Abstract
Here we report the functional assessment of two novel deafness-associated γ-actin mutants, K118N and E241K, in a spectrum of different situations with increasing biological complexity by combining biochemical and cell biological analysis in yeast and mammalian cells. Our in vivo experiments showed that while the K118N had a very mild effect on yeast behaviour, the phenotype caused by the E241K mutation was very severe and characterized by a highly compromised ability to grow on glycerol as a carbon source, an aberrant multi-vacuolar pattern and the deposition of thick F-actin bundles randomly in the cell. The latter feature is consistent with the highly unusual spontaneous tendency of the E241K mutant to form bundles in vitro, although this propensity to bundle was neutralized by tropomyosin and the E241K filament bundles were hypersensitive to severing in the presence of cofilin. In transiently transfected NIH3T3 cells both mutant actins were normally incorporated into cytoskeleton structures, although cytoplasmic aggregates were also observed indicating an element of abnormality caused by the mutations in vivo. Interestingly, gene-gun mediated expression of these mutants in cochlear hair cells results in no gross alteration in cytoskeletal structures or the morphology of stereocilia. Our results provide a more complete picture of the biological consequences of deafness-associated γ-actin mutants and support the hypothesis that the post-lingual and progressive nature of the DFNA20/26 hearing loss is the result of a progressive deterioration of the hair cell cytoskeleton over time.
INTRODUCTION
Hearing results from the conversion of the mechanical forces of sound waves into electrical signals, in a process called mechanotransduction (1). This process takes place in the cochlea where the sound-induced vibrations of the basilar membrane are detected and transduced by the hair cells, and the resultant electrical signals are transmitted to the brainstem via the cochlear ganglion. Mechanotransduction ultimately relies on highly specialized actin-based structures located at the apical surface of the cochlear hair cells, the stereocilia, which contain a core of parallel actin filaments that have identical polarity and are cross-linked by several actin-binding proteins (2). The stereocilia are anchored at their base into the cuticular plate, a dense gel-like network of randomly oriented actin filaments. Encircling each hair cell near its apical surface is the zona adherens junction, a circumferential belt of actin filaments that runs parallel to the plasma membrane (3). The integrity of the actin cytoskeleton in all these cellular structures is known to be crucial for hearing.
Actin filaments (F-actin) are formed through the polymerization of actin monomers, a globular 42 kDa protein (G-actin). G-actin is characterized by a cleft that runs through the protein separating it into two domains. The outer domain is comprised subdomains 1 and 2, whereas the inner larger domain contains subdomains 3 and 4. The monomer binds an adenine nucleotide and a divalent cation, usually Mg+2 in vivo, at the base of the cleft (4), and removal of either of these under physiological conditions leads to protein denaturation (5). During polymerization, the inherent slow ATPase activity of G-actin is greatly enhanced first producing ADP-Pi actin and then ADP actin following phosphate release. This behaviour of the nucleotide is important for actin filament dynamics since ATP and ADP-Pi F-actin are much more stable than ADP actin (6). On the basis of the Holmes model, the actin filament is a polar structure with a ‘barbed’ and a ‘pointed’ end, and this polarity is translated from the actin filament to the actin monomer (7). The barbed end (subdomains 1 and 3) is the preferred site for monomer addition during filament elongation, whereas the pointed end (subdomains 2 and 4) is the preferred site of monomer release. Also, the barbed end has been implicated in interactions with a number of proteins that regulate actin polymerization in the cell (8).
Non-muscle cells of birds and mammals, including cochlear hair cells, contain two very similar actin isoforms that are termed β- and γ-non-muscle isoactins (9). Filaments formed by non-muscle actins are involved in multiple dynamic processes including cell shape changes, cell spreading, motility, cytokinesis, polarity and vesicle trafficking (10). Although the protein sequences of γ-actin and β-actin differ by only four amino acids at their N-termini, the conservation of both genes in vertebrates and their different distribution suggests they may play different functions. In the mammalian cochlea, amounts of γ-actin increase and β-actin decrease in the order: outer pillar cells, inner pillar cells, Deiters’ cells and hair cells. In sensory cells, γ-actin appears to be the most prominent component with an apparent γ-actin:β-actin ratio of approximately 2:1 (11). β-actin is present in the cuticular plate but is more concentrated in the stereocilia, especially at the base (the rootlets and periphery of their shafts) where the stereocilia insert into the cuticular plate. The amount of γ-actin differs less between these structures, the stereocilia and cuticular plate, although its expression is apparently higher towards the tip of stereocilia and it is the predominant isoform of the hair cell's lateral wall (12).
Mutations in ACTG1, the gene encoding γ-actin, have been associated with DFNA20/26, a subtype of autosomal dominant non-syndromic sensorineural progressive hearing loss (ADNSHL). DFNA20/26 hearing impairment initially affects the high frequencies at onset and later progresses to affect the entire frequency range. Subtle differences in the onset and severity of the hearing loss have been observed in patients carrying the different mutations in ACTG1. At present, only seven missense mutations in ACTG1 have been identified causing ADNSHL, including four in American families (T89I, K118M, P264L, P332A), one in a Dutch family (T278I), one in a Norwegian family (V370A) and, more recently, one in a Chinese family (I122V) (13–16). All the mutations hitherto described are located in subdomains 1 and 3 of γ-actin.
The molecular basis for DFNA20/26 hearing loss has been a subject of discussion since the identification of the mutations. Recently, the biochemical effects of the mutations on actin structure/function have been investigated by cloning the original six deafness mutations into the yeast actin gene (8), thereby, allowing a direct comparison of the effects of the mutations in vivo and in vitro. In the present study, we report the identification and functional assessment of two novel deafness-causing mutations in ACTG1. We have used a combination of biochemical and cell biological analysis to delineate the effects of both mutations on actin function. Beside the in vivo experiments in yeast, we have also carried out transient expression of the mutants in NIH3T3 cells and gene-gun experiments in hair cells from mouse cochlear explants. Furthermore, we have characterized these mutant actins in vitro in terms of their polymerization properties and their interaction with two actin-regulatory proteins, tropomyosin and cofilin. This study provides, for the first time, a complete picture of the effect of deafness-associated γ-actin mutants in a spectrum of different situations with increasing biological complexity, from yeast to mammalian cells.
RESULTS
Identification of two novel pathogenic mutations (p.K118N and p.E241K) in human γ-actin
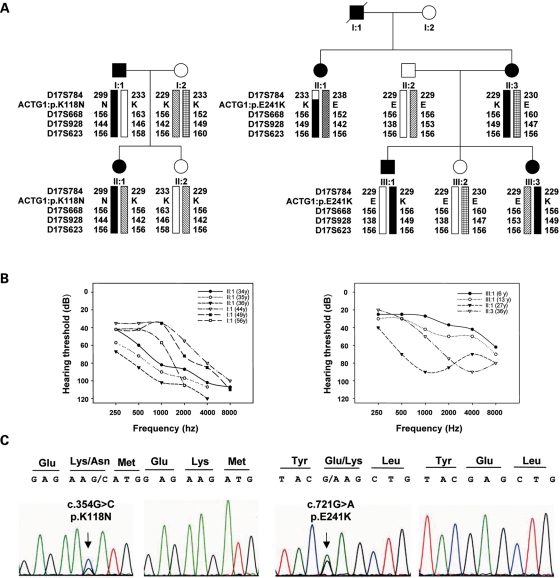
As affected individuals carrying mutations in ACTG1 display high-frequency hearing loss (DFNA20/26), we designed a phenotype-guided mutational screen in small families that were not suitable for linkage analysis. Thus, probands from 292 unrelated families displaying a mid-high frequency hearing loss in which segregation was compatible with the autosomal dominant pattern were screened for mutations in the ACTG1 gene. All exons and flanking intronic regions of ACTG1 were PCR amplified from genomic DNA isolated from each proband. The PCR amplimers were screened for mutations by DHPLC, and the different heteroduplex profiles obtained were characterized by sequencing. This approach led us to identify two novel pathogenic mutations in ACTG1 (Fig. 1C). The first mutation was identified in the subject II:1 of the family S840 and consists of the c.354G>C transversion (Fig. 1C, left) that replaces a lysine by an asparagine at position 118 (p.K118N). This lysine lies in subdomain 1 of the protein (Fig. 2) and was previously reported to be mutated to methionine in another family with DFNA20/26 hearing loss (13). The second mutation, identified in the subject II:3 of the family S582, is a c.721G>A transition (Fig. 1C, right) that replaces a glutamic acid by a lysine at position 241 (p.E241K) and is the first mutation reported to affect subdomain 4 of the protein (Fig. 2). Interestingly, an identical amino acid replacement has been reported in α-skeletal muscle actin, ACTA1, as a cause of nemaline myopathy, a subtype of inherited muscular disease (17). Both γ-actin mutations independently segregated with the hearing loss in the respective families (Fig. 1A), and they were not detected in 100 unrelated normal-hearing Spanish controls. Genotyping of microsatellite markers in the DFNA20/26 region and subsequent haplotype analysis confirmed segregation of the disease chromosome containing the mutation with the hearing loss in both families. Together, these data suggested that the identified mutations were the actual cause of the hearing loss in these families. This was further corroborated by the functional studies described below.
