SUMMARY
Expression profile analysis clusters Gpnmb with known pigment genes, Tyrp1, Dct, and Si. During development, Gpnmb is expressed in a pattern similar to Mitf, Dct and Si with expression vastly reduced in Mitf mutant animals. Unlike Dct and Si, Gpnmb remains expressed in a discrete population of caudal melanoblasts in Sox10-deficient embryos. To understand the transcriptional regulation of Gpnmb we performed a whole genome annotation of 2,460,048 consensus MITF binding sites, and cross-referenced this with evolutionarily conserved genomic sequences at the GPNMB locus. One conserved element, GPNMB-MCS3, contained two MITF consensus sites, significantly increased luciferase activity in melanocytes and was sufficient to drive expression in melanoblasts in vivo. Deletion of the 5’-most MITF consensus site dramatically reduced enhancer activity indicating a significant role for this site in Gpnmb transcriptional regulation. Future analysis of the Gpnmb locus will provide insight into the transcriptional regulation of melanocytes and Gpnmb expression can be used as a marker for analyzing melanocyte development and disease progression.
SIGNIFICANCE
Comparative analysis of gene expression profiles using melanocyte lines derived from mice provides a powerful resource to explore genetic components of melanocyte development and pigment cell function. Using expression data, we identified Gpnmb as a new marker for early melanoblast development. We show that Gpnmb is dependent on Mitf for in vivo expression and marks a unique set of Sox10-independent melanoblasts. We identified an 89 basepair evolutionarily conserved genomic sequence at the Gpnmb locus that can enhance expression in melanocytes and tested MITF E-box consensus sequences for their involvement in melanocyte-restricted expression. Gpnmb and the panel of genes identified in this study will be valuable resources for understanding the genetic components involved in melanocyte development and diseases.
Keywords: Gpnmb, Mitf, Sox10, melanoblast, melanocyte, melanoma
INTRODUCTION
Mammalian pigment-producing cells provide multiple functions for the organism including: color variation that may be used in camouflage or mating attraction, photo-protection of the skin from ultraviolet light, integrity of the stria vascularis of the cochlea needed for proper hearing, and prevention of intraocular light-scattering for visual acuity. The pigment-producing cells of the skin and cochlea are termed melanocytes and are originally derived from the neural crest (NC). Melanocyte precursor cells, termed melanoblasts, are specified to this lineage at the dorsal neural tube-ectoderm border. As fetal development continues, melanoblasts migrate dorsolaterally through the mesenchymal layer under the ectoderm where they proliferate, cross into the ectoderm, and differentiate into melanocytes in the epidermis and hair follicles. (Silver et al., 2006). Pigment producing cells are also found in the retinal pigmented epithelium (RPE) of the eye. While RPE cells have a different embryonic origin, being derived from an out-pocketing of the central nervous system, both melanocytes and RPE cells utilize many of the same genes to control synthesis and packaging of melanin pigment. In both NC and RPE, precise transcriptional control of these pigmentation genes is needed for proper development and function (Silver et al., 2006; Murisier and Beermann, 2006).
The microphthalmia-associated transcription factor (MITF) plays a major role in the transcriptional regulation of multiple pigmentation genes that modulate the balance between cell proliferation, survival and differentiation in melanocytes. MITF is a member of the basic helix loop-helix-leucine zipper family of proteins that preferentially binds to the E-box consensus CATGTG sequence flanked by either a 5’ or 3’ T nucleotide, in addition to the CACGTG E-box motif (Aksan and Goding, 1998). The importance of MITF function is apparent, as mutations in human MITF give rise to diseases associated with an absence of melanocytes (Waardenburg Syndrome (WS) 2a, OMIM# 193510_and Tietz syndrome, OMIM# 103500), MITF copy number variations are associated with melanoma (Garraway et al., 2005), and mutations of MITF transcriptional targets (TYR, OCA2 and TYRP1) are associated with oculocutaneous forms of albinism (OCA1, OMIM# 203100; OCA2, OMIM# 203200; and OCA3, OMIM# 203290). Therefore elucidating the transcriptional network involved in regulating melanocyte development and function as well as the role of MITF in this network is essential for understanding normal pigmentation and may lead to improved disease diagnostics and/or therapeutic interventions.
One powerful approach for understanding the dynamic nature of melanocyte cell regulation and function is through the use of immortalized melanocyte lines derived from mouse models of pigmentary diseases. Currently, mouse models exist for at least 70 defined genes that affect pigmentation, which model 37 distinct human disorders (Oetting et al., 2008), and multiple cell lines generated from a subset of these mutant mice are available at (http://www.sgul.ac.uk/depts/anatomy/pages/Dot/Cell%20bank%20holdings.htm#melanocytes). To increase the efficiency of generating immortal cell lines while maintaining the genetic integrity of the genome, Ink4a-Arf deletions were introduced into mice prior to explanting melanocytes, since cells of this genotype are immortal and so do not require immortalization by accumulation of random mutations in culture. Such Ink4a-Arf deletion melanocyte lines were generated from C57BL/6J mice (melan-a Ink4a-1) and from mice carrying classic coat color genes that disrupt melanocyte function. These include C57BL/6J lines carrying Tyrp1b/b (melan-Ink4a-b) (Sviderskaya et al., 2002) and Rab38cht/cht (melan-cht) genotypes (Wasmeier et al., 2006). By minimizing differences in genetic backgrounds through the use of C57BL/6J and the targeted Ink4a-Arf deletion, differences between cell lines are thus more indicative of the primary genetic defects.
Expression profiling allows for genome-wide evaluation of RNA expression within defined cell populations. Genome level evaluation of these large expression data sets, using clustering algorithms, involves the grouping of genes based upon similar patterns of expression. Such cluster analysis has been useful in demonstrating the coordinated transcriptional regulation of multiple genes in a common functional pathway or cell responses (Spellman et al., 1998). In this study, we utilized gene expression data from melanocyte cell lines to provide a repertoire of genes expressed in melanocytes. Profiling of these data revealed a collection of 258 genes that are enriched for expression in melanocyte lines relative to control cell populations. Further analysis demonstrated that one of these genes, Glycoprotein (transmembrane) nmb (Gpnmb), which encodes a type 1 transmembrane protein, is a novel melanoblast-expressed gene whose transcription is dependent on MITF.
