Abstract
The suprachiasmatic nuclei (SCN) of the mammalian hypothalamus function as the master circadian clock, coordinating the timing of diverse cell populations and organ systems. Dysregulation of clock timing is linked to a broad range of human conditions, including obesity, cardiovascular disease and a wide spectrum of neurological disorders. Aberrant regulation of expression of the PERIOD genes has been associated with improper cell division and human cancers, while the autosomal dominant disorder familial advanced sleep phase syndrome has been mapped to a single missense mutation within the critical clock gene hPERIOD2. An essential tool to begin to dissect the inherent molecular timing process is the clock gene reporter. Here, we functionally characterize two new mouse transgenic clock reporters, mPeriod1-Venus and mPeriod2-DsRED. Venus and DsRED are fluorescent proteins that can be used to monitor transcription in individual cells in real-time. Imaging of the SCN revealed oscillations, as well as light inducibility, in Venus and DsRED expression. Rhythmic Venus and DsRED expression was observed in distinct SCN cell populations, suggesting the existence of discrete cellular SCN clocks. Outside of the SCN, mPeriod1-Venus expression was broadly expressed in neuronal and non-neuronal populations. Conversely, mPeriod2-DsRED was expressed in glial populations and progenitor cells of the dentate gyrus; limited expression was detected in neurons. This distinct expression pattern of the two reporters reveals that the central nervous system possesses mechanistically distinct subpopulations of neuronal and non-neuronal cellular clocks. These novel mouse models will facilitate our understanding of clock timing and its role in human diseases.
INTRODUCTION
Most organisms possess an intrinsic time-keeping mechanism that drives rhythms in behavioral, physiological and cellular processes with nearly 24-h periodicity. The molecular basis that underlies these overt circadian (approximately 24 h) rhythms is a transcription–translation feedback loop within individual oscillator cells (1). In the mouse, the bHLH transcription factors Clock and Npas2 dimerize with Bmal1 and transactivate the expression of the Period (mPer1 and mPer2) and Cryptochrome (mCry1 and mCry2) genes via recognition of E-box motifs in the gene promoters (2–6). Period and Cryptochrome proteins translocate to the nucleus and negatively regulate their own expression via inhibition of E-box-mediated transcription. This inhibitory feedback loop takes approximately 24 h to complete. The negative impact of dysregulation of clock timing on human physiology is apparent in conditions such as cancer, obesity, cardiovascular disease, and a wide spectrum of neurological disorders. Moreover, mutations in specific clock genes have been implicated in several hereditary disorders: missense mutations in the human PERIOD2 and CASEIN KINASE I DELTA (CK1δ) genes have been attributed to the behavioral trait known as familial advanced sleep phase syndrome, a condition in which the subjects exhibit early sleep-onset followed by early morning awakenings (7,8). A related condition known as delayed sleep phase syndrome, in which the patient suffers from sleep-onset insomnia and an inability to awaken at a normal time, is associated with a polymorphism of the human PERIOD3 gene (9). Nevertheless, the processes by which clock timing influences physiological homeostasis remain poorly understood.
Clock genes are expressed in multiple tissues and cell types. However, the workings of the molecular clock are perhaps most striking in the suprachiasmatic nuclei (SCN), a bilateral hypothalamic structure above the optic chiasm that functions as the master circadian clock in mammals (10). As such, the SCN is a self-sustaining (i.e. autonomous) oscillator, and responds to external time cues such as light by resetting the clock phase (a process known as entrainment). With respect to the molecular clock, expression of mPer1, mPer2, mCry1 and mCry2 are strongly rhythmic but each peaks at a different circadian time (CT) (2,3,11). A light pulse administered in the early subjective night leads to induction of mPer1 and mPer2, whereas in the late subjective night mPer1 expression is preferentially activated (12).
Thus, one may use mPer1 and mPer2 expression not only as markers for the state of the molecular clock, but also to investigate mechanisms of circadian transcriptional regulation. Traditional in situ hybridization and immunohistochemical approaches are commonly employed, but they do not afford us the opportunity to monitor changes in gene expression in real time. A number of reporter transgenic mice that use luciferase or green fluorescent protein (GFP) have been constructed to circumvent this issue (13–16). Although the luciferase reporter mice have been widely used to study mPer1 or mPer2 rhythms on a macroscopic scale (e.g. SCN explants), their utility is somewhat limited for studies that seek to monitor timing in individual cells within tissue slices or in vivo. In addition, the use of luciferase as a marker of both mPer1 and mPer2 precludes dual, simultaneous monitoring of mPer1 and mPer2 expression in the same animal. With respect to the mPer1-GFP mice described by Kuhlman et al. (13), a short promoter fragment (approximately 3 kb) was used to drive transgene expression, and thus may not perfectly recapitulate endogenous mPer1 expression (14). Here, we describe the generation of two new transgenic clock reporters of endogenous mPer1- and mPer2-dependent transcription. These fluorescent protein reporters allow for simultaneous, single-cell detection of mPer1- and mPer2-dependent transcription in real time, and as such, provide a new level of resolution of the clock timing mechanism.
RESULTS
Generation of mPer1-Venus and mPer2-DsRED reporter mice by bacterial recombination
As an initial step to generating mPer1 and mPer2 reporter mice, we constructed Venus-NLS-PEST and DsRED.T3-NLS-PEST mammalian expression vectors. The DsRED.T3 variant, which carries eight point mutations relative to the parent DsRED protein, exhibits rapid maturation kinetics with a half-time of approximately 1.3 h, approximately 10 times faster than that of the wild-type protein (17). In the case of the YFP derivative Venus, five point mutations including a novel mutation, F46L, in the parent YFP not only enhances protein folding efficiency but also provide a 3-fold acceleration of chromophore oxidation, the rate-limiting step in maturation (18). Thus, the maturation kinetics of DsRED.T3 and Venus make them potentially well-suited for use as dynamic reporters. We further improved upon the kinetic properties of these fluorescent proteins by cloning the PEST domain of mouse ornithine decarboxylase in-frame with the penultimate or the last codon of Venus or DsRED.T3, respectively. Subsequently, a nuclear localization signal (NLS) sequence was cloned, in-frame, between the Venus/DsRED.T3 coding sequence and the PEST domain. The completed Venus-NLS-PEST and DsRED.T3-NLS-PEST expression vectors contained a human cytomegalovirus (hCMV) promoter and a SV40 polyadenylation (pA) site.
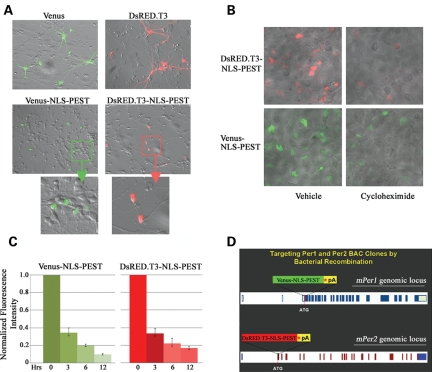
The functionality of the NLS and PEST sequences in the context of our expression vectors was examined in a series of cell culture-based overexpression experiments. Transfection of rat embryonic cortical neurons with vectors expressing the unmodified versions of Venus or DsRED.T3 yielded expression of the fluorescent proteins throughout the cell, including the soma and dendritic/axonal processes (Fig. 1A, top row). On the other hand, transfection with the Venus-NLS-PEST or DsRED.T3-NLS-PEST expression constructs resulted in fluorescence localized to the nucleus, with little detectable signal in the neuronal processes (Fig. 1A, middle and bottom rows). To examine protein turnover, HEK293 cells were transfected with Venus-NLS-PEST or DsRED.T3-NLS-PEST. Twenty-four hours post-transfection, cultures were treated with the protein synthesis inhibitor cycloheximide (CHX; 100 µg/ml). Compared with transfected cultures receiving mock (vehicle) treatment, the fluorescent intensity of CHX-treated HEK293 cells was significantly diminished 6 h following addition of CHX to the culture media (Fig. 1B and C). The mean fluorescent intensity per transfected cell, after 6 h of CHX treatment, was 19.5 ± 1.1% (for Venus-NLS-PEST) and 22.1 ± 2.9% (for DsRED.T3-NLS-PEST) relative to that of vehicle-treated controls (Fig. 1C; n = 50 cells per condition). Importantly, cell and nuclear morphological analyses by bright-field microscopy (Fig. 1B) and DAPI staining (data not shown), respectively, indicated that CHX treatment, at a concentration of 100 µg/ml for up to 12 h, did not compromise cell viability. Thus, the loss of fluorescent signal is directly attributed to the kinetics of protein turnover of the two modified reporters in viable cells, confirming the dynamic nature of the constructs.
Figure 1.
