Abstract
The development of soluble recombinant peptide-major histocompatibility complex class I (pMHCI) molecules conjugated in multimeric form to fluorescent labels has enabled the physical quantification and characterization of antigen-specific CD8+ T cell populations by flow cytometry. Several factors determine the binding threshold that enables visualization of cognate CD8+ T cells with these reagents; these include the affinity of the T cell receptor (TCR) for pMHCI antigen. Here, we show that multimers constructed from peptide-human leukocyte antigen (pHLA) A*0201 monomers engineered in the heavy chain α2 domain to enhance CD8 binding (KD≈85 µM) without impacting the TCR binding platform can detect cognate CD8+ T cells bearing low affinity TCRs that are not visible with the corresponding wild type pHLA A*0201 multimeric complexes. Mechanistically, this effect is mediated by a disproportionate enhancement of the TCR/pMHCI association rate. In direct ex vivo applications, these coreceptor-enhanced multimers exhibit faithful cognate binding properties; concomitant increases in background staining within the non-cognate CD8+ T cell population can be resolved phenotypically using polychromatic flow cytometry as a mixture of naïve and memory cells. These findings provide the first validation of a novel approach to the physical detection of low avidity antigen-specific CD8+ T cell populations; such coreceptor-enhanced multimeric reagents are likely to be useful in a multitude of settings for the detection of auto-immune, tumor-specific and cross-reactive CD8+ T cells.
Keywords: Peptide-major histocompatibility complex, class I tetramer, Polychromatic flow cytometry, T cell
1. Introduction
In contrast to antibodies, T cell receptors (TCRs) typically bind cognate antigens with low affinities and rapid kinetics. These biophysical properties preclude the use of soluble recombinant peptide-major histocompatibility complex class I (pMHCI) molecules in monomeric form for cell labeling purposes. However, ligand multimerization overcomes these limitations and enables stable binding and internalization of pMHCI complexes by antigen-specific CD8+ T cells (Laugel et al., 2005;Whelan et al., 1999), thereby providing the basis for flow cytometric applications (Altman et al.,1996). Indeed, the ability to quantify and characterize antigen-specific CD8+ T cells by flow cytometry using fluorochrome-conjugated multimeric pMHCI complexes has transformed our understanding of cellular immune responses over the past decade (Klenerman et al., 2002).
The primary requirement for pMHCI multimer binding is that the second moiety associates with a cell surface TCR before the first moiety dissociates; thus, regardless of valency, stable multimer binding is a function of TCR/pMHCI monomeric affinity. In addition to TCR engagement, cognate pMHCI is also bound by the CD8 coreceptor at a structurally discrete and largely invariant site (Gao et al., 1997). At the monomeric level, TCR/pMHCI and pMHCI/CD8 interactions are kinetically distinct and independent in three dimensions (Wyer et al., 1999). However, binding cooperativity does occur in membrane-constrained planar interactions at the cell surface. Thus, CD8 coreceptor binding stabilizes the TCR/pMHCI interaction by a factor of approximately two-fold by slowing the dissociation rate (Wooldridge et al., 2005). Furthermore, CD8 binding also enhances the TCR/pMHCI on-rate (Gakamsky et al., 2005; Laugel et al., 2007). These coreceptor-mediated effects have been exploited recently for the selective detection of high avidity antigen-specific CD8+ T cell populations using multimeric forms of α3 domain-mutated pMHCI molecules that fail to bind CD8 yet retain TCR binding fidelity (Laugel et al., 2007; Pittet et al., 2003; Choi et al., 2003; Price et al., 2005).
In this study, we characterized the binding properties of tetrameric pMHCI molecules mutated in the α2 domain to engage CD8 with enhanced affinity without impacting the integrity of TCR binding and examined the utility of these novel reagents for the detection of low avidity antigen-specific CD8+ T cell populations directly ex vivo.
2. Results
2.1. Coreceptor-enhanced pMHCI tetramers stain CD8+ T cells with avidities for cognate antigen that lie below the threshold of detection with corresponding wildtype reagents
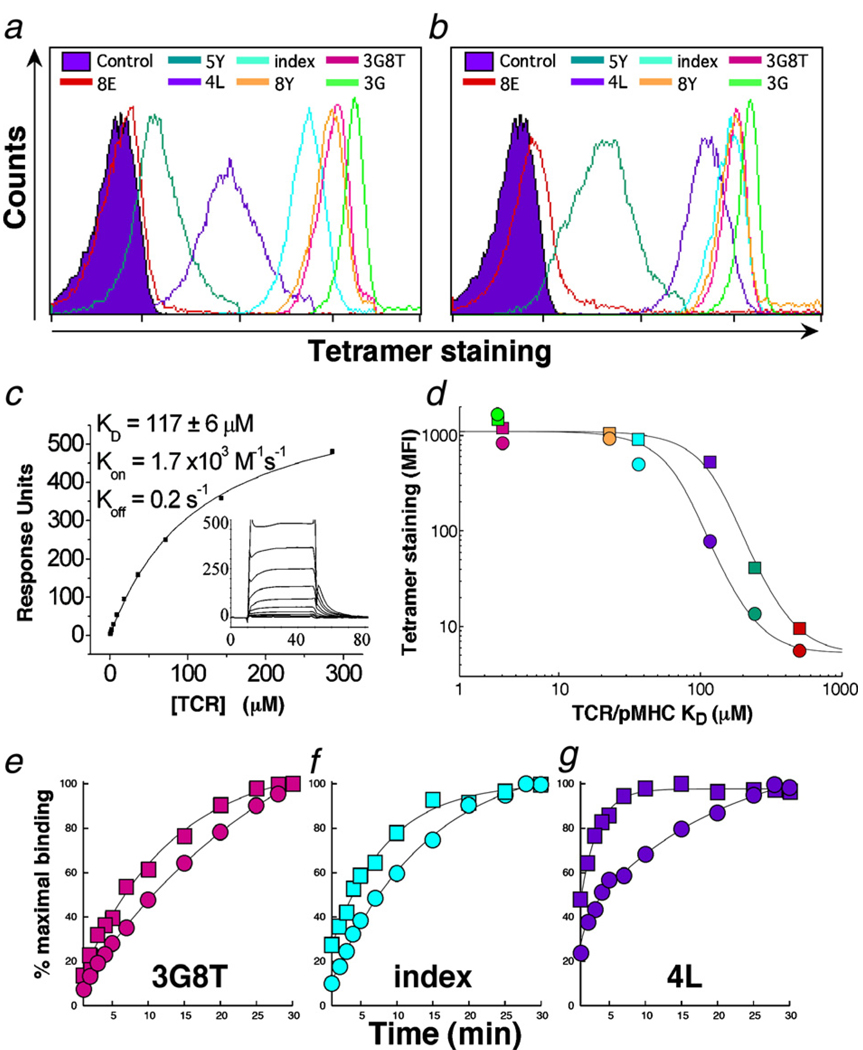