Figure 1.
(A) Pedigree of the Spanish families S840 (left) and S582 (right). Affected members are shown in black. Haplotypes are represented by bars, with the haplotype associated with hearing loss in black. The relative order of ACTG1 gene and the microsatellite markers within the DFNA20/A26 genetic interval is indicated. The numbers beside the bars are the allelic sizes for each microsatellite marker and the mutations identified in the families are notated as ACTG1: p.K118N and p.E241K, respectively. (B) Audiograms showing the air conduction values obtained from affected patients of S840 (I:1 and II:1) and S582 (II:1, II:3 and III:1) families. The age of the patients from which each audiometric record was obtained is indicated. Each graph point represents the average hearing loss for the right and left ears. (C) Electropherograms of the mutations c.354G>C and c.721G>A identified in exons 3 and 4 of ACTG1 in S840 and S582 families, respectively. The arrow points to the precise nucleotide that is mutated. An affected subject and a control are depicted.
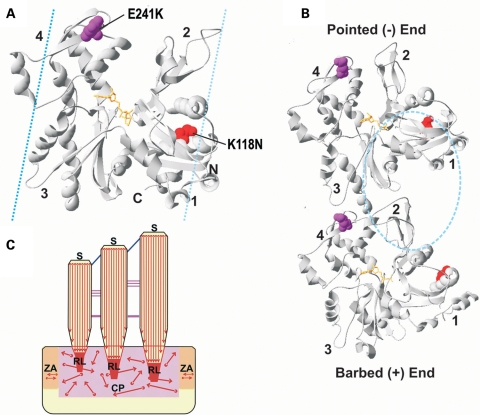
Figure 2.
Locations of the K118N and E241K γ-actin mutations in yeast actin. (A) Front view of the crystal structure of the yeast actin monomer (43), modified from Protein Data Bank code 1YAG using Swiss-PdbViewer, version 4.01. The positions of the mutations are modelled in space-fill and colour-highlighted as follows: K118N, red and E241K, purple. ATP is modelled in ball-stick and coloured orange. Numbers denote the actin subdomains. G-actin monomers are characterized by a cleft that divides it into two similar domains (4,7), the small outer domain (subdomains 1 and 2) and the larger inner domain (subdomains 3 and 4). The names relate to the position of each domain in the F-actin helix where the inner domain is closer to the helix axis. N and C mark the respective termini. Cyan dashed lines denote the general vicinity on the actin surface to which tropomyosin can occupy (29,30). (B) Model illustrating the longitudinal contacts between two neighbouring monomers within the same filament strand based on the Holmes filament model (7). The pointed (−) and barbed (+) ends are indicated. Each monomer is labelled as in (A) with regard to the mutations, ADP and subdomains. Cyan dashed circle denote the general vicinity on the actin surface to which cofilin binds (22). (C) Molecular architecture of the hair cell's (HC) actin cytoskeleton. Actin filaments are denoted by red arrows. The barbed (subdomains 1 and 3) and pointed (subdomains 2 and 4) ends of the filaments are represented by circles and arrowheads, respectively. CP, cuticular plate; S, stereocilia; ZA, zonula adherens; RL, rootlets.
In vivo experiments in yeast
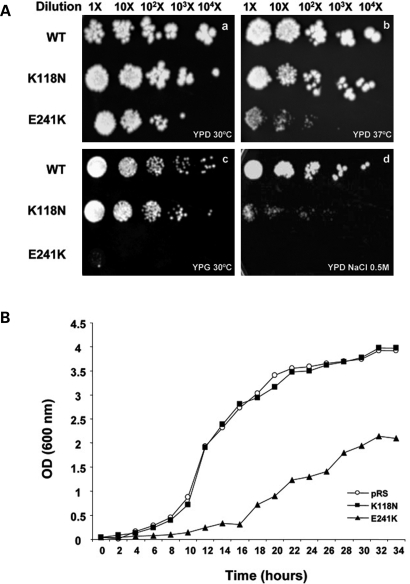
To assess the impact that p.K118N and p.E241K could exert on γ-actin function, we engineered both mutations into yeast actin and characterized their effects in vivo as previously reported (8). We first analyzed the effects of the mutations on the rate and extent of cell growth. The global effects of the mutations on cell behaviour were assessed by their ability to grow on YPD plates at two temperatures. While no significant growth differences are observed between the wild-type and the K118N mutant at 30°C, cells carrying the E241K mutation showed distinctly impaired growth (Fig. 3Aa). This growth deficiency was more evident for the E241K strain at 37°C (Fig. 3Ab). A more quantitative estimation of the growth defect was obtained after growing both mutants in yeast nitrogen base (YNB) liquid medium at 30°C. The K118N mutant exhibited a WT-like growth rate, whereas the E241K mutant exhibited severe growth retardation reaching a final density of ∼50% relative to the wild-type strain (Fig. 3B). As previously described for other ACTG1 mutations (8), one possible explanation for the extent of decreased growth is a defect in the mitochondrial function, as actin is required for mitochondrial inheritance, morphology and stabilization of mtDNA. This possibility was assessed by examining the ability of the cells to grow in glycerol as a sole carbon source, since the activity of the mitochondrial glycerol-3-phosphate dehydrogenase is often eliminated as a consequence of mitochondrial defects. This assay showed that, whereas K118N could normally grow in glycerol, the E241K mutant showed a dramatically decreased ability to grow, implying an impaired mitochondrial function (Fig. 3Ac). Additionally, the K118N mutation severely inhibited growth in high-salt YPD medium, whereas the E241K mutant strain could not grow in this environment (Fig. 3Ad), indicating that these mutations are differentially influencing the actin-mediated cell response to hyperosmotic stress.
Figure 3.
(A) Comparison of the growth of WT and mutant yeast strains on YPD medium at 30°C (a), YPD at 37°C (b), YPG medium with glycerol as a sole carbon source (c) and on YPD + NaCl 0.5 m at 30°C (d). The experiments were repeated three times with similar results. (B) Growth curves of cells expressing either Wt or mutant yeast actins. Cells were grown in YNB medium at 30°C on a shaking platform.
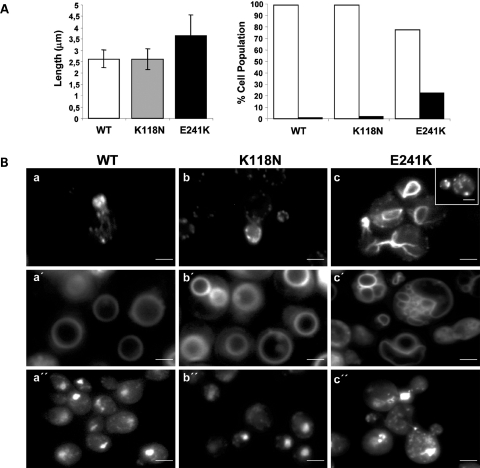
Next, the effects of the mutations on yeast cell morphology and actin cytoskeletal patterns were examined. Disruption of the actin cytoskeleton by actin mutations may lead to depolarized isotropic growth of the cell, resulting in larger and rounder cells. As previously reported (8), we estimated the effect of the mutations on cell size by measuring the cell diameter along its long axis using differential interference contrast microscopy. The E241K mutant cells exhibited a significant increase (P < 0.001) in comparison with the K118N or WT cells (Fig. 4A, left). Visualization of the actin cytoskeleton was achieved using Alexa-594-conjugated phalloidin. Actin in yeast forms two types of assemblies: cables and patches. Cables enable the movement of organelles from the mother cell to the bud during division. Actin patches localize at sites of endocytosis (18), and their distribution is cycle dependent. Figure 4B shows that the K118N mutation resulted in an apparent wild-type looking cytoskeleton (a and b). In the majority of E241K mutant cells that were compared at the similar cell-cycle stage (the bud was roughly one-third of the mother cell), abnormally randomly distributed thick-actin cables were observed in addition to patches (c) indicating a strong propensity for these actin filaments to bundle inappropriately. Furthermore, some E241K mutant cells displayed an abnormal cytoskeleton pattern in which the actin patches were randomly distributed, instead of being restricted to the growing bud as expected at this stage of the cell cycle (inset in c). Vacuole morphology in yeast is also regulated by the actin cytoskeleton. We therefore examined this parameter in the mutant yeast cells by staining the vacuoles with the dye FM4-64. In the majority of WT cells and K118N cells, one large vacuolar lobe per cell was observed (Fig. 4Ba′ and b′). Conversely, in the case of the E241K mutant, a percentage of cells displayed multiple vacuoles and two recognizable morphology patterns. Some cells showed one large lobe with multiple abnormally smaller lobes, whereas other mutant cells were filled with large numbers of small lobes only (Fig. 4Bc′). These patterns were displayed by nearly 25% of E241K mutant cells (Fig. 4A, right). Finally, to assess whether the mutations could affect the DNA distribution pattern, we visualized the mtDNA and genomic DNA with the DNA-intercalating dye Hoechst. Figure 4B shows that in both the wild-type and K118N mutant cells (a″ and b″), an apparently normal DNA pattern is observed in which, in addition to the large nuclear DNA spot, faint diffuse extra-nuclear spots representing mtDNA are observed. However, for the E241K mutant cells, larger and brighter mtDNA spots are numerous (c″) suggesting that the mtDNA may be aggregating in these cells. This phenotype was also observed with some of the other γ-actin mutant strains (K118M, T278I, P332A and V370A) that exhibited an inability to grow in glycerol as a sole carbon source (8).