RESULTS
Known MITF target genes reside within defined expression cluster nodes
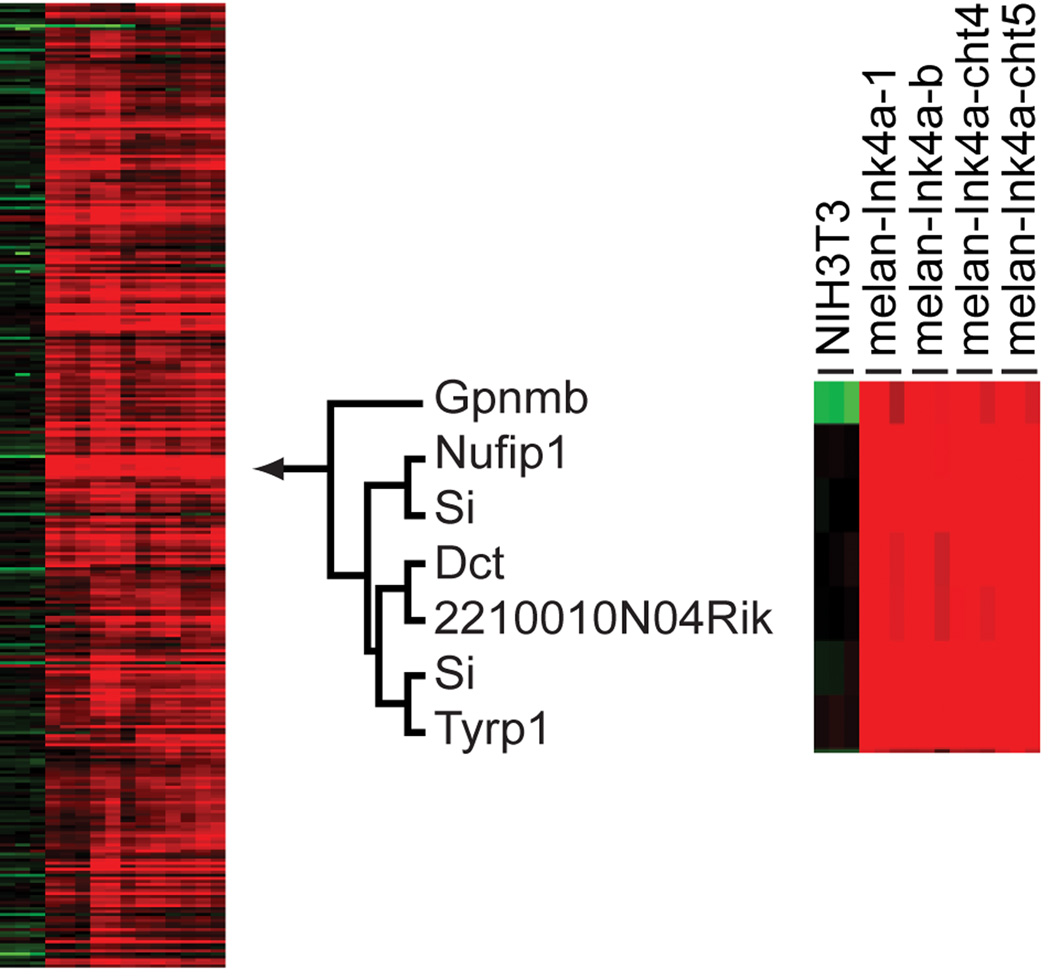
The transcriptional profiles of immortal melanocyte lines, melan-Ink4a-1, melan-Ink4a-b, melancht-4, melan-cht-5, and NIH3T3, were obtained by co-hybridization of labeled samples in triplicate with Universal RNA to a printed Operon Mv3.0 oligonucleotide probe set representing over 16,000 genes. Expression data from these 15 hybridizations were queried for genes that were: (1) highly expressed in melanocyte lines relative to Universal RNA and NIH3T3 levels; and (2) varied in expression using hierarchical clustering algorithms in a manner similar to previously identified pigmentation genes. A set of 318 microarray probes, representing 258 distinct genes, were identified that exhibited a calibrated ratio >2 for all melanocyte cell lines relative to the corresponding NIH3T3 control calibrated ratios, and had a calibrated ratio of >3 in any of the four melanocyte samples (Supplemental Figure 1, S1) relative to the Universal RNA control used for the hybridizations. This Universal RNA represented pooled expression from 11 different cell lines. Hierarchical clustering of the expression data for the 258 genes identified a distinct expression cluster node consisting of six genes, three of which, Si, Dct, and Tyrp1, are previously known pigmentation gene loci and MITF target genes (Figure 1).
Figure 1. Expression cluster gene set defined by Dct, Si, and Tyrp1 pigment genes.
Heat map of 318 array elements (258 genes) which are >2-fold over expressed in all melanocyte lines relative to NIH3T3 control calibrated ratios and also have expression increased >3 fold in any one cell line relative to the Universal control RNA. The location of the heirachical cluster node, defined by pattern of expression for Dct, Si, and Tyrp1 is denoted by the arrow, with the corresponding dendrogram and heatmap shown enlarged to the right.
Gpnmb expression is similar to known pigmentation genes in vivo
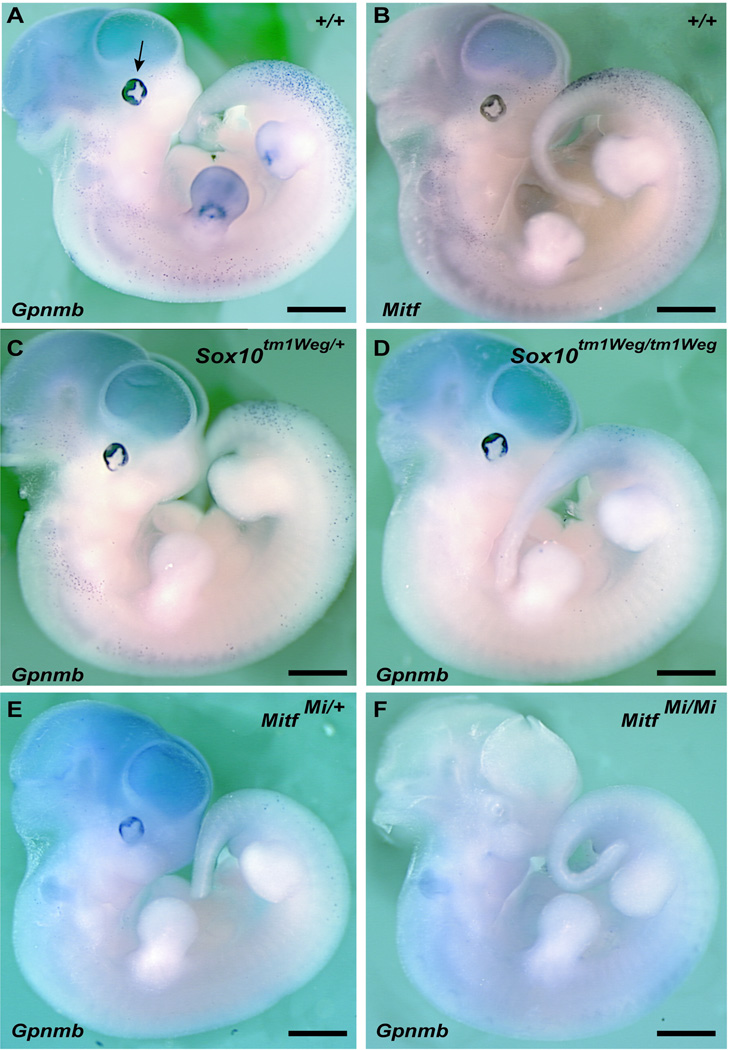
Previous work has demonstrated that three of the genes in this cluster node, Tyrp1, Dct and Si, are expressed in melanoblasts and/or the presumptive RPE during mouse embryonic development (Steel et al., 1992; Nakayama et al., 1998 ; Baxter and Pavan, 2003). Also of note within this defined cluster node was the gene Gpnmb. Although Gpnmb was known to be expressed in RPE (Bachner et al., 2002) and melanoma tissue samples (Weterman et al., 1995), representing cells from differentiated, adult pigment cell lineages, embryonic expression was unknown. Given the clustering of Gpnmb with these early markers for the developing pigment cell lineages we sought to evaluate if Gpnmb also exhibited an embryonic expression pattern similar to that of Tyrp1, Dct and Si. Whole-mount in situ hybridization was performed using wild-type murine embryos at embryonic day 11.5. Gpnmb was robustly expressed in both developing RPE and also in a punctate pattern throughout the embryo consistent with the expression of the early melanoblast marker Mitf (Figure 2A, B).
Figure 2. Developmental expression of Gpnmb is similar to known pigmentation gene expression.
Whole mount in-situ hybridization for (A, C–F) Gpnmb, and (B) Mitf, in mouse embryos at E11.5 (A, B). Expression pattern in wildtype embryos for Gpnmb and Mitf, respectively, demonstrates that Gpnmb is expressed in developing RPE (arrow) and in a punctuate cell population, similar to Mitf expression and consistent with melanoblast pattern of expression at this age. Gpnmb probe marks (C) a reduced number of melanoblasts in Sox10 LacZ/+ embryos and (D) only a small caudal population of melanoblasts that are Sox10 independent in Sox10 LacZ/LacZ embryos. Gpnmb is reduced in both melanoblast and RPE cells in (E) MitfMi/+ embryos, while expression is completely absent in (F) Mitf Mi/Mi embryos relative to wildtype embryos.
