Abstract
Protein tyrosine kinases are critical cell signaling enzymes. These enzymes have a highly conserved Arg residue in their catalytic loop which is present two residues or four residues downstream from an absolutely conserved Asp catalytic base. Prior studies on protein tyrosine kinases Csk and Src revealed the potential for chemical rescue of catalytically-deficient mutant kinases (Arg to Ala mutations) by small diamino compounds, particularly imidazole, however the potency and efficiency of rescue was greater for Src. This current study further examines the structural and kinetic basis of rescue for mutant Src as compared to mutant Abl tyrosine kinase. An X-ray crystal structure of R388A Src revealed the surprising finding that a histidine residue of the N-terminus of a symmetry-related kinase inserts into the active site of the adjacent Src and mimics the hydrogen bonding pattern seen in wild-type protein tyrosine kinases. Abl R367A shows potent and efficient rescue more comparable to Src, even though its catalytic loop is more like that of Csk. Various enzyme redesigns of the active sites indicate that the degree and specificity of rescue is somewhat flexible, but the overall properties of the enzymes and rescue agents play an overarching role. The newly discovered rescue agent 2-aminoimidazole is about as efficient as imidazole in rescuing R/A Src and Abl. Rate vs. pH studies with these imidazole analogs suggest that the protonated imidazolium is the preferred form for chemical rescue, consistent with structural models. The efficient rescue seen with mutant Abl points to the potential of this approach to be used effectively to analyze Abl phosphorylation pathways in cells.
Protein tyrosine kinases (PTKs1) are key enzymes in cell signaling and play roles in a wide range of diseases including cancer (1, 2). Several PTKs, including Abl, are targeted clinically with therapeutic agents, and others are the subject of pharmaceutical development (3). As members of the protein kinase superfamily, PTKs share conserved sequences and folds (2), and yet they show distinct substrate selectivity (4, 5) and modes of regulation (6). Evidence suggests that the mechanism of PTK-catalyzed phosphoryl transfer is dissociative, in which the bond to the leaving group ADP is largely broken prior to tyrosine phenol attack on the gamma phosphorus of ATP (7). The alignment, orientation, and spacing of the tyrosine and ATP substrates by key active site residues are believed to be crucial in facilitating catalysis based on structural (8), mutagenic (9), and kinetic (10) studies.
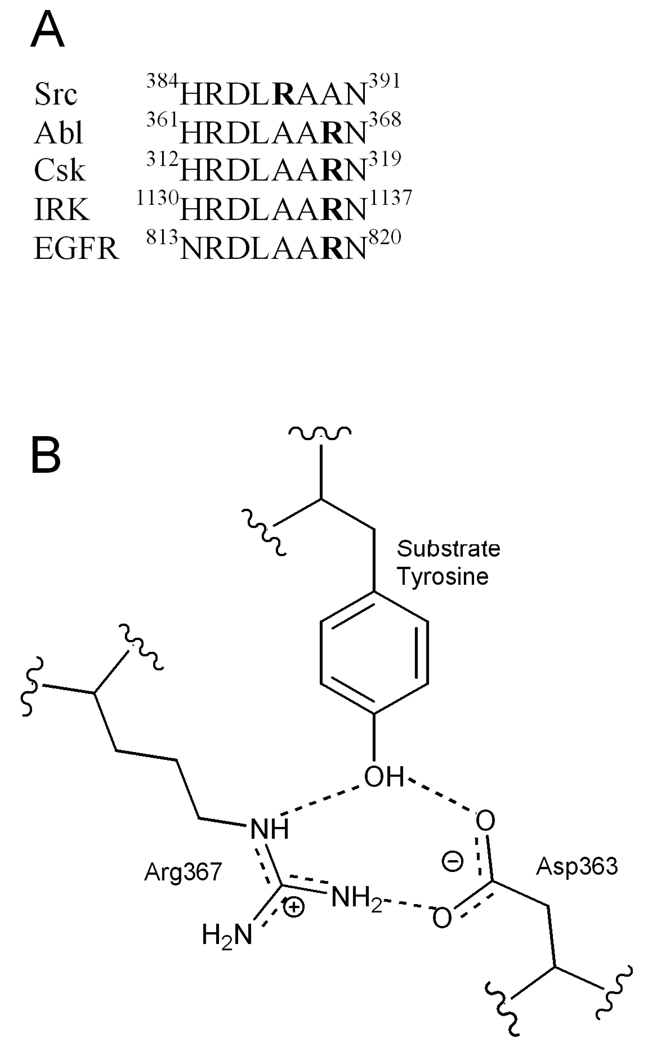
The catalytic loop sequence, DLAARN, is conserved throughout the vast majority of the 90 human PTKs, but differs in the nine members of the Src family, which have the sequence DLRAAN (Figure 1A) (2). This relocation of the Arg from the D+4 position in most PTKs to the D+2 position in the Src family suggests an unusual functional plasticity. A crystal structure of an insulin receptor tyrosine kinase-bisubstrate analog complex is believed to capture the active conformation of the enzyme and reveals the orientation of the active site residues (8). This structure shows that the side-chain of the catalytic loop Asp makes a hydrogen bond to the tyrosine hydroxyl and likely serves as the catalytic base. The catalytic loop Arg side-chain makes apparent hydrogen bonds to this Asp side-chain, as well as the substrate phenol oxygen. This triangle of hydrogen bonds (Figure 1B) appears to provide a scaffold that aligns the reactants. It is assumed that the Src Arg residue, located in the non-canonical D+2 position, fulfills a similar role to that of the catalytic loop Arg in the D+4 position in non-Src PTKs.
Figure 1. Catalytic Loop of Selected Kinases.
A) Amino acid sequence of the catalytic loop region for Src (residues 384–391), Abl (residues 361–368), Csk, insulin receptor tyrosine kinase (IRK), and epidermal growth factor receptor kinase (EGFR). The D+2 Arg and D+4 Arg that are examined here are in bold. B) Schematic of the interaction of the catalytic loop residues corresponding to Asp363 and Arg367 in Csk with the substrate tyrosine, as observed in the crystal structure of inhibited IRK.
Mutagenic analysis of the catalytic loop Arg has been performed previously on PTK Csk (11) and Src (12). Mutation of the D+4 Arg in PTK Csk to Ala (R318A Csk) resulted in a dramatic kcat drop of 3000-fold with relatively small (<10-fold) Km changes. A Csk double mutant was prepared to mimic the Src-like catalytic loop (D+2 Arg Csk), and this mutant showed a 150-fold increased kinase activity compared with R318A Csk (11). While still 20-fold less than wild type, the substantial kinase activity of D+2 Arg Csk mutant bolsters the argument for functional plasticity of the catalytic loop Arg residue.