Figure 4.
(A) Measure of the length of cells expressing Wt and mutants yeast actins (left). Difference was statistically significant between Wt and E241K mutant based upon Student t-test analysis with a P-value lower than 0.001. Error bars represent SD. Assessment of vacuole phenotype (right). White bars and black bars represent the percentage of cells with 1–4 vacuoles and with more than 4 vacuoles, respectively. Results based on the measurement of a population of at least 200 cells. (B) Fluorescence microscopy of cells expressing WT, K118N and E241K yeast actins. The cytoskeleton was visualized after staining fixed cells (at a stage of the cell cycle in which the bud represents roughly one-third of the mother cell) with rhodamine–phalloidin (a–c). At this stage, two abnormal cytoskeleton patterns were observed in the majority of E241K cells. They consisted of, in addition to patches, abnormally distributed thick-actin cables (c) or in randomly distributed actin patches that are not confined to the bud as expected at this stage of the cell cycle (inset). Vacuoles were observed following exposure the cells to dye FM4-64 (a′–c′). Nuclear and mitochondrial DNAs were visualized following staining of the cells with Hoechst (a″–c″). The largest bright spot in each cell is the nucleus and the cytoplasmic spots correspond to mtDNA. Scale bars = 2 µm.
In summary, our in vivo experiments show that the K118N had a very mild effect on yeast cell behaviour. Conversely, the phenotype caused by the E241K mutation was very severe and characterized by a highly compromised ability to grow on glycerol as a carbon source, an aberrant multi-vacuolar pattern and the deposition of thick F-actin bundles randomly in the cell.
Effects of the mutations on the actin monomer
To determine the molecular basis for the in vivo phenotypes described above, we first examined the effects of the mutations on the properties of the actin monomer. We thus assessed the effect of the mutations on the thermal stability of the mutant actin monomers and, because of the importance of the bound adenine nucleotide to actin behaviour; we determined their effects on nucleotide exchange rates (8). The E241K mutation had no significant effect on the overall conformation of the actin monomer as its melting temperature (Tm) is identical to that of WT actin (Table 1). In contrast, the K118N mutation resulted in a significant (P < 0.05) decrease of 4°C in the Tm relative to that of WT actin. The lower Tm with the K118N mutant is similar to that seen with its sister allele, K118M (8). Moreover, a change in the thermal stability of this magnitude suggests that the residue at position 118 is a significant determinant of monomer stability.
Table 1.
Effects of mutations on actin monomer thermostability and ATP exchange rates
| Strain | Thermostability average apparent Tm (°C) | Nucleotide exchange rate t1/2 (s) |
|---|---|---|
| WT | 58.5 ± 1 (4) | 44 ± 4.3 (4) |
| K118N | 54.2±0.3 (4)a | 44 ± 7.4 (4) |
| E241K | 58 ± 0.4 (4) | 35±5.1 (4)a |
The number of experiments performed is indicated in parentheses.
aDifference is statistically significant based upon Student t-test analysis with P-values of less than 0.002 and 0.03, respectively.
Next we measured the ability of the G-actin to exchange the fluorescently bound ATP analog (ε-ATP) in the presence of excess unlabeled ATP. Table 1 shows that the K118N mutation, as with its sister allele K118M (8), had no measurable effect on the monomer's nucleotide exchange rate. On the contrary, the t1/2 of exchange for the E241K was 35 ± 5 s (P < 0.03), ∼25% faster than the WT rate. This result suggests that the E241K mutant is more conformationally flexible or assumes a more open conformation than WT with respect to the interdomain cleft in which the nucleotide is bound. Interestingly, contrary to what might have been expected, there does not seem to be a correlation between lowered melting temperature and increased nucleotide exchange rate here, both potential indicators of a more conformationally dynamic molecule.
Effects of mutation on actin polymerization
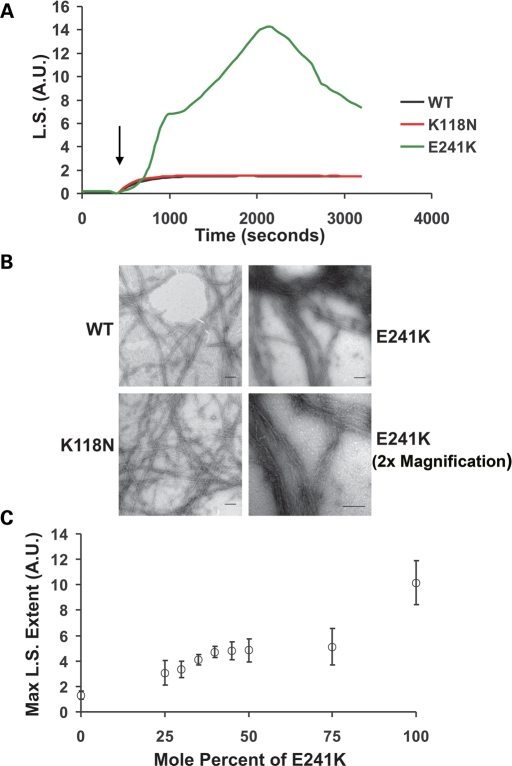
Proper function and structural integrity of both the stereociliary bundle and the cuticular plate depends on actin filaments. We thus wished to assess the effects of these two mutations on actin polymerization in vitro. Figure 5A indicates that both the kinetics and extent of polymerization of the K118N mutant are virtually identical to WT actin. In contrast, the polymerization of the E241K is highly unusual. Initially, polymerization lags behind that of WT actin. However, following a rapid elongation phase, the light scattering change reaches a level about 10-fold higher than that achieved by WT actin followed by a continual decrease in the light scattering signal over the next 1500 s. This decrease was accompanied by the appearance of what looked like an actin gel at the bottom of the cuvette. A possible explanation for this behaviour is the initial formation of highly scattering actin filament bundles followed by their coalescence and precipitation from solution.
Figure 5.
(A) Comparison of the polymerization kinetics of WT and mutant actins. Polymerization of 4.8 µm actin was initiated by the addition of magnesium and potassium chloride (denote by arrow) as described in Materials and Methods, and the increase in light scattering (LS) was monitored as a function of time at 25°C. Shown are representative plots from experiments performed at least three times with three independent actin preparations. AU stands for arbitrary units. (B) Effects of mutations on filament morphology. Samples of the actin were prepared as described in Materials and Methods. Shown are electron micrographs of WT, K118N and E241K filaments. Note the high degree of filament bundling of the E241K filaments. Scale bar = 100 nm. (C) Effect of increasing the mole percent of E241K on light scattering caused by actin polymerization. Shown is the relationship between the various percentages of E241K mutant actin in the samples and the maximum extent in light scattering signal value attained. The open circles represent the average light scattering value and the error bars are the standard deviation in the measurements from five independent trials using three different actin preparations.
