., Bruni C.B., Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crotzer V.L., Blum J.S. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic. 2008;9:10–16. doi: 10.1111/j.1600-0854.2007.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell. Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 37.Ding W.X., Ni H.M., Gao W., Hou Y.F., Melan M.A., Chen X., Stolz D.B., Shao Z.M., Yin X.M. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 38.Eskelinen E.L., Tanaka Y., Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 39.Whiteman P., Handford P.A. Defective secretion of recombinant fragments of fibrillin-1: implications of protein misfolding for the pathogenesis of Marfan syndrome and related disorders. Hum. Mol. Genet. 2003;12:727–737. doi: 10.1093/hmg/ddg081. [DOI] [PubMed] [Google Scholar]
- 40.Schaeffer C., Santambrogio S., Perucca S., Casari G., Rampoldi L. Analysis of uromodulin polymerization provides new insights into the mechanisms regulating ZP domain-mediated protein assembly. Mol. Biol. Cell. 2009;20:589–599. doi: 10.1091/mbc.E08-08-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesbit M.A., Bowl M.R., Harding B., Ali A., Ayala A., Crowe C., Dobbie A., Hampson G., Holdaway I., Levine M.A., et al. Characterization of GATA3 mutations in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. J. Biol. Chem. 2004;279:22624–22634. doi: 10.1074/jbc.M401797200. [DOI] [PubMed] [Google Scholar]
- 42.Nesbit M.A., Bowl M.R., Harding B., Schlessinger D., Whyte M.P., Thakker R.V. X-linked hypoparathyroidism region on Xq27 is evolutionarily conserved with regions on 3q26 and 13q34 and contains a novel P-type ATPase. Genomics. 2004;84:1060–1070. doi: 10.1016/j.ygeno.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Bradley K.J., Bowl M.R., Williams S.E., Ahmad B.N., Partridge C.J., Patmanidi A.L., Kennedy A.M., Loh N.Y., Thakker R.V. Parafibromin is a nuclear protein with a functional monopartite nuclear localization signal. Oncogene. 2007;26:1213–1221. doi: 10.1038/sj.onc.1209893. [DOI] [PubMed] [Google Scholar]
- 44.Wilcox W.D. Abnormal serum uric acid levels in children. J. Pediatr. 1996;128:731–741. doi: 10.1016/s0022-3476(96)70322-0. [DOI] [PubMed] [Google Scholar]
- 45.McBride M.B., Rigden S., Haycock G.B., Dalton N., Van't Hoff W., Rees L., Raman G.V., Moro F., Ogg C.S., Cameron J.S., et al. Presymptomatic detection of familial juvenile hyperuricaemic nephropathy in children. Pediatr. Nephrol. 1998;12:357–364. doi: 10.1007/s004670050466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.