In some cases, enzyme active site point mutants can be chemically rescued by small molecules that complement the deleted side-chains (13–15). Indeed, chemical rescue of R318A Csk with a series of diamino compounds (guanidiniums, amidiniums, imidazoles) could restore substantial activity to this mutant (11). Imidazole was the most efficient R318A Csk rescue agent, showed Km = 20 mM, and stimulated kinase activity up to the level of D+2 Arg Csk. Related studies were carried out on Src, and it was shown that mutation of the D+2 Arg in Src to Ala (R388A Src) is 200-fold less active than wild type (12). A Src double-mutant that mimics the canonical catalytic loop (D+4 Arg Src) was 2-fold more active than wild type. Imidazole rescued R388A Src to a level of 50% of wild type, with a Km = 5 mM.
The ability to rescue R318A Csk and R388A Src to 5–50% of wild-type kcat values with a non-toxic concentration of imidazole (< 50 mM) raised the interesting possibility that it might be feasible to complement these mutant tyrosine kinases in cells. Indeed, it was demonstrated that imidazole could restore Csk and Src functionality to mouse embryonic fibroblasts expressing replacement Arg → Ala mutant kinases (12, 16). Using this chemical rescue approach, new insights into the rapid signaling events have been obtained for Src and Csk (12, 17, 18).
Although progress on the chemical rescue of mutant Src and Csk has been made, questions remain. Does the imidazole occupy an active site position mimicking the position of the catalytic loop Arg side-chain? Are Arg → Ala mutant PTKs beyond Src and Csk likely to show more efficient chemical rescue, like Src, or less efficient rescue, like Csk? Can alternative potent rescue combinations of kinase mutations and small molecules be identified? Is the imidazole interacting with the mutant kinase as the imidazolium or the neutral species? Here we use a combination of X-ray crystallography, and extended mutagenic and kinetic experiments on Abl and Src, to address these questions.
Experimental Procedures
Rosetta Design Calculations
The crystal structure of the insulin receptor tyrosine kinase (PDB code 1IR3) (19) was used as the scaffold for design calculations. Because the binding site for the small molecules used for chemical rescue was unknown, atoms comprising the guanidinium group of the mutated arginine residue (corresponding to R388 in chicken c-Src) were converted into a fixed ligand. Alternate amino acids were selected at positions corresponding to W248 and E454 in chicken c-Src. Side-chain conformational freedom was modeled with a backbone-dependent rotamer library (20) supplemented by extra sample points with χ1 and χ2 values ±1 standard deviation in the observed torsional distributions. A Monte Carlo optimization algorithm was used to identify sequences and conformations that minimize the Rosetta full-atom energy potential of the complex (21).
Cloning, Expression and Purification
Src and Abl variants were generated following QuikChange protocols, with the addition of 2% DMSO to the reaction mixture. The targeted mutagenesis was confirmed by DNA sequencing.
Src variants used in enzymatic assays were expressed as previously described (12). These preparations consisted of residues 85–533 of chicken c-Src. Double and triple mutants of R388A Src plus mutations at position 454 to alanine, lysine, or arginine and/or at position 428 to glutamate or glutamine resulted in very low expression levels and were not purified. Double mutants of R388A Src plus mutations at 428 to tyrosine, phenylalanine, and alanine were purified as previously described (12).
For the purposes of crystallization, the R388A mutant of the kinase domain of chicken c-Src (R388A SrcKD; residues 251–533)(22) was expressed and purified in E. coli. R388A SrcKD was purified fused to an N-terminal TEV-cleavable His6-tag, which results in a Gly-His-Met sequence before the start of the kinase domain sequence. The identity of the purified protein was confirmed by mass spectrometry (David King, Howard Hughes Medical Institute, Berkeley) and were found to be of the calculated mass without any posttranslational modification.
The previously described protocol for expression and purification of the Abl kinase domain (AblKD; residues 229–511 of human c-Abl ) (23) was modified for the variants described here. By coexpressing AblKD with the catalytic domain of YopH phosphatase, instead of the full-length phosphatase, co-purification of YopH and AblKD was mitigated. Therefore, only a single purification step with a Ni affinity column was performed. DTT (2 mM) was added to all purification steps and the histidine-tag was not cleaved prior to kinetic measurements.
The catalytic domain of YopH phosphatase (corresponding to residues 164–468) was subcloned with Pfu Turbo polymerase and the primers 5'-AATTATCCATGGGCCGTGAACGACCACA-3' and 5'-ACCCGCGGATCCTTAGCTATTTAATAATGGTCGC-3'. pCDFDuet and the YopH PCR product were digested with NcoI and BamHI endonucleases. After dephosphorylation of digested pCDFDuet, both the vector and gene fragments were gel-purified (Qiaquick) and ligated together with T4 ligase. The final product was confirmed by DNA sequencing.
Synthesis of Biotin-Abltide
Abltide (EAIYAAPFAKKK) was synthesized using the Fmoc strategy on 0.2 mmol wang resin on a Rainin PS-3 peptide synthesizer. The DMF solvent was exchanged for NMP, and 60 mM of biotin-oNP was allowed to couple overnight in the presence of 20 mM HOBT at room temperature on a rotator. A second 2 hour coupling with 60 mM biotin-oNP and 20 mM HOBT was performed before cleavage of the peptide from the resin with 95% TFA and 2.5% triisopropylsilane. After ether precipitation, the crude peptide was purified by reverse-phase HPLC on a C18 column. The peptide was eluted with a linear gradient (acetonitrile/water + 0.05% TFA) and lyophilized to dryness. Pure biotin-Abltide was dissolved in water and the pH adjusted to neutral with NaOH. The mass was confirmed by MALDI-TOF and the concentration was determined by quanitative amino acid analysis at the Harvard Microchemistry and Proteomics Analysis Facility (Cambridge, MA).
Enzyme Kinetics
Three kinase assay methods were used to study the kinetics of the Src and Abl variants. All Src activity measurements were made using radiolabelled γ32P-ATP and poly[Glu,Tyr] as the peptide substrate, with separation on SDS-PAGE as described previously (12). Phosphorylation of biotin-Abltide by Abl was detected spectrophotometrically as described in (24, 25) or using radiolabelled ATP and selective binding to Avidin following the method in (26). The coupling enzymes used in the spectrophotometric assay, pyruvate kinase and lactate dehydrogenase, were supplied by Sigma as ammonium sulfate suspensions.
Michaelis constants (Km) and turnover numbers (kcat) were determined by fixing the concentration of one substrate at ≥ 3×Km and varying the concentration of the second substrate and fitting to equation 1.
| (1) |
Activators, such as imidazole, were treated as substrates, and activator-specific constants were derived from equation 1, as well.