To test this hypothesis and to examine the morphology of the filaments made by both actins, we examined negatively stained samples of the actins at the completion of polymerization by electron microscopy. Figure 5B shows normal appearing filaments for K118N actin. As predicted the E241K mutant actin forms thick F-actin bundles of normal appearing actin filaments. These data correlate with the tendency of this actin to form thick actin bars or bundles in yeast.
Since the mutant actin would account for ∼30% of the total actin content of the hair cells, we determined the effects of varying percentages of E241K mutant actin present versus WT actin when the two were copolymerized at a constant 4.8 µm total actin concentrations. Figure 5C clearly shows that between 25 and 50% mutant actin, the increase in light scattering increases roughly as does the percentage of E241K in the sample. Following a plateau region, a disproportionately sharp increase occurs between 75 and 100% mutant actin. This behaviour implies a concentration threshold above which smaller bundles may coalesce into larger bundles that ultimately drop out of solution. Bundles are observed by EM at 30% mutant concentration, the lowest concentration examined suggesting that filament formation and bundling occurred concurrently (data not shown).
Effects of tropomyosin on the bundling propensity of E241K mutant filaments
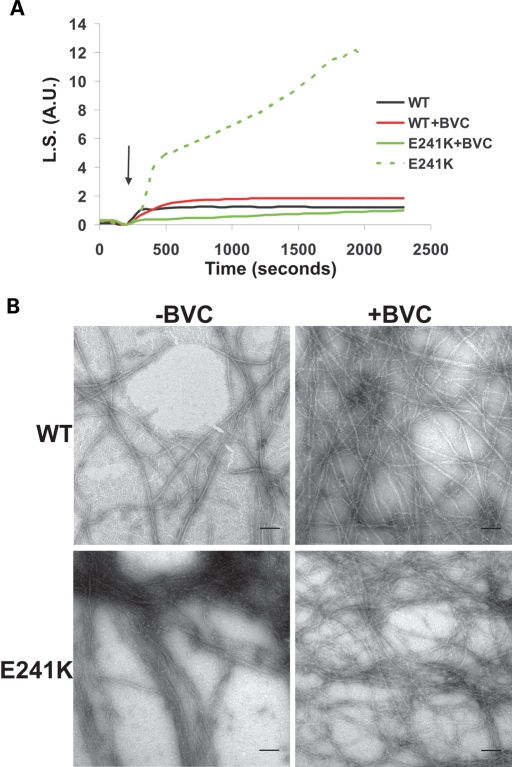
Although the E241K mutant exhibited a tendency to bundle in vivo, the compatibility of this actin with yeast viability indicated that many actin-dependent functions apparently still remained intact, probably as a result of the presence of actin-binding proteins. One such factor might be tropomyosin, also found in the auditory hair cell (19,20), which binds along the filament on both strands and might prevent bundling from occurring (21). Figure 6A shows that the addition of stoichiometric amounts of bovine cardiac tropomyosin (BVC) tropomyosin to the E241K mutant actin sample at the beginning of polymerization resulted in a decreased rate of polymerization and the elimination of the abnormally high light scattering caused by bundling. Examination of the sample by EM at the end of polymerization demonstrated that bundling is, in fact, greatly reduced as suggested by the kinetics (Fig. 6B).
Figure 6.
Effects of BVC on E241K mutant filament bundling. (A) Effects of the absence (dashed lines) or presence (solid lines) of bovine cardiac tropomyosin has on the polymerization kinetics and extents as measured by the light scattering of samples containing either WT and E241K actin when it is added prior to inducing polymerization at 25°C. Arrow denotes induction of polymerization by the addition of salts (Fig. 5). (B) Electron micrographs of WT and E241K filaments either in the absence (left) or presence of BVC (right). Scale bar = 100 nm.
Effects of mutations on filament sensitivity to cofilin
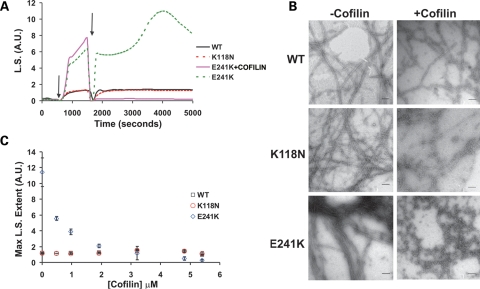
Auditory hair cell defects resulting from these two actin mutations might arise as a result of filament instability caused by the mutations. Both mutations lie in binding sites for the actin filament severing protein cofilin (22), a protein also found in the cochlea (www.brighamandwomens.org/bwh_hearing/InnerEarcDNAArrays.aspx#). To determine if either mutation might result in enhanced cofilin sensitivity, we examined the effect that stoichiometric amounts of cofilin had on the extent of light scattering when added at the end of polymerization. This amount of yeast cofilin combined with the yeast actin will actually coat WT filaments (23) without causing appreciable severing under experimental conditions. Figure 7A shows that K118N, which is in a weak binding site for cofilin (24), caused a response to this protein similar to that of WT actin: filament decoration with minimal severing based on the recovery after a sharp drop in light scattering. In contrast, the addition of cofilin to the E241K filaments prior to the time they would normally fall out of solution resulted in a dramatic decrease in light scattering to approximately the same level observed with G-actin. A control sample in which buffer was added in place of the cofilin showed this change does not result simply from mixing the sample. Electron micrographs of the cofilin-treated E241K actin verified the destruction of bundles and an extreme shortening of filaments consistent with enhanced sensitivity to cofilin severing (Fig. 7B). We then repeated the experiment as a function of cofilin concentration to determine thresholds for bundle disappearance. Figure 7C shows an exponential decrease in light scattering as a function of cofilin concentration resulting in the elimination of most of the bundling by the time the cofilin reaches 40% of the total actin concentration. Because of the enhanced sensitivity of E241K F-actin to cofilin, we wanted to determine the effect of increasing the mole fraction of E241K actin at a constant actin concentration on cofilin sensitivity. The cofilin sensitivity appeared to be roughly proportional to the amount of E241K in the samples (data not shown).
Figure 7.
Effects of mutations on filament sensitivity to cofilin severing. (A) The change in light scattering as a function of time was monitored for samples containing 4.8 µm of either WT, K118N or E241K actin. Polymerization was induced by the addition of salts (denoted by first arrow) and once the samples reached steady state a stoichiometric amount of yeast cofilin or an equivalent amount of buffer was added to the samples (denoted by the second arrow). Dashed green line represents an E241K control sample to which buffer was added instead of cofilin. (B) Electron micrograph which shows the effect the post-polymerization addition of a stoichiometric amount of cofilin has on E241K filaments. Scale bar = 100 nm. (C) Each point represents the average maximum light scattering value obtained for samples containing either 4.8 µm WT, K118N or E241K actin and the following concentrations of yeast cofilin: 0, 0.48, 0.96, 1.92, 3.2, 4.8 and 5.4 µm, respectively. Error bars denote range determined from two independent trials with two different preparations of actin.
Expression of the actin mutants in NIH-3T3 fibroblast
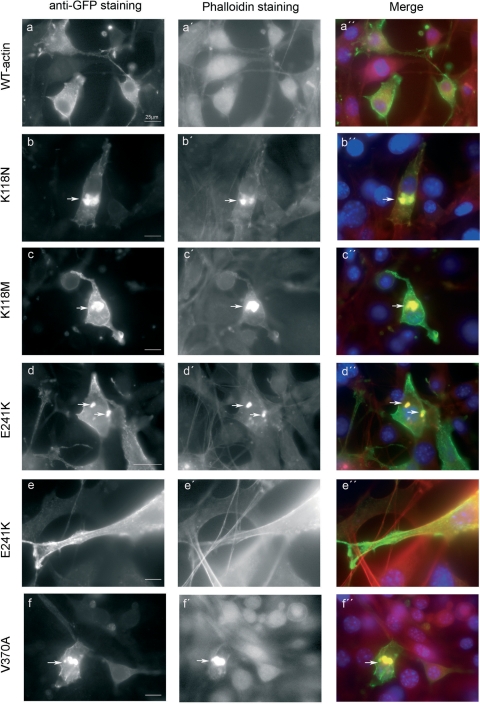
Next we wanted to examine the ability of the actual mammalian K118N and E241K γ-actin mutants on actin dynamics in cultured mammalian cells, a model system with increased biological complexity. We also assayed the K118M mutant since it has a more drastic amino acid substitution at this position, and the V370A mutant since this was the only deafness mutant previously shown to have altered polymerization properties. A modified expression plasmid EGFP-γ-actin was mutagenized to introduce each mutation and then used to transiently transfect NIH3T3 fibroblasts, a cell line previously used as a model to evaluate the effects of actin mutations (25). Wild-type γ-actin is incorporated into stress fibres, filamentous actin in ruffles and lamellipodia, and into the actin network beneath the plasma membrane (Fig. 8A). All structures that stained with phalloidin in these cells also showed co-staining with anti-EGFP antibody, indicating a good incorporation into the cellular F-actin pools of the plasmid-derived γ-actin. The K118N mutant, however, localized more weakly to stress fibres and lamellipodia and accumulated in a large cytoplasmic aggregate that intensely stained with phalloidin (Fig. 8B). This pattern was observed in nearly 50% of cells expressing the K118N mutant. With the K118M mutant, the aggregate was observed in roughly a 30% of cells, and its incorporation to stress fibres was similar to that of wild-type γ-actin (Fig. 8C). E241K mutant actin was also incorporated into stress fibres and filamentous actin in lamellipodia and ruffles, but in roughly 20% of cells this mutant also localized to small aggregates or a large phalloidin-staining clump in the cytoplasm (Fig. 8D and E). Similar to E241K, the V370A mutant incorporated into stress fibres, lamellipodia and ruffles, and a large clump that localized with phalloidin was observed in 20% of cells. Small aggregates, however, were not seen in the V370 mutant cells (Fig. 8F).