Construction of mPer1-Venus and mPer2-DsRED reporters by bacterial recombination. (A) Subcellular distribution of Venus and DsRED.T3 proteins. Rat embryonic cortical neurons were transfected with constructs expressing Venus, Venus-NLS-PEST, DsRED.T3 and DsRED.T3-NLS-PEST, and the distribution of the fluorescent signals was determined 48 h later. (B) Protein stability of DsRED.T3-NLS-PEST and Venus-NLS-PEST. HEK293 cells were transfected with the reporter gene expression constructs. After 24 h, cultures were treated with the protein synthesis inhibitor cycloheximide (CHX: 100 µg/ml) or vehicle (dimethyl sulfoxide) and the intensity of DsRED and Venus fluorescence was determined 6 h after addition of CHX (or vehicle) to the culture media. Representative data are an overlay of the bright-field and fluorescence micrographs. (C) Quantitative analysis of Venus or DsRED fluorescence in Venus-NLS-PEST- or DsRED.T3-NLS-PEST-transfected HEK293 cells at 0–12 h following CHX (or vehicle) treatment. For each time point, the fluorescence intensity of the CHX-treated cells is expressed relative to the fluorescence of vehicle-treated cells, which were normalized to a value of 1. Error bars denote the SEM. (D) Diagram of the mPer1 and mPer2 loci, and the Venus- and DsRED-encoding gene cassettes. The Venus and DsRED.T3 genes were modified by an in-frame insertion of a NLS and PEST domain at the 3′ end. Asterisk (*) represents in-frame STOP codon. pA denotes SV40 pA signal. The Venus and DsRED gene cassettes were inserted in-frame with the starting ATG codon of the mPer1 and mPer2 genes, respectively. Amino acid-encoding exons are denoted as dark blue (for mPer1) and red (for mPer2) blocks. Exons encoding 5′ and 3′ untranslated regions are represented by light blue (for mPer1) and blue (for mPer2) blocks.
To generate the mPer1 and mPer2 reporter constructs, we introduced the Venus-NLS-PEST and DsRED.T3-NLS-PEST mini-gene cassettes into the mPer1 and mPer2 loci, respectively, by bacterial recombination (19) and the complete bacterial artificial chromosome (BAC) transgenic constructs were then used to generate mPer1-Venus and mPer2-DsRED reporter mice by pronuclear injection (Fig. 1D).
Circadian and light-inducible expression of mPer1-Venus in the suprachiasmatic nuclei
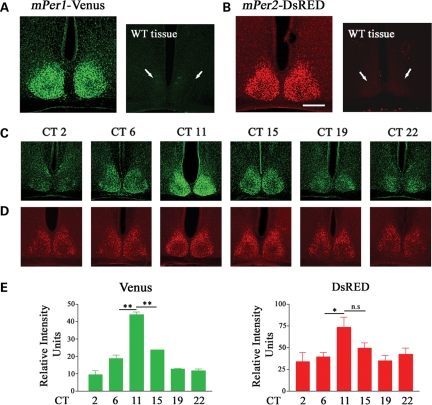
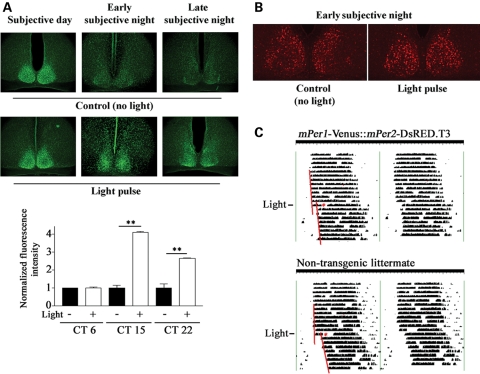
In order to confirm that mPer1-Venus transgenic mice are bona fide reporters of mPer1-dependent transcription, we characterized the expression of mPer1-Venus in the SCN. To this end, mPer1-Venus transgenic mice were housed under a standard 12 h:12 h light:dark (LD) cycle for 14 days followed by dark-adaptation (DD) for 2 days; subsequently, SCN tissue was harvested at various CTs spanning both the subjective day- and night-time domains. Using a polyclonal GFP antiserum that cross-reacts with Venus, we detected strong Venus immunoreactivity (IR) in the transgenic SCN at CT 12, in contrast with wild-type (WT) SCN (Fig. 2A). Furthermore, Venus expression exhibited a robust circadian rhythm, reaching a peak at CT 11 and declining by CT 15 (Fig. 2C and E, left graph). In a second set of experiments, mPer1-Venus transgenic mice were dark-adapted for 2 days, and subsequently exposed to a brief light pulse (100 lux, 15 min) in the subjective day (projected time: CT 6), or early or late subjective night (projected times: CT 15 and 22, respectively); Venus expression was determined 4 h later. A light pulse in the subjective night, but not the subjective day, strongly induced the expression of Venus in the ventrolateral region of the SCN (Fig. 3A). Thus, two key properties of the mPer1 gene, i.e. rhythmic and light-inducible expression in the SCN, are reflected faithfully by the expression of Venus in the SCN of mPer1-Venus transgenic mice.
Figure 2.
mPeriod-driven transgene expression in the SCN. (A) Tissue from an mPer1-Venus transgenic mouse and a non-transgenic (wild-type; WT) littermate was immunolabeled for Venus expression. (B) Tissue from an mPer2-DsRED transgenic mouse and a non-transgenic (WT) littermate was immunolabeled for DsRED expression. Brain tissue was harvested between circadian time (CT) 11 and 12 and processed for Venus or DsRED expression by immunofluorescent labeling. Immunolabeling was specific to the transgenic mice. Arrows denote the approximate locations of the suprachiasmatic nuclei (SCN) in the non-transgenic littermates. Scale bar = 200 µm. Circadian expression of (C) mPer1-Venus and (D) mPer2-DsRED in the SCN. Mice were killed at the indicated circadian times (CT) and SCN tissues were processed for Venus or DsRED expression by immunofluorescent labeling. (E) Quantitative circadian profile of Venus and DsRED expression in the SCN. SCN fluorescence intensity values were averaged from two to three animals per time point, and are representative of triplicate determinations. Error bars = SEM. **P < 0.001; *P < 0.01; n.s. denotes not significant.
Figure 3.
Light-induced clock-resetting of mPer1-Venus and mPer2-DsRED Mice. (A) Top panels: Light-induced expression of mPer1-Venus in the suprachiasmatic nuclei (SCN). Mice received a brief light pulse (100 lux, 15 min) during the mid-subjective day (CT 6), early subjective night (CT 15) or late subjective night (CT 22), and killed 4 h after light treatment; brain sections were immunolabeled for Venus expression. Dark control animals were not exposed to light but were killed at the same circadian times (CTs). Bottom graph: Quantitation of light-evoked Venus expression at the three CT points. SCN fluorescence intensity values were averaged from two to three animals per time point, and are representative of triplicate determinations. For each time point, the mean fluorescence intensity value of the no light condition is normalized to a value of 1, and the light pulse condition is expressed as a relative fold-value. Error bars = SEM. **P < 0.001 (B) Light-induced expression of mPer2-DsRED in the SCN. Mice received a brief light pulse (100 lux, 15 min) at CT 15 and were sacrificed 6 h later. Dark control animals were not exposed to light but were sacrificed at the same CT (CT 21). Scale bar = 100 µm. (C) Representative wheel running activity in a dark-adapted double hemizygotic transgenic mPer1-Venus::mPer2-DsRED mouse and a non-transgenic littermate (WT). After 13 days of free-running, mice were exposed (asterisks) to light (100 lux, 15 min) at CT 15, and maintained under constant darkness for an additional 8 days. Note the phase-delaying effect of light in both the double transgenic and non-transgenic mice. Regression lines approximate the phase-shifting effects of light.
Circadian and light-inducible expression of mPer2-DsRED in the suprachiasmatic nuclei
In a parallel series of experiments, we validated the utility of the mPer2-DsRED transgenic mice. Using the entrainment and dark-adaptation paradigms described above, brain tissues from mPer2-DsRED mice were harvested and processed for DsRED immunoreactivity. As expected, DsRED was strongly expressed in the transgenic SCN at CT12, in contrast with WT SCN (Fig. 2B). Furthermore, DsRED-IR was rhythmic, reaching a peak at CT 11–15 (Fig. 2D and E, right graph). Previous studies indicated that light-induced expression of the mPer2 gene, unlike that of mPer1, exhibits delayed kinetics and is observed only following a light pulse in the early, and not the late, night (2,12). Therefore, to examine the inducibility of DsRED expression in the SCN, we exposed mice to a brief light pulse (100 lux, 15 min) at CT 15 and determined transgene expression 6 h later. We found that light triggered a modest increase in DsRED expression in the SCN relative to dark (no light) control animals (Fig. 3B). Induction was observed in the central and dorsal regions of the SCN, analogous to the early night light-induced mPer2 protein expression pattern reported by Yan and Silver (20). Using the no-light control condition, we applied a digital threshold filter to eliminate all but the highest 10% of DsRED-immunoreactive cells in the SCN. Counting the remaining cells in both the control and light-pulse condition revealed that light triggered a significant increase (P = 0.02 Student's t-test) in the number of DsRED-immunopositive cells (43.2 ± 4.6 SEM; n = 6 mice) versus control condition (26.8 ± 0.8 SEM; n = 4 mice). These data indicate that the characteristics of DsRED expression in mPer2-DsRED transgenic mice are similar to those of the mPer2 protein in terms of its rhythmic and light-inducible nature.