Crystallization of R388A SrcKD
R388A SrcKD was crystallized using the hanging drop vapor diffusion method by mixing 1 µL of protein at 6 mg/mL in 20 mM Tris pH 8.0, 100 mM NaCl, 5% glycerol, 1 mM DTT, 50 mM imidazole, 500 µM PP2 (4-Amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo[3,4-d]pyrimidine; CalBiochem) with 1 µL of 50 mM imidazole, 500 µM PP2, 20 % PEG4000, 50 mM NH4Ac and 100 mM Bis-Tris pH 5.5 over a 500 µL reservoir of precipitant buffer. Rectangular crystals grew at room temperature within 8 hours to 350µm × 100µm × 50µm. Crystals were cryoprotected in mother liquor plus 20% glycerol and flash frozen in liquid nitrogen.
Crystal Data Collection and Refinement
X-ray diffraction data were collected over 180° in 180 frames with 1° oscillation at beamline 8.3.1 at the Advanced Light Source (ALS, Lawrence Berkeley National Laboratory). Reflections were indexed and merged in P21 with unit cell dimensions of a = 59.65 Å b = 63.46 Å c = 83.86 Å α = 90° β = 91.45 ° γ = 90° using DENZO and data were scaled with scalepack via the HKL2000 interface (27). The structure was solved by molecular replacement using the kinase domain of human c-Src of PDB entry 1Y57 (28) (residue 260–520) minus the C-helix (residues 298–310) and the activation loop (residues 400–425) as the template in Phaser (29) of ccp4i (30). The structure was built in O (31) and Coot (32) and refinement of the structure was straightforward in CNS (33).
Results
Overall structure of the mutant kinase is conserved
Protein kinases have been shown to yield crystals more easily and form better ordered crystals when complexed with ligands that bind to the kinase active site. Therefore we decided to use the non-specific Src kinase family inhibitor, PP2, which inhibits Src with nanomolar affinity, to improve crystallization. The protein crystallized readily in a variety of conditions: for some conditions within minutes of mixing the mother liquor with the protein solution. However, the fast crystallization yielded unusable groups of clustered crystals, and we screened further buffer conditions to control crystallization onset and growth speed (see Experimental Procedures). Data were recorded at ALS beamline 8.3.1 (Lawrence Berkeley National Lab) and crystals diffracted X-rays to 2.2 Å The structure was solved by molecular replacement (Table 1 and Experimental Procedures) with two molecules of R388A SrcKD per asymmetric unit and each kinase in complex with one molecule of PP2.
Table 1.
Data Collection and Refinement Statistics for R388A SrcKD Structure Determination
| Data Collection | Average (highest resolution shell) |
|---|---|
| Beamline | ALS 8.3.1 |
| Resolution (Å) | 50 − 2.2 (2.28 − 2.2) |
| Space group | P21 |
| Unit cell parameters (Å) | a = 59.65, b = 63.46, c = 83.86 |
| α = γ = 90° β = 91.45° | |
| Content of the assymteric unit | two kinase domains, two molecules of PP2 |
| Measured reflections (#) | 110199 |
| Unique reflections (#) | 31912 |
| Data Redundancy | 3.5 |
| Data completeness (%) | 98.7 (93.9) |
| Rsym (%)a | 9.4 (48.9) |
| I/sig | 13.5 (2.3) |
| Refinement | |
| R factor / R free | 22.6 % / 27.3 % |
| Number of protein atoms | 4356 |
| Number of non-protein atoms | 282 |
| Rmsd bond length (Å) | 0.0076 |
| Rmsd bond angle | 1.2° |
| Rmsd B factors (Å2) (mainchain / side-chain) | 1.5 / 2.1 |
| PDB entry | 3GEQ |
Rsym = Σ|I-<I>|/ ΣI, where I is the observed intensity of a reflection, and <I> is the average intensity of all the symmetry related reflections
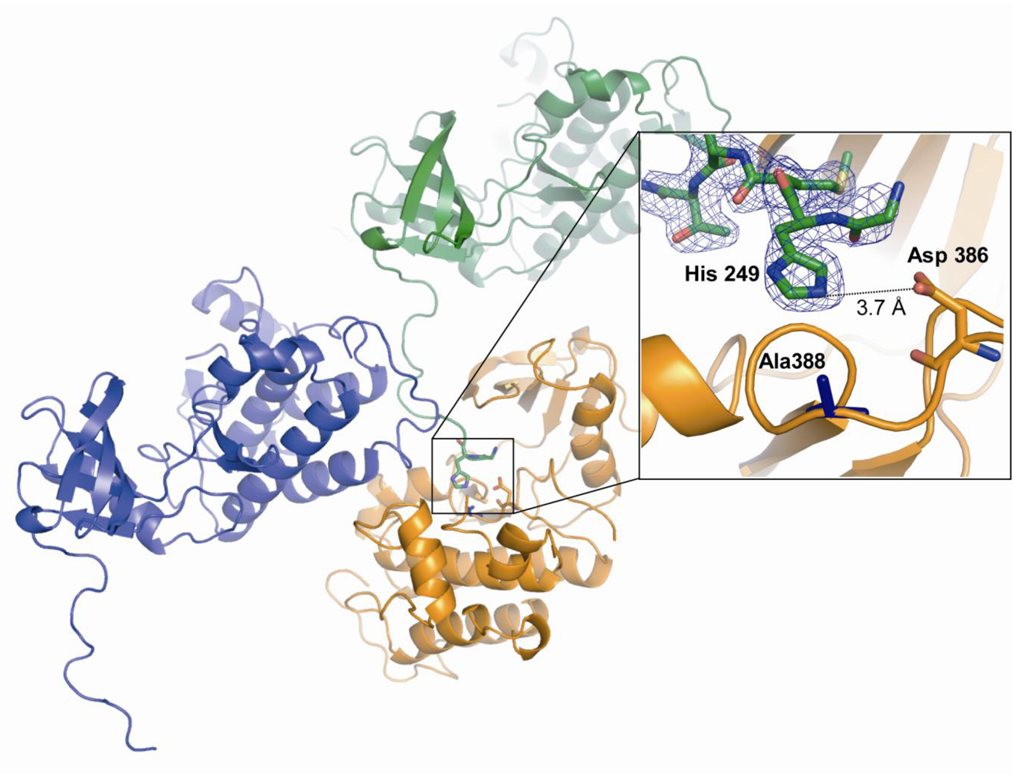
The kinase domains show the canonical fold of the α/β-N-lobe and the α-helical C-terminal lobe (Figure 2) with the high affinity ligand PP2 bound to the nucleotide binding pocket as seen before for Lck kinase (34). The two kinase domains in the asymmetric unit are virtually identical and align onto each other with a Cα RMSD of 0.241 Å R388A SrcKD and the kinase domain of the molecular replacement search template, human Src (PDB entry 1Y57) align with a Cα RMSD of 0.8 Å when the residues N-terminal of W260 are omitted from the RMSD calculation. The salt bridge between D310 of the C-helix and K295 is formed in R388A SrcKD, which is the hallmark of the catalytically active kinase. The activation loop in R388A SrcKD is disordered between residues 412 and 424 as is often seen in tyrosine kinases without activation loop phosphorylation.
