Abstract
Ribosome-targeting antibiotics block protein synthesis by binding at functionally important regions of the bacterial rRNA. Resistance is often conferred by addition of a methyl group at the antibiotic binding site within an rRNA region that is already highly modified with several nucleotide methylations. In bacterial rRNA, each methylation requires its own specific methyltransferase enzyme, and this raises the question as to how an extra methyltransferase conferring antibiotic resistance can be accommodated and how it can gain access to its nucleotide target within a short and functionally crowded stretch of the rRNA sequence. Here, we show that the Sgm methyltransferase confers resistance to 4,6-disubstituted deoxystreptamine aminoglycosides by introducing the 16S rRNA modification m7G1405 within the ribosomal A site. This region of Escherichia coli 16S rRNA already contains several methylated nucleotides including m4Cm1402 and m5C1407. Modification at m5C1407 by the methyltransferase RsmF is impeded as Sgm gains access to its adjacent G1405 target on the 30S ribosomal subunit. An Sgm mutant (G135A), which is impaired in S-adenosylmethionine binding and confers lower resistance, is less able to interfere with RsmF methylation on the 30S subunit. The two methylations at 16S rRNA nucleotide m4Cm1402 are unaffected by both the wild-type and the mutant versions of Sgm. The data indicate that interplay between resistance methyltransferases and the cell's own indigenous methyltransferases can play an important role in determining resistance levels.
Keywords: aminoglycoside resistance, 16S rRNA methylation, Sgm, YebU (RsmF), RNA mass spectrometry
INTRODUCTION
The sisomicin-producing actinomycete Micromonospora zionensis protects its own ribosomes from the toxic effects of the aminoglycoside product by expression of the sisomicin–gentamicin resistance methyltransferase, Sgm (Kojic et al. 1992). Sgm confers resistance to 4,6-disubstituted deoxystreptamine aminoglycosides such as kanamycin, and thus functions similarly to its orthologs KgmB (Holmes and Cundliffe 1991), GrmA (Cundliffe 1992), and FmrO (Ohta and Hasegawa 1993) from other aminoglycoside-producing microorganisms, and also to ArmA (Galimand et al. 2003), RmtA (Yokoyama et al. 2003), RmtB (Doi et al. 2004), RmtC (Wachino et al. 2006), and RmtD (Doi et al. 2007) that have recently been identified in clinical strains. The methylation target of KgmB (Holmes and Cundliffe 1991), ArmA (Liou et al. 2006), and RmtB (Perichon et al. 2007) is the N7-position of nucleotide G1405 in 16S rRNA (Escherichia coli numbering). Sgm is also thought to modify at G1405 (Holmes and Cundliffe 1991; Tomic et al. 2008), although this has not previously been verified experimentally. Nucleotide G1405 is located within a highly conserved and heavily modified rRNA region at the decoding site (A site) of the ribosome (Fig. 1). The 4,6-disubstituted deoxystreptamine aminoglycosides disrupt mRNA decoding by binding to this region through contact with nucleotide G1405 (Yoshizawa et al. 1998; Vicens and Westhof 2002). This contact is hindered by methylation of G1405 and results in drug resistance.
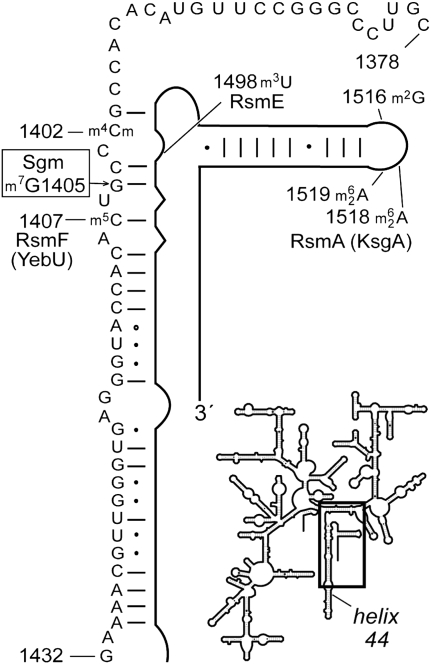
FIGURE 1.