We previously demonstrated that the human leukocyte antigen (HLA) A*0201 heavy chain α2 domain Q115E mutation enhances equilibrium CD8 coreceptor binding (KD≈85 µM) without altering the cognate TCR binding platform(Wooldridge et al., 2005). To determine the binding threshold of wildtype and coreceptor-enhanced pHLA A*0201 tetramers with respect to monomeric TCR/pMHCI ligand affinity, we used a CD8+ T cell clone (ILA1) specific for the HLA A*0201-restricted human telomerase reverse transcriptase (hTERT) epitope ILAKFLHWL (residues 540–548) and studied TCR interactions with altered peptide ligand (APL) pHLA A*0201 complexes at both the molecular and cellular level (Laugel et al., 2007). Importantly, cellular factors that can influence pMHCI tetramer binding, such as TCR and coreceptor density, membrane flexibility and cell surface molecular topography, are standardized in this monoclonal system. As shown in Fig. 1, coreceptor-enhanced pHLA A*0201 tetramers refolded with APL that exhibited low monomeric TCR/pMHCI binding affinities in surface plasmon resonance (SPR) experiments (KD100 µM) stained ILA1 CD8+ T cells with greater intensities compared to the corresponding wildtype pHLA A*0201 tetramers. This effect was most marked with the 4L (KD=117 µM; Fig. 1c) and 5Y (KD=242 µM (Laugel et al., 2007)) APL/HLA A*0201 tetramers; no significant staining differences between wildtype and coreceptor-enhanced tetramers were observed with lower monomeric TCR/pMHCI affinities (KD500 µM) (Fig. 1a–d).
Fig. 1.
Tetrameric complexes of pHLA A*0201 containing the heavy chain α2 domain Q115E substitution bind low avidity CD8+ T cells. The ILA1 CD8+ T cell clone was stained with wildtype (a) and Q115E-substituted (b) pHLA*0201-PE tetramers bearing different APL at a final concentration of 10 µg/ml with respect to the monomeric pMHCI component. The staining protocol and APL binding/activation properties were described previously (Laugel et al., 2007). As reported, the TCR/pMHCI affinities measured by SPR for the APL interactions shown are as follows: 3G, 3.7 µM; 3G8T, 4.0 µM; 8Y, 22.6 µM; index, 36.6 µM; 5Y, 242.1 µM; 8E,>500 µM. The 3G, 3G8T and 8Y ligands act as superagonists in functional assays relative to index; 5Y and 8E are weak agonists relative to index. (c) Affinity and binding kinetics of the ILA1 TCR/4L HLA A*0201 interaction; SPR experiments were performed as described previously (Laugel et al., 2007). Standard deviation from the mean dissociation constant (KD) of 3 separate experiments is shown. (d) Staining intensities shown in (a) and (b) are plotted against TCR/pMHCI binding affinity (KD) for wildtype (circles) and Q115E (squares) pHLA A*0201 tetramers; APL color codes match those shown in a and b. Curves were fitted as described previously (Laugel et al., 2007). (e–g) Tetramer capture from solution by ILA1 CTL. Concentrations of tetramer were titrated in an attempt to achieve a maximum fluorescence intensity of ~200. This level of staining could not be achieved for the wildtype 4L ligand, and staining only reached an MFI of ~60 in this case. From left to right, panels show % maximal binding over 30 min for ILA1 CTL stained with 0.2 µg/ml 3G8T (e), 1 µg/ml ILAKFLHWL index (f) and 5 µg/ml 4L (g) wildtype (circles) and Q115E (squares) tetramers. Curves in panels (e–g) represent least squares fit to Eq. (20) in van den Berg et al. (2007).
2.2. The extended avidity threshold for coreceptor-enhanced pMHCI tetramer staining is mediated primarily by a disproportionate increase in TCR/pMHCI association rate
The CD8 coreceptor stabilizes TCR/pMHCI interactions at the cell surface by approximately two-fold through an effect on dissociation rate that is consistent across a range of ligand affinities (Wooldridge et al., 2005). Interestingly, the Q115E mutation enhances this stabilization effect at the level of TCR/pMHCI dissociation only minimally (2–5%) and exhibits only subtle effects in tetramer dissociation assays (Wooldridge et al., 2005). Thus, the observation that Q115E pHLA A*0201 tetramers extend the avidity threshold of detection for cognate CD8+ T cells by a factor of approximately two-fold at the level of the corresponding monomeric TCR/pMHCI affinity (Fig. 1a–d) indicates an effect mediated via the association rate. To test this possibility, we compared the rate at which ILA1 CD8+ T cells bound identical concentrations of wildtype and Q115E pHLA A*0201 tetramers from solution as described previously (Laugel et al., 2007). In all cases, Q115E pHLA A*0201 tetramers exhibited more rapid uptake in these association assays (Fig. 1e–g). Notably, however, this effect was more pronounced for APL/HLA A*0201 complexes with relatively low monomeric affinities for the ILA1 TCR (Fig. 1c,g). Thus, mechanistically, the enhanced detection of low avidity CD8+ T cells afforded by Q115E pHLA A*0201 tetramers is mediated primarily by a disproportionate increase in the TCR/pMHCI association rate.