Figure 2. Extension of N-terminus of One R388A SrcKD Molecule into the Active Site of Another.
The relative orientation of three kinase molecules are shown as observed in the crystal. The N-terminal kinase domain-SH2 domain linker extends for 27 Å inserting the side-chain of His249 (green) deep into the active site of another molecule (orange), complementing the cavity created by the R388A mutation (pop-out).
Structure of the site of mutation shows binding of N-terminal histidine
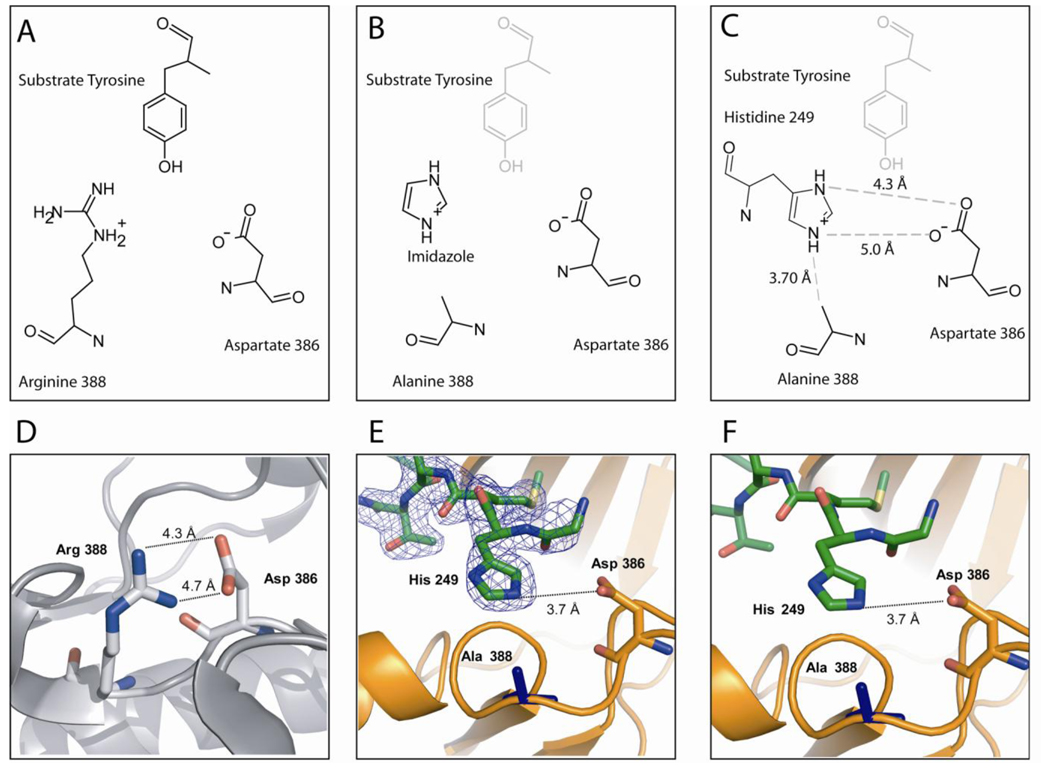
Based on the model described previously for the chemical rescue of R388A Src (Figure 3B) (12), we expected to see electron density for imidazole in the vicinity of the side-chain of residue 388 since it was present as 50 mM in the crystallization solution. Surprisingly, there was not only density to accommodate imidazole but a continuous stretch of electron density extended from the expected imidazole binding site to the N-terminus of a symmetry-related molecule (Figure 3E and Figure 2). We were able to build the N-terminus of the kinase up to the very first residue of the expressed and purified construct and found that the histidine side-chain of the second residue in our construct occupies the expected position of the imidazole (Figure 3C). Whereas the N-terminal SH2-kinase domain linker in 1Y57 (residues 245–260) is bound to the SH3 domain, it is extended in the R388A SrcKD mutant (Figure 2). This is all the more surprising as the first two residues of our construct (Gly and His) are non-native; they are part of the TEV recognition sequence for the cleavable affinity tag.
Figure 3. Occupation of Histidine in R388A SrcKD Active Site.
(A) Orientation of the catalytic residues and substrate tyrosine in the structure of insulin receptor kinase (PDB entry 1IR3; Src numbering) (19). (B) Model for chemical rescue of R388A Src by imidazole as proposed by Qiao et al.(12). The protonated imidazolium fulfills the hydrogen bonding network between the catalytic residues and the susbtrate tyrosine. (C) Schematic drawing of the coordination of His249 by the active site residues. (D) Distances between the active site residues of wild-type Src in the absence of substrate peptide (PDB entry 1Y57). (E, F) Structure of R388A SrcKD with electron density of the 2F0-Fc contoured to 1σ. The His249 residue of a symmetry related molecule (green) inserts into the active site of a kinase (orange). The distances for this structure are virtually identically to the distances in the wild-type Src structure, which is also free of susbstrate peptide.
We were surprised that the kinase domain would be able to accommodate the N-terminal peptide in a mode that has not been seen for substrate peptides before and which is almost certainly not physiological for a number of reasons: (1) the intruding N-terminus occludes the ATP binding site which is only partially occupied by PP2; (2) the kinase domain construct used in this study is preceded by bulky SH3 and SH2 domains in the full length Src kinase; and (3) dynamic light scattering and size exclusion chromatography showed that R338A SrcKD is monomeric in solution at concentrations of up to 1 mg/ml, a concentration that almost certainly exceeds the cellular concentration of Src.
This cloning and crystallization artifact is very helpful in determining the binding mode of imidazole. The fact that histidine takes the position of the imidazole reduces the number possible rotations of the imidazole ring from 5 for imidazole to 2 for the histidine (180° rotation around the Cα − Cβ bond). We found that the most favorable side-chain rotation of His 249 that fit the electron density yielded distances between the imidazolium nitrogens and the oxygen atoms of the catalytic aspartate side-chain D386 comparable to R388 – D386 distances in wild-type Src structures (Figure 3). Note that these distances are longer than those in the IRK ternary complex and presumably tighten up when peptide substrate binds. In any case, this structure implicitly suggests a natural affinity for the imidazole in the R388A mutant of Src.