Representation of the 16S rRNA secondary structure showing the sequence in helix 44 (boxed) that was isolated for analysis by mass spectrometry. The sites of post-transcriptional housekeeping modification in this E. coli rRNA region are indicated, and the methyltransferases that introduce these modifications are given (where known); the site of the m7G1405 modification added by the Sgm resistance methyltransferase is also shown.
Drug resistance methylations in bacterial rRNAs, such as that at nucleotide G1405, have the sole function of providing protection against a specific group of antibiotics and offer no known physiological advantage in the absence of the drug. In this sense they contrast with the numerous indigenous methylations (also termed housekeeping methylations) that contribute to the overall structure and general function of the rRNA. In many other respects, however, the resistance and housekeeping methylations are similar in that they cluster in phylogenetically conserved and functionally essential regions of the rRNA, and each nucleotide methylation is introduced by its own distinct methyltransferase enzyme (Ofengand and Del Campo 2004; Andersen and Douthwaite 2006). At the ribosomal A site of E. coli 16S rRNA, several nucleotides including C1402, C1407, and U1498 (Fig. 1) are methylated by housekeeping methyltransferases. It seems that the interactions of these methyltransferases have evolved to be sterically compatible and/or temporally displaced such that each of the enzymes is able to gain access to and modify its own nucleotide target. This raises the question as to how well an additional methyltransferase modifying at nucleotide G1405 is accommodated in this functionally crowded region of the ribosome.
In this study, we have introduced a recombinant version of the Sgm methyltransferase gene into an E. coli strain that has a full complement of housekeeping methyltransferases. Analyses of the 16S rRNA by primer extension and MALDI mass spectrometry show that the m5C1407-specific housekeeping methyltransferase RsmF (previously termed YebU) is outcompeted by Sgm as the resistance methyltransferase gains access to its own m7G1405 target on the 30S ribosomal subunit. A single amino acid change in Sgm, which lowers the level of conferred resistance, reduces the ability of Sgm to interfere with RsmF methylation on the 30S subunit. The two housekeeping methylations at 16S rRNA nucleotide m4Cm1402 are unaffected by both the wild-type and the mutant versions of Sgm.
RESULTS AND DISCUSSION
Sgm modifies m7G1405 in 16S rRNA
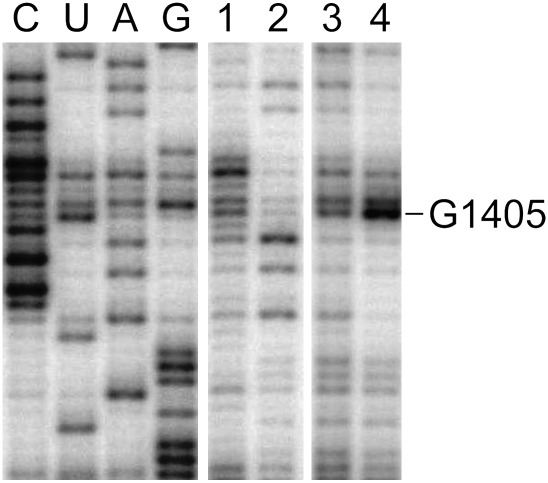
We identified the site of Sgm methylation by two independent approaches. Total rRNA was extracted from ribosomal subunits isolated from E. coli cells harboring an expression vector encoding the sgm gene. In the first approach, sites of m7G methylation in the rRNAs were identified by borohydride/aniline treatment to induce β-elimination. Analysis of the rRNAs by primer extension with reverse transcriptase showed that one reverse transcriptase stop (Fig. 2) was dependent on Sgm and borohydride/aniline treatment and indicated that Sgm methylates at the N7-position of nucleotide G1405.
FIGURE 2.
Gel autoradiogram of primer extension through the G1405 region of 16S rRNA. The rRNAs were from E. coli cells without sgm (lanes 1,2) or from cells expressing wild-type sgm (lanes 3,4); samples were either untreated (lanes 1,3) or were treated with NaBH4/aniline prior to primer extension. In lane 4, rRNA scission stops reverse transcriptase immediately 3′ to the site of Sgm methylation at G1405.