To test whether the random integration of the transgenes affected SCN clock timing or entrainment, the wheel running activity of mPer1-Venus and mPer2-DsRED double hemizygotic transgenic mice was analyzed. Under DD conditions, double transgenic mice exhibited a free-running period of 23.91 ± 0.06 h, which was not statistically different (P = 0.50) from the 23.98 ± 0.18 h free-running period of non-transgenic littermates (n = 6 per genotype; Fig. 3C). Photic stimulation (100 lux, 15 min) at CT 15 triggered a 1.90 ± 0.25 h phase delay in double transgenic mice, and a 2.28 ± 0.39 h phase delay in non-transgenic littermates (n = 6 per genotype, Fig. 3C). Student's t-test revealed a non-significant difference (P = 0.43) in light-induced phase shifts during the early subjective night between double transgenic mice and non-transgenic littermates. These data indicate that clock timing processes as well as light-induced clock resetting of the SCN in transgenic mice are indistinguishable from those in non-transgenic littermates. Thus, the SCN clock timing system has not been altered by the integration of the BAC transgene constructs into the mouse genome or by Venus or DsRED transgene expression.
mPer1-Venus is expressed in neurons in multiple brain regions outside the suprachiasmatic nuclei
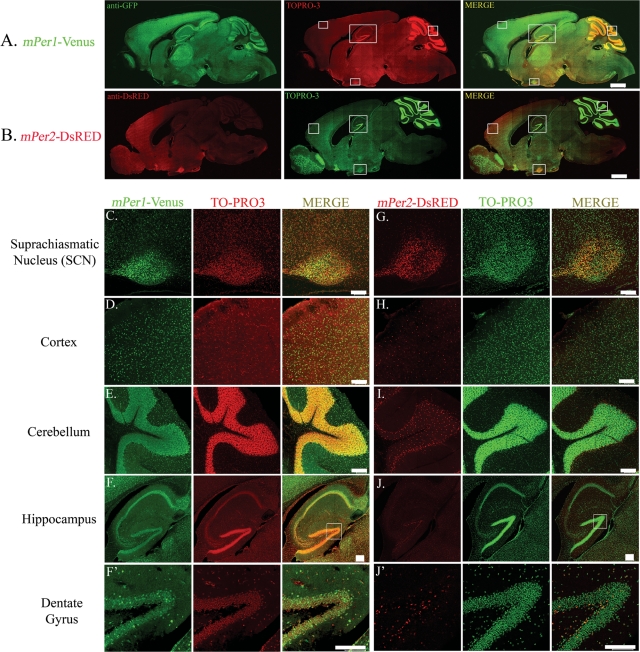
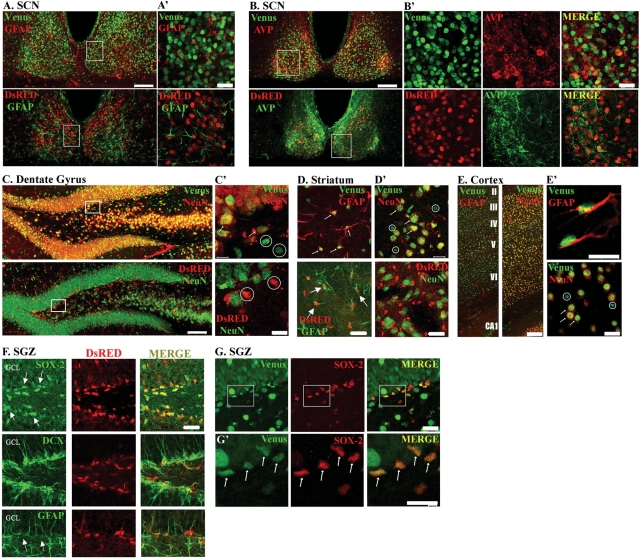
To examine mPer1 transgene expression in the central nervous system (CNS), we analyzed Venus-IR in mid-sagittal as well as coronal brain sections. Venus-IR was detected in multiple brain regions (Fig. 4A), and paralleled the distribution of mPer1 gene expression as reported previously (2,21). Levels of Venus-IR were particularly high in the SCN (Fig. 4C), Purkinje cells and the internal granule layer of the cerebellum (Fig. 4E), the cerebral cortex (Fig. 4D), and the CA regions of the hippocampus (Fig. 4F). Expression was also found in the dentate gyrus of the hippocampus (Fig. 4F'), striatum, thalamus, piriform cortex, brain stem and olfactory bulb, among other regions (Fig. 4A and data not shown). The expression of Venus co-localized with that of the post-mitotic neuron-specific antigen, neuronal nuclei (NeuN), in multiple brain regions including the dentate gyrus of the hippocampus (Fig. 5C and C' top panels, arrows), striatum (Fig. 5D' top panel, arrows), and cortex [Fig. 5E (right panel) and E' (bottom panel) arrows]. In addition, Venus-IR was detected in some NeuN-negative cells with small cellular nuclei of approximately 3 µm in diameter [Fig. 5C' and D' (top panels) and E' (bottom panel) circles], suggestive of a glial population. Indeed, double-labeling revealed co-localized expression of Venus and glial fibrillary acidic protein (GFAP), a marker of glial cells, in the striatum (Fig. 5D top panel, arrows) and cortex [Fig. 5E (left panel) and E' (top panel)]. Venus also broadly co-localizes with GFAP throughout the dentate gyrus (data not shown), and is expressed in a subset of GFAP-positive cells within the subgranular zone (SGZ) that co-expresses the SRY-related transcription factor SOX-2 (Fig. 5G and G', arrows). Within the SCN, Venus did not co-localize with GFAP, indicating that SCN expression is neuron-specific (Fig. 5A and A', top panels). In addition, double-labeling for Venus and arginine vasopressin (AVP), a neuropeptide that is expressed in a subset of SCN neurons, revealed co-localized expression within the SCN (Fig. 5B and B', top panels). Quantitative analysis revealed that 94.4 ± 2.02% of AVP-positive cells in the SCN co-expressed Venus (152 cells analyzed from three mice).
Figure 4.
Transgene expression in the brain. Expression of (A) mPer1-Venus and (B) mPer2-DsRED in mid-sagittal sections of single transgenic mice. Brain tissue was harvested between ZT 12 and 13 and processed for Venus or DsRED expression by immunofluorescent labeling and confocal microscopy. Sections were counterstained with TO-PRO3 [pseudocolored in red (for A) or green (for B)] to visualize nuclei. Scale bar = 1 mm. Boxed regions are provided in higher magnification (C–J'). (C–J') Higher magnification images of (C–F') mPer1-Venus and (G–J') mPer2-DsRED expression in the (C, G) SCN, (D, H) cortex, (E, I) cerebellum, (F, J) hippocampus and (F', J') dentate gyrus. TO-PRO3 is pseudocolored in red (for C–F', middle column) or green (for G–J', middle column). Scale bars = 100 µm. Note the distinct expression patterns of Venus and DsRED in all brain regions shown with the exception of the SCN.
Figure 5.
Distinct mPer1-Venus and mPer2-DsRED expression patterns. (A, B) Double-labeling for Venus or DsRED and (A) the glial marker glial fibrillary acidic protein (GFAP) or (B) the neuronal marker arginine vasopressin (AVP) in the suprachiasmatic nuclei of mPer1-Venus or mPer2-DsRED single transgenic mice. Scale bar = 100 µm. (A', B') Higher magnification images of the boxed regions in (A, B). Scale bar = 20 µm. (C) Double-labeling for Venus or DsRED and the neuronal marker NeuN in the dentate gyrus. Scale bar = 100 µm. (C') Higher magnification images of the boxed regions in (C). Scale bar = 10 µm. (D, D') Double-labeling for Venus or DsRED and (D) GFAP or (D') NeuN in the striatum. Scale bar = 20 µm. (E) Double-labeling for Venus and GFAP or NeuN in the cortex. Roman numerals denote mouse cortical cell layers. Scale bar = 100 µm. (E') Higher magnification images of (E). Scale bar = 10 µm. For (C'–E'), arrows indicate co-localized expression, whereas circles indicate lack of co-localization (F) mPer2-DsRED expression in the subgranular zone (SGZ) of the dentate gyrus. Top panels: Within the SGZ (arrows), mPer2-DsRED expression was observed in SOX-2-positive progenitor cells. Scale bar = 30 µm. Middle panels: Tissue was labeled for doublecortin (DCX) and DsRED. The merged image indicates that the nuclear DsRED signal was not localized to doublecortin-positive neuronal precursor cell nuclei. Bottom panels: mPer2-DsRED expression was observed in GFAP-positive cells in the SGZ. GCL: granule cell layer. (G) Venus expression in SOX-2 progenitor cells of SGZ. Top panels: mPer1-Venus expression was detected in SOX-2-positive progenitor cells. Scale bar = 20 µm. (G') Higher magnification images of the boxed regions in (G). Arrows denote double-labeled progenitor cells. Scale bar = 10 µm. All animals were sacrificed between ZT 11–12 and images were captured by confocal microscopy.