Investigation of the catalytic loop Arg367 residue in Abl
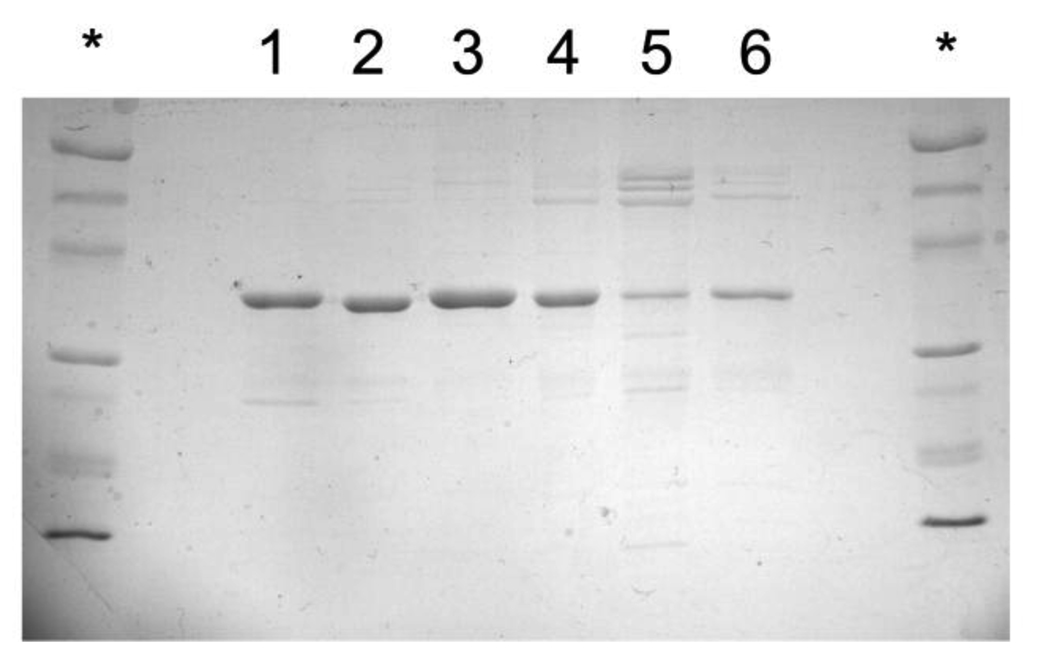
Generation of wild type and mutant Abl kinase domains was performed using a modified E. coli expression system which overcomes host toxicity by co-expressing a tyrosine phosphatase (YopH) (23). One technical improvement over the original method was to use a truncated YopH construct that shows reduced affinity to Ni-resin, and is therefore more readily separated during this purification step. Purity of Abl kinase mutants is shown in Figure 4.
Figure 4. SDS-PAGE of Purified AblKD Enzyme Variants.
Molecular weight marker indicated by asterisk. Numbered lanes are 1) wild-type AblKD, 2) R367A, 3) A365R + R367A, 4) R367A + W428Y, 5) R367G, 6) A365G + R367A.
Kinetic analyses of wild type Abl and Abl mutants were performed with an efficient Abl peptide substrate (Abltide, biotin-EAIYAAPFAKKK) and a direct, 32P-ATP incorporation quenched assay (26) as well as a continuous (spectrophotometric) coupled assay (25). Both assay methods gave similar catalytic parameters. As shown in Table 2, the mutation of the D+4 Arg to Ala (R367A AblKD) showed a reduction in catalytic efficiency (kcat/Km = 35 M−1s−1) of about 5000-fold compared with the wild-type enzyme (kcat/Km 1.9×104 M−1s−1). Given the slow kinase rate of R367A AblKD, it was not possible to accurately determine the Km values for this mutant. Interestingly, the R367A/A365R AblKD double mutant showed a kcat/Km (80,000 M−1s−1) value within two-fold of wild-type AblKD, suggesting excellent compensation by moving the Arg to the D+2 position seen in Src.
Table 2.
Kinetic Constants of Abl variantsa
| [Imidazole] (mM) |
Abltideb | ATPc | |||||
|---|---|---|---|---|---|---|---|
| kcat(s−1) | Km(µM) | kcat/Km(M−1 s−1) | kcat(s−1) | Km(µM) | kcat/Km(M−1 s−1) | ||
| wild-type AblKD | 0 | 12 ± 1 | 60 ± 20 | 190,000 ± 50,000 | 9.9 ± 0.2 | 45 ± 3 | 220,000 ± 20,000 |
| 40 | 12.7 ± 0.9 | 38 ± 9 | 330,000 ± 80,000 | 12.5 ± 0.2 | 41 ± 2 | 300,000 ± 20,000 | |
| R367A AblKD | 0 | NS (> 0.04) | NS (> 500) | 35 ± 9 | |||
| 40 | 8.9 ± 0.3 | 130 ± 20 | 68,000 ± 8,000 | 5.6 ± 0.1 | 47 ± 5 | 120,000 ± 10,000 | |
| A365R/R367A AblKD | 0 | 22 ± 1 | 280 ± 30 | 80,000 ± 10,000 | 13.2 ± 0.4 | 114 ± 9 | 115,000 ± 9,000 |
Reactions were performed at 30 ° in 100 mM Tris, pH 7.4 and 10 mM MgCl2. NS: No saturation. If no value is given, then the constants were not measured.
For wild-type AblKD and R367A AblKD reactions without imidazole: [γ−32P]ATP = 0.15 mCi/mL and ATP = 0.25 mM ATP. For wild-type AblKD with imidazole: [γ−32P]ATP = 0.15 mCi/mL and ATP = 1 mM ATP. Otherwise, reaction mixtures contained 1 mM ATP, 0.1 U/µL pyruvate kinase, 0.05 U/µL lactate dehydrogenase, 1 mM phosphoenolpyruvate, 150 µM NADH, and 0.5 mM Na3VO4
Reaction mixtures included 0.1 U/µL pyruvate kinase, 0.05 U/µL lactate dehydrogenase, 1 mM phosphoenolpyruvate, 150 µM NADH, and 0.5 mM Na3VO4. For wild-type AblKD, [Abltide] = 190 µM; for R367A AblKD, [Abltide] = 440 µM; for A365R/R367A AblKD, [Abltide] = 840 µM.