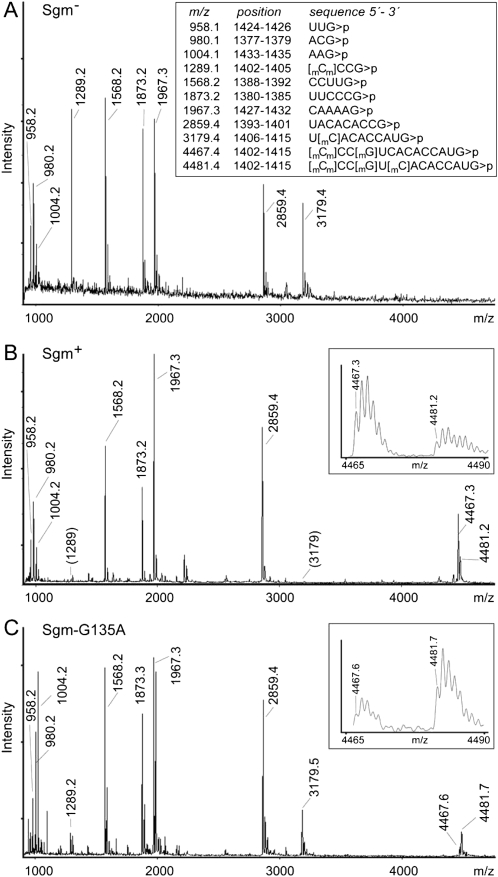
Using a second approach, the 16S rRNA sequence 1378 to 1432 (Fig. 1) was isolated by hybridization to a complementary deoxyoligonucleotide and the rest of the rRNA was removed with nucleases. The 1378–1432 sequence was then digested with RNase T1 and the fragments were analyzed by MALDI mass spectrometry. The rRNA from Sgm− cells produced a distinct MS spectrum that included the two peaks at m/z 1289 and 3179, which correspond to the fragments C1402–G1405 and U1406–G1415, respectively (Fig. 3A).
FIGURE 3.
MALDI-MS spectra of RNase T1 oligonucleotides from the E. coli 16S rRNA sequence C1378–G1432. (A) Spectrum of rRNA from cells without the sgm gene. The theoretical monoisotopic masses of the RNase digestion products with 3′-end cyclic phosphates (>p) are shown in the box. Fragments smaller than trinucleotides are not shown. The theoretical and empirical masses (given above the peaks) match to within 0.2 Da. (B) The corresponding spectrum from Sgm+ cells showing that the fragments at m/z 1289 and 3179 are missing and remain combined in the longer sequence at m/z 4467 and 4481. (Inset) Enlargement of this spectral region; the multiple tops in these fragments reflect the natural distribution of 12C and 13C isotopes. (C) The rRNA spectrum from Sgm-G135A cells; this differs from the Sgm+ rRNA in the relative amplitudes of the m/z 4467 and 4481 peaks, and also peaks at m/z 1289 and 3179 are evident.
After expression of the Sgm methyltransferase (Sgm+ cells), the rRNA could no longer be cleaved at G1405 by RNase T1, resulting in the disappearance of the m/z 1289 and 3179 peaks (Fig. 3B) and the appearance of a new, larger peak at m/z 4481 (and at m/z 4467, see below). The m/z 4481 peak corresponds to sequence C1402–G1415 in which the two smaller fragments remain combined and include an extra methyl group. The resistance to RNase T1 cleavage at nucleotide G1405 and the addition of 14 Da extra mass clearly establish that Sgm methylates this nucleotide position. The complete loss of the m/z 1289 and 3179 peaks from the spectrum (Fig. 3B) indicates that Sgm methylation at G1405 is stoichiometric.
In addition to the Sgm site at G1405, this sequence of the 16S rRNA contains three housekeeping methylations (Fig. 1). Two of these modifications are at nucleotide m4Cm1402 and are added by presently uncharacterized methyltransferases; the third is at nucleotide m5C1407 and is added by the RsmF (formerly YebU) methyltransferase (Andersen and Douthwaite 2006). These methylations are evident in Figure 3A as the m/z 1289 peak (containing m4Cm1402) and the 3179 peak (containing m5C1407). The lack of unmodified fragments, which would run at m/z 1261, m/z 1275, or m/z 3165 in the Figure 3A spectrum, indicates that m4Cm1402 and m5C1407 are normally stoichiometrically methylated in E. coli rRNA.