2.3. Coreceptor-enhanced pMHCI tetramers retain cognate binding properties in mononuclear cell samples directly ex vivo
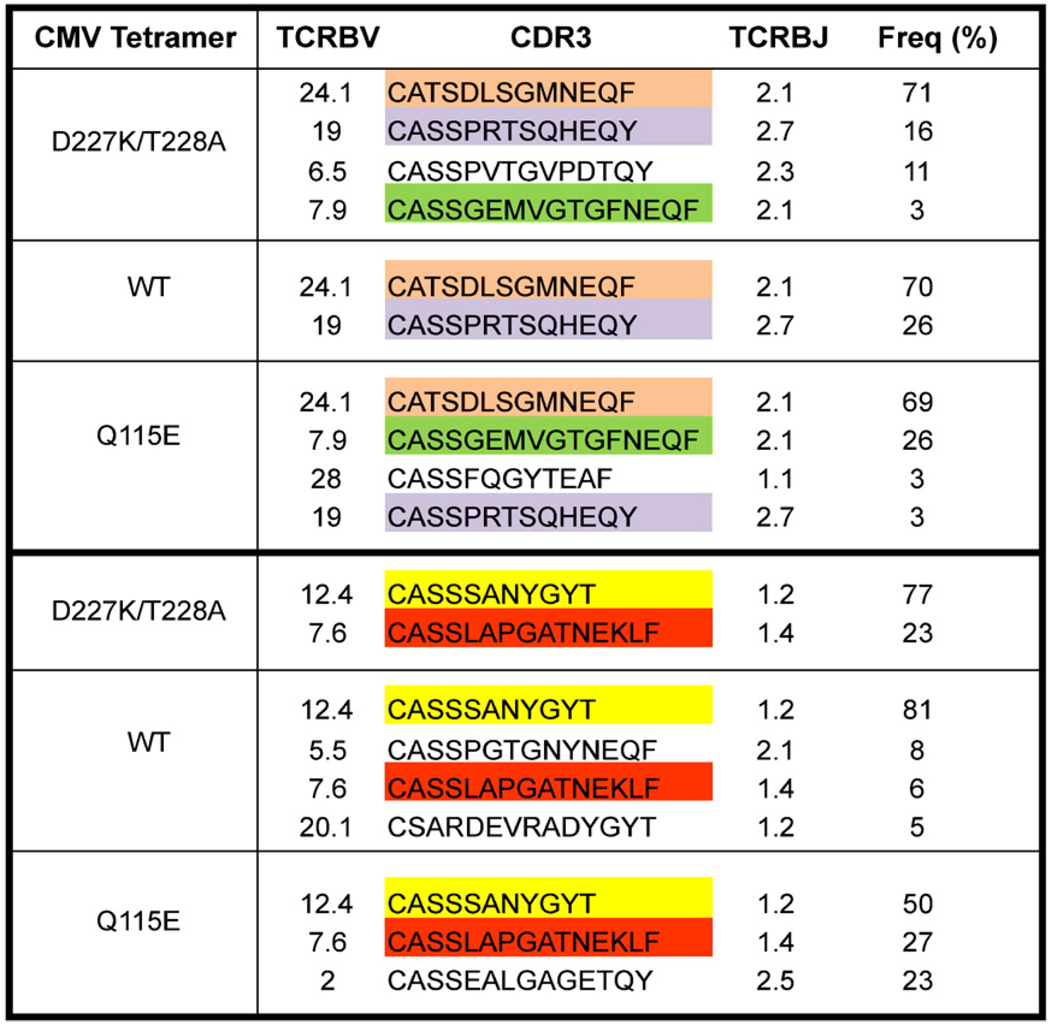
To test the validity of coreceptor-enhanced pMHCI tetramers in direct ex vivo applications, we produced D227K/T228A, wildtype and Q115E pHLA A*0201 tetramers refolded with viral and leukemia antigen-associated (LAA) peptide epitopes as described in the Materials and methods. Experiments were performed on mononuclear cell preparations isolated from both healthy donors (n=12) and individuals with myeloid leukemias (n=14); representative polychromatic flow cytometric data are shown in Fig. 2. In the vast majority of cases, CD8+ T cell populations specific for the CMV pp65495–503 epitope were defined in mononuclear cell preparations at comparable magnitudes with all three forms of the corresponding cognate tetramer (data not shown). This suggests that aberrant binding events are not introduced in mixed cell populations by the Q115E mutation and is consistent with previous studies showing that interclonal competition drives the selection of high avidity CD8+ T cell populations specific for persistent viral antigens (Price et al., 2005). To confirm this, we sorted equivalent CD8+ T cell populations specific for CMV pp65495–503 that were identified with the D227K/T228A, wildtype and Q115E tetramers directly ex vivo and conducted a molecular analysis of all expressed TCRB genes using an unbiased template-switch anchored RT-PCR as described previously (Douek et al., 2002; Price et al., 2004). In each of two donors studied, the constituent clonotypes within the respective tetramer-binding CD8+ T cell populations were almost identical (Fig. 3). Thus, Q115E pHLA A*0201 tetramers retain TCR binding specificity directly ex vivo in mixed cell populations.