Extra-suprachiasmatic nuclei expression of mPer2-DsRED is distinct from mPer1-Venus: restricted and non-neuronal
An examination of mPer2-DsRED expression in mid-sagittal and coronal brain sections (Fig. 4B) revealed a distribution pattern that was largely distinct from mPer1-Venus. Intense DsRED immunoreactivity was observed predominantly in the SCN (Fig. 4G), with other brain regions exhibiting more modest expression levels. DsRED expression was present in scattered cells of the cerebral cortex (Fig. 4H), striatum, internal granule layer of the cerebellum (Fig. 4I) and dentate gyrus (Fig. 4J and J'). Low or no DsRED-IR was detected in the CA1 pyramidal region of the hippocampus (Fig. 4J), the Purkinje cell layer of the cerebellum, or the piriform cortex (data not shown).
Next, we examined the expression of DsRED in different cell populations. DsRED-IR was not detected in NeuN-positive neurons in all brain regions examined, including the dentate gyrus (Fig. 4J', 5C and C' bottom panels, circles) and striatum (Fig. 5D', bottom panel). As shown in Fig. 5D (bottom panel, arrows), a subset of DsRED-expressing striatal cells was immunoreactive for GFAP, indicating a glial expression origin. In contrast, within the SCN, DsRED expression was not detected in glial cells, as assessed by GFAP immunolabeling (Fig. 5A and A', bottom panels). In other words, whereas DsRED is conspicuously absent from neurons in all other brain regions examined, SCN neurons uniquely express DsRED. Double-labeling for DsRED and AVP demonstrated co-localized expression within the SCN (Fig. 5B and B', bottom panels). Quantitative analysis revealed that 84.3 ± 1.45% of AVP-positive cells (218 cells analyzed from three mice) co-expressed DsRED.
In the dentate gyrus, we noted strong DsRED-IR within the SGZ of the dentate gyrus, the locus of neural stem cells (Fig. 4J' and 5F). Double-labeling for DsRED and SOX-2, which is expressed by pluripotent, self-renewing stem cells (22), revealed co-localization, indicating that mPer2 transcription occurs in progenitor cells (Fig. 5F top row, arrows). Interestingly, within the SGZ, DsRED-IR was not detected in precursor cells that had been committed to a neuronal lineage, as assessed by double-labeling with doublecortin (Fig. 5F middle row), whereas we noted co-localization with GFAP, which is expressed in stem cell progenitors (Fig. 5F bottom row, arrows). Together, these data suggest that mPer2-mediated transcription occurs in stem cell progenitors of the SGZ, and that following commitment, mPer2 transcription is limited to the glial lineage.
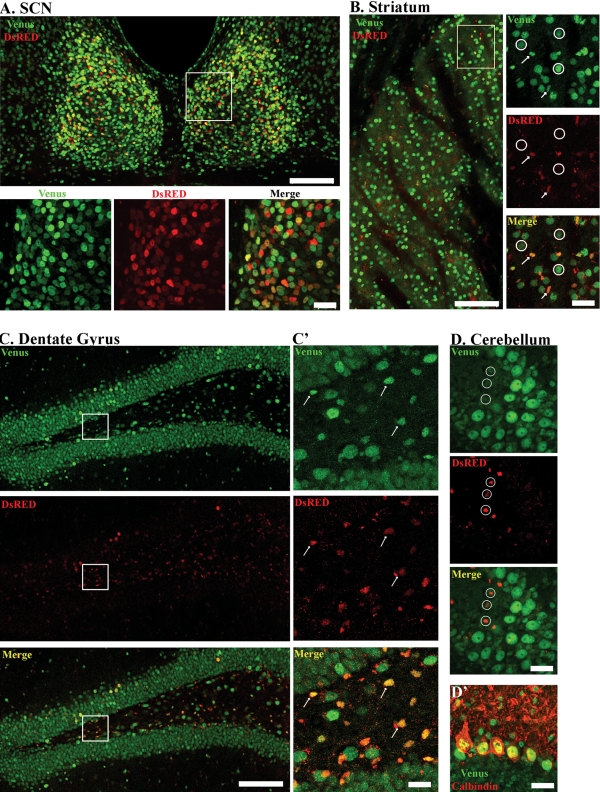
The expression profile of Venus and DsRED in the brains of single hemizygous transgenic mice, as well as the results from the double-labeling experiments using neuron- and glial-specific markers, suggest overlapping, but not identical, patterns of mPer1 and mPer2 expression in the brain. To better illustrate this, we analyzed the distribution of Venus-IR and DsRED-IR in coronal sections from double-hemizygous transgenic mice (Fig. 6). Using a goat polyclonal GFP antiserum and a rabbit polyclonal DsRED antiserum, we found a similar pattern of Venus-IR in the SCN (Fig. 6A), striatum (Fig. 6B), dentate gyrus (Fig. 6C and C') and cerebellum (Fig. 6D) as described in the previous sections. Within the SCN, we observed overlapping expression of Venus and DsRED at CT 12 (Fig. 6A): 67.2 ± 4.3% of Venus-positive SCN cells also expressed DsRED (234 cells analyzed from four mice).
Figure 6.
Brain-specific colocalization of mPer1-Venus and mPer2-DsRED in double transgenic mice. Expression of mPer1-Venus and mPer2-DsRED in the (A) suprachiasmatic nuclei, (B) striatum, (C) dentate gyrus and (D) cerebellum. Boxed regions (in A and B) are provided as higher magnification images in the bottom and right-most panels, respectively. Low magnifications images: scale bar = 100 µm. High magnification images: scale bar = 20 µm. Arrows indicate co-localized expression of Venus and DsRED, whereas circles indicate lack of co-localization. Panel D' represents double-labeling for Venus and Calbindin in the cerebellum. Note that DsRED is not expressed in Purkinje cells of the cerebellum. All animals were sacrificed between ZT 11and 12 and images were captured by confocal microscopy.
Double-labeling revealed that DsRED-IR cells in the striatum (Fig. 6B) and dentate gyrus (Fig. 6C and C') overlap with a subset of Venus-IR cells (arrows), which have a nuclear size of approximately 3 µm, suggestive of the glial lineage. Quantitative analysis showed that 10.6 ± 1.9% of Venus-positive cells in the striatum co-expressed DsRED (259 cells analyzed from four mice). In the cerebellum, Venus-IR and DsRED-IR cells represent distinct subpopulations of cells (Fig. 6D, circles) and do not co-localize. We find co-localized expression of Venus with the Purkinje cell marker, Calbindin (Fig. 6D'), but not with DsRED-positive cells (data not shown). Thus, whereas Venus is broadly expressed in the brains of our transgenic mice, the expression of DsRED is restricted to a much sparser population of cells outside of the SCN.
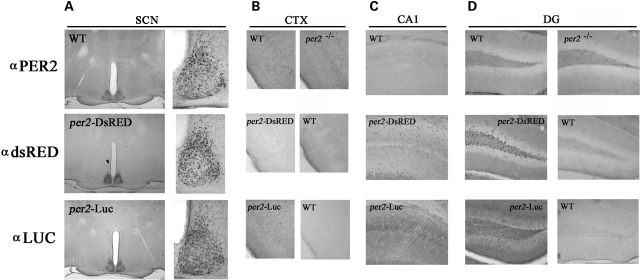
Comparative analysis of DsRED, endogenous mPer2 and mPer2::Luc protein expression in the mouse brain
To further test the utility of mPer2-DsRED as a bona fide reporter of endogenous mPer2 expression, we used an immunohistochemical approach to compare the expression pattern of DsRED in the brains of mPer2-DsRED hemizygous transgenic mice with that of mPer2 in WT (C57Bl/6) mice and mPer2::Luc fusion protein in mPer2-Luciferase knock-in mice (16). The distribution of DsRED-IR in the SCN at Zeitgeber time (ZT) 11–12 paralleled mPer2-IR as well as Luc-IR (Fig. 7A). Specifically, DsRED-/mPer2-/Luc-IR was highly expressed in the dorsal and lateral regions of the SCN at this time point. Under the DAB development conditions used to detect robust expression of all three antigens in the SCN, immunoreactivity in other brain regions was negligible. Increasing the development time of the DAB substrate revealed DsRED-IR in the SGZ of the dentate gyrus (Fig. 7D middle row, left panel), consistent with the results of the immunofluorescence experiments; sparse expression in the GCL and CA1 region was detected (Fig. 7C and D, middle row, left panel). Furthermore, low DsRED expression was detected in the piriform cortex (Fig. 7B, middle row, left panel). These signals appeared to be specific, since this labeling pattern was absent from WT tissue processed with the DsRED antibody (Fig. 7B and D, middle row, right panels). In comparison, there was no detectable mPer2-IR in the dentate gyrus or CA1 in WT mice (Fig. 7C and D, top row, left panel), and very sparse and weak expression of Luc in the GCL and CA1 (Fig. 7C and D, bottom row, left panel) in mPer2-Luciferase knock-in mice. In the piriform cortex, very limited endogenous mPer2 protein was detected in WT mice (Fig. 7B, top row, left panel); this signal appeared to be slightly higher than the signal detected in mPer2−/− tissue (Fig. 7B, top row, right panel). In the CA1 region of the hippocampus, only faint Luc expression slightly above background was observed in mPer2-Luciferase knock-in mice (Fig. 7C, bottom panel). However, this signal appeared to be specific, since Luc staining was not detected in wild-type CA1 that was immunolabeled with a Luc antiserum (data not shown). Together these data indicate that, during the late subjective day, mPer2 is not strongly expressed in neuronal populations outside of the SCN. The discord in DsRED versus mPer2 and Luc expression in non-neuronal cells may be attributed to the relative sensitivities of the primary antibodies as well as the subcellular concentration of the antigen (e.g. DsRED is forcibly localized to the nucleus, whereas mPer2 and Luc are not). Together, these data are not inconsistent with the notion that DsRED is a bona fide reporter of mPer2 expression (see Discussion), as strongly suggested by the comparative expression data in the SCN.