Gpnmb expression in RPE and melanoblasts is dependent on MITF
Given the striking expression pattern of Gpnmb in both the RPE and putative melanoblasts in wild type embryos, we next determined if the intensity and pattern of Gpnmb expression is dependent upon levels of Mitf or Sox10 transcription factors. For this, Gpnmb mRNA expression was examined by whole mount in situ hybridization in mouse embryos mutated for either Mitf or Sox10. No change in Gpnmb signal intensity was observed in the developing RPE of Sox10tm1Weg/+ or Sox10tm1Weg/tm1Weg embryos (Figure 2C, D). This was expected, as Sox10 is neither expressed in nor needed for optic cup/RPE development and function. However, Gpnmb signal intensity was substantially reduced in the developing RPE of MitfMi/+ embryos, and was completely absent in the RPE of MitfMi/Mi embryos (Figure 2E, F). This lack of RNA expression in the developing RPE of MitfMi/Mi embryos has also been observed for the MITF target genes, Tyrp1, Tyr and Si (Baxter and Pavan, 2002; Baxter and Pavan, 2003; Nakayama et al., 1998). These results demonstrate that Gpnmb expression is dependent on MITF function for expression in the developing RPE. Examination of the punctate pattern of Gpnmb expression in these embryos showed that Gpnmb positive cell numbers were reduced in Sox10tm1Weg/+, Sox10tm1Weg/tm1Weg and MitfMi/+embryos, and in MitfMi/Mi embryos there was a complete lack of Gpnmb expression over the entire embryo (Figure 2, 3). This is consistent with the punctate, non-RPE, Gpnmb-positive cells being putative melanoblasts, as SOX10 and MITF are necessary for melanoblast development, and suggests normal Gpnmb expression is dependent on SOX10 and MITF function in melanoblast cells.
Figure 3. Gpnmb is expressed in melanoblasts and marks a distinct caudal population of cells in Sox10 null embryos.
Dorsal view of caudal melanoblast population marked by (A–C) Gpnmb and (D–F) Mitf in (A, C) wildtype, (B, E) Sox10LacZ/LacZ, and (C,F) MitfMi/Miembryos.
Gpnmb marks a novel Sox10-independent cell population
In Sox10tm1Weg/tm1Weg embryos, while there was a complete absence of Gpnmb expressing melanoblasts along the head and trunk, we observed a small, discrete population of Gpnmb-positive cells present at the tail-hindlimb region along the dorsal neural tube (Figure 3B). This is in contrast to the complete loss of Gpnmb expression in MitfMi/Mi embryos (Figure 3C) and also distinct from expression of the Mitf target gene Si, where there is a complete loss of melanoblast expression in MitfMi/Mi embryos (Baxter and Pavan, 2003), and in Sox10tm1Weg/tm1Weg (data not shown). Given these results we then evaluated Mitf RNA expression by whole mount in situ hybridization in both Sox10tm1Weg/tm1Weg and MitfMi/Mi embryos to assess if these caudal cells were also Mitf positive. This analysis revealed that in both Sox10tm1Weg/tm1Weg and MitfMi/Mi embryos, head and trunk melanoblasts were absent, yet an Mitf-positive population of punctate cells remained at the tail-hindlimb, dorsal neural tube region (Figure 3D–F). Taken together these results indicate that in Sox10tm1Weg/tm1Weg embryos a cell population that is positive for Mitf and for Gpnmb is present, while in MitfMi/Mi embryos this cell population is positive for MitfMi RNA but negative for Gpnmb expression. This demonstrates that Gpnmb expression in this discrete population of caudal melanoblasts at E11.5 requires expression of functional MITF protein but not SOX10.
GPNMB harbors highly conserved non-coding sequences
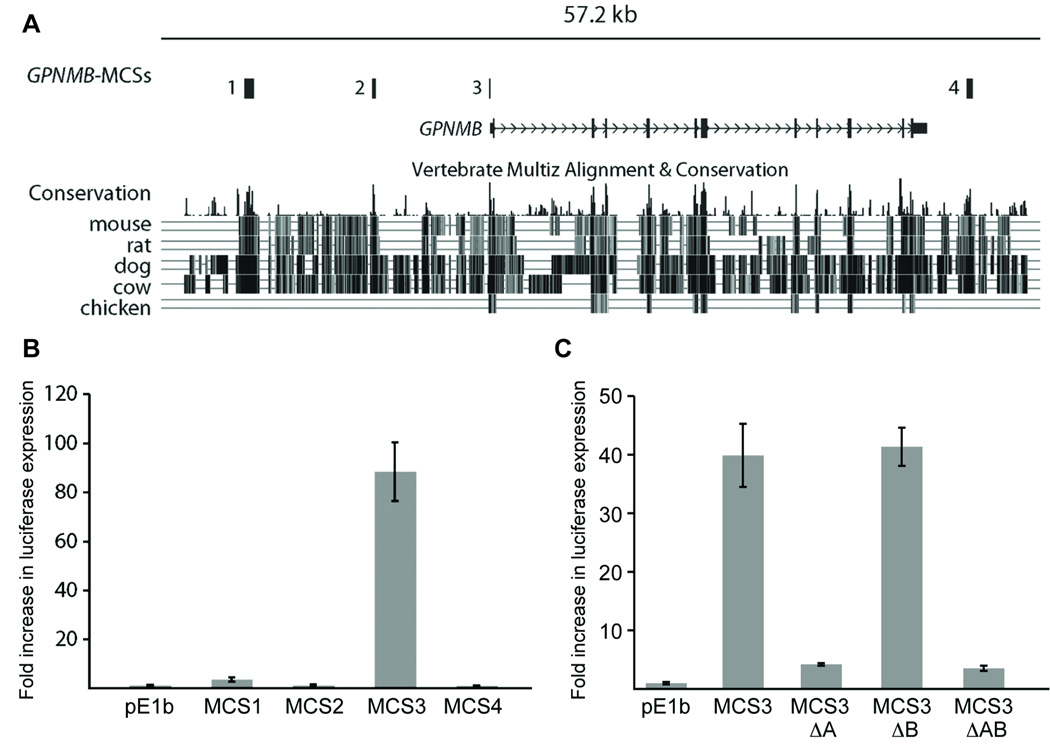
To identify candidate genomic sequences that regulate GPNMB transcription, we identified highly conserved non-coding sequences surrounding the human GPNMB gene using MultiPipMaker program and PhastCons scores. Both analyses revealed four multiple-species conserved sequences (MCSs): GPNMB-MCS1, GPNMB-MCS2, GPNMB-MCS3, and GPNMB-MCS4 (Figure 4A).
Figure 4. Comparative and functional analysis of conserved sequences at GPNMB.
(A) The interval of human chromosome 7 harboring the GPNMB locus as depicted by the UCSC Human Genome Browser (March 2006 build, chr7:23207149–23305465), (genome.ucsc.edu). Comparative sequence analysis revealed four highly-conserved sequences (bars across top): GPNMB-MCS1, GPNMB-MCS2, GPNMB-MCS3, and GPNMB-MCS4. (B) Enhancer activity of each GPNMB-MCS. Each MCS region indicated in (A) was cloned upstream of a minimal promoter driving luciferase expression. Melan-a cells were transfected with each construct along with an internal-control vector expressing renilla, and incubated for 48 hours. The ability of each MCS to enhance luciferase expression was determined by calculating the ratio of luciferase expression to renilla expression, and then determining the fold increase (y-axis) over the ratio calculated for pE1b with no insert. (C) Similar experiments as in (B) for MCS3 deletion constructs in which MITF(A) and MITF(B) sites were independently deleted, (MCS3-ΔA) and (MCS3-ΔB) respectively, and in combination (MCS3-ΔAB).