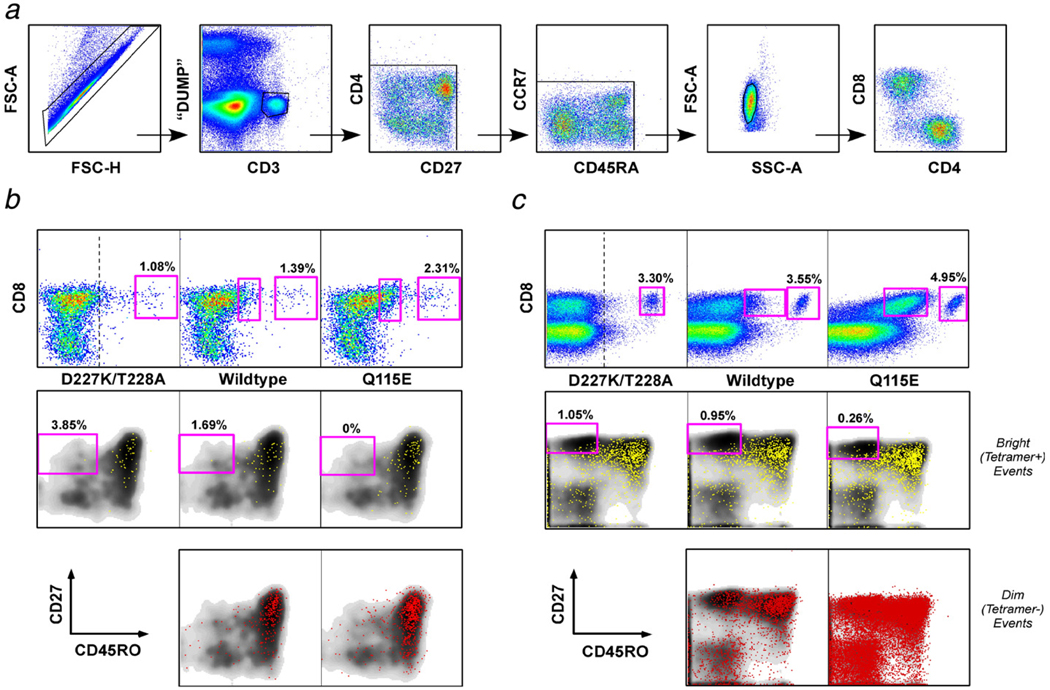
Fig. 2.
Coreceptor-enhanced pHLA A*0201 tetramers exhibit distinct staining characteristics with preservation of binding specificity directly ex vivo. (a) Successive panels show the gating strategy used to identify antigen-specific CD8+ T cells. First, doublets were distinguished from single cell events on the basis of their forward scatter-area vs. forward scatter-height profile. Live CD3+ T cells were then distinguished from dead cells (ViViD+), monocytes (CD14+) and B cells (CD19+). Subsequently, fluorochrome aggregates were gated out in two consecutive bivariate plots and lymphocytes were further identified according to forward scatter vs. side scatter profile. Histograms were then transformed at the level of the CD4 vs. CD8 plot. (b) Shown is the frequency (top panels) and phenotype (middle and bottom panels) of leukemia-associated antigen PR1-specific CD8+ T cells using D227K/T228A, wildtype and Q115E pHLA A*0201 tetramer analysis in a bone marrow sample from a patient with chronic myeloid leukemia 12 months after hematopoietic stem cell transplantation. Live CD3+ T cells were gated as shown in (a), and tetramer-positive events are displayed in a bivariate plot with CD8. The vertical bar in the D227K/T228A tetramer vs. CD8 plot serves as an orientation point for the positioning of the gate on tetramer-dim cells in subsequent panels. Tetramer-bright (yellow; middle panels) and tetramer-dim (red; bottom panels) CD8+ T cells are overlaid on a CD45RO vs. CD27 density plot for the total CD8+ T cell population; tetramer-bright CD8+ T cells lack naïve cells, whereas tetramer-dim CD8+ T cells are distributed across naïve and antigen-experienced phenotypes. This is most obvious for the Q115E pHLA A*0201 tetramer. (c) As for (b), except that the analysis shows CD8+ T cells specific for CMV pp65495–503 in the peripheral blood of a patient with acute myeloid leukemia. Staining was not observed with any of the three cognate pHLA A*0201 tetrameric forms in CMV seronegative controls (data not shown). In all experiments, tetrameric preparations were standardized for concentration and volume, such that conditions differed solely with respect to the pHLA A*0201/CD8 interaction.
Fig. 3.
Coreceptor-enhanced Q115E pHLA A*0201 tetramers retain cognate binding integrity directly ex vivo. PBMC from two CMV seropositive patients with acute myeloid leukemia were stained with D227K/T228A, wildtype (WT) and Q115E pHLA A*0201 tetramers refolded around the pp65495–503 peptide. Tetramer-positive cells gated as shown in Fig. 2a were sorted by flow cytometry to >98% purity and analyzed for TCRB gene expression as described in the Materials and methods. Shown are the CDR3 amino acid sequences, TCRBV and TCRBJ usage, and the relative frequency of CD8+ T cell clonotypes specific for CMV pp65495–503. Colored boxes denote identical clonotypes.
2.4. Modulation of CD8 coreceptor binding properties differentially influences inter-individual pMHCI tetramer staining patterns directly ex vivo
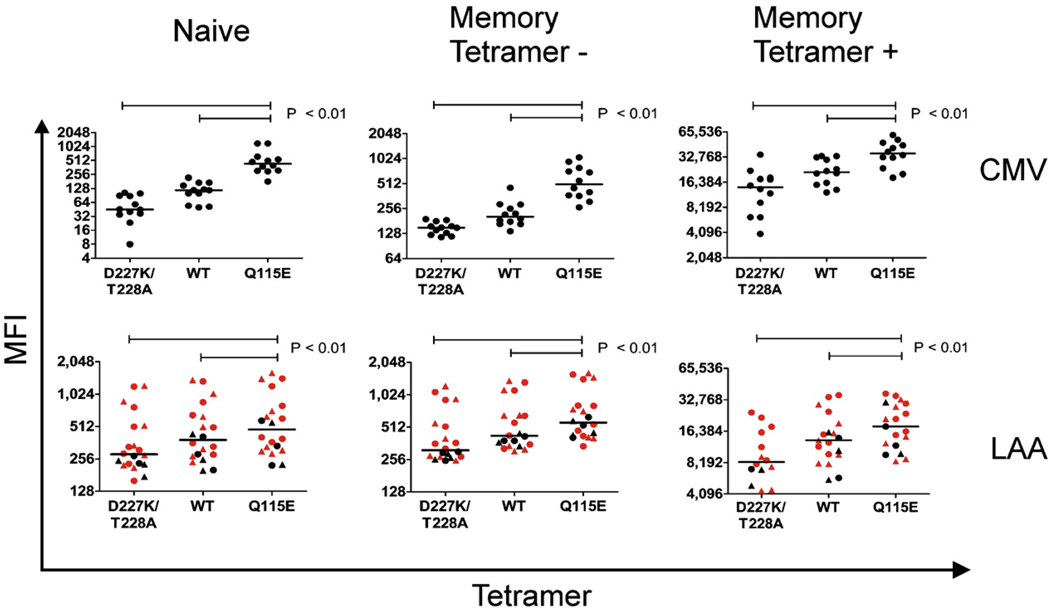
In further direct ex vivo experiments, we characterized differential tetramer staining patterns in multiple individuals according to both the affinity of the pMHCI/CD8 interaction and the nature of the cognate antigen; tetramer concentrations and volumes were standardized in all cases to enable direct comparisons limited solely to differences in CD8 binding properties within antigen specificities. Generically, Q115E pHLA A*0201 tetramers stained cognate antigen-specific CD8+ T cell populations with greater intensity compared to their wildtype and D227K/T228A counterparts (Fig. 2 and Fig.4). Enhanced tetramer uptake that corresponded with the affinity of the pMHCI/CD8 interaction was also observed within the non-cognate CD8+ T cell population as described previously (Wooldridge et al., 2005); however, this increased background staining comprised