Sgm reduces RsmF housekeeping methylation at m7C1407
After expression of the Sgm methyltransferase, one of these housekeeping modifications is no longer stoichiometric; this is apparent from the presence of the m/z 4467 peak (containing three methyl groups) in addition to the m/z 4481 peak (with four methyl groups). Tandem MS analysis of the m/z 4467 peak (not shown) showed that both methylations on nucleotide m4Cm1402 were present, and this fits with our previous observation that these methylations occur as a pair and have not been seen individually (Andersen and Douthwaite 2006). The m7G1405 modification must be present in the m/z 4467 peak to prevent its cleavage by RNase T1; thus, the rRNA is missing the RsmF methylation at C1407.
At a first glance, it might seem that the relative heights of the m/z 4467 and 4481 peaks indicate the extent to which RsmF methylation is suppressed by Sgm. However, any such quantitative interpretation of MALDI-MS data should be accompanied with a great deal of caution. The MALDI technique is notoriously inconsistent in the degree to which it ionizes molecules, and consequently the amplitudes of the resultant spectra signals are variable. This is illustrated in the heights of the m/z 1004, m/z 1568, and m/z 1967 peaks corresponding to the AAG, CCUUG, and CAAAAG fragments. Despite the fact that these fragments are equimolar (as a consequence of being derived from the same RNA chain), they produce signals of different heights, and these relative heights vary from spectrum to spectrum (Fig. 3). That being said, our analyses of dozens of spectra for the Sgm+ rRNA (as in Fig. 3B) showed that the heights of the m/z 4467 and 4481 peaks were remarkably constant relative to each other (although not to the other spectral peaks). The consistency between the m/z 4467 and 4481 peaks undoubtedly reflects the near identical composition of the two ions, and, in this particular case, we believe that some guarded inferences can be made from the relative heights (see below).
Sgm and RsmF recognize the same 30S subunit substrate
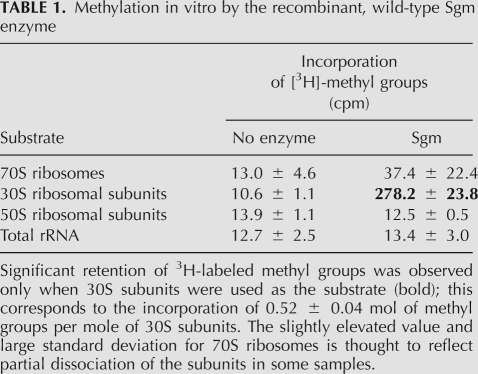
We determined the substrate that is recognized by Sgm by following the methylation reaction in vitro using purified histidine-tagged Sgm and a range of potential substrates including 70S ribosomes, separated 30S and 50S subunits, and naked 16S rRNA. The methylation reaction was monitored by the incorporation of radiolabeled S-adenosylmethionine into the rRNA (Table 1). Sgm methylation is dependent on the presence of the S-adenosylmethionine cofactor, and only the 30S ribosomal subunit functions as a valid substrate. In vitro methylation of 30S subunits at nucleotide G1405 was confirmed in parallel MS experiments using nonlabeled cofactor (not shown). Substrate recognition by Sgm is therefore similar to that of RsmF, which also uses 30S subunits as its substrate and is unable to modify naked 16S rRNA or 70S ribosomes (Andersen and Douthwaite 2006). Docking simulations with the RsmF crystal structure indicate contact with ribosomal components including r-protein S12 at the interface of the 30S subunit; in the 70S ribosome, access of the methyltransferase to these contact sites is blocked by the 50S subunit (Hallberg et al. 2006).
TABLE 1.
Methylation in vitro by the recombinant, wild-type Sgm enzyme
Sgm activity affects its ability to hinder C1407 methylation by RsmF
The in vitro methylation studies indicate that Sgm and RsmF attempt to gain access to the same region of the 16S rRNA and at the same time—immediately after (and possibly during) 30S subunit assembly, but before association with the 50S subunit. The apparent competition between the two methyltransferases was studied further in vivo using a Sgm variant with impaired enzymatic activity. Several Sgm mutants have previously been shown to confer greatly reduced aminoglycoside resistance (Maravić Vlahoviček et al. 2008; Savic et al. 2008). We chose a representative mutant, Sgm-G135A, which is a poor binder of the S-adenosylmethionine cofactor (Savic et al. 2008), to determine whether the resistance level correlates with a reduced ability to methylate at the G1405 and to compete with RsmF. In E. coli, this mutant enzyme confers resistance to kanamycin at up to 32 μg/mL, compared with resistance at >1280 μg/mL for E. coli expressing wild-type Sgm, and <1 μg/mL for Sgm− cells (Maravić Vlahoviček et al. 2008).