The human genomic version of each GPNMB-MCS was cloned upstream of a minimal promoter driving luciferase expression. The resulting constructs were then transfected into cultured melanocytes (melan-a cells) and NIH3T3 cells. Subsequently, luciferase expression was assessed in cells carrying each vector, and compared to cells transfected with a construct bearing only the minimal promoter driving luciferase expression. These analyses revealed that while GPNMB-MCS1, GPNMB-MCS2 and GPNMB-MCS4 were associated with modest increases in luciferase expression (3.5, 1.1 and 0.9-fold respectively), GPNMB-MCS3 was associated with an 88.3-fold increase in expression (Figure 4B). Similar results were seen in both melan-a and NIH3T3 cells (Supplemental Figure 1, S2) indicating that factors necessary to activate GPNMB-MCS3 are present in both cell types.
In order to determine if MITF consensus binding sites were contained within the GPNMB-MCS sequences, we took a genome scale approach to map and annotate E-box motifs throughout the genome in multiple species. The E-box motif (CACGTG) and the two reciprocal complementary sequences CATGTG and CACATG, were each annotated with respect to flanking nucleotides 5’ T/A or 3’ A/T. Results from this genome wide analysis are available at http://research.nhgri.nih.gov/manuscripts/Loftus/June2008. This dataset was then queried to determine if any MITF motifs reside within the regions GPNMB-MCS1-4. This analysis revealed that two of the MCS regions harbor a total of three MITF consensus sequences. Human GPNMB-MCS1 harbors one CACATGT, however when evaluated for each of the other organisms only the human sequence contains the intact CACATGT element (data not shown).
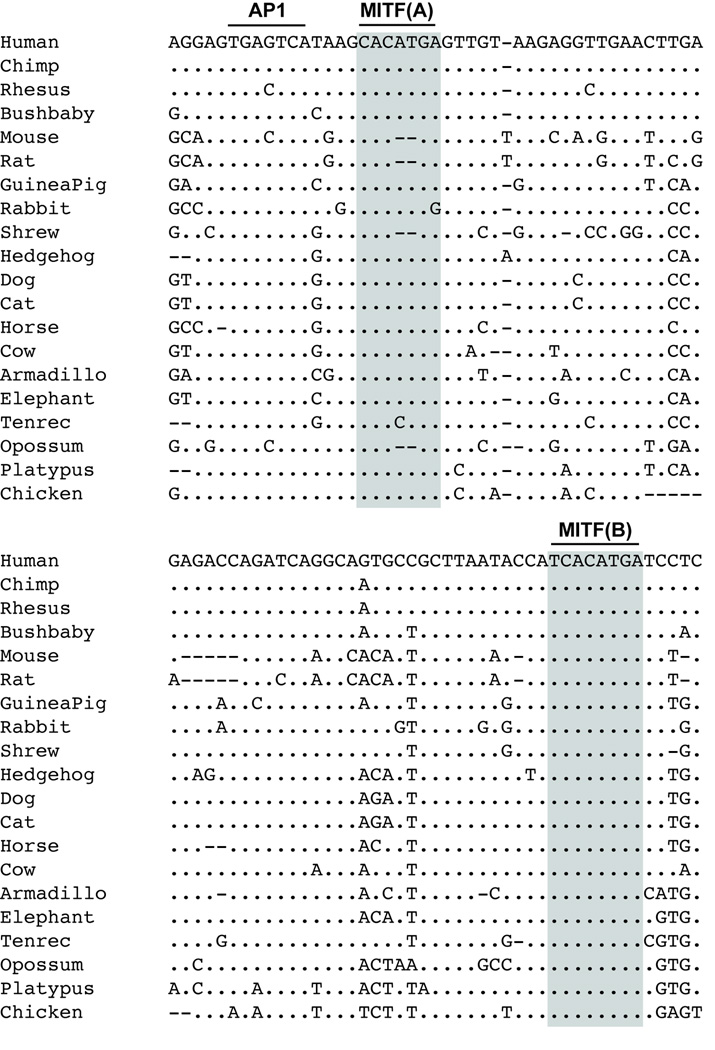
Two MITF consensus motifs are contained within human GPNMB-MCS3 (Figure 5). The first element MITF(A), (CACATGA) is fully conserved among 14 species, but 3 separate sequence variations are observed within 6 additional species. The second MITF consensus motif, MITF(B), (TCACATGA), is also located within the same MCS3 element, is fully conserved among all 20 species evaluated, and resides within the predicted 5’UTR of the human GPNMB transcript. Further analysis of 5 kb of DNA upstream of the transcription start site revealed no other conserved MITF consensus motifs other than those in MCS3.
Figure 5. Sequence conservation for 89 bp MCS3 element across 20 species.
The location of AP-1 and two MITF consensus binding sequences MITF(A) and MITF(B) are indicated by bars and sequences corresponding to MITF(A) and MITF(B) in 20 species are indicated in grey. Identical basepairs are indicated by dots and insertion/deletion events are indicated by dashes. For MITF(A) note the conservation between human and 15 additional species including chicken, and the 2 bp deletion/insertion (dashes) in rat, mouse, opossum and shrew while MITF(B) is conserved in all 20 species.
We next assessed the degree to which the two MITF motifs contribute to the observed enhancer activity of MCS3 in vitro. Site-directed mutagenesis was performed to delete both elements, independently and together, and luciferase expression assays were repeated in melan-a (Figure 4C) and NIH3T3 cells, (Supplemental Figure 2, S2). Deletion of MITF(A) motif results in a dramatic ~30-fold reduction in activity in melan-a cells, while deletion of MITF(B) exhibits expression comparable to that of the wild type MCS3 element. Deletion of both MITF(A) and MITF(B) motifs does not change the activity from that observed for deletion of MITF(A) alone. This analysis revealed that of the two MITF motifs, MITF(A) is the major contributor for expression in both melan-a and in NIH3T3 cells.
Given the striking expression pattern of Gpnmb in developing pigment cells and the significant enhancement of expression we observed from MCS3 in melan-a cells, we assayed whether MCS3 was sufficient to provide in vivo lineage restricted expression utilizing a zebrafish developmental transgenic assay (Fisher et al., 2006a; Fisher et al., 2006b) and if the corresponding MITF motif deletion constructs abrogated this pattern. We found that the 89 basepair MCS3 element was sufficient to drive GFP expression in a restricted population of cells emerging from the dorsal neural tube at 24 hpf, consistent with the pattern, morphology, and location of pre-migratory neural crest cells, melanoblast precursors (Figure 6A). At 48 hpf, GFP positive cells were found to colocalize with pigment melanocytes, demonstrating that the sequences within this 89 basepair MCS3 element are capable of directing expression to the melanocyte lineage in zebrafish (Figure 6B). Interestingly, this same GFP expression pattern was also observed for constructs in which either MITF(A), MITF(B), or both MITF(A) and MITF(B) were deleted (data not shown). This indicates that additional sequences contained with the MCS3 element are sufficient for expression in melanocytes in vivo.