a mixture of memory and naïve CD8+ T cells, thereby providing a phenotypic basis for the accurate discrimination of true binding events (Fig. 2, Fig.4 and Fig.5). Interestingly, although these staining characteristics applied almost invariably, differences were apparent within individuals according to the nature of the antigen. Specifically, Q115E pHLA A*0201 tetramers refolded with LAA peptide antigens, especially PR1, exhibited substantially reduced background staining in some individuals compared to the corresponding CMV pp65495–503 tetramers (Fig. 4; data not shown). Further-more, substantial differences in background staining with the same Q115E pHLA A*0201 tetramer were apparent between individuals. Thus, while coreceptor binding properties clearly dictate tetramer staining patterns within both the cognate and non-cognate CD8+ T cell populations, both host factors and the nature of the TCR/pMHCI interaction also appear to play determinative roles.
Fig. 4.
Modulation of CD8 coreceptor binding properties differentially influences pMHCI tetramer staining patterns directly ex vivo according to antigen origin. Mean fluorescence intensity (MFI) of distinct populations identified in PBMC (black) and bone marrow mononuclear cells (BMMC; red) with D227K/T228A, wildtype (WT) and Q115E pHLA A*0201 tetramers refolded around CMV (pp65495–503) and LAA (PR1, circles; WT1, triangles) peptides. PBMC from 12 healthy donors were stained with the three CMV pp65495–503/HLA A*0201 tetramers; PBMC from 10 patients and BMMC from 8 patients with acute or chronic myeloid leukemias were stained with the six LAA/HLA A*0201 tetramers (only data from samples with detectable tetramer binding events are shown). The MFIs for three populations are shown in each case: (i) naïve CD8+ T cells (CD27+CD45RO−); (ii) memory CD8+ T cells (CD27+CD45RO+; CD27−CD45RO+; CD27−CD45RO−) that did not bind tetramer (tetramer −); and, (iii) memory CD8+ T cells that did bind tetramer (tetramer +). Higher MFI values are apparent in all three populations of CD8+ T cells stained with the various Q115E pHLA A*0201 tetramers compared to the corresponding D227K/T228A and wildtype tetramers. These effects applied to all antigen-specific CD8+ T cell populations, but were generally more pronounced with the CMV pp65495–503 tetramers. In all experiments, tetrameric preparations were standardized for concentration and volume, such that conditions differed solely with respect to the pHLA A*0201/CD8 interaction. Statistical comparisons were conducted using the Wilcoxon signed rank test.
Fig. 5.
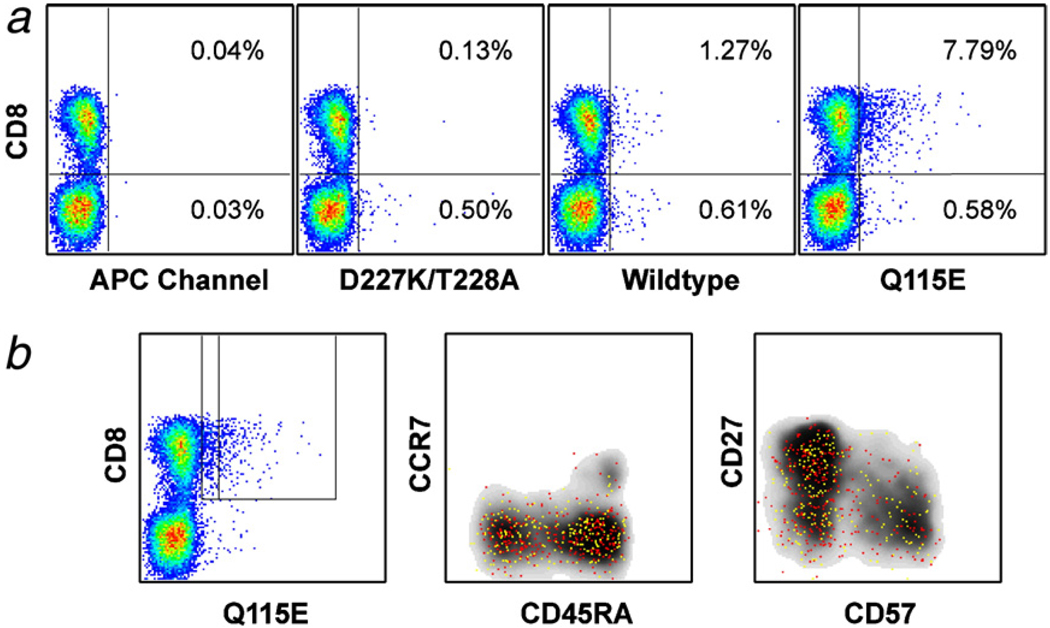
Coreceptor-enhanced pHLA A*0201 tetramers can identify low avidity antigen-specific CD8+ T cells that are not visible with the corresponding wildtype tetramer directly ex vivo. (a) Bone marrow mononuclear cells from a patient with acute myeloid leukemia were stained with PR1/HLA A*0201 tetramers. The frequencies of D227K/T228A, wildtype and Q115E PR1/HLA A*0201 tetramer-positive and background events, calculated within the CD8+ and CD8− populations respectively after gating as shown in Fig. 2a, are shown as a percentage of the total live CD3+ lymphocyte population. (b) Cells staining brightly (yellow) and dimly (red) with the Q115E PR1/HLA A*0201 tetramer in the experiment shown in (a) exhibit similar phenotypes based on expression of CCR7, CD27, CD45RA and CD57; the tetramer−positive events are almost exclusively CCR7− memory cells, although a few naïve events consistent with background staining are apparent as the gate is extended towards the CD8+ tetramer-negative population. The phenotypic distribution of these PR1-specific CD8+ T cells differs considerably from that of the overall CD8+ T cell population (gray density plot). In all experiments, tetrameric preparations were standardized for concentration and volume, such that conditions differed solely with respect to the pHLA A*0201/CD8 interaction.