The methylation pattern of the 16S rRNA from cells with Sgm-G135A was analyzed by MALDI-MS (Fig. 3C). The spectral peaks at m/z 4467 and 4481 show that the Sgm-G135A mutant is still capable of methylating G1405; however, the reappearance of the m/z 1289 and m/z 3179 peaks indicates that G1405 methylation is no longer complete. There is no peak at m/z 1365, and therefore all the rRNA molecules are methylated at C1407 and/or G1405. The molecules left unmethylated by RsmF congregate exclusively in the m/z 4467 peak, which is now reproducibly smaller than the m/z 4481 peak (Fig. 3C). Thus, the impaired cofactor binding of Sgm-G135A reduces its efficiency in methylating the G1405 target, which in turn enables RsmF to partially re-establish its claim to the region and methylate at nucleotide C1407.
Modeling of the Sgm structure (Maravić Vlahoviček et al. 2008) and of RsmF on the 30S subunit (Hallberg et al. 2006) is consistent with a mechanism whereby the bulky methyltransferases sterically clash in their attempts to gain access to their respective targets at G1405 and C1407. In this scenario, the wild-type Sgm interacts more effectively with the 30S subunit and physically obstructs the binding of RsmF; this advantage of Sgm is then markedly reduced when the G135A mutation lowers its activity. It should be noted, however, that our observations fit equally well with an alternative explanation where modification at m7G1405 obscures an essential element for RsmF recognition and, in this model, inhibition would persist after Sgm has dissociated from the substrate.
Interference between methyltransferases and variance in resistance levels
The rRNA target sites of resistance methyltransferases tend to be close to those of the indigenous housekeeping modifications. Loss of an individual housekeeping modification generally has a measurable fitness cost, although this is often quite small. In the case of RsmF, the growth rate of E. coli is reduced by ∼20% after loss of m5C1407 methylation (Andersen and Douthwaite 2006). In E. coli, the consequences of Sgm affecting RsmF function and cell fitness would naturally be offset in the presence of aminoglycoside antibiotics, although it might be expected that there would be a selection against methyltransferases such as Sgm in the absence of antibiotics.
Sgm has evolved as a natural component in its actinomycete host, Micromonospora zionensis, and as such it would not be expected to clash with other indigenous methyltransferase. In fact, no ortholog of rsmF is apparent in any actinomycete genome that has been sequenced to date. We have analyzed the rRNA from three actinomycetes: Mycobacterium smegmatis, Mycobacterium tuberculosis (Johansen et al. 2006), and Streptomyces coelicolor (S Johansen and S Douthwaite, unpubl.) by MALDI-MS, and all lack modification at 16S rRNA nucleotide C1407. Although the genome and rRNAs from M. zionensis have not yet been analyzed, it seems unlikely based on the available evidence that this species possesses an RsmF homolog.
Competition in accessing rRNA targets is presumably by no means limited to RsmF and Sgm, and might involve the nearby modification at nucleotide U1498 (Basturea et al. 2006), or C1404 in thermophiles (Guymon et al. 2006), or C1409 and G1410 in mycobacterial actinomycetes (Johansen et al. 2006). Other G1405 methyltransferases would presumably also clash with housekeeping modification enzymes, and the outcome of these confrontations might explain why, despite modifying the same site, they do not always confer identical resistance phenotypes. For instance, the resistance conferred by the G1405 methyltransferases RmtC (Wachino et al. 2006) and RmtA (Yokoyama et al. 2003) appears particularly potent. Differences in resistance phenotypes can be due to many factors, although one aspect that until now has been overlooked is how effectively a resistance methyltransferase that has been acquired as an exogenous genetic element can elbow aside the cell's own housekeeping methyltransferases to gain access to its target.