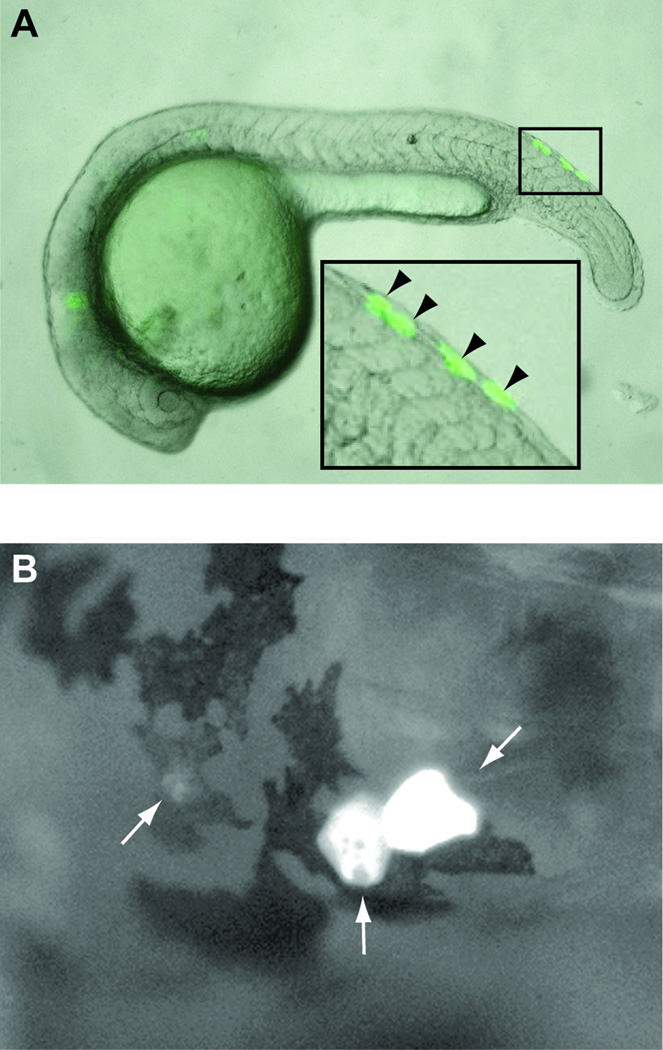
Figure 6. MCS3 is sufficient to drive expression to zebrafish melanocyte lineage.
(A) MCS3-directed GFP localizes within dorsally positioned cells (open triangles) consistent with the premigratory/migratory neural crest population of melanoblasts at 24 hours post fertilization (24 hpf). (B) GFP signal co-localizes with maturing melanocyte populations at 48 hpf (white arrows) as they begin to form pigment and obliterate the GFP signal therein.
Taken together, this study demonstrates that GPNMB is expressed in early pigment cell lineages, RPE and melanoblast, and its expression in these structures is dependent on MITF. We have identified an 89 basepair conserved sequence element MCS3 contained with the GPNMB promoter that is capable of directing expression to the melanocyte lineage in vivo. Contained within MCS3 are located two MITF binding sequence consensus elements, of which only one, annotated MITF(A), is needed for robust expression in melan-a and NIH3T3 cells, however neither of these sites is required for melanocyte expression in vivo.
DISCUSSION
In this study we have queried the expression network of melanocytes using cell lines derived from mice where mutant alleles of the pigmentation genes Tyrp1 and Rab38 were crossed onto a C57BL/6 background. Using criteria based on consistently higher gene expression in multiple melanocyte lines relative to other tissue sources, we identified a set of 258 genes. Contained within this 258 gene set are nine genes that have been shown to either directly affect MITF expression or are downstream MITF target genes. These nine genes include the transcription factor Sox10, mutated in WSIV, Peripheral Demyelinating Neuropathy, Central Dysmyelinating Leukodystrophy, and Hirschsprung Disease (PCWH) (Southard-Smith et al., 1998; Pingault et al., 1998; Inoue et al., 1999) and known to directly regulate Mitf expression (Verastegui et al., 2000; (Potterf et al., 2000; (Lee et al., 2000), and the gene Mitf itself (mutated in WSI and III). The remaining seven genes represent additional MITF targets including: Cdk2 (Du et al., 2004), Met (McGill et al., 2006), Tyr (Bentley et al., 1994; Yasumoto et al., 1997) (Murisier et al., 2007), Mlana (Du et al., 2003), Si (Du et al., 2003), Dct (Bertolotto et al., 1998; Jiao et al., 2004; Ludwig et al., 2004), and Tyrp1 (Fang et al., 2002; Yasumoto et al., 1997; Bertolotto et al., 1998). A tenth gene, Slc45a2, demonstrates reduced gene expression in Mitf mutant mice (Baxter and Pavan, 2002). However, promoter analysis by Du et al. could not find supporting evidence for direct regulation of the proximal promoter sequence by MITF suggesting that Slc45a2 is either regulated by MITF indirectly or through yet unidentified distal regulatory elements (Du and Fisher, 2002)
As three of these genes, Tyrp1, Si and Dct were contained within a defined expression cluster node, and each are dependent on MITF function for appropriate expression, we sought to assess whether the gene Gpnmb, also present within the cluster defined by Tyrp1, Dct and Si, had the capacity to mark the early melanoblast and RPE lineage. Our expression data in wild type embryos demonstrate that during early embryonic development, Gpnmb is robustly expressed in RPE and also in melanoblasts, displaying a pattern similar to Mitf, Dct and Si. This is consistent with Gpnmbexpression in the developing RPE and iris (Bachner et al., 2002) and also with the glaucoma phenotype observed in DBA/2J mice, which harbor mutations in both Gpnmb (GPNMB R150X) and Tyrp1 (Tyrp1b/b genotype) (Anderson et al., 2002; Howell et al., 2007). The robust expression of Gpnmb in melanoblasts extends the list of markers for melanocyte development in mouse embryogenesis and will be useful to further analyze mouse mutations that disrupt melanoblast development. For example, extension of the expression analysis of Gpnmb to embryos containing mutations in Mitf and Sox10 allowed us to detect differences in Gpnmb expression from that of Si and Dct, specifically in a population of melanoblasts in the caudal region of mouse embryos. In Sox10 null embryos a population of putative melanoblasts exists that express both Mitf and Gpnmb, even within the context of a Sox10 null background. Previous studies have determined that Mitf expression is dependent on SOX10 (Potterf et al., 2000; (Verastegui et al., 2000; Lee et al., 2000). However our results show that Mitf and Gpnmb, but not Si or Dct, can be expressed in melanoblasts at this time and site even in the absence of SOX10 protein. These results suggest that at this time point and location, Mitf gene expression is not solely dependent on Sox10 expression, and that melanocyte-expressed genes have differing requirements for Sox10 and Mitf. Understanding how the transcriptional regulation of these two classes of genes is controlled in these caudal melanoblasts will provide interesting insights into the transcription networks coordinating melanocyte development and how they are altered in disease states.
From the results of gene expression profiling and in situ hybridization analyses on mutant embryos, we hypothesized that GPNMB expression is dependent on MITF function in melanoblasts and RPE. To test if this regulation was direct, we directly examined the GPNMB locus for regulatory elements that might be candidates for regulation by MITF. We computationally identified four MCS non-coding regions at the GPNMB locus that may act as transcriptional regulatory elements, and found that two of these sequences harbor consensus MITF binding motifs by cross referencing with our whole genome annotation of E-box, MITF binding consensus sequences. One of these elements, GPNMB-MCS3, was able to enhance luciferase reporter expression in melan-a cells, and robust expression was dependent on one of the MITF binding consensus sequences, MITF(A), but not the other, MITF(B). Consistent with MCS3 enhancing pigment cell expression in vivo, transgenic zebrafish exhibit GFP expression in putative melanoblasts in response to sequences contained within MCS3. Although it is clear that Gpnmb expression is dependent on MITF function in vivo as its expression is absent in MITFMi mutant melanoblasts, it is unclear if MCS3 is directly regulated by MITF. MCS3 is a strong enhancer in both melan-a and a non-melanocyte derivative NIH3T3, and importantly deletion of MITF(A) sequence abrogates enhancement in both lines. However, zebrafish transgenics still express GFP even when MITF(A), MITF(B), or both are deleted. These results suggest that MCS3 is a melanoblast enhancer and that the MITF(A) site is needed for enhancement of expression in cell line based assays, but additional regulatory factors interact with this element to facilitate the lineage restricted expression observed in vivo.