Figure 7.
Comparative analysis of endogenous mPer2, mPer2::Luc and mPer2-DsRED expression in the brain. Brain tissue was harvested between ZT 11 and ZT 12 from wild-type (WT) mice or mPer2−/− null mice, mPer2-DsRED transgenic mice and mPer2::Luc heterozygous mice, and processed for endogenous mPer2 (top), DsRED (middle), and Luciferase (LUC, bottom) expression by immunohistochemical labeling. Protein expression was examined in the (A) SCN, (B) piriform cortex (CTX), (C) CA1 region of the hippocampus and (D) dentate gyrus (DG). For (A), the SCN are magnified and shown to the right for each panel. The genotype of the representative tissue is noted for each panel. Data are representative of at least six animals of each genotype.
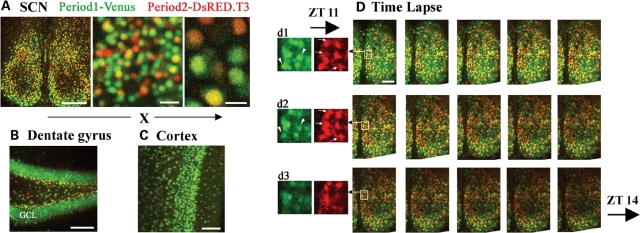
Dual-imaging of mPer1-Venus and mPer2-DsRED in the suprachiasmatic nuclei: in real time and with single-cell resolution
While various mPer1 and mPer2 reporter mice are currently available, it has not been possible to combine different transgenic lines to monitor both transcriptional processes simultaneously. The spectral properties of Venus and DsRED are sufficiently distinct that they could be resolved easily under standard fluorescence microscopic conditions. Furthermore, as a result of localization of the fluorescent signals to the nuclei, it should be possible to resolve individual mPer1-Venus- and mPer2-DsRED-expressing cells in live tissue. To illustrate this point, tissues from double hemizygotic transgenic mice were isolated at ZT 11 and live images from the SCN, hippocampus and cortex were captured. In the SCN, under low and high magnification, we were able to clearly identify transgenic cells that express Venus, DsRED or both transgenes (Fig. 8A). In the dentate gyrus (Fig. 8B), the broad expression of Venus was in contrast with the largely restricted expression of DsRED to the SGZ; however, it should be noted that the majority of DsRED-positive cells within the SGZ co-expressed Venus (Figs 8B and 6C'). Expression of both Venus and DsRED was detected in the cortex (Fig. 8C).
Figure 8.
Real-time monitoring of dynamic changes in mPer1-Venus and mPer2-DsRED expression in live brain slices of double transgenic mice. Venus and DsRED fluorescence in the (A) suprachiasmatic nuclei (SCN), (B) dentate gyrus and (C) cortex from live brain slices of double transgenic mice. Brain tissue (a 300 µm slice) was prepared from mice sacrificed at ZT 11, and maintained at 34°C in oxygenated physiological media during image acquisition using a confocal microscope. A subset of SCN neurons co-expresses Venus and DsRED, although cells which express only one of the markers are also detected. Scale bars in (A) = 20 µm (middle panel) and 10 µm (right panel). (B and C) Note the distinct expression patterns of Venus in the granule cell layer (GCL) of the dentate gyrus and cortex. (D) Time-lapse recording from mPer1-Venus::mPer2-DsRED double hemizygotic transgenic mice. Animals were sacrificed at ZT 9 and images of brain slices containing the SCN were captured on a confocal microscope at 4-min intervals. Representative images are presented between ZT 11 and ZT 14, with an approximate time interval of 12 min. (d1–d3) DsRED and Venus signals are separated and magnified from the boxed regions in the montage. Arrows denote cells showing decreased Venus expression and arrowheads denote cells showing elevated DsRED expression as a function of time.
Next, we asked whether changes in the expression levels of both mPer1- and mPer2-regulated transgenes could be detected in real time in live SCN slices. To this end, mPer1-Venus::mPer2-DsRED double hemizygotic transgenic mice were entrained to a fixed light–dark cycle, and SCN tissue was dissected at ZT 9 and placed in a live imaging/perfusion chamber with oxygenated physiological media at 34°C. Confocal images were acquired at 4-min intervals from ZT 9 to ZT 14. As shown in Figure 8D, we were able to detect and discriminate between Venus and DsRED fluorescence on a live SCN slice. Importantly, within a 4-h interval (ZT 11–14), we were able to observe a decline in both the number of Venus-expressing cells and the intensity of Venus fluorescence, from its peak in expression at ZT 11. Furthermore, within the dorsal SCN we detected a transient increase in the number of DsRED-positive cells from ZT 11 to approximately ZT 12.5, followed by a decline in DsRED expression by ZT 14 (Fig. 8D).
DISCUSSION
The data presented here reveal the utility of these new clock gene transgenic mouse strains. As expected, the highest levels of expression for both Venus and DsRED were detected in the SCN from the late day to early night. Outside of the SCN, Venus was expressed in neuronal and non-neuronal cell populations throughout the CNS. This expression pattern is consistent with the expression pattern of the mPer1 transcript, as assessed by in situ hybridization (2,3). In contrast, DsRED expression outside of the SCN was largely restricted to non-neuronal cell populations. This finding was surprising, given that several studies have detected inducible mPer2 (23,24) outside of the SCN. Although these studies did not verify the phenotype of the cells that exhibit mPer2 antigenicity, it was suggested that the cells in question were neurons. The discrepancy between our data set and the noted studies is not clear. One possibility is that DsRED expression does not faithfully recapitulate mPer2 expression. However, several pieces of data provided here argue against this possibility. First, as with the endogenous gene, expression of DsRED was neuronal and rhythmic within the SCN. Second, consistent with our DsRED expression pattern, we detected minimal mPer2 or mPer2-Luc immunoreactivity in neurons outside of the SCN. Compared with mPer1-Venus, mPer2-DsRED exhibited a lower amplitude rhythm. One reason for this attenuated oscillation was the relatively high level of DsRED detected at the anticipated nadir (CT 22–CT 6). This result contrasts with the marked variation of mPer2 protein expression observed over the circadian cycle in WT mice. Indeed, immunohistochemical labeling has shown a near complete loss of mPer2 expression from the late subjective night to mid-subjective day (25,26). One potential reason for this difference between the reporter and the endogenous gene product is that, in addition to transcriptional regulation, the circadian expression of endogenous mPer2 may be influenced by post-transcriptional as well as post-translational processes. Support for post-translational circadian regulation comes from a number of studies showing that cycling of mPer2 protein occurs even in the absence of rhythmic mPer2 mRNA expression (27–29). More recently, post-transcriptional mRNA decay has been implicated in the circadian oscillation of mPer2 mRNA (30). As the expression of the reporter only captures the transcriptional component of the mPer2 rhythm, the oscillation amplitude may thus be dampened relative to the endogenous gene product.