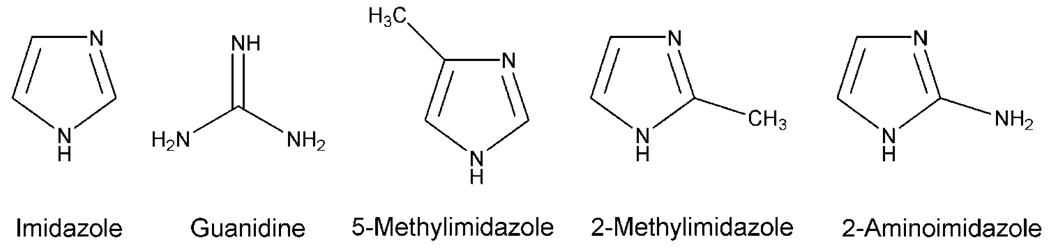
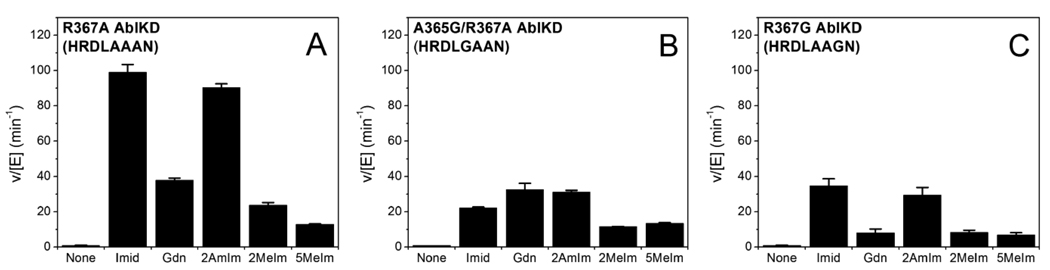
The catalytic rescue of R367A AblKD was examined with a series of diamino analogs that have been tested previously with Csk (11) and Src (12) including guanidinium, imidazole, 2-methylimidazole, and 5-methylimidazole (Figure 5). All showed the ability to stimulate R367A AblKD at least 5-fold, but maximal stimulation was achieved with imidazole (100-fold, Figure 6A). More extensive analysis of imidazole rescue of R367A AblKD revealed that it could afford kcat/Km (68,000 M−1s−1) within 3-fold of the wild-type level (Table 2), with minor changes in kcat and Km. Imidazole had minimal impact on the wild-type AblKD kinase parameters, as shown in Table 2. The Km of imidazole is 8.5 mM, similar to the Km for rescue of R388A Src (5 mM) (12). We also examined the rescue of two other Abl mutants that were analogous to those studied previously in Src: R367G AblKD and R367A/A365G AblKD, which have the potential to expand the cavity surrounding the deleted Arg side-chain. These mutants were also quite catalytically impaired compared to wild type, as expected, and complementation by imidazole was less efficient (Figure 6B–C). It is noteworthy that the methylimidazoles showed a similar efficacy (within 3-fold) for R367A/A365G AblKD, suggesting that the larger hole could be filled more effectively by the larger imidazole analog. Related behavior was observed with Src rescue (12). Taken together, the Abl chemical rescue data are quantitatively more similar to that of Src than Csk.
Figure 5. Diamino Analogs.
Structures of the chemical rescue agents used in this study.
Figure 6. Rescue of AblKD Variants with Diamino Analogs.
Relative activation of AblKD variants by imidazole (Imid), guanidine (Gdn), 2-methylimidazole (2MeIm), or 5-methylimidazole (5MeIm) compared to basal activity (None). The letters in parentheses indicate the catalytic loop sequence. Activity was measured at 30 ° using 40 mM of the indicated activator in 100 mM Tris, pH 7.4, 10 mM MgCl2, 500 µM ATP, 2 mM Abltide, and 0.15 mCi/mL [γ−32P]ATP.
Second sphere mutation and amino-imidazole complementation in Src and Abl
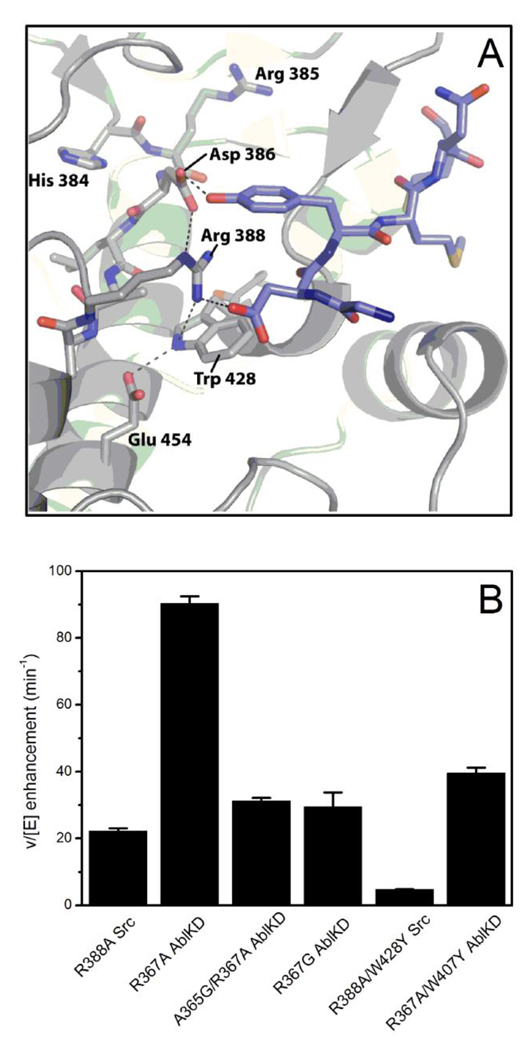
We investigated the interactions of kinase residues that might influence chemical rescue efficiency and small molecule selectivity. A computational analysis using Rosetta (21) was undertaken to select "second-sphere" residues that might be most important. The mutations that were selected fell into two types: 1) mutations that filled in the space vacated by the Arg→ Ala mutation and 2) substitutions of different interactions for the Glu454-Tyr428 hydrogen bond. We pursued these studies on the well characterized Src enzyme initially. The mutants made to address the former case were not activatable by imidazole (data not shown). This may be due to uncertainty in the placement of the guanidinium ligand, and the additional atoms in the mutant side-chain may occlude the ligand binding site. For the second class of mutations selected by Rosetta, Trp428 and Glu454, were replaced by a series of amino acids. These mutations were carried out in a background of R388A. In Figure 7A, we highlight the observed interactions of Trp428 and Glu454 with active site residues. Many of the double (and triple) mutants expressed poorly in E. coli, or appeared to be very unstable in kinase assays, making characterization very difficult. These problems may indicate the importance of a cation-pi interaction between the guanidinium/imidazolium and the residue at that position.
Figure 7. Modulation of Chemical Rescue.
A) Interactions of Trp428 and Glu454 (Src numbering) as seen in the crystal structure for IRK (PDB entry 1IR3). Peptide inhibitor is shown in blue, while the kinase backbone is shown in gray. Stick representations are used for side-chains of the active site loop (HRDLAAR) as well as the conserved tryptophan and glutamate. Hydrogen bonding interactions are shown as dashed lines. B) Relative activation of AblKD and Src variants by 2-aminoimidazole compared to no activator. Note that due to the different substrate preferences, AblKD activity is significantly higher than Src activity. Trp407 in Abl corresponds to Trp428 in Src. Src activity was measured as in Table 2 using 50 mM of 2-aminoimidazole; Abl assays are as described for Figure 6 (using 40 mM of 2-aminoimidazole).