Given the striking inability of deletion of MITF(B) to reduce luciferase expression enhancement in melan-a cells, it is of interest to note that the MITF(B) site is fully conserved in all 20 species analyzed, while the MITF(A) site is fully conserved in only a subset (14 of 20) species evaluated. The MITF(A) site exhibits classes of alterations: tenrec contains an A to C substitution at base pair 4, rabbit an A to G substitution at basepair 7, and rat, mouse, shrew and opossum a two basepair deletion corresponding to base pairs five and six of the E-box consensus. This two basepair deletion changes CACATGA to CACGAGT, and the resulting T to A nucleotide change at position 5 of the deletion element would theoretically no longer be recognized by MITF. It is interesting that the same two basepair deletion has arisen within these four species (rat, mouse, shrew and opossum) representing three different evolutionary branches. This sequence difference also suggests an intriguing possible evolutionary divergence in the regulation of Gpnmb among different species.
Gpnmb has previously been referred to under the name Osteoactivin, and as such has been implicated in osteoblast development as a mediator of BMP-2 signaling (Abdelmagid et al., 2007). However, Ripoll et al. have recently shown that previous expression analysis for Gpnmb utilizing osteoblast cultures most likely measured GPNMB expression found in macrophages co-purified in the cultures, rather than in osteoblasts (Ripoll 2008). Ripoll et al. then evaluated the ability of MITF to regulate the proximal promoter sequence of mouse Gpnmb in the osteoclast lineage. They focused on a 323 basepair fragment encompassing the corresponding mouse region of GPNMB-MCS3 (and additional sequence). This fragment exhibited enhancer activity, as 6-fold activation was observed in the macrophage/osteoclast cell line RAW/C4 when co-transfected with MITF. Their analysis of this enhancer (based on the mouse genome) only identified one conserved MITF consensus binding site located within the proximal promoter of mouse Gpnmb (UCSC Mouse Genome Browser, Feb. 2006 build), which corresponds to MITF(B) in our study. When Ripoll and colleagues mutated MITF(B) from the corresponding mouse sequence, the enhancer lost the ability to be activated upon co-transfection with MITF in RAW/C4 cells.
Both our analysis and the Ripoll study implicate MITF to be important for regulation of GPNMB expression. However, each study identified different MITF consensus sequence elements as being the major sequence element responsible for the observed enhancer activity assayed, and when each respective MITF motif sequence was deleted, both studies observed significant loss of enhancer activity. There are several notable differences between the studies. In our analysis of enhancer sequence activity our construct was shorter, only 89 basepair in length, and we utilized the human GPNMB sequence, not mouse. This difference in species used for the analysis is relevant as the mouse lineage has a two basepair deletion, relative to the human MITF(A) consensus binding site, although MITF(A) is fully conserved between 14 other different species. Therefore, while the mouse enhancer in the Ripoll study has assayed the corresponding genomic sequence to our human MCS3 element, the Ripoll wild-type mouse construct would more appropriately correspond to our MCS3-A construct. Also distinct were the cell lineages in which the proximal GPNMB/Gpnmb promoter activities were evaluated. Our analysis was performed in the melan-a melanocyte line with endogenous MITF while Ripoll and colleagues utilized the corresponding mouse sequence in RAW/C4 cells with co-transfection of Mitf. The differences in functional activity observed in our and Ripoll's study may be attributed to other sequence differences in the constructs themselves, species-specific differences, or cell lineage (macrophage/osteoclast vs. melanocyte) specific factors.
While Gpnmb exhibits 33% amino acid identity to the melanosomal protein Si (Weterman et al., 1995) and proteomic analysis has revealed that GPNMB is present in all stages (I–IV) of melanosomes (Chi et al., 2006), a functional role for Gpnmb has yet to be determined. GPNMB may be a useful disease marker, as higher GPNMB expression has been correlated with lower metastasis in melanoma (Weterman et al., 1995) and the GPNMB locus has been associated with translocation breakpoints in a small number of melanomas (Okamoto et al., 2005). In addition to MITF binding sites, the GPNMB proximal promoter sequence also contains a highly conserved AP-1 binding site sequence directly upstream of the MITF(A) site. Given that a physical interaction between MITF and AP-1 has been previously demonstrated (Ogihara et al., 2001), the proximity of the AP-1 site to MITF(A) suggests a possible direct interaction between MITF and AP-1 in regulation of GPNMB expression. Both AP-1 and MITF have been implicated in melanoma progression. AP-1 activation through the MAP kinase signaling cascade has been correlated with malignant transformation of melanocytes to melanoma (Govindarajan et al., 2003), while amplifications of the MITF locus have been correlated with poor melanoma prognosis (Garraway et al., 2005). This taken together with our results highlight that further analysis of GPNMB with respect to melanoma is warranted.
The observation of a hierarchical cluster node, enriched with known melanocyte-expressed genes, allowed for the identification of Gpnmb as a new Mitf-dependent melanoblast marker and the identification of a novel, caudal, Sox10-deficient population of melanoblasts. Expression data generated from melanocyte derived cell lines can be useful in providing information to both identify and validate transcription factor target genes (Hoek et al., 2008). Further analysis of Gpnmb and the additional genes identified in this study will be a valuable resource for understanding the genetic components involved in melanocyte development and diseases.
MATERIALS AND METHODS
Cell growth conditions and RNA isolation
C57BL/6J Ink4a-Arf−/− (melan-Ink4a-1), C57BL/6J Ink4a-Arf−/−; Tyrp1b/b (melan-Ink4a-b) (Sviderskaya et al., 2002) and two separate C57BL/6J Ink4a-Arf−/−; Rab38cht/cht lines (melan-cht4 and melan-cht5) (Wasmeier et al., 2006) were grown at 37°C, 10% CO2 in RPMI 1640 medium containing 10% FCS, 2 mM L-glutamine, 100 units/ ml each penicillin and streptomycin (Invitrogen, Carlsbad, CA), 200 nM TPA (12-O-Tetradecanoylphorbol 13-acetate) Sigma-Aldrich and 200 pM cholera toxin (Sigma-Aldrich), (Sviderskaya et al., 1997). NIH3T3 cells were grown at 37°C, 5% CO2 in DMEM medium containing 10% FCS, 2 mM L-glutamine, and 100 units each penicillin and streptomycin. All cell lines were grown to 90% confluence and RNA was isolated as reported by Loftus et al. (Loftus et al., 2002), with one modification: following Trizol reagent (Invitrogen, Carlsbad, CA) lysis of cells, samples were incubated for 5 min at 65°C prior to proceeding with the RNA isolation protocol.
Microarray analysis
Operon Mv3.0 oligonucleotides (Operon, Huntsville, Al) containing over ~16,000 unique known genes were printed to slides by the National Human Genome Research Institute Genomics Core facility and probes hybridized with RNA isolated from the above melanocyte lines. All slide hybridizations were performed in triplicate, with one Cy3-Cy5 dye swap, and were co-hybridized with Universal RNA reference control (Stratagene, La Jolla, CA) and calibrated intensity ratios between the sample cell line derived RNA and the reference control were obtained. Analysis was performed using the MATLAB program (Natick, MA, USA) for log transformed calibrated ratio values. Anova Pattern Navigation Analysis was performed to identify probes with calibrated ratios > 2 fold higher than that of NIH 3T3 in all melanocyte lines and > 3 fold higher in any one of the four melanocyte lines relative to Universal controls using a correlation coefficient = 0.98. Significance thresholds of p-value < 0.05, and Benjamini and Hochberg false discovery rate (FDR) = 0.05 to account for multiple testing were used for all analyses performed. Hierarchical clustering was performed using the program Cluster 3.0 (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/software.htm#ctv) (University of Tokyo, Japan) using Spearman Rank correlation and average linkage with visualization of the heatmap and dendrogram generated by Java TreeView v1.1.3 (Saldanha, 2004). Microarray data was provided to the GEO database under accession number GSE10163.