Figure 8.
Expression of wild-type and γ-actin mutants in NIH3T3 fibroblasts. Wild-type actin (A) was used as a control and was incorporated into stress fibres, ruffles, lamellipodia and into the cortical network beneath the plasma membrane and generally co-localized with the endogenous filamentous actin, as visualized with phalloidin (A′, A″). The mutant K118N localized less to stress fibres and lamellipodia (B′, B″). The mutant protein accumulated in a big cytoplasmic clump (arrow) in the cytoplasm that stained with phalloidin in 50% of cells, (B). The incorporation of the E241K into the stress fibres, ruffles and lamellipodia appears normal (D and E), but in 20% of cells, a big or several small aggregates were visualized that co-localized with phalloidin (D′, D″). Both the K118M and the V370A were incorporated into stress fibres, ruffles and lamellipodia (C and F). However, in 30% of cells expressing K118M and in 20% expressing V370A, the mutant proteins accumulated in a big cytoplasmic clump that co-localized with phalloidin (C′, C″ and F′, F″). Scale bar = 25 µm.
Gene-gun experiments in cochlear hair cells
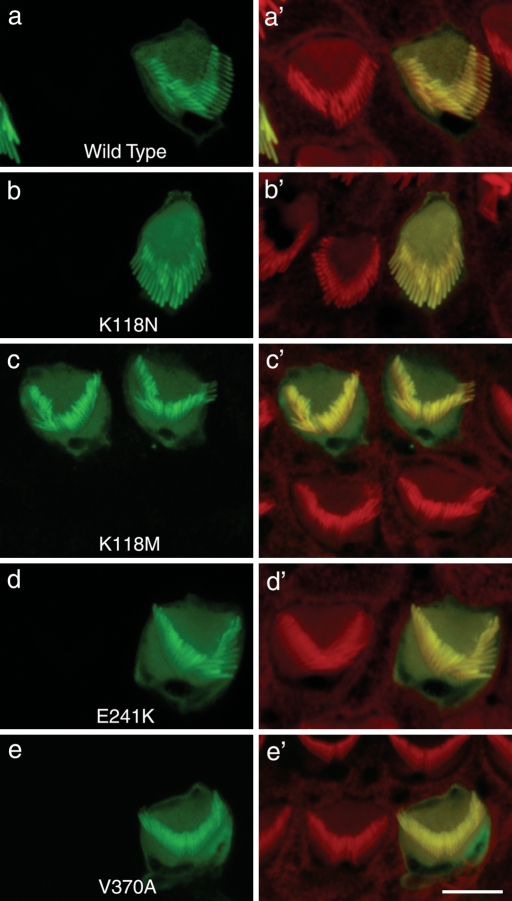
Finally, we wanted to determine whether the mutations affected either the incorporation of the mutant monomers into filaments in hair cells or the morphology and functional architecture of their stereocilia. We therefore performed gene-gun mediated expression of wild-type and the four mutant EGFP-actins, K118N, K118M, E241K and V370A, in hair cells of mouse cochlear explants obtained at postnatal days 2–3 (P2–P3). We found no differences between cells transfected with either the wild-type or any of the mutants tested (Fig. 9A–E). There was no impairment of incorporation of the mutant actin into the hair bundle, and EGFP-tagged protein was observed along the entire length of each stereocilium. Similarly, incorporation into the cuticular plate appeared normal. In all these structures, the mutant actins co-localized with phalloidin indicating a good incorporation of the exogenous γ-actin into cellular F-actin assemblies. Confocal analysis performed on the soma of the hair cells did not reveal aberrant accumulation of any of the mutant forms tested (data not shown). Thus, in both NIH3T3 cells and cultured hair cells, the mutant actins appeared to show no aberrant behaviour in terms of incorporation into any of the normal actin structures, although they did form aggregates in the NIH3T3 cells.
Figure 9.
Gene-gun mediated expression of γ-actin mutants into mouse cochlear hair cells. Cochlear explants of mice sacrificed at post-natal day 2–3 (P2–P3) were transfected with the same constructs as in Figure 8, maintained in medium and fixed 24 h after transfection. Cells expressing any of the four γ-actin mutants assayed, K118N, K118M, E241K and V370A, displayed a normal wild-type appearance, with no appreciable alteration in the incorporation of actin or the morphology of the stereocilia. Confocal microscopy revealed that at 24 h after transfection, the wild-type γ-actin-GFP (green) and mutant actins were incorporated along the entire length of the stereocilia bundle and into the randomly organized filaments of the cuticular plate. In all cases, co-localization with rhodamine–phalloidin staining (red) was observed. The stereocilia of the actin-GFP transfected cells maintained their lengths as judged by comparing with the stereocilia of neighbouring non-transfected cells (red) in each panel. Scale bars = 2.5 µm.
DISCUSSION
In this work, we report the identification and functional assessment of two novel γ-actin mutations, K118N and E241K, responsible for DFNA20/26. Our analysis of the functional effects of the mutations combines in vivo experiments in yeast and biochemical analysis in vitro, as previously reported (8), with those based on expression of the mutant actins in NIH3T3 and mouse cochlear hair cells. It therefore represents the first study to show the effects of deafness-associated γ-actin mutants in mammalian cells.
Our results using yeast actin reveal that both mutants have allele-specific effects on yeast actin behaviour that are not simply caused by a wholesale disassembly of the actin cytoskeleton. This fact further strengthens yeast as an appropriate model for delineating the aspects of actin function that are affected as a consequence of a given mutation in the absence of other actin isoforms. The identification of the K118N allele is the second deafness-causing amino acid substitution to be identified at the same site (K118M) and it allows a comparison of the relative effects of the two mutations on actin behaviour. No abnormal phenotype was observed for the K118N in terms of growth in rich medium or ability to use glycerol as a sole carbon source, although the K118N mutation severely impaired growth in hyperosmolar medium. Additionally, no obvious effects of the mutation were seen on cell size, shape, cytoskeletal structure or vacuole morphology, the latter of which is a sensitive indicant of cytoskeletal behaviour. In comparison, the K118M mutation resulted in severely compromised growth in rich medium and an inability to grow on glycerol indicating mitochondrial dysfunction. Moreover, cells expressing K118M showed altered cytoskeleton and vacuolar patterns (8). Our in vitro results do not show any appreciable differences in monomer behaviour between K118N and K118M, both with decreased thermostability, or in polymerizability that was similar to that of the wild-type. K118 is located on the outside of subdomain 1 where it is exposed to solvent and can potentially interact with different actin-binding proteins. Its position suggests that it is not directly involved in a monomer–monomer interface within the actin filament. Thus, unless a substitution here caused a propagated conformational change throughout the protein, it would not be expected to affect polymerization per se. The more drastic effects shown by the K118M versus the K118N mutation in vivo may reflect the fact that asparagine has a much more polar side chain than methionine and is therefore closer to the parent lysine in its ability to engage in potential hydrogen-bonding interactions affecting actin function. In terms of deafness, however, differences between these alleles are not so apparent. Subjects with these same mutations exhibit practically identical audiometric profiles (13). Although, the later onset of the hearing loss in the family carrying the K118N mutation correlates with the milder phenotype of this mutation in vivo in yeast, this correlation could also be coincidental.