2.5. Coreceptor-enhanced pMHCI tetramers enable direct ex vivo detection of low avidity antigen-specific CD8+ T cells that cannot be visualized under identical conditions with wildtype tetramers
In contrast to pathogen-specific responses, CD8+ T cell populations directed against tumor-derived epitopes tend to exhibit low avidities for antigen and bear TCRs with weaker monomeric affinities for pMHCI (Molldrem et al., 2003; Cole et al., 2007). Consistent with these properties, LAA-specific CD8+ T cell populations showed variable staining with different tetrameric reagents (Fig. 2, Fig.4 and Fig.5). Thus, while high avidity LAA-specific CD8+ T cells could be identified with D227K/T228A pHLA A*0201 tetramers in some individuals with myeloid leukemias, larger populations were generally defined with the corresponding wildtype and Q115E tetramers; a similar pattern was observed with CD8+ Tcells specific for CMV pp65495–503 in one donor (Fig. 2c). In some cases, this effect was most marked with the Q115E pHLA A*0201 tetramers, thereby indicating the existence of cognate CD8+ T cell populations below the threshold of detection with wildtype tetramers (Fig. 5).
3. Discussion
In this study, we investigated the properties of tetrameric pHLA A*0201 complexes mutated in the α2 domain of the heavy chain to engage CD8 with enhanced affinity without affecting the integrity of the TCR binding platform. The principal findings were: (i) coreceptor-enhanced Q115E pHLA A*0201 tetramers bind antigen-specific CD8+ T cells with avidities that lie below the detection threshold for wildtype tetramers through a disproportionate enhancement of the TCR/pMHCI association rate; (ii) these reagents retain their cognate binding properties in direct ex vivo applications and exhibit faithful identification of antigen-specific CD8+ T cell clonotypes within the memory pool; (iii) background staining of the CD8+ T cell subset is a function of coreceptor binding affinity and can be distinguished phenotypically as a mixture of naïve and memory cells; and, (iv) staining patterns are influenced both by host factors and by the nature of the antigen. These results provide validation of a novel approach to the physical detection of low avidity antigen-specific CD8+ T cell populations by polychromatic flow cytometry.
The general utility of coreceptor-enhanced pMHCI tetramers for the detection, quantification and characterization of low avidity antigen-specific CD8+ T cells is difficult to predict. Thus, while these reagents clearly lower the corresponding monomeric TCR/pMHCI affinity threshold for tetramer binding at the clonal level (Fig. 1), contemporaneous background effects in direct ex vivo specimens can mitigate the improved detection of low avidity antigen-specific CD8+ T cells. However, not all individuals and not all antigen specificities are equally affected by these increases in non-cognate staining with coreceptor-enhanced tetramers (Fig. 2 and Fig.4); such variability might reflect differences in the way that individual antigens interface with the available TCR repertoire. Furthermore, background staining can be reliably distinguished from cognate staining on the basis of phenotypic characteristics; thus, CD8+ T cells that exhibit non-specific uptake, which always occurs at lower fluorescence intensities compared to cognate binding, can be identified as a mixed population that comprises both naïve and memory cells. For these reasons, polychromatic flow cytometric platforms are desirable for the optimal use of coreceptor-enhanced pMHCI tetramers. Even with access to such technology, however, separation of true antigen-specific CD8+ T cells from back-ground events can be subtle (Fig. 5). In addition, although low affinity pMHCI ligands can elicit functional consequences in cognate CD8+ T cells (Laugel et al., 2007; Gagnon et al., 2006), low avidity CD8+ T cells that bind coreceptor-enhanced tetramers in an antigen-specific manner might not be readily amenable to functional verification. Indeed, in this study, the almost exclusively low avidity PR1-specific CD8+ T cell population shown in Fig. 5 exhibited some evidence of CD107a mobilization but failed to produce detectable levels of four distinct soluble factors, including IFNγ (data not shown); it remains to be determined to what extent such functional inertia reflects the nature of the targeted antigen and the conditions of encounter, as suggested previously (Lee et al., 1999; Zippelius et al., 2004), in relation to TCR/pMHCI binding properties. Nevertheless, it has also been established that CD8+ T cells can exhibit effector functions in the absence of detectable wildtype pMHCI tetramer binding (Laugel et al., 2007; Rubio-Godoy et al., 2001; Lyons et al., 2006), and it is conceivable that coreceptor-enhanced pMHCI tetramers might enable the physical identification of such CD8+ T cell populations.
In summary, we have demonstrated that coreceptor-enhanced pMHCI tetramers can be used in conjunction with polychromatic flow cytometric platforms for the identification, quantification and characterization of low avidity antigen-specific CD8+ T cells. Together with coreceptor-null pMHCI tetramers (Laugel et al., 2007; Pittet et al., 2003; Choi et al., 2003; Price et al., 2005; Purbhoo et al., 2001), these reagents allow for the rigorous physical assessment of antigen avidities within CD8+ T cell populations of defined specificity. Thus, despite the caveats elucidated above, it seems likely that the ability to detect low avidity CD8+ T cells will prove useful for the full evaluation of antigen-specific immune responses in several settings; these include auto-immune conditions, tumor immunity, heterologous phenomena and lymphopenic situations in which low avidity T cells expand during the process of homeostatic reconstitution (Goldrath and Bevan, 1999; Cho et al., 2000; Krupica et al., 2006; Selin et al., 2006; Kasprowicz et al., 2008).