In-situ hybridization
Riken cDNA clones for Gpnmb (G370004B14; Accession # BY474724), Mitf (G370008D06; Accession # BB852578), and Si (G370069C13; Accession # BB766987) were each digested with Kpn1 and transcribed with T3 polymerase to generate in situ probes. In situ hybridizations were performed on three embryos for each genotype as described previously (Loftus et al., 2002).
Mouse genotyping
E11.5 embryos were obtained from Sox10tm1Weg/+(Britsch et al., 2001) heterozygous intercrosses and MitfMi/+(B6C3Fe a/a-MitfMi/J strain #001573, Jackson Laboratories) heterozygous intercrosses. Following mating, the morning of positive plug was established as day 0.5. Genotyping for Sox10tm1Weg was performed as described previously (Britsch et al., 2001).Genotyping for MitfMi/+ was performed using ABI Taqman primers: mitfabiF - CCTTTCCCATGCTCTTTTCTTGAAG; mitfabiR – GCTCCTTAATGCGGTCGTTTATGT; vic-labeled Mitf+/+ probe –AACGAAGAAGAAGATTT; fam-labeled MitfMi probe – TTGAACGAAGAAGATTT.
Multiple-species comparative sequence analysis at GPNMB
We used two methods to identify conserved, non-coding sequences at GPNMB. First, genomic sequences at GPMNB were collected from human (UCSC Human Genome Browser, March 2006 build, chr7:23207149-23305465), rat (UCSC Rat Genome Browser, November 2004 build, chr4:77065632-77278341), mouse (UCSC Mouse Genome Browser, July 2007 build, chr6:48889957-49104776), cow (GenBank Accession No. AC157864.2), dog (UCSC Dog Genome Browser, May 2005 build, chr14:39752800-40030599), armadillo (GenBank Accession No. AC157452.2), hedgehog (GenBank Accession No. AC158438.2), chicken (UCSC Chicken Genome Browser, May 2006 build, chr2:30977810-31144369), and zebrafish (UCSC Zebrafish Genome Browser, July 2007, chr19:10878976–11271285). Sequences were aligned with MultiPipMaker software to obtain pip and acgt files. The ExactPlus program was then used to identify non-coding regions that harbor greater than four six-basepair fragments, 100% identical in at least seven of the nine species (Antonellis et al., 2006). Second, the GPNMB locus was examined at the UCSC Genome Browser (March 2006 build) to identify non-coding regions with a PhastCons conservation score of 340 or higher. Four MCS regions were identified corresponding to UCSC Human Genome Browser, March 2006 build as follows: MCS1- chr7: 23236842–23237413 (571 bp), MCS2 -chr7: 23245111–23245367 (256 bp), MCS3-chr7: 23252780–23252868 (88 bp) and MCS4- chr7: 23283864–23284187 (323 bp) and cloned as indicated below. The corresponding starting location for GPNMB MCS3 sequence in 20 species with associated genome builds are as follows: Human/chr7:23252780, UCSC Mar 2006; Chimp/chr7:23543405,UCSC: Mar 2006; Rhesus/chr3:102992177,UCSC: Jan 2006; Bushbaby/GeneScaffold_2348:67548, Ensembl:otoGar1, May 2006; Mouse/chr6:48986520, UCSC: July 2007; Rat/chr4:77161260, UCSC: Nov 2004; GuineaPig/scaffold_44:453683UCSC: Feb 2008; Rabbit/scaffold_153544:16001,UCSC TEST: May 2005; Shrew/scaffold_253863:51730, Ensembl: sorAra1, Oct 2005; Hedgehog/GeneScaffold_4005:4899,Ensembl: eriEur1, Jun 2006; Dog/chr14:39877714, UCSC: May 2005; Cat/scaffold_216322:112896,UCSC: Mar. 2006; Horse/chr4:54368849, UCSC: Jan. 2007; Cow/chr4:33202350, UCSC: Oct. 2007; Armadillo/scaffold_20519:46487, UCSC TEST: May 2005; Elephant/scaffold_149215:4649, UCSC TEST: May 2005; Tenrec/GeneScaffold_3894:6178, Ensembl: TENREC, July 2005; Opossum/chr8:296389507, UCSC: Jan. 2006; Platypus/Ultra65:618004, UCSC: Mar 2007; Chicken/chr2:31055481, UCSC: May 2006. UCSC TEST sequences are located at http://genome-test.cse.ucsc.edu/.
Identification of whole genome MITF motif consensus sequences
To identify potential MITF binding sites in the human genome, we first determined the genomic coordinates of all E-box consensus sequences (CAYRTG) in the human genome (UCSC Human Genome Browser, March 2006 build). Next, we marked those that could be extended by one nucleotide on either side to include the MITF motif consensus nucleotides (T)CAYRTG(A) and recorded their coordinates. Each of these steps was coded with Perl scripts and used flatfiles downloaded from the UCSC Human Genome Browser. The UCSC Genome Browser's 'RefSeq Genes' track was used to determine gene coding and non-coding regions. These data can be viewed on a per chromosome basis as a UCSC Custom Track at http://research.nhgri.nih.gov/manuscripts/Loftus/June2008/.
Luciferase reporter constructs
Luciferase expression constructs were generated using Gateway technology (Invitrogen, Carlsbad, CA). Briefly, PCR primers containing flanking attB sites were designed to amplify each region of interest at the human GPNMB locus. Following PCR reactions, products were separated on 1% low-melt agarose gels, purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), and recombined into the pDONR 221 vector according to the manufacturer’s specifications (Invitrogen Carlsbad, CA). Each recombination reaction was transformed into E. coli, and colonies selected for resistance to 25 µg/ml kanamycin. Each insert was sequenced to ensure the absence of PCR-induced errors. Subsequently, plasmid DNA isolated from each entry clone was recombined with the pLGF-E1b destination vector according to the manufacturer’s specifications (Invitrogen, Carlsbad, CA). Each reaction was transformed into E. coli, and colonies selected for resistance to 100 µg/ml ampicillin. Each construct underwent restriction enzyme digest with BsrGI to confirm the presence of the appropriately sized insert. A negative control vector bearing only the Gateway sequences upstream of the E1b promoter was generated in a similar manner. Removal of the MITF motif sequences, MITF(A) - (CACATGA) and MITF(B) - (TCACATGA) from GPNMB-MCS3 was performed using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and appropriate mutation-bearing oligonucleotides.
Cell culture, transfections, and luciferase assays
NIH 3T3 and melan-a cells were cultured as indicated above. Evaluation of MCS elements 1–4 were performed in 96-well culture plates using 16 replicates and MCS3 deletion constructs were performed in 48 well culture plates using 8 replicates, transfected with luciferase reporter vectors (see above) using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For each reaction 96-well assay, 0.25 µl of lipofectamine 2000 and 25 µl of OptiMEM I minimal growth medium (Invitrogen, Carlsbad, CA) were combined and incubated at room temperature for 10 minutes. Purified luciferase reporter vector (200 ng) and 2 ng of the internal control pCMV-RL renilla expression vector (Promega, Madison, WI) were diluted in 25 µl of OptiMEM I and combined with the lipofectamine-OptiMEM I mixture. The ~50µl reactions were incubated at room temperature for 20 minutes and then added to a single well of the 96-well culture plate containing melan-a cells. For 48 well assays tranfections were proportionally scaled 4-fold. After a 4-hour incubation at 37°C, the medium was aspirated, the slides washed with 1X PBS, and normal growth medium added. After a 48-hour incubation at 37°C, cells were washed with 1X PBS and lysed at room temperature using 1X Passive Lysis Buffer (Promega, Madison, WI). A total of 4 µl of the resulting cell lysate were transferred to a white polystyrene 96-well assay plate (Corning Inc., Corning, NY). Luciferase and renilla activities were determined using the Dual Luciferase Reporter 1000 Assay System (Promega, Madison, WI) and a model Centro LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany). For each experiment the ratio of luciferase to renilla activity and the fold increase in this ratio over that observed for pLGF-E1b with no insert were calculated. The mean (bar height in figures) and standard deviation (error bars in figures) were determined using standard calculations.