One mutant, R388A/W428Y Src, showed sufficient stability and baseline kinase activity to determine the kinetic parameters. It showed modestly less efficient rescue (4.6-fold) than the parent R388A Src itself. We also assessed this and other mutants with an additional, novel rescue agent, 2-aminoimidazole, which in the course of this work was found to efficiently rescue R388A Src and R367A AblKD (about 30–40% more potent than imidazole in terms of Km). As shown in Table 3, 2-aminoimidazole was about equally effective (within 2-fold) at rescuing R388A/W428Y Src as the parent R388A Src, suggesting a relative compensation compared to imidazole. These findings support the idea that Trp428 can subtly influence the preference for rescue agent. However, the results with Abl were quite different. R367A/W407Y AblKD was greatly catalytically impaired and was only poorly rescuable by imidazole (35-fold down in kcat/Km compared to parent R367A AblKD). Despite the generally conserved chemical rescue properties of Abl and Src, Trp407 in Abl appears to be more crucial to facilitating imidazole rescue than the corresponding Trp428 residue in Src.
Table 3.
Comparison of Imidazole and 2-Amino-imidazole as Activatorsa
| Imidazole | 2-Aminoimidazole | |||||
|---|---|---|---|---|---|---|
| kcat(min−1) | Km (mM) | kcat/Km (M−1 s−1) | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) | |
| R388A Srcb | 21.5 ± 0.7 | 9.3 ± 0.8 | 40 ± 10 | 11.0 ± 0.3 | 6.8 ± 0.5 | 27 ± 2 |
| R388A/W428Y Srcb | 8.30 ± 0.02 | 16 ± 1 | 8.7 ± 0.8 | 5.56 ± 0.03 | 6 ± 1 | 16 ± 3 |
| R367A AblKDc | 460 ± 20 | 8.5 ± 0.9 | 900 ± 100 | 350 ± 20 | 5.3 ± 0.9 | 1100 ± 200 |
| R367A/W407Y AblKDd | NS (> 60) | NS (> 30) | 26 ± 7 | |||
Reactions were performed at 30° at pH 7.4 with 10 mM MgCl2. NS: No Saturation. If no value is given, then the constants were not measured.
50 mM Tris, 10 mM DTT, 200 µg/mL BSA, 400 µM ATP, 1 mg/mL PGT, 0.15 mCi/mL [γ−32P]ATP
100 mM Tris, 1 mM ATP, 440 µM Abltide, 0.1 U/µL pyruvate kinase, 0.05 U/µL lactate dehydrogenase, 1 mM phosphoenolpyruvate, 150 µM NADH, and 0.5 mM Na3VO4
100 mM Tris, 1 mM ATP, 1.5 mM Abltide, 0.1 U/µL pyruvate kinase, 0.05 U/µL lactate dehydrogenase, 1 mM phosphoenolpyruvate, 150 µM NADH, and 0.5 mM Na3VO4
Analysis of the imidazole protonation state in chemical rescue
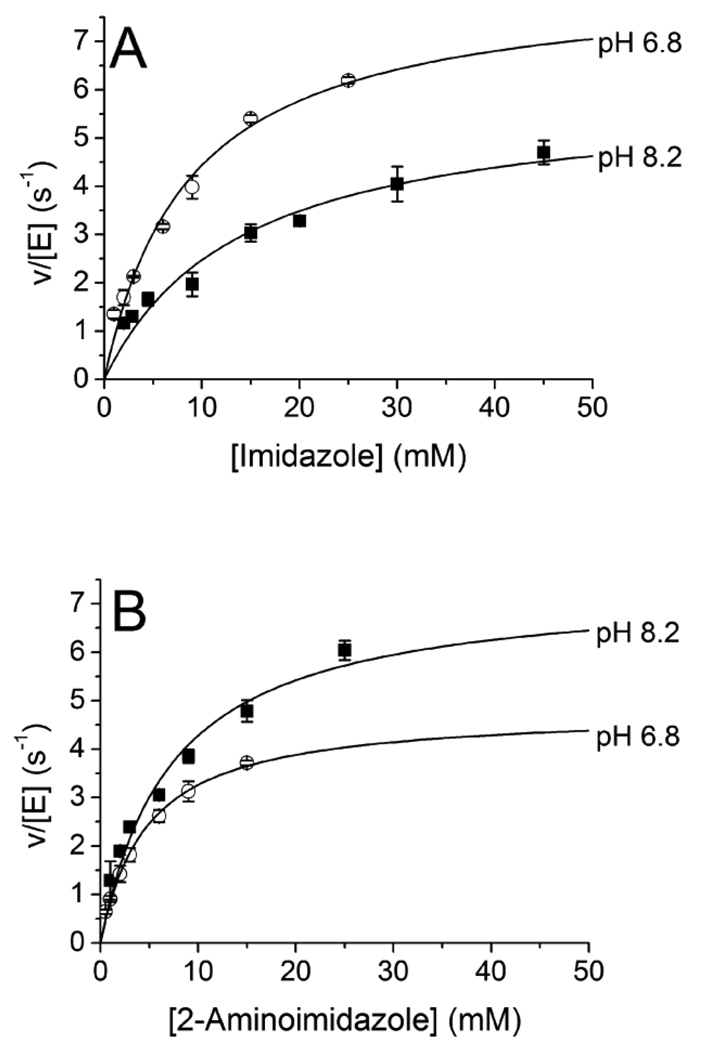
More extensive analysis of Src and Abl mutants (Figures 6 and 7) revealed similar trends for chemical rescue by imidazole and 2-aminoimidazole, suggesting that the general interactions of these compounds with these PTKs are similar. Because imidazole has pKa = 7.0 and 2-aminoimidazole has pKa = 8.5 (35), we hypothesized that an analysis of the pH effects of chemical rescue by these compounds might provide information about the protonation state of the analog employed for chemical rescue. As shown in Figure 8, imidazole effects greater rescue of R367A AblKD than 2-aminoimidazole when both are studied at pH 6.8 and each is > 50% protonated. In contrast, chemical rescue by 2-aminoimidazole is superior to imidazole at pH 8.2, when only the 2-aminoimidazole is greater than 50% protonated. These data are consistent with the model that the imidazolium is a more potent rescue agent than the neutral imidazole form of these compounds.
Figure 8. Effect of pH on Rescue of Abl R367A by Imidazole and 2-Aminoimidazole.