Unlike the mPer2-DsRED mice, immunohistochemical labeling failed to detect marked mPer2 and mPer2-Luc expression in glial cells outside the SCN. There may be several reasons for this difference. As noted above, one possibility may be the relative sensitivity of the primary antibodies. Hence, the mPer2 and Luc antibodies may not be sufficiently sensitive to detect a relatively modest level of expression in glia. In addition, glial-specific binding partners may limit the ability to use immunohistochemical labeling techniques to detect endogenous mPer2. On a related note, our construct is designed to drive the DsRED reporter into the nucleus. This high concentration of antigen may be required to detect an appreciable signal above the baseline/non-specific signal. As a final note, Prolo et al. (31) reported rhythmic mPer2 expression in mouse forebrain astrocytes, and Suh et al. (32) reported glial rhythmicity in Drosophila. These findings do fit with our work showing mPer2-driven reporter gene expression in astrocytes. Finally, it should also be noted that the insertion of the DsRED coding sequence into the mPer2 translation start site did not interfere with any of the recently characterized sequence motifs shown to regulate rhythmic expression of mPer2. For example, neither the non-canonical E-box (a target of Clock and Bmal1) located 20 base pairs upstream from the transcription start site (33), nor the E4BP4 element which functions as a negative regulator of mPer2 rhythm amplitude (34), were disrupted by the insertion of the transgene. Our data noting divergent mPer1 and mPer2 expression in extra-SCN tissue/cells raises the interesting possibility that there are distinct cellular oscillators outside of the SCN: one that is neural-specific and largely mPer1-dependent, and a second glial oscillator that is driven by both mPer1 and mPer2. Additional studies will be required to test this possibility.
Our data showing mPer2-DsRED expression in SGZ progenitor cells is consistent with studies showing that mPer2 is expressed in stem cells (35). Interestingly, mPer2 expression was reported to be antiphasic to the peak in the circadian rhythm of DNA synthesis, and the disruption of mPer2 expression was found to increase the DNA synthesis rate of neural stem cells. Kochman et al. (36) reported a circadian variation in the rate of cell proliferation in the dentate gyrus, which was ascribed to gliogenesis, rather than neurogenesis. It is also interesting to note that deregulated expression of the PERIOD genes is observed in various human cancers, and that in animal models mPer2 controls the temporal expression profile of several key cell cycle genes such as c-myc, Cyclin D1 and Gadd45α (37–39). These results and others showing that mPer2 functions as a potent regulator of cell proliferation (40) raises the possibility that the relatively high level of mPer2-dependent transcription we observed in glial cells serves as a suppressor of mitogenesis.
Our double-immunolabeling and live cell imaging of the SCN revealed that DsRED and Venus expression were colocalized in a subset of neurons. Although the peak in mPer1- and mPer2-dependent transcription within the SCN is offset by several hours (21,41), the observation that a subset of the cells express either high levels of DsRED or Venus raises the interesting possibility that SCN timing is influenced by discrete mPer1- and mPer2-dependent cellular clocks. One possibility is that within the SCN, there is a subpopulation of cellular oscillators in which either mPer1 or mPer2 has emerged as the dominant player in clock timing, and this subset of cells is distinct from the majority of SCN oscillators in which mPer1 and mPer2 are acting as co-regulators of clock timing. A number of reports have shown that mPER1 and mPER2 have both complementary and distinct roles in clock timing and output. For example, with respect to the molecular clock, mPer1 has been shown to function at a post-translational level, and that it is dispensable for rhythmic clock gene transcription (42). Conversely, mPer2 functions at a transcriptional level, positively regulating clock gene expression (42). Furthermore, with respect to clock output, microarray analysis has revealed that mPer1 and mPer2 regulate distinct populations of clock-controlled genes (43). Additional work will be required to determine whether this partitioning of functionality occurs at the level of autonomous cellular oscillators within the SCN.
In conclusion, these new mouse strains can be used to address research questions that are fundamental to the timing field (e.g. photic input, intercellular coupling, cell-autonomous oscillations), and to examine the role of circadian timing in diverse processes, such as cell proliferation, sleep and learning, and memory (44–46). The mPer1-Venus and mPer2-DsRED mice will be a valuable addition to the ‘circadian toolbox’ of mouse models that will help to advance our understanding of the complex interplay between biological timing and human physiological processes.
MATERIALS AND METHODS
Vector construction
BAC constructs encompassing either the mPer1 gene (Clone ID number RP24-277K16) or the mPer2 gene (Clone ID number RP24-391L2) were obtained from BACPAC Resources Center (Oakland, CA, USA). To construct the Venus-NLS-PEST mammalian expression vector, the mouse ornithine decarboxylase PEST sequence was amplified by PCR from the pTAL-d2EGFP vector (Clontech, Mountain View, CA, USA) using the following primers (forward: 5′-GTG TAC AGC CAT GGC TTC CCG CCG GAG-3′; reverse: 5′-CGA ATT CCT ACA CAT TGA TCC TAG CAG AAG-3′), and cloned into the BsrGI/NotI site of the pCS2-Venus expression plasmid (gift of A. Miyawaki). Complementary oligonucleotides encoding the NLS of SV40 T antigen (forward: 5′-GTA CCC TCC AAA AAA GAA GAG AAA GGT AGA AGA CCC CTT-3′; reverse: 5′-GTA CAA GGG GTC TTC TAC CTT TCT CTT CTT TTT TGG AGG-3′) were annealed and subsequently inserted into the BsrGI site. To construct the DsRED.T3-NLS-PEST mammalian expression vector, the DsRED.T3 coding sequence was amplified by PCR from the Z/RED vector (gift of A. Nagy) using the following primers (forward: 5′-GCG TCG ACA TGG CCT CCT CCG AGG ACG TC-3′; reverse: 5′-GTG TAC AAC AGG AAC AGG TGG TGG CGG C-3′), and cloned into the pcDNA3.1 Hygro+ vector (Invitrogen, Carlsbad, CA, USA). The PEST and NLS domains were cloned downstream of the DsRED.T3 sequence essentially as described above. All vectors were confirmed by sequencing.
The Venus-NLS-PEST fusion gene was introduced into the mPer1-containing BAC construct by BAC recombineering using galK selection (47). Recombineering reagents were kindly provided by Neal Copeland (National Cancer Institute Frederick, MD) and detailed protocols can be found at the website http://recombineering.ncifcrf.gov/. The galK cassette was amplified by PCR using primers containing flanking sequences with 56–60 bp homology to the mPer1 gene, at the desired site of Venus-NLS-PEST insertion (mPer1-galK forward: 5′-CCG TGG TGG CTT CTT CAC CTT CCC TGT TTC GTC CTC CAC TGT ATG GCC CAG ACA TGC CTG TTG ACA ATT AAT CAT CGG CA-3′; mPer1-galK reverse: 5′-GGG GAT GGG ACT CCT CCA GGA CAA AAA GGT TCT CCG GGC CTG GGG TCT CCT CCC CCA TCG TCA GCA CTG TCC TGC TCC TT-3′). The galK cassette was transformed into electrocompetent SW102 cells containing the BAC, and galK+ colonies were selected on M63 minimal media containing galactose, leucine, biotin and chloramphenicol. The Venus-NLS-PEST cassette was generated by PCR amplification using primers containing homology arms to mPer1 (mPer1-Venus forward: 5′-CCG TGG TGG CTT CTT CAC CTT CCC TGT TTC GTC CTC CAC TGT ATG GCC CAG ACA TGG TGA GCA AGG GCG AGG AGC TG-3′; mPer1-Venus reverse: 5′-GGG GAT GGG ACT CCT CCA GGA CAA AAA GGT TCT CCG GGC CTG GGG TCT CCT CCC CCA TCG GGG CCC AAT GCA TTG GCG CC-3′). To replace the galK cassette with the Venus-NLS-PEST cassette, the latter was transformed into electrocompetent SW102 cells derived from a single galK+ clone. Cells were selected on M63 minimal media containing glycerol, leucine, biotin, 2-deoxy-galactose and chloramphenicol. Correct recombinants were identified by SpeI digestion (comparing the pattern with parental BAC DNA) and PCR analysis using the genotyping primers indicated below.
To introduce the DsRED cassette into the BAC, we first amplified the DsRED.T3-NLS-PEST cassette by PCR using the following primers (forward: 5′-CCC GAT ATC ATG GCC TCC TCC GAG GAC GTC A-3′; reverse: 5′-CCC GAA TTC CTC AGA AGC CAT AGA GCC CAC C-3′), and subcloned it into the EcoRV/EcoRI sites of pBluescript KS (Stratagene, La Jolla, CA, USA). mPer2 homology arms were amplified from the BAC clone using the following PCR primers (5′ mPer2 homology arm forward: 5′-CCC GGT ACC GTG ACC CGA CTC AGC TTA AGT GAC-3′; 5′ mPer2 homology arm reverse: 5′-CCC GAT ATC GTC GGG CTC TGG AAT AAG CTG G-3′; 3′ mPer2 homology arm forward: 5′-CCC GAA TTC AGT CCC ACC AGT CCC ACC AAG G-3′; 3′ mPer2 homology arm reverse: 5′-CCC ACT AGT CTG ACC AGG GAC ATC ACA CCA G-3′). The 5′ mPer2 homology arm was inserted upstream of the DsRED.T3-NLS-PEST sequence at the KpnI/EcoRV sites, and the 3′ mPer2 homology arm was inserted downstream of the cassette at the EcoRI/SpeI sites. Following confirmation by DNA sequencing, the mPer2-flanked DsRED cassette was excised by KpnI/SpeI digestion and gel-purified to remove the backbone vector. Electrocompetent SW102 cells containing the BAC were then co-transformed with this DsRED fragment (100 ng) and pBluescript KS (1 ng), and selected on media containing chloramphenicol, ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal: Sigma–Aldrich, St Louis, MO, USA). Four out of 96 colonies screened were true recombinants containing the DsRED cassette. The positive clones were subsequently grown under chloramphenicol (no ampicillin) selection to eliminate the pBluescript KS plasmid. Targeted BAC clones were verified by DNA sequencing prior to use for pronuclear injections. All primers were obtained from Integrated DNA Technologies (Coralville, IA, USA).