4. Materials and methods
4.1. Cells
Leukapheresis and bone marrow samples were collected under protocols for allogeneic stem cell transplantation approved by the Institutional Review Board of the National Heart, Lung and Blood Institute. Mononuclear cells were isolated by standard Ficoll–Hypaque density gradient centrifugation prior to cryopreservation. Cells were thawed and rested for a minimum of 2 h at 37 °C/5% CO2 in Iscove's Modified Dulbecco's Medium (IMDM; Cambrex, Walkersville, MD) supplemented with 10% heat-inactivated human AB serum (Gemini Bio-Product, Woodland, CA), 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen, Carlsbad, CA) prior to staining. The ILA1 CD8+ T cell clone was generated and maintained as described previously (Laugel et al., 2007; Purbhoo et al., 2007).
4.2. Tetrameric pMHCI complexes
The compound α3 domain mutation D227K/T228A and the α2 domain mutation Q115E in the HLA A*0201 heavy chain have been described previously; these mutations abrogate and enhance CD8 binding respectively, without affecting the integrity of the TCR binding platform (Wooldridge et al., 2005; Purbhoo et al., 2001; Wooldridge et al., 2007). Coreceptor-null (D227K/T228A), wildtype and coreceptor-enhanced (Q115E) biotinylated monomeric pHLA A*0201 complexes were produced for the following epitopes as described previously (Wooldridge et al., 2005; Hutchinson et al., 2003): (i) CMV pp65495–503 (NLVPMVATV) (Diamond et al., 1997); (ii)WT1126–134 (RMFPNAPYL) (Gao et al., 2000; Oka et al., 2000); and, (iii) PR1 (proteinase 3, residues 169–177; VLQELNVTV) (Molldremet al.,1996). In addition, wildtype and coreceptor-enhanced pHLA A*0201 monomers were produced for hTERT540–548 (ILAKFLHWL) and APL variants thereof as listed in Fig. 1. To eliminate effects related to differential protein stability, all tetramers were prepared afresh from cryopreserved monomers by conjugation to fluorochrome-labeled streptavidin as described previously (Hutchinson et al., 2003). Tetramer concentration and volume were standardized for all experiments to enable direct comparisons limited solely to differences in CD8 binding properties.
4.3. Antibodies
The following directly conjugated monoclonal antibodies (mAbs) were purchased from commercial vendors: (i) αCD3 Cy7-allophycocyanin (Cy7APC) and αCD3 Cy7-phycoerytherin (Cy7PE) (BD Biosciences, San Diego, CA); (ii) αCD4 Cy5.5PE, αCD4 Pacific Blue, αCD8 Cy7APC, αCD8 Pacific Orange, αCD14 Pacific Blue, αCD19 Pacific Blue and αCD57 fluorescein isothiocyanate (FITC) (Caltag/Invitrogen, Burlingame, CA); (iii) αCD27 Cy5PE and αCD45RO PE (Beckman Coulter, Miami, FL). In-house conjugates were prepared from commercially available unconjugated mAbs (BD Biosciences) and fluorophores (Invitrogen) as follows: (i) αCD33 Pacific Blue and αCD34 Pacific Blue; (ii) αCD8 Quantum Dot (QD) 585, αCD45RA QD705 and αCD57 QD545. Further, αCCR7 (R&D Systems, Minneapolis, MN) was conjugated to Alexa 594 (Invitrogen/Molecular Probes, Eugene, OR). The amine reactive viability dye ViViD (Invitrogen/Molecular Probes) was used as a dead cell exclusion marker (Perfetto et al., 2006).
4.4. Flow cytometry
For experiments conducted directly ex vivo, thawed cells (1–2×106/experiment) were washed with phosphate-buffered saline (PBS) and stained with ViViD (Invitrogen/Molecular Probes) as described previously (Perfetto et al., 2006). After a further wash, the cells were stained with pre-titered APC-labeled pHLA A*0201 tetrameric complexes (typically 1 µg/test with respect to the monomeric pMHCI component) according to previously optimized protocols (Whelan et al., 1999). Cells were then washed again, stained for surface markers with pretitered mAbs for 15 min at room temperature, washed and fixed in 4% paraformaldehyde (Tousimis, Rockville, MD) for 10 min at room temperature prior to resuspension in 200 µl FACS buffer (PBS containing 2% FCS and 0.05% sodium azide) and acquisition using either a Canto II or a modified LSR II flow cytometer (BD Biosciences). A minimum of 2×105 events was collected for each condition. Compensation for spectral overlap was performed electronically using antibody capture beads (BD Biosciences) stained separately with the individual mAbs used in each experiment. Data were analyzed according to the gating scheme shown in Fig. 2a using FlowJo software (Treestar, San Carlos, CA). Tetramer binding experiments conducted with the ILA1 CD8+ T cell clone were acquired using a FACSCalibur flow cytometer and analyzed with Cell Quest software.
4.5. Clonotype analysis
Antigen-specific CD8+ T cells identified on the basis of pHLA A*0201 tetramer binding as described above were sorted to >98% purity using a modified FACSAria (BD Biosciences). A minimum of 1500 live CD8+ tetramer+ T cells was sorted for each experimental condition directly into collection tubes containing RNAlater (Ambion, Austin, TX). Clonotype analysis was performed using an unbiased template-switch anchored RT-PCR as described previously (Douek et al., 2002; Price et al., 2004).
4.6. Production of soluble recombinant ILA1 TCR, surface plasmon resonance and pMHCI tetramer association assays
The ILA1 TCR was produced in soluble recombinant form as described previously (Laugel et al., 2007; Purbhoo et al., 2007; Boulter et al., 2003). Surface plasmon resonance experiments and pHLA A*0201 tetramer association assays were conducted and analyzed as described previously (Laugel et al., 2007; van den Berg et al., 2007).
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, Vaccine Research Center, National Institute of Allergy and Infectious Diseases and the National Heart, Lung and Blood Institute. DAP is a Medical Research Council (UK) Senior Clinical Fellow. LW is a Wellcome Trust (UK) Clinical Intermediate Fellow.