MATERIALS AND METHODS
Strains and plasmids
The sgm gene was expressed from the plasmid vector pET-25b(+) in E. coli C41(DE3) and purified as described previously (Maravić Vlahoviček et al. 2008). Mutations including G135A were introduced into sgm based on the tertiary structure predictions for the methyltransferase (Maravić Vlahoviček et al. 2008). Measurements of resistance levels expressed as minimal inhibitory concentrations of kanamycin (Sigma) were described previously (Maravić Vlahoviček et al. 2008).
Analysis of rRNA by primer extension
Total rRNA was extracted from ribosomal particles isolated from E. coli C41(DE3) cells harboring the expression vector pET-25b(+) with and without recombinant versions of the sgm gene. Cells were grown in the presence of 1 mM IPTG and were harvested in early log phase at an OD450 of 0.4–0.5. Sites of m7G methylation in the rRNAs were reduced with sodium borohydride (NaBH4) and cleaved via aniline-induced β-elimination (Peattie 1979). This reaction is generally incomplete (Douthwaite et al. 1983) but was improved by inclusion of hypermethylated tRNA carrier in the reactions (Zueva et al. 1985). A 5′-32P-end-labeled deoxynucleotide primer, complementary to 16S rRNA nucleotides 1459–1479, was hybridized to the rRNA and extended with AMV reverse transcriptase (Finnzymes) (Stern et al. 1988). Sequencing reactions were preformed on rRNA from the E. coli cells lacking Sgm. Extension products were separated on denaturing polyacrylamide gels and were visualized by phosphorimaging (Typhoon, Amersham Biosciences).
Analysis of rRNA by MALDI mass spectrometry
The 16S rRNA sequence from C1378–G1432 was isolated by hybridization to complementary deoxyoligonucleotides, and 40 pmol of rRNA were heated with 400 pmol of deoxyoligonucleotide (complementary to the sequence shown in Fig. 1) for 1 min at 85°C, followed by slow cooling over 2 h to 45°C. Unhybridized regions of the rRNAs were digested with mung bean nuclease (NE Biolabs) and RNase A (Sigma), and the rRNA fragment protected by the deoxyoligonucleotide was isolated by gel electrophoresis (Andersen et al. 2004). The rRNA fragment was digested with 20 units of RNase T1 (Ambion) for 3 h at 37°C in 2 μL of H2O containing 0.25 μL of 0.5 M 3-hydroxypicolinic acid. Samples were dried and resuspended in 1 μL of H2O prior to analysis by MALDI-MS (Voyager Elite, Perseptive Biosystems) recording in reflector and positive ion mode (Kirpekar et al. 2000). Spectra were analyzed using the program m/z (Proteometrics, Inc).
In vitro methylation reactions
Ribosome 70S couples, separated 30S and 50S subunits, and naked rRNAs were tested as substrates for Sgm-mediated methylation. Reactions for in vitro methylation contained 5 pmol of substrate and 1 pmol of Sgm enzyme with a mixture of 2 nmol of nonradioactive AdoMet (Sigma) and 1 pmol of [methyl-3H] AdoMet (82 Ci/mmol, Amersham Biosciences/GE Healthcare) as the methyl donor. All reactions were carried out at 37°C in 100 μL of 20 mM Tris, pH 7.5, 40 mM NH4Cl, 7.5 mM MgCl2, 6 mM β-mercaptoethanol with 10% glycerol. Reactions were stopped by the addition of 10% (w/v) trichloroacetic acid, and the precipitated substrates were pelleted by centrifugation and counted for radioactivity. In control experiments, the Sgm enzyme and the substrates were incubated separately with AdoMet. Duplicate samples were measured in three independent experiments in all cases.
ACKNOWLEDGMENTS
We thank Anette Rasmussen and Finn Kirpekar (University of Southern Denmark) for invaluable help with the collection and interpretation of the MALDI data. G.M.V. thanks the Croatian Ministry of Science (grant 006-0982913-1219), ICGEB (grant CRP/CRO08-02), and FP6 (grant #043682 “EuroPharm” as support to F.B.) for support. S.D. thanks the Danish Research Agency (FNU-rammebevilling 272-07-0613), the Nucleic Acid Center of the Danish Grundforskningsfond, and NAC-DRUG under the FP6 Marie Curie Initial Training Networks (to S.Č.) for support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1618809.