Generation of transgenic mice and animal husbandry
The DNA of the mPer1-Venus and mPer2-DsRED BAC constructs was purified using NucleoBond Plasmid Maxi EF Kit (Clontech), and injected into the pronuclei of oocytes from FVB/N mice at the Ohio State University Transgenic Facility. Founders were identified by PCR genotyping using the following primers: mPer1-Venus transgenic mice (Venus forward: 5′-ACA ACC ACT ACC TGA GCT ACC AGT-3′; mPer1 reverse: 5′-CAG GCT GGG GCC TGG ACA AG-3′); mPer2-DsRED transgenic mice (DsRED forward: 5′-ATC TAC ATG GCC AAG AAG CCC GT-3′; mPer2 reverse: 5′-GCT GGA GCC ACT GCT CAT GTC-3′). Four out of seven mPer1-Venus transgenic lines (nos. 4126, 7, 13, 33) exhibited robust expression of Venus in the SCN as well as an identical expression pattern in other brain regions. The data presented herein were derived from mPer1-Venus founder line no. 33, and are consistent with results obtained from the other three transgenic lines. The remaining three transgenic lines showed no expression in the brain and other tissues analyzed. Three out of five mPer2-DsRED transgenic lines (nos. 6, 8, 12) exhibited an identical distribution of DsRED expression in the SCN and other brain regions. The other two transgenic lines showed no expression in the brain. The three independent mPer2-DsRED transgenic lines exhibited strong neuronal expression within the SCN and glial-specific expression outside of the SCN. The only line-specific differences that were noted pertained to the relative intensities of transgene expression. Variations in transgene expression levels are commonly observed and to be expected, and are likely attributed to the chromosomal location of the transgene insertion for each of the founder lines. Data presented in this manuscript were obtained from all three mPer2-DsRED founder lines. For experiments requiring quantitative comparison, the founder line that exhibited the strongest expression intensity, mPer2-DsRED founder line no. 12, was used. All mice used in this study were hemizygotic for mPer1-Venus, mPer2-DsRED, or both transgenes (i.e. double hemizygotic).
Data presented were obtained from mPer1-Venus and mPer2-DsRED mice that had been backcrossed onto a C57Bl/6J background for three (minimum) to six (maximum) generations. Mice carrying a targeted deletion of the mPer2 gene (mper2−/−) and those that express the mPer2::Luciferase fusion protein (mPer2::Luc) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Mice were maintained at the animal facilities of the University of Ottawa and the Ohio State University in accordance with institutional guidelines. All animal handling and experimental procedures were approved by the Animal Welfare Committees of the University of Ottawa and the Ohio State University.
Cell culture and transfection
Embryonic day 19–20 Sprague–Dawley rat cortical cultures were prepared and maintained using the methods described elsewhere (26). After 7–10 days in culture, neurons were transfected using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. HEK293 cells were obtained from The American Type Culture Collection and grown according to supplier's instructions. For CHX kinetics, HEK293 cells were transfected at approximately 90% confluency using Lipofectamine 2000 (Invitrogen). Twenty-four hours post-transfection, cells were incubated with media containing CHX at 100 µg/ml in DMSO (Sigma) and vehicle (DMSO alone). Graphed data represents normalized fluorescence intensity units for three independent experiments.
Behavioral and photic stimulation paradigms
Mice were individually housed in polycarbonate cages equipped with a running wheel in ventilated cabinets with timer-controlled lighting (white fluorescent tubular bulbs approximately 400 lux at cage level). To monitor wheel running activity, cages were equipped with magnetic switches, and switch closures were recorded and analyzed with VitalView and Actiview software (MiniMitter Co. Inc., Sunriver, OR, USA). To examine free-running rhythms and the phase-delaying effect of light, mice were stably entrained to a 12 h:12 h light:dark (LD) cycle for 14 days prior to dark adaptation (DD). Period length and the phase-shifting effects of light were determined using the linear regression method described in Cheng et al. (48). Data are presented as the mean ± SEM. Statistical significance was measured using the Student's t-test: significance was accepted for P < 0.05. For the photic stimulation paradigm, cages were removed from the cabinet, placed in a secondary containment chamber and exposed to fluorescent white light (100 lux) for 15 min at CT 15. Immediately after the light pulse (LF), cages were returned to the behavioral cabinet and remained in DD for at least 7 days post-LF. For all other photic stimulation assays, mice were entrained to a 12:12 LD cycle (400 lux), dark-adapted for 2 days, exposed to light (100 lux, 15 min) at designated CTs and processed as described below. To profile circadian transgene expression, mice were entrained and dark-adapted for 2 days, and then sacrificed at the designated CTs. For these experiments, CT was estimated based on the prior light–dark cycle, where ZT 12 (i.e. lights off) was used to define CT 12. As noted in the Results section, for time-lapse monitoring of transgene expression brain slices and double-immunolabeling experiments, tissue was prepared from mice maintained under a standard 12:12 LD cycle, where time-of-day is denoted by ZT.
Mice were killed via cervical dislocation, and their brains were removed rapidly under red dim light. Brains were immersed in chilled oxygenated physiological saline, cut into 600-µm coronal sections with an oscillating tissue slicer, and fixed (6 h, room temperature) in a 4% (w/v) formaldehyde/phosphate-buffered saline (PBS) solution, pH 7.4. Tissue was cryoprotected in 30% sucrose (w/v) overnight, and then cut into thin (40 µm) sections using a freezing microtome.
Immunofluorescent and immunohistochemical detection
For immunofluorescent labeling, thin (40 µm), free-floating brain sections were washed five times in PBST (PBS with 0.1% Triton X-100), blocked for 1 h in 5% horse serum/PBST, and incubated (overnight, 4°C) in primary antibodies. The following primary antibodies were used: rabbit polyclonal anti-GFP (1:2000; Eusera, Edmonton, Alberta, Canada), goat polyclonal anti-GFP (1:2000; Eusera), rabbit polyclonal anti-DsRED (1:200; Clontech), mouse monoclonal anti-NeuN (1:1000; Chemicon, Temecula, CA, USA), mouse monoclonal anti-GFAP (1:500; Molecular Probes, Eugene, OR, USA), goat polyclonal anti-SOX-2 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat polyclonal anti-doublecortin (1:1000; Santa Cruz Biotechnology), guinea pig polyclonal anti-AVP (1:4000; Peninsula Laboratories, San Carlos, CA, USA) and rabbit polyclonal anti-calbindin (1:400; Millipore, Billerica, MA, USA). After 12–18 h at 4°C, sections were washed five times in PBST and incubated (2 h, room temperature) with Alexa Fluor® 488- and/or Alexa Fluor® 594-conjugated secondary antibodies (1:1000; Invitrogen) against the IgG domains of the primary antibodies. In some experiments, sections were counterstained with the nuclear marker TO-PRO®-3 (1:1000; Invitrogen). Sections were mounted on glass slides with DAKO fluorescent mounting media (Glostrup, Denmark) and sealed with nail polish.
For immunohistochemical labeling, thin (40 µm), free-floating brain sections were pre-treated with 0.3% hydrogen peroxide in PBST for 15 min prior to blocking. The following primary antibodies were used: rabbit polyclonal anti-DsRED (1:1000), rabbit polyclonal anti-mPer2 (1:2500; Alpha Diagnostic International, San Antonio, TX, USA), and goat polyclonal anti-Luciferase (1:1000; Promega, Madison, WI, USA). The following day, sections were washed and incubated in biotinylated anti-goat or anti-rabbit IgG (1:300; Vector Laboratories, Burlingame, CA, USA), and further processed using the VECTASTAIN ABC System (Vector Laboratories). Signal detection was performed using the DAB/Nickel Substrate Kit (Vector Laboratories). After staining, sections were washed and mounted on gelatin-coated slides with Permount media (Fisher Scientific, Pittsburgh, PA, USA).