Zebrafish Reporter Expression Assays
Zebrafish were raised and bred in accordance with standard conditions (Kimmel et al., 1995); (Westerfield, 2000). Embryos were maintained at 28 C and staged in accordance with standard methods (Kimmel et al., 1995); (Westerfield, 2000). EGFP expression constructs (see above) were injected into AB background G0 embryos (n≥200), as previously described (Fisher et al., 2006a), Injected embryos were evaluated for reporter expression at 24 and 48 hpf. Embryos were analyzed and imaged using a Carl Zeiss Lumar V12 Stereo microscope with AxioVision version 4.5 software.
Supplementary Material
Hierarchical clustering of 318 array elements (258 genes) which are >2-fold over expressed in all melanocyte lines relative to NIH3T3 control calibrated ratios and also have expression increased >3 fold in any one cell line relative to the Universal control RNA.
Enhancer Assay for MCS3 and MCS3 deletion constructs performed in NIH3T3 cells. The ability of each MCS to enhance luciferase expression was determined by calculating the ratio of luciferase expression to renilla expression, and then determining the fold increase (y-axis) over the ratio calculated for pE1b with no insert.
ACKNOWLEDEGMENTS
We would like to acknowledge Shih-Queen Lee-Lin for expert technical assistance, Abdel Elkahloun for technical assistance with microarray hybridizations, Julia Feckes for graphics assistance and NIH Intramural Sequencing Center (NISC) for generating sequence data. Handling of mice was performed in accordance with NIH guidelines under animal care and use mouse protocol G94-7. All zebrafish work was performed under protocols approved by the Johns Hopkins University Animal Care and Use Committee. This work was supported in part by the National Human Genome Research Institute’s (NHGRI) Intramural Research Program.
REFERENCES
- Abdelmagid SM, Barbe MF, Arango-Hisijara I, Owen TA, Popoff SN, Safadi FF. Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol. 2007;210:26–37. doi: 10.1002/jcp.20841. [DOI] [PubMed] [Google Scholar]
- Aksan I, Goding CR. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, John SW. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet. 2006;15:259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- Bachner D, Schroder D, Gross G. mRNA expression of the murine glycoprotein (transmembrane) nmb (Gpnmb) gene is linked to the developing retinal pigment epithelium and iris. Brain Res Gene Expr Patterns. 2002;1:159–165. doi: 10.1016/s1567-133x(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Pavan WJ. The oculocutaneous albinism type IV gene Matp is a new marker of pigment cell precursors during mouse embryonic development. Mech Dev. 2002;116:209–212. doi: 10.1016/s0925-4773(02)00130-2. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Pavan WJ. Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns. 2003;3:703–707. doi: 10.1016/j.modgep.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Busca R, Abbe P, Bille K, Aberdam E, Ortonne JP, Ballotti R. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol Cell Biol. 1998;18:694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, Huang H, Canfield VA, Cheng KC, Yang F, Abe R, Yamagishi S, Shabanowitz J, Hearing VJ, Wu C, Appella E, Hunt DF. Proteomic and bioinformatics characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- Du J, Fisher DE. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem. 2002;277:402–406. doi: 10.1074/jbc.M110229200. [DOI] [PubMed] [Google Scholar]
- Du J, Miller AJ, Widlund HR, Horstmann MA, Ramaswamy S, Fisher DE. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol. 2003;163:333–343. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fang D, Tsuji Y, Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. doi: 10.1093/nar/gkf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006a;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006b;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Govindarajan B, Bai X, Cohen C, Zhong H, Kilroy S, Louis G, Moses M, Arbiser JL. Malignant transformation of melanocytes to melanoma by constitutive activation of mitogen-activated protein kinase kinase (MAPKK) signaling. J Biol Chem. 2003;278:9790–9795. doi: 10.1074/jbc.M212929200. [DOI] [PubMed] [Google Scholar]
- Hoek HS, Schlegell NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008 doi: 10.1111/j.1755-148X.2008.00505.x. In press. [DOI] [PubMed] [Google Scholar]
- Howell GR, Libby RT, Marchant JK, Wilson LA, Cosma IM, Smith RS, Anderson MG, John SW. Absence of glaucoma in DBA/2J mice homozygous for wild-type versions of Gpnmb and Tyrp1. BMC Genet. 2007;8:45. doi: 10.1186/1471-2156-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Tanabe Y, Lupski JR. Myelin deficiencies in both the central and the peripheral nervous systems associated with a SOX10 mutation. Ann Neurol. 1999;46:313–318. doi: 10.1002/1531-8249(199909)46:3<313::aid-ana6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ. Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res. 2004;17:352–362. doi: 10.1111/j.1600-0749.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)- associated hypopigmentation and deafness to WS2. J Biol Chem. 2000;275:37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA, 3rd, Pavan WJ. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci U S A. 2002;99:4471–4476. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 2004;556:236–244. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem. 2006;281:10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- Murisier F, Beermann F. Genetics of pigment cells: lessons from the tyrosinase gene family. Histol Histopathol. 2006;21:567–578. doi: 10.14670/HH-21.567. [DOI] [PubMed] [Google Scholar]
- Murisier F, Guichard S, Beermann F. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res. 2007;20:173–184. doi: 10.1111/j.1600-0749.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Oetting WS, L M, Bennett DC Color Genes. European Society for Pigment Cell Research. 2008 September; World Wide Web (URL: http://www.espcr.org/micemut.
- Ogihara H, Morii E, Kim DK, Oboki K, Kitamura Y. Inhibitory effect of the transcription factor encoded by the mutant mi microphthalmia allele on transactivation of mouse mast cell protease 7 gene. Blood. 2001;97:645–651. doi: 10.1182/blood.v97.3.645. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Pirker C, Bilban M, Berger W, Losert D, Marosi C, Haas OA, Wolff K, Pehamberger H. Seven novel and stable translocations associated with oncogenic gene expression in malignant melanoma. Neoplasia. 2005;7:303–311. doi: 10.1593/neo.04514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Silver DL, Hou L, Pavan WJ. The genetic regulation of pigment cell development. Adv Exp Med Biol. 2006;589:155–169. doi: 10.1007/978-0-387-46954-6_9. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Davidson DR, Jackson IJ. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development. 1992;115:1111–1119. doi: 10.1242/dev.115.4.1111. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Bennett DC, Ho L, Bailin T, Lee ST, Spritz RA. Complementation of hypopigmentation in p-mutant (pink-eyed dilution) mouse melanocytes by normal human P cDNA, and defective complementation by OCA2 mutant sequences. J Invest Dermatol. 1997;108:30–34. doi: 10.1111/1523-1747.ep12285621. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC. p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem. 2000;275:30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, Seabra MC. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish(Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Weterman MA, Ajubi N, van Dinter IM, Degen WG, van Muijen GN, Ruitter DJ, Bloemers HP. nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. Int J Cancer. 1995;60:73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hierarchical clustering of 318 array elements (258 genes) which are >2-fold over expressed in all melanocyte lines relative to NIH3T3 control calibrated ratios and also have expression increased >3 fold in any one cell line relative to the Universal control RNA.
Enhancer Assay for MCS3 and MCS3 deletion constructs performed in NIH3T3 cells. The ability of each MCS to enhance luciferase expression was determined by calculating the ratio of luciferase expression to renilla expression, and then determining the fold increase (y-axis) over the ratio calculated for pE1b with no insert.