E241K is the first deafness-causing actin mutation identified in subdomain 4 of the protein. The previous mutations have all been in subdomains 1 and 3. The severe phenotypes in vivo in yeast caused by the E241K mutation were remarkable in that the mutation also led to the abnormal formation of thick randomly oriented actin filament bundles. This phenotype was not seen with any of the previously reported γ-actin mutations (8). This behaviour in vivo correlates with the polymerization defect of E241K in vitro that leads to the formation of massive actin filament bundles in the absence of ancillary actin-bundling proteins. The E241 occupies a position that could be involved in dictating actin filament behaviour in a number of ways. First, it is very close to an actin-binding site (243–245) involved in inter-molecular contacts along the F-actin helix (7) and could produce changes in monomer interactions that alter filament topology leading to an association of actin filaments. Secondly, this residue, at the top and inside of subdomain 4, is also in a position where it could affect the relationship of the two large domains of the protein across the inter-domain cleft in which the nucleotide binds, as our results in vitro indicate. Several filament conformations have been identified based on the size of this cleft ranging from a relatively open to a closed conformation that can affect filament behaviour (26–28), and such a change in cleft conformation might result from this mutation. Both of these explanations are consistent with our demonstrated hypersensitivity of the E241K filament bundles to the severing effects of yeast cofilin suggesting an alteration in monomer–monomer contacts along the filament length.
It is difficult to understand how the pronounced bundling tendency of the E241K mutant actin is compatible with yeast cell viability since the dynamic properties needed for cytoskeletal function would be severely compromised due to the bundles acting as an actin sink. However, our finding that tropomyosin prevents bundling of E241K filaments provides a clue as to why this actin still functions in vivo. EM reconstructions of tropomyosin–actin complexes show that tropomyosin can lie across the actin surface very near residue 241 (29,30). With the E241K substitution, as suggested for the E241K, Q246K and Q246R mutations in the alpha-skeletal muscle actin that are associated with nemaline myopathy (17), the introduction of a positive charge in place of a negative charge at this site might result in an attractive interaction with the anionic face of a second actin filament leading to bundle formation. Tropomyosin as well as other filament stabilizing proteins is present in both yeast and mammalian cells. Such proteins may interfere with inherent bundling enough to allow the limited cytoskeletal function commensurate with the severely affected phenotypes associated with this mutation.
In transiently transfected NIH3T3 cells, all the tested mutants, K118M, K118N, E241K and V370A were able to incorporate normally into cytoskeletal structures. However, with the mutants abnormal aggregates of actin were also observed indicating an element of abnormality caused by the mutations in vivo. The clumping we observed with the actin mutants might be due to the effects of the mutations on the actin structure combined with over expression, therefore resulting in abnormal aggregation. The results of the gene-gun experiments indicate that no gross alteration is observed in the cytoskeleton structures and morphology of stereocilia for any of the mutants tested.
There are a number of possible explanations for the differences in the behaviour of these mutant actins in yeast versus mammalian cells. The most likely is the fact that in yeast, the mutant actin is the only actin in the cell, whereas in the mammalian cells, the full effects of the mutations are diluted by the presence of the β- and γ-actins normally in the cell (12,31). In hair cells, the stereociliary actin cytoskeleton is part of a complex protein network in which different proteins are interacting with the actin core, either directly or mediated by scaffolding proteins (32), which may mask the underlying structural deficiencies. The absence of abnormal effects with the gene-gun experiments could also reflect the fact that the stereociliary actin bundle is a much more stable cytoskeletal structure than the highly spatially and temporally dynamic yeast cell cytoskeleton.
This stability could underlie the post-lingual and progressive character of DFNA20/26 hearing loss for which a congenital defect of the inner ear function is not expected. Indeed our results are consistent with those obtained in the Actg1-null mice in which only β-actin is present. These mice initially have normal hearing but develop progressive hearing loss during adulthood that is characterized by stereocilia actin core disruptions and stereocilia degradation (33).
In summary, this study highlights the importance of using different experimental approaches to obtain a complete picture of the various functional aspects of γ-actin that are caused by a given mutation. Our results show that these two mutations have biochemical consequences on actin function and provide support for the hypothesis that the post-lingual and progressive character of the DFNA20/A26 hearing loss due to mutations in ACTG1 could be the result of a progressive deterioration of the hair cells’ cytoskeletal structures over time.
MATERIALS AND METHODS
The protocols of this study were approved by the Institutional Review Board of Hospital Ramón y Cajal (Madrid, Spain).
Clinical data
Appropriate informed consent was obtained from all study participants; peripheral blood samples were collected from all members of generations I and II for family S840 (Fig. 1A, left) and II and III for family S582 (Fig. 1A, right) and DNA extraction was performed following standard methods. Clinical history ruled out environmental factors as the cause of the hearing loss in both families and physical examination did not reveal any evidence of syndromic features. Tympanometry indicated proper functioning of the middle ear. Pure-tone audiometry was performed to test for air conduction (125–8000 Hz) and bone conduction (250–4000 Hz). Patients of family S840 show bilateral, symmetrical and progressive sensorineural hearing loss at mid- and high frequencies (a down-sloping audiometric profile) of post-lingual onset (Fig. 1B, left). The earliest clinical evidence of hearing loss was obtained from patient II:1 in the third decade of life. Affected subjects of family S582 also show a post-lingual, bilateral, symmetrical and progressive sensorineural hearing impairment that affect the mid- and high frequencies. All the patients of this family were referred with hearing problems at school age and the earliest record of hearing loss in the family was obtained from individual III:1 at the age of 6 years (Fig. 1B, right).
Molecular genetic methods
The microsatellite markers, D17S784, D17S668, D17S928 and D17S623, from the DFNA20/A26 region were used to genotype the families S582 and S840. The fluorescently labelled alleles were analyzed in an ABI PRISM 3100 automated DNA genetic analyzer (Applied Biosystems, Foster City, CA, USA). Primers to PCR-amplify all the exons and intron–exon boundaries of the human ACTG1 gene were designed using Oligo 4.0 (Molecular Biology Insights, West Cascade, CO, USA). PCR reactions were carried out according to the conditions described in Supplementary Material, Table S2. PCR amplimers were screened for mutations by denaturing high performance liquid chromatography (DHPLC) on a Wave™ DNA fragment analysis system (Transgenomic™, USA) according to manufacturer's protocol. The different DHPLC heteroduplex profiles were characterized by sequencing a product of a second PCR amplification in an automated DNA sequencer ABI PRISM 3100 (Applied Biosystems). For the detection of the c.721G>A mutation in the members of family S582 and control subjects, we developed a RFLP screening test in which a fragment of the ACTG 1 exon 4 is amplified with a pair of primers in which the reverse primer was modified to create a SacI site when the mutation is present. After digestion the mutant allele is 21 nt shorter than the wild-type one and it could be detected by running the digested amplimers in a 3% agarose gel. Members of family S840 and control subjects were screened for the c.354G>C mutation by digesting exon 3 amplimers with NlaIII restriction enzyme and running the digestion products in 2% agarose gels. Wild-type amplimers yield five fragments of 178, 79, 59, 26 and 9 bp. When the mutation is present, the 178 bp fragment is cleaved in two of 111 and 67 bp.
To exclude the involvement of other ACTG1 mutations in the hearing loss of the S840 and S582 families, we sequenced the remaining exons in the probands of both families. No other mutations were found apart from two synonymous changes (rs1139405, c.918C>T, p.Tyr306Tyr and rs1135989, c.930C>T; p.Ala310Ala) previously reported as non-pathogenic (15) in the subject II:1 of family S840, and two intronic changes (IVS1 + 15G>C, IVS3 + 13C>A) in the member III:1 of family S582.
Construction of the mutant yeast strains and determination of the growth characteristics
Mutations were introduced into the centromeric plasmid pRS314 (34) containing the yeast actin coding sequence driven by ACT1 promoter using QuickChange® mutagenesis kit according to the manufacturer's instructions. Plasmids containing the desired mutations were introduced into a recipient yeast strain expressing WT actin (pCENWT) as described previously (35). Plasmid shuffling yielded viable haploid strains for both mutations. The plasmids containing the mutations were re-isolated from these strains and sequenced to confirm the presence of the desired mutation. Growth characteristics of the mutant yeast strains were determined as previously described (8). YNB is composed by YNB without amino acids 0.67% (BD 291940) and glucose 0.3%. YPD is yeast peptone/2% dextrose medium. YPG is YPD with 2% glycerol substituted for dextrose as sole carbon source.