A) Activation by imidazole (pKa = 7.0) and B) 2-aminoimidazole (pKa = 8.5) of R367A AblKD at pH 6.8 and 8.2 were fit to equation 1. Reactions were performed at 30 ° in 100 mM HEPES, 10 mM MgCl2, 1 mM ATP, 440 µM Abltide, 0.1 U/µL pyruvate kinase, 0.05 U/µL lactate dehydrogenase, 1 mM phosphoenolpyruvate, 150 µM NADH, and 0.5 mM Na3VO4 at the indicated pH.
Discussion
Kinetic experiments done on Csk, Src, and now, Abl, in which complementation of various single and double Arg mutant PTKs by several compounds (guanidinium, imidazole, methylimidazoles) argues for localization of the rescue agent in the mutant cavity. The surprising findings from X-ray crystallography of R388A SrcKD provides independent evidence that chemical rescue of mutant PTKs involves the positioning of an imidazole in place of the native Arg. Electron density for a His residue from one kinase molecule in the active site of a second was unexpected for two reasons. First, Src is a monomer in solution so that pre-existing dimeric interactions were not known. Second, it has been shown previously that the catalytic-loop Arg→His mutant of Csk is about as dead as the Arg→Ala mutant, and free His in solution is a rather poor rescue agent compared with imidazole (11). We can rationalize the inactivity of Arg→His Csk mutant as due to inability of His to extend far enough into the active site. The fact that free His is a poor rescue agent could be due to the added bulk of the alanyl moiety of the residue, creating unfavorable interactions. Additionally, in the R388A SrcKD crystal structure, the backbone atoms of His seem to prevent effective ATP binding, thus free His may have the same effect and serve to inactivate the kinase. For these reasons, rescue of AblKD variants with free His was not attempted.
The relatively high efficiency of R387A AblKD chemical rescue bodes well for future applications in cell signaling studies. Abl rescue efficiency is more similar to Src's than Csk's. This is significant because Abl's catalytic loop sequence is identical to that of Csk and different from that of Src. This suggests that residues outside the catalytic loop are more influential in governing rescue efficiency. However, Csk is, in many ways, an outlier PTK because of its high substrate selectivity for one Tyr site in the Src protein and low rate of phosphorylation of peptides (36). Recent studies on Csk's naturally truncated activation loop suggest that its unique activation loop may contribute to its unique properties (37).
The discovery of 2-aminoimidazole as a new and potent rescue agent for mutant PTKs may have practical importance for cell signaling studies. Its structure can be viewed as a hybrid molecule of guanidinium and imidazole, perhaps accounting for its improved properties over 2-methylimidazole. In this study, it provided a useful tool for establishing the preference for the imidazolium form of the rescue agent to complement mutant Src and Abl. This protonation state is consistent with expectation since the Arg pKa is above 10 and likely to be protonated in the active site. Furthermore, the enhanced electrostatic interaction of the catalytic base Asp carboxylate with the imidazolium is likely to be catalytically important in substrate alignment.
Acknowledgements
We thank Steve Zimmerman and members of the Cole lab for helpful ideas and discussions. Crystallographic data was collected at the Advanced Light Source, supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
This work was supported by National Institutes of Health Grant CA74305 (P.A.C.), the Johnson & Johnson Fellowship of Life Science Research (M.A.S.), and the Jane Coffin Childs Memorial Fund for Medical Research (J.J.H.)
AblKD, human c-Abl kinase domain; D+2, PTK with active loop sequence DLRAAN; D+4, PTK with active loop sequence DLAARN; PP2, 4-amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo[3,4-d]pyrimidine; PTK, protein tyrosine kinase; SrcKD, chicken c-Src kinase domain
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 3.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 4.Miller WT. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc Chem Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Songyang Z, Carraway KL, 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 6.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 7.Parang K, Cole PA. Designing bisubstrate analog inhibitors for protein kinases. Pharmacol Ther. 2002;93:145–157. doi: 10.1016/s0163-7258(02)00184-5. [DOI] [PubMed] [Google Scholar]
- 8.Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Mechanism-based design of a protein kinase inhibitor. Nat Struct Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- 9.Williams DM, Cole PA. Proton demand inversion in a mutant protein tyrosine kinase reaction. J Am Chem Soc. 2002;124:5956–5957. doi: 10.1021/ja025993a. [DOI] [PubMed] [Google Scholar]
- 10.Kim K, Cole PA. Kinetic Analysis of a Protein Tyrosine Kinase Reaction Transition State in the Forward and Reverse Directions. Journal of the American Chemical Society. 1998;120:6851–6858. [Google Scholar]
- 11.Williams DM, Wang D, Cole PA. Chemical rescue of a mutant protein-tyrosine kinase. J Biol Chem. 2000;275:38127–38130. doi: 10.1074/jbc.C000606200. [DOI] [PubMed] [Google Scholar]
- 12.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 13.Toney MD, Kirsch JF. Direct Bronsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989;243:1485–1488. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- 14.Craik CS, Roczniak S, Largman C, Rutter WJ. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987;237:909–913. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- 15.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 16.Lowry WE, Huang J, Ma YC, Ali S, Wang D, Williams DM, Okada M, Cole PA, Huang XY. Csk, a critical link of g protein signals to actin cytoskeletal reorganization. Dev Cell. 2002;2:733–744. doi: 10.1016/s1534-5807(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 17.Madhusoodanan KS, Guo D, McGarrigle DK, Maack T, Huang XY. Csk mediates G-protein-coupled lysophosphatidic acid receptor-induced inhibition of membrane-bound guanylyl cyclase activity. Biochemistry. 2006;45:3396–3403. doi: 10.1021/bi052513u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Im E, Kazlauskas A. Src family kinases promote vessel stability by antagonizing the Rho/ROCK pathway. J Biol Chem. 2007;282:29122–29129. doi: 10.1074/jbc.M702637200. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 22.Takeya T, Hanafusa H. Structure and sequence of the cellular gene homologous to the RSV src gene and the mechanism for generating the transforming virus. Cell. 1983;32:881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 23.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook PF, Neville ME, Jr, Vrana KE, Hartl FT, Roskoski R., Jr Adenosine cyclic 3',5'-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 25.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 26.Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, Leahy DJ. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography. 1997;276(Pt A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D-Biological Crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X, Kim JL, Newcomb JR, Rose PE, Stover DR, Toledo LM, Zhao H, Morgenstern KA. Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors. Structure. 1999;7:651–661. doi: 10.1016/s0969-2126(99)80086-0. [DOI] [PubMed] [Google Scholar]
- 35.Storey BT, Sullivan WW, Moyer CL. The pKa Values of Some 2-Aminomidazolium Ions. The Journal of organic chemistry. 1964;29:3118–3120. [Google Scholar]
- 36.Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Peptide and protein phosphorylation by protein tyrosine kinase Csk: insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 37.Levinson NM, Seeliger MA, Cole PA, Kuriyan J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]