Image acquisition and processing
Cell culture images (Fig. 1) were captured using an inverted microscope (Leica DMIRB) with a 20X objective and with a Zeiss Axiovert Observer Z1 epifluorescent microscope equipped with an AxioCam MRm cooled-color camera (Oberkochen, Germany). Fluorescent and bright-field images were merged using Metamorph software (Molecular Devices Inc.). Immunofluorescent tissue sections were examined using a Zeiss 510 laser scanning confocal microscope with argon (488 nm), helium/neon (546 nm) and helium/neon (633 nm) lasers. For the rhythmic Venus intensity analysis, images (10X) of the SCN were acquired and a digital 150 (x-axis) × 200 (y axis)-pixel oval was placed in the central SCN and the mean intensity values were determined. A digital circle (Φ 200 pixels) was placed in the lateral hypothalamus and the mean value was subtracted from the adjacent SCN, thus generating a normalized SCN intensity value. The same approach was used to examine light-induced Venus expression. For the rhythmic DsRED intensity analysis, a digital crescent, as described in (49), was placed in the dorsal SCN and the mean labeling intensity was determined. Data were normalized using the same methods as those described for Venus. Quantitation of light-induced DsRED expression is described in the Results section. Group statistical analysis was performed via the two-tailed Student's t-test. P < 0.05 was accepted as statistically significant. The values are presented as the mean ± SEM.
For live cell and time-lapse imaging of SCN slices, mice were sacrificed at ZT 9 and SCN-containing coronal brain sections were isolated using the techniques described above. Sections were then gradually brought up to 34°C and placed in a laminar flow perfusion chamber (50) mounted on a Zeiss 510 scanning confocal microscope. Both the stage and perfusion media were maintained at 34°C. Two-micron thick sections were acquired at 4-min intervals using a 20X objective.
FUNDING
This work is supported by National Institutes of Health Grants MH62335 and NS47176 to K.O., and the Ohio State University Neuroscience Center Core Grant 5P30NS045758, as well as an operating grant from the Canadian Institute of Health Research (CIHR) no. 086549 to H.Y.M.C.
ACKNOWLEDGEMENTS
We thank A. Miyawaki for the generous gift of the pCS-Venus construct, and A. Nagy for the gift of Z/RED. Many thanks also to the following individuals: N. Copeland for sharing the BAC recombineering reagents; X.-A. Pu and S. Kharzai for performing the pronuclear injections; and J. Curfman for assistance in animal husbandry. H.Y.M.C. is a Canada Research Chair in the Genetics of Biological Timing. All requests for reagents must be made jointly to H.Y.M.C. and K.O.
Conflict of Interest statement. The authors declare that there are no competing interests.
REFERENCES
- 1.Okamura H. Clock genes in cell clocks: roles, actions, and mysteries. J. Biol. Rhythms. 2004;19:388–399. doi: 10.1177/0748730404269169. [DOI] [PubMed] [Google Scholar]
- 2.Shearman L.P., Zylka M.J., Weaver D.R., Kolakowski L.F., Reppert S.M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 3.Tei H., Okamura H., Shigeyoshi Y., Fukuhara C., Ozawa R., Hirose M., Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 4.Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Griffin E.A., Jr, Staknis D., Weitz C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 6.Bunger M.K., Wilsbacher L.D., Moran S.M., Clendenin C., Radcliffe L.A., Hogenesch J.B., Simon M.C., Takahashi J.S., Bradfield C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A., Virshup D.M., Ptácek L.J., Fu Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y., Padiath Q.S., Shapiro R.E., Jones C.R., Wu S.C., Saigoh N., Saigoh K., Ptácek L.J., Fu Y.H. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 9.Ebisawa T., Uchiyama M., Kajimura N., Mishima K., Kamei Y., Katoh M., Watanabe T., Sekimoto M., Shibui K., Kim K., et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralph M.R., Foster R.G., Davis F.C., Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto Y., Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc. Natl Acad. Sci. USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L., Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur. J. Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhlman S.J., Quintero J.E., McMahon D.G. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport. 2000;11:1479–1482. [PubMed] [Google Scholar]
- 14.Yamaguchi S., Mitsui S., Miyake S., Yan L., Onishi H., Yagita K., Suzuki M., Shibata S., Kobayashi M., Okamura H. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr. Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 15.Wilsbacher L.D., Yamazaki S., Herzog E.D., Song E.J., Radcliffe L.A., Abe M., Block G., Spitznagel E., Menaker M., Takahashi J.S. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc. Natl Acad. Sci. USA. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo S.H., Yamazaki S., Lowrey P.L., Shimomura K., Ko C.H., Buhr E.D., Siepka S.M., Hong H.K., Oh W.J., Yoo O.J., Menaker M., Takahashi J.S. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevis B.J., Glick B.S. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 18.Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 19.Copeland N.G., Jenkins N.A., Court D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 20.Yan L., Silver R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur. J. Neurosci. 2004;19:1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z.S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., Lee C.C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 22.Komitova M., Eriksson P.S. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neurosci. Lett. 2004;369:24–27. doi: 10.1016/j.neulet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 23.Verwey M., Khoja Z., Stewart J., Amir S. Differential regulation of the expression of Period2 protein in the limbic forebrain and dorsomedial hypothalamus by daily limited access to highly palatable food in food-deprived and free-fed rats. Neuroscience. 2007;147:277–285. doi: 10.1016/j.neuroscience.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 24.Feillet C.A., Mendoza J., Albrecht U., Pévet P., Challet E. Forebrain oscillators ticking with different clock hands. Mol. Cell. Neurosci. 2008;37:209–221. doi: 10.1016/j.mcn.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H.Y., Papp J.W., Varlamova O., Dziema H., Russell B., Curfman J.P., Nakazawa T., Shimizu K., Okamura H., Impey S., Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H.Y., Dziema H., Papp J., Mathur D.P., Koletar M., Ralph M.R., Penninger J.M., Obrietan K. The molecular gatekeeper Dexras1 sculpts the photic responsiveness of the mammalian circadian clock. J. Neurosci. 2006;26:12984–12995. doi: 10.1523/JNEUROSCI.4253-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishii K., Yamanaka I., Yasuda M., Kiyohara Y.B., Kitayama Y., Kondo T., Yagita K. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: a real-time analysis. Neurosci. Lett. 2006;401:44–48. doi: 10.1016/j.neulet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y., Yagita K., Okamura H. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol. Cell. Biol. 2005;25:1912–1921. doi: 10.1128/MCB.25.5.1912-1921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto Y., Yagita K., Okamura H. Does mPER2 protein oscillate without its coding mRNA cycling?: post-transcriptional regulation by cell clock. Genes Cells. 2006;11:525–530. doi: 10.1111/j.1365-2443.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- 30.Woo K.C., Kim T.D., Lee K.H., Kim D.Y., Kim W., Lee K.Y., Kim K.T. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prolo L.M., Takahashi J.S., Herzog E.D. Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh J., Jackson F.R. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:335–347. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo S.H., Ko C.H., Lowrey P.L., Buhr E.D., Song E.J., Chang S., Yoo O.J., Yamazaki S., Lee C., Takahashi J.S. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno T., Onishi Y., Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriya T., Hiraishi K., Horie N., Mitome M., Shinohara K. Correlative association between circadian expression of mouse Per2 gene and the proliferation of the neural stem cells. Neuroscience. 2007;146:494–498. doi: 10.1016/j.neuroscience.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 36.Kochman L.J., Weber E.T., Fornal C.A., Jacobs B.L. Circadian variation in mouse hippocampal cell proliferation. Neurosci. Lett. 2006;406:256–259. doi: 10.1016/j.neulet.2006.07.058. [DOI] [PubMed] [Google Scholar]
- 37.Gery S., Gombart A.F., Yi W.S., Koeffler C., Hofmann W.K., Koeffler H.P. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S.T., Choo K.B., Hou M.F., Yeh K.T., Kuo S.J., Chang J.G. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 39.Fu L., Pelicano H., Liu J., Huang P., Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 40.Lee C.C. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 41.Albrecht U., Sun Z.S., Eichele G., Lee C.C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 42.Bae K., Jin X., Maywood E.S., Hastings M.H., Reppert S.M., Weaver D.R. Differential functions of mPer1, mPer2 and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 43.Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z.S., Eichele G., Bradley A., Lee C.C. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 44.Eckel-Mahan K.L., Phan T., Han S., Wang H., Chan G.C., Scheiner Z.S., Storm D.R. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat. Neurosci. 2008;11:1074–1082. doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 46.Wisor J.P., O'Hara B.F., Terao A., Selby C.P., Kilduff T.S., Sancar A., Edgar D.M., Franken P. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng H.Y., Obrietan K., Cain S.W., Lee B.Y., Agostino P.V., Joza N.A., Harrington M.E., Ralph M.R., Penninger J.M. Dexras1 potentiates photic and suppresses nonphotic responses of the circadian clock. Neuron. 2004;43:715–728. doi: 10.1016/j.neuron.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Butcher G.Q., Lee B., Obrietan K. Temporal regulation of light-induced extracellular signal-regulated kinase activation in the suprachiasmatic nucleus. J. Neurophysiol. 2003;90:3854–3863. doi: 10.1152/jn.00524.2003. [DOI] [PubMed] [Google Scholar]
- 50.Forscher P., Kaczmarek L.K., Buchanan J.A., Smith S.J. Cyclic AMP induces changes in distribution and transport of organelles within growth cones of Aplysia bag cell neurons. J. Neurosci. 1987;7:3600–3611. doi: 10.1523/JNEUROSCI.07-11-03600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]