The actin cytoskeleton was visualized after staining fixed cells with rhodamine–phalloidin as described previously (36). Vacuoles were observed following exposure of the cells to the dye FM 4–64 (Molecular Probes) as reported elsewhere (37). The genomic and mitochondrial DNA was visualized after staining the cells with Hoechst (Sigma, Hoechst 33342). To measure cell size, mounted samples were visualized, and the long axis of the cells was measured. The average length for each strain was determined by measuring more than 100 cells of each sample. In all cases, the cells were examined with a fluorescence inverted microscope (Olympus IX81) equipped with a UPLFLN40X, PLAPON60XO and UPLSAPO100XO objectives. Images were recorded with a cooled CCD-F View II (Soft Imaging System) camera and processed using Cell R software (Olympus).
Actin biochemistry
DNase I (grade D) was purchased from Worthington. DE52 DEAE-cellulose was obtained from Whatman. Micro Bio-Spin P-30 Tris columns and Affi-Gel 10-activated resin were purchased from BioRad. ATP was from Sigma. N6-ethenoadenosine 5′-triphosphate (ϵ-ATP) was purchased from Molecular Probes. Yeast cakes for WT actin preparations were purchased from a local bakery. All other chemicals were reagent-grade quality.
Purification of WT and mutant actins
WT and mutant actins were purified from lysates of frozen cells via a combination of DNase I-agarose affinity chromatography, DEAE-cellulose chromatography and polymerization/depolymerization cycling as described previously (38). The concentration of G-actin was determined from the absorbance at 290 nm using an extinction coefficient of 0.63 ml mg−1 cm−1. All actins were used within 4 days following completion of purification.
Thermal denaturation
The apparent melting temperatures of WT and mutant G-actins were determined using circular dichroism. A continuously stirred sample containing 1 µm G-actin was heated at 1°C per minute between 25 and 90°C while the change in ellipticity of the G-actin at 222 nm was recorded as a function of temperature using an Aviv 62 DS spectropolarimeter. Data were fit to a two-state model, and the apparent Tm value was determined by fitting the data in Excel to the Gibbs–Helmholtz equation to approximate the temperature at which 50% of the actin was denatured.
Nucleotide exchange
The ability of G-actin to exchange its bound nucleotide was assessed by first loading the actin with fluorescent ϵ-ATP as described previously (39) and following its displacement from the actin in the presence of a 33-molar excess of ATP via monitoring the resulting decrease in fluorescence using a Fluorolog-3 fluorescence spectrometer outfitted with a computer controlled thermostatted sample chamber (HORIBA Jobin Yvon Inc.). Exchange rates were determined by fitting the data to a single exponential expression using BioKine Version 3.1 using the Simplex algorithm.
Actin polymerization
Polymerization of a sample containing 4.8 µm G-actin in a total volume of 120 µl was induced by the addition of MgCl2 and KCl to a final concentration of 2 and 50 mm, respectively. Polymerization was monitored at 25°C by following the increase in light scattering of the sample in a FluoroMax-3 fluorescence spectrometer outfitted with a computer controlled thermostatted multi-sample exchanger (HORIBA Jobin Yvon Inc.). The excitation and emission wavelengths and the slit widths were both set to 360 and 1 nm, respectively. For the polymerization experiments involving tropomyosin, BVC was purified from rabbit cardiac muscle acetone powder according to Butters et al. (40), and stoichiometric amounts were added to samples of actin prior to triggering polymerization. For the polymerization experiments involving yeast cofilin, the cofilin was purified from E. coli according to Lappalainen et al. (41), and the protein concentration was determined by BCA. Cofilin listed in each experiment was added post-polymerization to the actin sample once the light scattering signal reached steady state. All polymerization experiments were performed at least three times with different actin preparations. For the experiments where we examined the effects of varying the mole percentage of E241K relative to WT actin on either bundling or cofilin sensitivity, working stocks of 4.8 µm were prepared for both WT and E241K samples before they were mixed together to yield the listed percentage E241K mutant relative to WT. The polymerization kinetics and extents were followed by the increase in light scattering as a function of time at 25°C.
Electron microscopy
Actin filaments were visualized by depositing 2 µl of a sample containing 4.8 µm F-actin onto carbon-coated Formvar grids. The grids were negatively stained with 1% uranyl acetate, and observed using a JOEL 1230 transmission electron microscope (University of Iowa Central Electron Microscopy Facility). Image J was used to process the images.
Expression of actin mutants in NIH-3T3 and gene-gun transfections in cochlear hair cells
The human cytoplasmic β-actin cloned into the pEGFP-Actin plasmid (Clontech) was converted to γ-actin, and the mutations K118N, K118M, E241K and V370A were introduced in steps using the QuickChange® mutagenesis system. NIH-3T3 cells were cultured in DMEM medium supplemented with 10% fetal calf serum (FCS), 100 U/ml of penicillin and 100 µg/ml of streptomycin on 12 mm glass coverslips (Menzel-Glaser, GmbH, Braunschweig, Germany) in flat-bottomed 24-well multititer plates (Falcon; BD Biosciences, San Jose, CA, USA) for immunocytochemistry studies. Cells were transiently transfected with 800 ng of the wild-type γ-actin gene or each of the four mutated constructs using Lipofectamine TM 2000 (111668, Invitrogen) according to the manufacturer's directions. For immunocytochemistry, cells on coverslips were fixed 48 h after transfection in 4% paraformaldehyde in phosphate buffer (0.1 m NaH2PO4 pH 7.04) for 10 min and then washed three times briefly in PBS. To study intracellular protein localization, cells were permeabilized for 10 min at room temperature with 0.5% Triton X-100 in PBS and blocked with 3% BSA in PBS for 30 min, washed twice with PBS and incubated for 1 h at room temperature with a 1:100 dilution of the rat monoclonal antibody to EGFP (Roche). Cells were washed three times with PBS, and then incubated for 1 h at room temperature with a 1:500 dilution of the secondary antibody, Alexa Fluor™ 498 conjugated goat anti-rat IgG (Molecular Probes A-11007; Invitrogen). Total actin was stained with Alexa 594 phalloidin (Invitrogen, A-12381). After washing three times in PBS, slides were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) containing Hoechst stain (Sigma, Hoechst 33342). Percentages accounting for the aberrant distribution of γ-actin in the mutant transfected cells were obtained by scoring by eye, 100 randomly chosen green-positive cells per coverslip by an observer blinded to experimental design. Values represent the mean ± SD of cells judged to display γ-actin aggregates of three coverslips from four independent transfections.
For gene-gun experiments, cochlear cultures were prepared on collagen-coated round glass coverslips from the inner ears of P2–P3 mice as described previously (42) and maintained in Maximow slide assemblies in a medium containing 93% DMEM/F12, 7% fetal bovine serum, 10 µg/ml ampicillin at 37°C for 1 day prior to gene-gun transfection. Gene-gun bullets were prepared by precipitating plasmid DNAs onto 1 µm diameter gold particles with CaCl2, and cultures were transfected using a BioRad Helios gene-gun. Medium was drained from the culture, the coverslip was placed at a distance of 12 mm from the end of the gene-gun barrel, and plasmid-coated gold particles were delivered using a helium pressure of 120–140 psi. The cultures were then immediately placed in a 35 mm diameter plastic petri dish, fed with 3 ml of medium and maintained for a further 24 h at 37°C in a 5% CO2 incubator. Following fixation for 1 h in 4% formaldehyde in 0.1 m sodium phosphate buffer pH 7.4, cultures were pre-blocked and permeabilized with TBS/10% horse serum/0.1% Triton X-100, stained with rabbit antibodies to GFP followed by Alexa 488-conjugated goat anti-rabbit IgG and Texas Red conjugated phalloidin, mounted in Vectashield and examined with a Zeiss LSM 510 confocal microscope using a ×100 oil immersion lens, NA 1.4.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants from the Spanish Ministerio de Ciencia y Tecnología [grant numbers SAF2005-06355, SAF2008-03216 to F.M.], Spanish Fondo de Investigaciones Sanitarias [grant numbers CP03/00014 and PI08/0045 to M.A.M.P.], the European Commission [grant numbers FP6 Integrated Project EUROHEAR, LSHG-CT-2004-512063], the Wellcome Trust [grant number #071394/Z/03/Z], the National Institutes of Health Grants and Institute on Deafness and other Communication disorders [DC008803 to P.A.R.].