REFERENCES
- Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J Mol Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E. Resistance to macrolides and lincosamides in Streptomyces lividans and to aminoglycosides in Micromonospora purpurea. Gene. 1992;115:75–84. doi: 10.1016/0378-1119(92)90543-x. [DOI] [PubMed] [Google Scholar]
- Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother. 2004;48:491–496. doi: 10.1128/AAC.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother. 2007;51:852–856. doi: 10.1128/AAC.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S, Garrett RA, Wagner R. Comparison of Escherichia coli tRNAPhe in the free state, in the ternary complex and in the ribosomal A and P sites by chemical probing. Eur J Biochem. 1983;131:261–269. doi: 10.1111/j.1432-1033.1983.tb07258.x. [DOI] [PubMed] [Google Scholar]
- Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003;47:2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: The complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- Hallberg BM, Ericsson UB, Johnson KA, Andersen NM, Douthwaite S, Nordlund P, Beuscher AE, Erlandsen H. The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J Mol Biol. 2006;360:774–787. doi: 10.1016/j.jmb.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Holmes DJ, Cundliffe E. Analysis of a ribosomal RNA methylase gene from Streptomyces tenebrarius which confers resistance to gentamicin. Mol Gen Genet. 1991;229:229–237. doi: 10.1007/BF00272160. [DOI] [PubMed] [Google Scholar]
- Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Kirpekar F, Douthwaite S, Roepstorff P. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA. 2000;6:296–306. doi: 10.1017/s1355838200992148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Topisirovic L, Vasiljevic B. Cloning and characterization of an aminoglycoside resistance determinant from Micromonospora zionensis. J Bacteriol. 1992;174:7868–7872. doi: 10.1128/jb.174.23.7868-7872.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GF, Yoshizawa S, Courvalin P, Galimand M. Aminoglycoside resistance by ArmA-mediated ribosomal 16S methylation in human bacterial pathogens. J Mol Biol. 2006;359:358–364. doi: 10.1016/j.jmb.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Maravić Vlahoviček G, Čubrilo S, Tkaczuk KL, Bujnicki JM. Modeling and experimental analyses reveal a two-domain structure and amino acids important for the activity of aminoglycoside resistance methyltransferase Sgm. Biochim Biophys Acta. 2008;1784:582–590. doi: 10.1016/j.bbapap.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M. Modified nucleosides in Escherichia coli ribosomal RNA. EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology. In: Curtiss R, editor. ASM Press; Washington, DC: 2004. [DOI] [PubMed] [Google Scholar]
- Ohta T, Hasegawa M. Analysis of the self-defense gene (fmrO) of a fortimicin A (astromicin) producer, Micromonospora olivasterospora: Comparison with other aminoglycoside-resistance-encoding genes. Gene. 1993;127:63–69. doi: 10.1016/0378-1119(93)90617-c. [DOI] [PubMed] [Google Scholar]
- Peattie DA. Direct chemical method for sequencing RNA. Proc Natl Acad Sci. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perichon B, Courvalin P, Galimand M. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother. 2007;51:2464–2469. doi: 10.1128/AAC.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic M, Ilic-Tomic T, Macmaster R, Vasiljevic B, Conn GL. Critical residues for cofactor binding and catalytic activity in the aminoglycoside resistance methyltransferase Sgm. J Bacteriol. 2008;190:5855–5861. doi: 10.1128/JB.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Tomic TI, Moric I, Conn GL, Vasiljevic B. Aminoglycoside resistance genes sgm and kgmB protect bacterial but not yeast small ribosomal subunits in vitro despite high conservation of the rRNA A-site. Res Microbiol. 2008;159:658–662. doi: 10.1016/j.resmic.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother. 2006;50:178–184. doi: 10.1128/AAC.50.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet. 2003;362:1888–1893. doi: 10.1016/S0140-6736(03)14959-8. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zueva VS, Mankin AS, Bogdanov AA, Baratova LA. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur J Biochem. 1985;146:679–687. doi: 10.1111/j.1432-1033.1985.tb08704.x. [DOI] [PubMed] [Google Scholar]