Abbreviations
- APC
allophycocyanin
- APL
altered peptide ligand
- FITC
fluorescein isothiocyanate
- HLA
human leukocyte antigen
- MHC
major histocompatibility complex
- PE
phycoerythrin
- TCR
T cell receptor
References
- Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94. [PubMed] [Google Scholar]
- Boulter JM, et al. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003;16:707. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- Cho BK, Rao VP, Ge Q, Eisen HN, Chen J. Homeostasis-stimulated proliferation drives naive T cells to differentiate directly into memory T cells. J. Exp. Med. 2000;192:549. doi: 10.1084/jem.192.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EM, et al. High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J. Immunol. 2003;171:5116. doi: 10.4049/jimmunol.171.10.5116. [DOI] [PubMed] [Google Scholar]
- Cole DK, et al. Human TCR-binding affinity is governed by MHC class restriction. J. Immunol. 2007;178:5727. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- Diamond DJ, York J, Sun JY, Wright CL, Forman SJ. Development of a candidate HLA A0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751. [PubMed] [Google Scholar]
- Douek DC, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 2002;168:3099. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Gagnon SJ, et al. T cell receptor recognition via cooperative conformational plasticity. J. Mol. Biol. 2006;363:228. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Gakamsky DM, et al. CD8 kinetically promotes ligand binding to the T-cell antigen receptor. Biophys. J. 2005;89:2121. doi: 10.1529/biophysj.105.061671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GF, et al. Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature. 1997;387:630. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- Gao L, et al. Selective elimination of leukemic CD34(+) progenitor cells by cytotoxic T lymphocytes specific for WT1. Blood. 2000;95:2198. [PubMed] [Google Scholar]
- Goldrath AW, Bevan MJ. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity. 1999;11:183. doi: 10.1016/s1074-7613(00)80093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson SL, et al. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J. Biol. Chem. 2003;278:24285. doi: 10.1074/jbc.M300633200. [DOI] [PubMed] [Google Scholar]
- Kasprowicz V, et al. Defining the directionality and quality of influenza virus-specific CD8+ T cell cross-reactivity in individuals infected with hepatitis C virus. J. Clin. Invest. 2008;118:1143. doi: 10.1172/JCI33082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenerman P, Cerundolo V, Dunbar PR. Tracking T cells with tetramers: new tales from new tools. Nat. Rev. Immunol. 2002;2:263. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- Krupica T, Jr, Fry TJ, Mackall CL. Autoimmunity during lymphopenia: a two-hit model. Clin. Immunol. 2006;120:121. doi: 10.1016/j.clim.2006.04.569. [DOI] [PubMed] [Google Scholar]
- Laugel B, et al. Design of soluble recombinant T cell receptors for antigen targeting and T cell inhibition. J. Biol. Chem. 2005;280:1882. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- Laugel B, et al. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J. Biol. Chem. 2007;282:23799. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- Lee PP, et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat. Med. 1999;5:677. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- Lyons GE, et al. T-cell receptor tetramer binding or the lack there of does not necessitate antigen reactivity in T-cell receptor transduced T cells. Cancer Immunol. Immunother. 2006;55:1142. doi: 10.1007/s00262-005-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molldrem J, et al. Targeted T-cell therapy for human leukemia: cytotoxic T lymphocytes specific for a peptide derived from proteinase 3 preferentially lyse human myeloid leukemia cells. Blood. 1996;88:2450. [PubMed] [Google Scholar]
- Molldrem JJ, et al. Chronic myelogenous leukemia shapes host immunity by selective deletion of high-avidity leukemia-specific T cells. J. Clin. Invest. 2003;111:639. doi: 10.1172/JCI16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, et al. Human cytotoxic T-lymphocyte responses specific for peptides of the wild-type Wilms' tumor gene (WT1) product. Immunogenetics. 2000;51:99. doi: 10.1007/s002510050018. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, et al. Amine reactive dyes: an effective tool to discriminatelive and dead cells in polychromatic flowcytometry. J. Immunol. Methods. 2006;313:199. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pittet MJ, et al. alpha3 domain mutants of peptide/MHC class I multimers allow the selective isolation of high avidity tumor-reactive CD8 T cells. J. Immunol. 2003;171:1844. doi: 10.4049/jimmunol.171.4.1844. [DOI] [PubMed] [Google Scholar]
- Price DA, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Price DA, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2005;202:1349. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbhoo MA, et al. The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor zeta chain. J. Biol. Chem. 2001;276:32786. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- Purbhoo MA, et al. The HLA A*0201-restricted hTERT(540–548) peptide is not detected on tumor cells by a CTL clone or a high-affinity T-cell receptor. Mol. Cancer Ther. 2007;6:2081. doi: 10.1158/1535-7163.MCT-07-0092. [DOI] [PubMed] [Google Scholar]
- Rubio-Godoy V, et al. Discrepancy between ELISPOT IFN-gamma secretion and binding of A2/peptide multimers to TCR reveals interclonal dissociation of CTL effector function from TCR-peptide/MHC complexes half-life. Proc. Natl. Acad. Sci. U S A. 2001;98:10302. doi: 10.1073/pnas.181348898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin LK, et al. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol. Rev. 2006;211:164. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg HA, Wooldridge L, Laugel B, Sewell AK. Coreceptor CD8-driven modulation of T cell antigen receptor specificity. J. Theor. Biol. 2007;249:395. doi: 10.1016/j.jtbi.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan JA, et al. Specificity of CTL interactions with peptide-MHC class I tetrameric complexes is temperature dependent. J. Immunol. 1999;163:4342. [PubMed] [Google Scholar]
- Wooldridge L, et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J. Biol. Chem. 2005;280:27491. doi: 10.1074/jbc.M500555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, et al. Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur. J. Immunol. 2007;37:1323. doi: 10.1002/eji.200636765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyer JR, et al. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity. 1999;10:219. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- Zippelius A, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Res. 2004;64:2865. doi: 10.1158/0008-5472.can-03-3066. [DOI] [PubMed] [Google Scholar]