ACKNOWLEDGEMENTS
We thank the family members for their participation in this study. We also express our gratitude to J. L. Rodriguez-Barbancho for kindly donating the anti-GFP antibody and to I. Rodriguez-Escudero for advice in immunocytochemistry in yeasts. K.E.B, S.M.H, F.M.-M. are recipients of a fellowship from NRSA (DC008913); from L'Oréal-UNESCO ‘for women in Science’ and from the Spanish Ministerio de Ciencia y Tecnología, respectively. A.M.R is recipient of a contract from the Centre for Biomedical Research on Rare Diseases (CIBERER).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hudspeth A.J. How the ear's works work. Nature. 1989;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- 2.Tilney L.G., Egelman E.H., DeRosier D.J., Saunder J.C. Actin filaments, stereocilia, and hair cells of the bird cochlea. II. Packing of actin filaments in the stereocilia and in the cuticular plate and what happens to the organization when the stereocilia are bent. J. Cell Biol. 1983;96:822–834. doi: 10.1083/jcb.96.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirokawa N., Tilney L.G. Interactions between actin filaments and between actin filaments and membranes in quick-frozen and deeply etched hair cells of the chick ear. J. Cell Biol. 1982;95:249–261. doi: 10.1083/jcb.95.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabsch W., Mannherz H.G., Suck D., Pai E.F., Holmes K.C. Atomic structure of the actin: DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 5.De La Cruz E.M. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol. Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 6.Isambert H., Venier P., Maggs A.C., Fattoum A., Kassab R., Pantaloni D., Carlier M.F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 7.Holmes K.C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 8.Bryan K.E., Wen K.K., Zhu M., Rendtorff N.D., Feldkamp M., Tranebjaerg L., Friderici K.H., Rubenstein P.A. Effects of human deafness gamma-actin mutations (DFNA20/26) on actin function. J. Biol. Chem. 2006;281:20129–20129. doi: 10.1074/jbc.M601514200. [DOI] [PubMed] [Google Scholar]
- 9.Vandekerckhove J., Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 1978;126:783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- 10.Pollard T.D., Blanchoin L., Mullins R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 11.Höfer D., Ness W., Drenckhahn D. Sorting of actin isoforms in chicken auditory hair cells. J. Cell Sci. 1997;110:765–770. doi: 10.1242/jcs.110.6.765. [DOI] [PubMed] [Google Scholar]
- 12.Furness D.N., Katori Y., Mahendrasingam S., Hackney C.M. Differential distribution of beta- and gamma-actin in guinea-pig cochlear sensory and supporting cells. Hear. Res. 2005;207:22–34. doi: 10.1016/j.heares.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhu M., Yang T., Wei S., DeWan A.T., Morell R.J., Elfenbein J.L., Fisher R.A., Leal S.M., Smith R.J., Friderici K.H. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am. J. Hum. Genet. 2003;73:1082–1091. doi: 10.1086/379286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Wijk E., Krieger E., Kemperman M.H., De Leenheer E.M., Huygen P.L., Cremers C.W., Cremers F.P., Kremer H. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26) J. Med. Genet. 2003;40:879–884. doi: 10.1136/jmg.40.12.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rendtorff N.D., Zhu M., Fagerheim T., Antal T.L., Jones M., Teslovich T.M., Gillanders E.M., Barmada M., Teig E., Trent J.M., et al. A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur. J. Hum. Genet. 2006;14:1097–1105. doi: 10.1038/sj.ejhg.5201670. [DOI] [PubMed] [Google Scholar]
- 16.Liu P., Li H., Ren X., Mao H., Zhu Q., Zhu Z., Yang R., Yuan W., Liu J., Wang Q., Liu M. Novel ACTG1 mutation causing autosomal dominant non-syndromic hearing impairment in a Chinese family. J. Genet. Genomics. 2008;35:553–558. doi: 10.1016/S1673-8527(08)60075-2. [DOI] [PubMed] [Google Scholar]
- 17.Sparrow J.C., Nowak K.J., Durling H.J., Beggs A.H., Wallgren-Pettersson C., Romero N., Nonaka I., Laing N.G. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1) Neuromuscul. Disord. 2003;13:519–531. doi: 10.1016/s0960-8966(03)00101-9. [DOI] [PubMed] [Google Scholar]
- 18.Engqvist-Goldstein A.E., Drubin D.G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 19.Slepecky N., Chamberlain S.C. Immunoelectron microscopic and immunofluorescent localization of cytoskeletal and muscle-like contractile proteins in inner ear sensory hair cells. Hear. Res. 1985;20:245–260. doi: 10.1016/0378-5955(85)90029-2. [DOI] [PubMed] [Google Scholar]
- 20.Slepecky N., Chamberlain S.C. Tropomyosin co-localizes with actin microfilaments and microtubules within supporting cells of the inner ear. Cell Tissue Res. 1987;248:63–66. doi: 10.1007/BF01239963. [DOI] [PubMed] [Google Scholar]
- 21.Gunning P., O'Neill G., Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 22.Galkin V.E., Orlova A., VanLoock M.S., Shvetsov A., Reisler E., Egelman E.H. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrianantoandro E., Pollard T.D. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Galkin V.E., Orlova A., Lukoyanova N., Wriggers W., Egelman E.H. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa C.F., Rommelaere H., Waterschoot D., Sethi K.K., Nowak K.J., Laing N.G., Ampe C., Machesky L.M. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J. Cell Sci. 2004;117:3367–3377. doi: 10.1242/jcs.01172. [DOI] [PubMed] [Google Scholar]
- 26.Orlova A., Shvetsov A., Galkin V.E., Kudryashov D.S., Rubenstein P.A., Egelman E.H., Reisler E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl Acad. Sci. USA. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orlova A., Galkin V.E., VanLoock M.S., Kim E., Shvetsov A., Reisler E., Egelman E.H. Probing the structure of F-actin: cross-links constrain atomic models and modify actin dynamics. J. Mol. Biol. 2001;312:95–106. doi: 10.1006/jmbi.2001.4945. [DOI] [PubMed] [Google Scholar]
- 28.Belmont L.D., Orlova A., Drubin D.G., Egelman E.H. A change in actin conformation associated with filament instability after Pi release. Proc. Natl Acad. Sci. USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C., Craig R., Tobacman L., Horowitz R., Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys. J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman W., Hatch V., Korman V., Rosol M., Thomas L., Maytum R., Geeves M.A., Van Eyk J.E., Tobacman L.S., Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y., Sakiyama S. Analysis of isoactin expression in cultured mouse cells by in situ hybridization. Biol. Cell. 1990;69:211–213. doi: 10.1016/0248-4900(90)90347-6. [DOI] [PubMed] [Google Scholar]
- 32.Kremer H., van Wijk E., Märker T., Wolfrum U., Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet. 2006;2:R262–R270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- 33.Belyantseva I.A., Perrin B.J., Sonnemann K.J., Zhu M., Stepanyan R., McGee J., Frolenkov G.I., Walsh E.J., Friderici K.H., Friedman T.B., et al. γ-actin is required for cytoskeletal maintenance but not development. Proc. Natl Acad. Sci. USA. 2009;106:9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook R.K., Sheff D.R., Rubenstein P.A. Unusual metabolism of the yeast actin amino terminus. J. Biol. Chem. 1991;266:16825–16833. [PubMed] [Google Scholar]
- 36.McKane M., Wen K.K., Boldogh I.R., Ramcharan S., Pon L.A., Rubenstein P.A. Mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J. Biol. Chem. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- 37.Vida T.A., Emr S.D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook R.K., Blake W.T., Rubenstein P.A. Removal of the amino-terminal acidic residues of yeast actin. Studies in vitro and in vivo. J. Biol. Chem. 1992;267:9430–9436. [PubMed] [Google Scholar]
- 39.Yao X., Nguyen V., Wriggers W., Rubenstein P.A. Regulation of yeast actin behavior by interaction of charged residues across the interdomain cleft. J. Biol. Chem. 2002;277:22875–22882. doi: 10.1074/jbc.M201685200. [DOI] [PubMed] [Google Scholar]
- 40.Butters C.A., Willadsen K.A., Tobacman L.S. Cooperative interactions between adjacent troponin–tropomyosin complexes may be transmitted through the actin filament. J. Biol. Chem. 1993;268:15565–15570. [PubMed] [Google Scholar]
- 41.Lappalainen P., Fedorov E.V., Fedorov A.A., Almo S.C., Drubin D.G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell I.J., Richardson G.P. The morphology and physiology of hair cells in organotypic cultures of the mouse cochlea. Hear. Res. 1987;31:9–24. doi: 10.1016/0378-5955(87)90210-3. [DOI] [PubMed] [Google Scholar]
- 43.Vorobiev S., Strokopytov B., Drubin D.G., Frieden C., Ono S., Condeelis J., Rubenstein P.A., Almo S.C. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl Acad. Sci. USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.